Abstract
Motivation: Many published manuscripts contain experiment protocols which are poorly described or deficient in information. This means that the published results are very hard or impossible to repeat. This problem is being made worse by the increasing complexity of high-throughput/automated methods. There is therefore a growing need to represent experiment protocols in an efficient and unambiguous way.
Results: We have developed the Experiment ACTions (EXACT) ontology as the basis of a method of representing biological laboratory protocols. We provide example protocols that have been formalized using EXACT, and demonstrate the advantages and opportunities created by using this formalization. We argue that the use of EXACT will result in the publication of protocols with increased clarity and usefulness to the scientific community.
Availability: The ontology, examples and code can be downloaded from http://www.aber.ac.uk/compsci/Research/bio/dss/EXACT/
Contact: Larisa Soldatova lss@aber.ac.uk
1 INTRODUCTION
‘Everything is vague to a degree you do not realize till you have tried to make it precise.’
Bertrand Russell
The ability to repeat a published experiment protocol is the foundation stone of laboratory science. It is widely accepted that for new knowledge to be published in a scientific journal the protocols used to derive that new knowledge must also be published. This is essential to validate that the process by which the knowledge was inferred was not flawed in any fundamental way, and to ensure that the result was not caused by some chance event. In order to repeat a protocol it must necessarily be described in sufficient and unambiguous detail to enable another agent (human or machine) to be able to replicate the original experiment actions. With the increasing complexity of experiment methods, the description of laboratory protocols is becoming correspondingly more complicated and intricate. This means that there is a growing technological need to be able to represent experiment protocols in an efficient and unambiguous way.
We propose the Experiment ACTions (EXACT) ontology as the basis of a method of representing biological laboratory protocols. EXACT provides a model for the description of experiment actions and it can be used for the fully formalized representation of protocols. It can also be combined with other formalisms for the description of bio-medical investigations.
The rest of this article is organized as follows: Section 2 provides the background to this work, Section 3 provides a detailed description of the proposed ontology of experiment actions and Section 4 demonstrates the application of the ontology to the formalized description of two protocols: for creating competent cells and for compound library replication. In Section 5 we describe the opportunities for the application/implementation of EXACT and finally Section 6 provides discussion and conclusion.
2 BACKGROUND
2.1 Current problems
The degree of information granularity present in many published protocols is often insufficient to allow the method to be repeated successfully. An optimization period is then necessary to bridge the gap in knowledge between the published protocol and one which works reliably. Knowledge of how best to implement an existing method is regarded as a group's intellectual property and is often not included in published manuscripts. Each and every time research results are published with insufficient information in the materials and methods section this duplication of labour is repeated, adding to inconvenience and cost.
A manuscript published by Akada et al. 2006 illustrates the difficulty in repeating another researcher's protocol when not all the necessary information is provided. The manuscript outlines a novel protocol for gene deletion in Saccharomyces cerevisiae. The protocol focusses on the generation of a deletion cassette through the fusion of two DNA fragments. The manuscript provides ample information on strains, media, primer sequence and polymerase chain reaction (PCR) conditions. However, when repeated in our laboratory the deletion cassette could not be generated. Personal communication with the author revealed that the two DNA fragments require gel purification before a successful PCR fusion can occur. This proved to be a vital step yet was not included in the published manuscript.
The excerpt shown in Table 1 describes the first stages of making yeast cells competent and was taken from the High Efficiency Transformation of Yeast protocol published in Methods in Yeast Genetics Amberg et al., 2005, a text book routinely cited in published papers.
Table 1.
High-efficiency transformation of yeast, methods in yeast genetics
| 1. Inoculate 4 ml of liquid YPAD or 10 ml of SC and incubate with shaking overnight at 30○C |
| 2. Count overnight culture and inoculate 50 ml of YPAD to a cell density of 5 × 106/ml culture |
| 3. … |
The protocol is summarized in point form using natural language. This can lead to ambiguous statements with unclear objectives. Point 1: does the word inoculate mean using a single yeast colony from a solid media plate or can the liquid YPAD (Yeast extract, Peptone, Adenine hemisulfate, Dextrose) be inoculated from another previously inoculated YPAD liquid culture? The statement incubate with shaking, is this at 20 rpm or 400 rpm? Does overnight mean 12 or 24 h? Point 2: it states count overnight culture, which only suggests that the person executing the protocol needs to calculate approximately how many cells are present in the overnight culture. How this estimate is achieved is not stated. Having statements which can be interpreted in different ways introduce inconsistencies in how the protocol is executed. This can introduce noise and contribute to inaccurate findings. In this instance the author has nothing to gain from omitting information from the method. The target audience of this protocol are yeast biologists and therefore the author assumes that the reader has a degree of prior knowledge. However, this is not always the case. A researcher new to this field may mis-interpret a statement resulting in an inaccurate objective. It is also possible to envisage information being deliberately left out of published protocols, particularly if omitted information reduces the impact of the findings. This information could suggest that the findings cannot be reliably reproduced or could reflect poorly on the success rate of a novel technique.
2.2 Existing approaches
The requirement for an efficient representation of experiment protocols is recognized as a pressing problem, and several other projects are applying ontologies to the formalization of knowledge about experiment data. The Microarray Gene Expression Data (MGED)1 ontology is one of the pioneering attempts to use an ontology to record information about experiment data (Whetzel et al., 2006b). The MGED ontology was designed to formalize the descriptors required by the Minimum Information About a Microarray Experiment (MIAME)2 standard for capturing core information about microarray experiments (Brazma et al., 2001). Many journals (∼50 thus far3) require {MIAME} compliant data as a condition for publishing microarray-based papers. This is a trend that looks set to continue for other accepted ontological standards. The Minimum Information About Proteomics Experiment (MIAPE) standard supports proteomic experiments (Taylor et al., 2007). The Metabolomics Standards Initiative (MSI) ontology working group is building an ontology to facilitate the consistent annotation of metabolomic experiments (Sansone et al., 2007).
Minimum Information for Biological and Biomedical Investigation (MIBBI) is a web-based resource designed to act as a one-stop-shop for those seeking for or looking to contribute to Minimum Information (MI) checklists.4 According to MIBBI's website these checklists are intended to promote transparency in experiment reporting, enhance accessibility to data and support effective-quality assessment, thereby increasing the value of a body of work. The MIBBI project maintains a web-based resource for extant checklist projects, complementary data formats, tools, controlled vocabulary and databases. MIBBI aims to provide guidelines for checklist development, both by increasing connectivity between MI checklist development projects, and by disseminating best practise both in relation to process (such as open mechanisms to receive and respond to public comment) and presentation (e.g. use of shared language, documentation style and structure, production of user-friendly summaries). Checklists developed using MIBBI guidelines focus largely on capturing the minimum information needed to usefully annotate data generated by biological/biomedical investigations. Information pertaining to wet-lab processes such as experiment execution(s) is described using natural language with the aid of some controlled vocabulary. However, the degree of information granularity in these descriptions is thus far at the authors discretion. These descriptions may be sufficient to make effective use of reported data but are likely to be insufficient to independently repeat the experiment actions used to generate the date.
PRoteomics IDEntification (PRIDE)5 is a data repository, supported by a combination of tools, standards and infrastructure for the description of proteomic data (Martens et al., 2005). PRIDE's schema presents a minimum of information about protein identifications. PRIDE's top level structure contains the part <protocol description> annotated with key words used to mark the type of method used to generate the proteomic data.
MGED, MIAME, MIAPE and PRIDE are ontologies primarily focused on developing controlled vocabulary and descriptors for high-throughput strategies such as mass spectroscopy and array-based comparative binding assays. These ontologies are centred around the annotation of data. Protocol information is only present at a level of detail, which is sufficient to describe the data. None of these projects provides a detailed enough formalism for the representation of experiment actions.
The Functional Genomics Investigation Ontology (FuGO) project Whetzel et al., 2006a), and its successor the Ontology for Biomedical Investigations6 (OBI) project, are developing an integrated ontology for the description of biological and medical experiments and investigations. This ontology aims to model the design of an investigation, including the protocols, instrumentation, materials used and the data generated. OBI has not yet been released, but it already has the key classes for the description of protocols: <OBI: investigator>, <OBI: instrument>, <OBI: biomaterial entity>. The generic ontology of scientific experiments (EXPO) aims to formalize domain-independent knowledge about the organization, execution and analysis of scientific experiments (Soldatova and King 2006). This ontology has the class <EXPO: experiment action> and defines some of its properties: has Goal, has Object, has Instrument, but there are no subclasses specified. Our proposal differs from the existing ontology-based approaches for the description of experiment protocols by suggesting a meta-language for the description of experiment actions and their properties. EXACT provides a formalized representation of the domain that is not sufficiently covered by any other ontology.
In theoretical computer science, process algebras have been used to specify and reason about descriptions of processes and actions. Process algebras are algebraic systems for the manipulation of elements of processes (the individual elements being actions or events). They define laws governing the sequencing, composition and synchronization of actions. Leading examples of process algebras are the Communicating Sequential Processes (CSP), the Calculus of Communicating Systems (CCS) and the Algebra of Communicating Processes (ACP) (Bergstra and Klop 1984; Hoare 1985; Milner 1980). For example, a process algebra would provide laws stating that:
is equivalent to the choice of
Process algebras in biology have generally been used as modelling languages for biological systems rather than as a way to specify experiment actions. The ontology we propose provides much more detail than process algebras are usually designed to give, but could be used together with a suitable process algebra for verification and other algebraic reasoning over protocols. The process algebra for biological protocols would need to represent parameterized actions (e.g. to incubate at 30○C). It would also require the ability to represent state changes (e.g. CSP∥B, Treharne and Schneider 2002, which combines the CSP representation of processes with the B formal language to represent changes in state).
Logics for agency have also considered some of the issues that we deal with in this work. In particular agents are described by their actions and goals (desires/intentions). There are many logics for agency, each allowing different expressiveness and covering areas such as belief, knowledge, possibility, time, branching, the relationships between actions and goals and the distinction between understanding what must be done and why it must be done.
In EXACT we first provide a vocabulary and ontology, and then begin to look at the grammatical aspects of describing experiment actions.
2.3 Our proposed solution
To develop EXACT, we first analysed protocols from several bio-medical domains, including functional genomics, metabolomics and drug screening, as well as protocols published in Nature Protocols7. We then consulted with biologists, microbiologists, biochemists and chemists with experience in the execution of these protocols to clarify ambiguous statements and to enrich the protocols with as much information as possible. This helped to capture the precise meaning of each experiment action performed. General concepts were abstracted from these experiments actions and were used to develop the ontological classes. The scientific experts then used these classes to try and represent their own protocols. After many painstaking rounds of consultation, classes were added and removed or changed in the ontology to help better represent the actions performed in various protocols. The EXACT hierarchy of experiments is sufficient to formalize many of the protocols used in our labs. However, as we formalize more and more protocols using EXACT, its class structure will grow and evolve to meet the needs of new methods and techniques.
3 AN ONTOLOGY OF EXACT
An ontology of EXACT aims to provide a structured vocabulary of concepts for the description of protocols in bio-medical domains. Our ontology intends to be compatible with other formalisms, to share and reuse already formalized knowledge. For example it reuses classes from the phenotypic qualities ontology PATO,8 OBI and the W3C Time Ontology (OWL-Time).9 EXACT is expressed in OWL–DL and was developed using the Protégé ontology editor.10
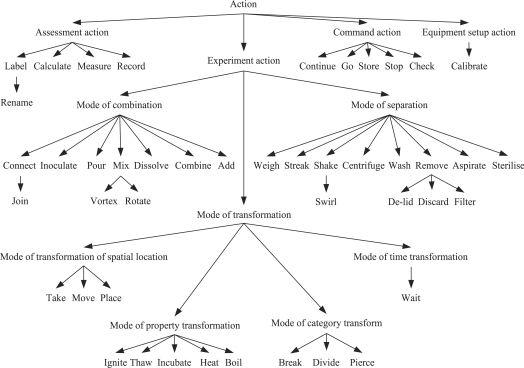
The main part of EXACT is a hierarchy of experiment actions. This hierarchy was created using a classification based on goals of actions. The experiment actions are divided into three groups according to their goals:
separation;
transformation;
combination.
In defining these groups of actions we follow the classes of elementary processes used by Noy (1997). Our approach differs by separating ‘what is done’ from ‘how it is done’. The same goal can be achieved by many different actions. For example, the goal <separation> may be achieved in various ways: by the experiment action <centrifuge>, by the experiment action <filter> or by other actions.
Experiment actions that provide a mode of transformation are classified into the following subclasses:
a mode of property transformation, with such experiment actions as <incubate>, <heat>, <thaw>;
a mode of transformation of spatial location, for example <move>;
a mode of transformation of time, for example <wait>;
a mode of category transformation, with such experiment actions as <break>, <pierce>, <divide>.
Figure 1 shows our classification of experiment actions according to their goals.
Fig. 1.
EXACT classification of experiment actions.
Many experiment procedures have experiment actions that are executed only if a certain condition is valid. For example in our experiments with yeast, the optical density (OD) of yeast cells should be between 0.6 and 1.0 before processing. If the OD is too low, the culture must be incubated for longer. In order to represent conditions, EXACT defines the class <condition> with the sub-classes <if−condition>, <pre−condition>, <post−condition> and <store−condition>. Each condition has a ‘boolean expression’, a ‘yes-command’ for execution if the value of the expression is true and a ‘no-command’ for execution if the value of the expression is false. Pre-conditions and post-conditions can be used to check whether materials and instruments are ready for execution of experiment actions, whether final volumes of solutions are correct, or whether objects are in the correct locations. Such checks during running of experiments are important to prevent errors. Store conditions are used to indicate when it is possible to temporarily stop execution of the protocol and put materials in a store under the defined storage requirements.
EXACT also defines a set of command actions, which control the flow of execution of the protocol: <continue>, <stop>, <check>, <store> and <go>. <Continue> is the null action that does nothing at all (but is useful as the yes-command for pre- and post-conditions, and the no-command for store-conditions). <Stop> terminates the flow of execution immediately and is used as the no-command for pre- and post-conditions. <Check> is used in conjunction with all four conditions described above to test the expression and execute the yes-command or no-command as appropriate. <Store> is the action of storage and is used as the yes-command of the Store-condition. <Go> is a command that moves the flow of execution to an action elsewhere in the protocol. The command actions currently defined by EXACT are minimal and are likely to be enhanced in the future by more complex constructs such as loops.
The class <role> is used in the ontology to describe that some entities can play a certain role. A location can be a start or end location, a piece of equipment can be a start or end container; a location can be a lid location, etc.
Apart from experiment actions that are performed by manipulating dependent variables of an experiment, EXACT defines the class <equipment setup action> with instances that have the goal of preparing for experiment actions. These actions are considered as preliminary to later actions. EXACT also defines the class <data action> with instances for recording measurements and observations that have the goal of preservation of information. EXACT includes a hierarchy of instructions with the classes <warning> e.g. <flammable> and <caution> e.g. <critical step> (taken from Nature Protocols) that can be ignored by automated agents executing protocols, but warn human users to take extra care.
EXACT is available in two versions: EXACT/EXPO is compliant with EXPO (Soldatova and King 2006) and more suitable for automated laboratories; EXACT/OBI is more suitable for using within OBO communities. EXACT/OBI provides an explicit mapping to OBI (the current draft March, 2008).
The principal difference between these two versions is in philosophical foundations. OBO ontologies are based on a philosophy of reality and do not include abstract entities. This does not put considerable restrictions on the description of the existing protocols as most protocols are designed for execution of experiments in the real physical world by manipulating real physical objects. The results of our research show (King et al., 2004; Soldatova et al., 2006; Whelan and King 2008) that the representation of logical and mathematical objects (i.e. sets, relations, facts) and other entities within a computer system as abstract entities provides a clearer description of computational experiments and experiments executed in automated laboratories.
Philosophers have argued about fundamental ontological questions for at least two and a half thousand years, and we do not wish to enter these debates. What we need to do is to make practical decisions about how best to describe protocols. We believe that supplying different versions of EXACT is the best way to deal with conflicting upper ontologies. Hopefully the two versions of EXACT can be merged when a philosophical solution is found that is suitable for all needs.
EXACT/EXPO has only two abstract entities <true value> and <false value>, which are defined as the value of a statement that corresponds/does not correspond to reality. These classes are used in pre- and post-conditions of actions. EXACT/EXPO is designed to be compliant with an ontology for automated laboratories, which we are developing at Aberystwyth, UK. In EXACT/OBI these classes are defined as subclasses of the class <information entity>, which was recently introduced into OBI (January, 2008). The class <OBI: information entity> is a subclass of the class <BFO: generically dependent continuant>. Table 2 shows an explicit mapping between the top classes of the two EXACT versions.
Table 2.
EXACT/EXPO – EXACT/OBI mapping of the top classes
| EXACT/EXPO | EXACT/OBI |
|---|---|
| <process> | <BFO: occurent> |
| <object> | <BFO: continuant> |
| <proposition> | <OBI: information entity> |
| <quality> | <BFO: quality> |
| <role> | <BFO: role> |
| <abstract entity> | <OBI: information entity> |
EXACT/OBI defines a mapping of the EXACT/EXPO top classes to the leaf classes of Basic Formal Ontology (BFO)11 (Grenon and Smith 2004) without considerable loss of semantics and can be reused within OBO ontologies.
EXACT aims to follow OBO Foundry principles:12 ‘the ontology is open and available to use by all’, ‘is in a common formal language’, ‘includes textual definitions of all terms’, ‘uses relations which are unambiguously defined’, it is orthogonal to OBO ontologies and it follows the naming convention of (Schober et al., 2007).
The current version of EXACT does not yet include axioms. We are collecting statements about experiment actions and plan to include them in the form of axioms in the next version. Here are some examples of such statements:
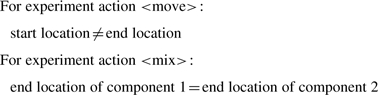 |
The second statement tells us that in order to mix components, the components must be moved to the same location.
Apart from the well-defined foundational relations is_a and part_of, EXACT includes the relations from the OBO Relational Ontology (RO) (Smith et al., 2005) located_in, has_participant and has_agent, the relations has_role and has_quality that are used in OBI, DOLCE (Gangemi et al., 2003) and HOZO (Kozaki et al., 2002), and a relation has_proposition (or has_information for the EXACT/OBI version).
The specialization of BFO in representing real world entities is reflected in the set of RO relations. RO relations are not suitable for linking physical entities to information entities. The set of RO does not allow to easily represent such knowledge as ‘an experiment action has a goal’, ‘an action is conditional’, ‘an investigator has a plan’. EXACT includes the relation has_proposition to fill this gap. The relation allows the connection of an agent of a process with a certain portion of information that is essential for participating in the process. We define this relation following the methodology suggested in Smith et al. (2005). First, we add one more relation to the ‘pain of infinite regress’ of primitive instance-level relations:
There is a primitive relation between an agent of a process, a proposition and a time, where a is an agent, p is a process, i is a proposition and t is time.
Second, we define a class-level relation using this primitive instance-level relation:
 |
where A and I are classes of agents and propositions. We can express ‘an experiment action has a goal’ with this relation as follows: an agent of an <experiment action> has_proposition <goal>.
EXACT is a modular ontology. It defines a conceptual scheme for describing experiment actions and their properties. To represent individual experiment actions it is necessary to import individuals of the classes <object>, <equipment>, <location> and <method>. These classes are part of EXACT. Individuals of these classes are stored in the corresponding knowledge bases. An example of a protocol with a sequence of particular experiment actions in OWL-DL can be found on the EXACT website: http://www.aber.ac.uk/compsci/Research/bio/dss/EXACT/.
4 EXAMPLES OF FORMALIZED PROTOCOLS
4.1 Example 1: competent-cells protocol
The excerpt in Table 3 from the competent-cells protocol is structured as a series of experiment actions explicitly stating what the user must do step by step. All objects used in the experiment actions for example YPD media bottle, yeast culture flask are defined as instances of the class <object>. All locations for example laminar flow hood, cold room are defined as instances of the class <location> (more precisely as objects playing a role of <location>). Each particular instance of an experiment action has to specify values of all parameters. The action move 12 is an instance of the class <move>, which is defined in EXACT as ‘an experiment action to change a spatial location of an entity from a start location to an end location’. In order to specify an instance move 12, it is required to specify a start location (=store), end location (=laminar flow hood) and an object of the action—the entity that is changing location (=YPD media bottle).
Table 3.
EXACT competent-cells protocol (a fragment)
| Operating procedure: grow yeast culture | |
| pre-condition: sealed yeast colonies plate located_in cold room | |
| pre-condition: YPD media bottle located_in cold room | |
| experiment action: | move 12 |
| object: | YPD media bottle |
| start location: | in store |
| end location: | in laminar flow hood |
| experiment action: | move 13 |
| object: | 500ml conical flask |
| start location: | in store |
| end location: | in laminar flow hood |
| experiment action: | move 14 |
| object: | sealed yeast colonies plate |
| start location: | in cold room |
| end location : | in laminar flow hood |
| experiment action: | add 15 |
| component 1: | YPD medium |
| volume: | 50ml |
| start container: | YPD media bottle |
| end container: | 500ml conical flask |
| equipment: | pipette |
| experiment action: | rename 16 |
| old name: | 500ml conical flask |
| new name: | YPD conical flask |
| experiment action: | add 17 |
| component 1: | single yeast colony |
| volume: | small volume |
| start container: | sealed yeast single colonies plate |
| end container: | YPD conical flask |
| equipment: | inoculating loop |
| experiment action: | rename 18 |
| old name: | YPD conical flask |
| new name: | yeast culture flask |
| experiment action: | move 19 |
| object: | yeast culture flask |
| start location: | in laminar flow hood |
| end location: | in incubator |
| experiment action: | incubate 20 |
| object: | yeast culture flask |
| equipment: | shaking incubator |
| rpm: | 200 |
| temp: | 30○C |
| time interval: | 12–24h |
| goal: | grow yeast until medium becomes cloudy |
| Post condition: | yeast culture located_in incubator |
In the EXACT formalism laboratory protocols are divided into many operating procedures. Prerequisite objects for each operating procedure are represented in <pre−condition> and objects created as a result of executing an operating procedure are represented in <post−condition>. In the above operating procedure grow yeast culture, pre-conditions include sealed yeast colonies plate located_in cold room and YPD media bottle located_in cold room, where sealed yeast colonies plate, YPD media bottle are instances of the class <object>, cold room is an instance of the class <location>, and located_in is a defined relation. Therefore in order to execute the operating procedure grow yeast culture the user must have first executed one or more operating procedures where the post-conditions include sealed yeast colonies plate located_in cold room and YPD media bottle located_in cold room. This provides the protocol user with the knowledge of exactly what he/she needs to have in place before commencing. The <move> action ensures that each object is in the correct location. For example, when adding YPD to a conical flask, first both objects are moved to the laminar flow hood. Similarly, a yeast culture flask cannot be incubated if it is not first moved to an incubator.
The action <rename> was used to represent a change in an object's state. A 500ml conical flask changes to YPD conical flask when YPD is added to the flask. The <rename> action was put in place to make the protocol easier to follow when being executed by a human. It has no significance when the protocol is being executed by laboratory robotics.
Figure 2 illustrates the difference between the original text book representation of a portion of this protocol, the detailed EXACT representation, and a basic text representation generated automatically from the EXACT representation.
Fig. 2.
(1) Protocol from Methods in Yeast Genetics. (2) Protocol represented using EXACT. (3) Text generated automatically from EXACT representation.
4.2 Example 2: Formalized protocols for commissioning of equipment
EXACT has also been used to formalize protocols to assist with the commission of laboratory-automation equipment. The Computational Biology group at Aberystwyth (UK) is in the process of purchasing robotic equipment for the automated screening and design of drugs. As part of this procedure, protocols were created describing the work that the robotic equipment would need to perform. If a robotic system is to automate a protocol it will need every temperature, every movement and every decision fully specified. Explicitly describing protocols that were always intended to be automated forced us to be precise and this helped the development of EXACT.
We applied EXACT to define which experiment actions, with which properties, were necessary to achieve the planned goals. This enabled us to specify what type of equipment, and what functionality, was required to execute the planned investigations. The level of detail that can be expressed in EXACT corresponds to the level usually represented by the control software for managing integrated laboratory-automation systems. This is the level at which the protocols become concrete, well-defined and implementable. We sent these protocols to companies that sell laboratory automation equipment (such as Tecan, Beckman, Hamilton, FluidX, Matrix and many others), as specifications of what we wanted to achieve. Several of these companies then obliged us with demonstrations of how their equipment could meet the protocols.
As an example, one of our compound library replication protocols is available on the EXACT website.
The strict specification of protocol elements helped us to recognize inconsistencies and potential problems with equipment. For example: a lack of space for a lid location. Equipment demos are often done using plates without lids, causing de-lidding operations to be skipped. The action of de-lidding must involve a lid location property. The lid location must be available, reachable by the robot and must not obstruct other operations. As another example, experiment actions such as <discard> may not seem important for demos, but inefficient execution can cause serious problems in future investigations. The strict description of all experiment actions forces one to pay attention to all operations.
Currently there is no standard language for programming the protocols for automated-laboratory systems, no single language that all laboratory equipment understands. Each device has a proprietary driver, and these are generally linked into an overarching software system by a laboratory integration specialist, who will provide a domain specific language for end users to represent the protocol they need to run on the system. Each of these languages provides its own functionality and vocabulary. EXACT provides a vocabulary at a particular level of detail useful for specification: for example we have an experiment action: <incubate> which has properties describing the temperature, shaking speed and duration, but does not specify the lower level of which serial commands to use, which location in the incubator should be used, or what to do if the incubator should raise an error.
5 APPLICATION
5.1 Validation
EXACT is extremely valuable as a language for the initial specification of an automated system, because it forces the removal of ambiguity and can be used for validation. As an example, we have implemented EXACT in the programming language Haskell. This allows an EXACT specification to be executed and tested. An example of a part of the competent-cells protocol is implemented in Figure 3.
Fig. 3.
An example of part of the competent-cells protocol, implemented in Haskell, using EXACT.
The implementation in Haskell allows actions to be combined with other actions to create an ‘operating procedure’ which is itself a (complex) action to be combined with others. Each action may modify the state of the equipment and write a description to a log. This description can be used as a simple text representation of the formalized protocol. The state updates and existence of equipment and locations can be validated during the execution of the protocol, and the protocol must typecheck in order to be a valid Haskell program (all necessary properties of actions must be defined).
The other benefit of a Haskell implementation of EXACT is as a tool to test the validity of the ontology itself. The semantics of conditions and command actions can be examined and refined. The type system enforces and makes clear the distinction between materials, equipment and locations, but also demonstrates that some locations are created from equipment, that equipment can contain materials, and that when materials are combined in a container, the container may hold a new material that has been created from the combination.
Other approaches also exist that can assist in the validation of protocols. The use of agency logics may be able to provide proof by axioms or by model checking that the protocols give the correct results and are achievable. However we would need efficient and practical implementations of such logic-based reasoning. The relation between logic theory and practical approaches is still unclear (de Boer et al., 2007). Some logics do not allow the expression of how an action is achieved, only what is achieved. Several examples of families of logics that may be suitable to enhance EXACT in the future include Belief-Desire-Intention (BDI), Knowledge, Actions, Results and Opportunities (KARO) and ‘Sees To It That’ (STIT) (Troquard et al., 2006; van der Hoek and Wooldrige 2003). However, logics for agency have a different emphasis than the work of EXACT, namely that they describe the underlying causes of agent behaviour rather than provide a language for precise description of actions.
5.2 Tools
Good tools are vital to the adoption of standards. If we expect biomedical scientists to unambiguously define their protocols we must give them tools that are easy to use. Fully formalized protocols will span many pages of text. Generating such descriptions by hand is labour-intensive, error-prone and uninspiring. We need tools for generating, validating, viewing and reasoning with protocols.
Protocol-generation tools should:
provide an intuitive graphical user interface;
automatically enforce the vocabulary of EXACT;
supply default values and allow reuse of existing protocols.
Protocols that are formally defined should be validated before being accepted for publication. Tools for validation should ensure that:
all equipment and objects have defined initial locations and properties;
names for equipment and objects are consistently used;
locations of objects and equipment are consistent (a flask cannot be moved from the bench to the cold store and then from the incubator to the laminar flow hood);
properties of equipment are valid (if you have only one incubator then it cannot be used at two different temperatures at the same time);
biological materials exist and are available (a plate cannot be used as a source of yeast culture if yeast has not been added to it previously);
stated pre-conditions/post-conditions for each subpart of the protocol can be met by the protocol as a whole.
Text generation tools are also needed. Usually a biologist will not require a full description of a protocol, and will prefer a much higher level summary, but may require clarification of certain steps. For this we would like a tool that can translate from a fully specified EXACT protocol into a summarized human-friendly readable format, with the option of expansion of any instruction for more detailed information.
Given a formalized protocol, useful tools would generate equipment lists and their necessary range of settings, calculate timings and storage points that are friendly to a biologist's working-hours, and compare two or more published protocols and state how and where they differ.
6 DISCUSSION AND CONCLUSION
Laboratory-based scientific experiments must, by definition, be repeatable. However, many, perhaps most, scientific protocols in the literature are so poorly described and deficient in information that their exact repetition is impossible. Indeed, many experiment protocols bear more resemblance to recipes in cook books than to detailed scientific methodologies. And even in bioinformatics, where experiments may be wholly computational, it is often very hard to obtain enough information to fully repeat an experiment. This unhappy situation is being made worse by the unfortunate trend in scientific journals to downplay the ‘Methods’ section, moving it from its traditional place after the introduction to the end, reduce its font, move it into ‘Further information’, etc.
This careless/vague description of experiment protocols was perhaps viable when molecular biology focused on qualitative experiments: the correct result being indicated by a band on a gel in the correct place, a colony growing, etc. Such experiments were routinely executed in batches of 10 or 20. However, with the ever increasing importance of quantitative methods such as microarrays (where numerical values have to be interpreted as biological observations and tens of thousands of experiments are executed simultaneously) the precise and unambiguous description of experiment protocols is essential.
We propose the EXACT ontology as the basis for the description of protocols. We followed the current best practice in ontology development by not allowing multiple inheritance, providing definitions for all classes and relations, using top-level classes (Rosse and Mejino Jr. 2003; Smith et al., 2005; Soldatova and King 2005). We have demonstrated the utility of EXACT to represent drug screening and functional-genomic protocols.
The EXACT hierarchy of the experiment actions is currently sufficient to formalize many of the protocols used in our Computational Biology Group. However, more work is required before it is sufficiently comprehensive to be able to represent all protocols in laboratory biology. We are currently working on the development of tools that will make the generation of these protocols easy for biologists. We also have to define a consistent language for specifying the flow of execution through the protocols and the relationship of our work to process algebras.
It is intrinsically valuable to describe one's own experiments in a precise and unambiguous way as it provides a clear record of what one has achieved. However, the value of describing protocols clearly is greatly amplified by being able to exchange and compare protocols. Ontologies provide a basis for such a shared understanding. We therefore envisage developing an EXACT repository as a place where investigators and practitioners can accumulate their knowledge about representing protocol actions. We invite researchers from all areas to participate in the development of an ontology of experiment actions and to contribute to an Open Source project for the formalized representation of protocols.
ACKNOWLEDGEMENTS
We would like to acknowledge RC UK, RAEng/EPSRC, and BBSRC for providing funding to accomplish this work.
Conflict of Interest: none declared.
Footnotes
3MIAME journals: http://www.mged.org/Workgroups/MIAME/journals.html
4MIBBI: http://mibbi.sourceforge.net/
5PRIDE: http://www.ebi.ac.uk/pride/
7Nature Protocols: http://www.nature.com/nprot/
9OWL-Time: http://www.w3.org/TR/owl-time/
10Protégé: http://protege.stanford.edu
11BFO: http://www.ifomis.org/bfo
12OBO Foundry: http://ontoworld.org/wiki/OBO_foundry
REFERENCES
- Akada R, et al. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast. 2006;15;23:399–405. doi: 10.1002/yea.1365. [DOI] [PubMed] [Google Scholar]
- Amberg DC, et al. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Bergstra JA, Klop JW. Process algebra for synchronous communication. Inform. Control. 1984;60:109–137. [Google Scholar]
- Brazma A, et al. Minimum information about a microarray experiment MIAME-toward standards for microarray data. Nat. Genet. 2001;4:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- de Boer FS, et al. A verification framework for agent programming with declarative goals. J. Appl. Logic. 2007;5:277–302. [Google Scholar]
- Gangemi A, et al. Sweetening ontologies with DOLCE. AI Magazine. 2003;24:13–24. [Google Scholar]
- Grenon P, Smith B. SNAP and SPAN: towards dynamic spatial ontology. Spat. Cogn. Comput. 2004;4:69–103. [Google Scholar]
- Hoare CAR. Communicating Sequential Processes. Prentice Hall; 1985. [Google Scholar]
- King RD, et al. Functional genomics hypothesis generation by a Robot Scientist. Nature. 2004;427:247–252. doi: 10.1038/nature02236. [DOI] [PubMed] [Google Scholar]
- Kozaki K, et al. Hozo: an environment for building/using ontologies based on a fundamental consideration of ‘role’ and ‘relationship’. In. Knowledge Engineering and Knowledge Management. 2002:213–218. [Google Scholar]
- Martens L, et al. Pride: the proteomics identifications database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- Milner R. A Calculus of Communicating Systems. Springer Verlag; 1980. [Google Scholar]
- Noy N. Ph.D. thesis. USA: College of Computer Science, Northeastern University; 1997. Knowledge Representation for Intelligent Information Retrieval in Experimental Sciences. [Google Scholar]
- Rosse C, Mejino JLV., Jr A reference ontology for bioinformatics: the Foundational Model of Anatomy. J. Biomed. Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Sansone S, et al. Metabolomics standards initiative – ontology working group. work in progress. Metabolomics. 2007;3:249–256. [Google Scholar]
- Schober D, et al. Towards naming conventions for use in controlled vocabulary and ontology engineering. In. Proceedings of BioOntologies SIG, ISMB07. 2007:29–32. [Google Scholar]
- Smith B, et al. Relations in biomedical ontologies. Genome Biology. 2005;6:R46:1–15. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatova LN, King RD. Are the current ontologies used in biology good ontologies? Nat. Biotechnol. 2005;9/23:1096–1098. doi: 10.1038/nbt0905-1095. [DOI] [PubMed] [Google Scholar]
- Soldatova LN, King RD. An ontology of scientific experiments. J. R. Soc. Interface. 2006;3/11:795–803. doi: 10.1098/rsif.2006.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatova L, et al. An ontology for a Robot Scientist. Bioinformatics (Special issue for ISMB) 2006;22/14:e464–e471. doi: 10.1093/bioinformatics/btl207. [DOI] [PubMed] [Google Scholar]
- Taylor CF, et al. The minimum information about a proteomics experiment (MIAPE) Nat. Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- Treharne HE, Schneider S. Communicating B machines. In ZB2002: International Conference of Z and B Users. 2002 [Google Scholar]
- Troquard N, et al. Towards an ontology of agency and action : from STIT to OntoSTIT+. In. 2006:179–190. International Conference on Formal Ontology in Information Systems (FOIS), Baltimore, Maryland, USA. [Google Scholar]
- van der Hoek W, Wooldrige M. Towards a logic of rational agency. Logic Journal of the IGPL. 2003;11:133–157. [Google Scholar]
- Whelan KE, King RD. Using a logical model to predict the growth of yeast. BMC Bioinformatics. 2008;9(97) doi: 10.1186/1471-2105-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetzel PL, et al. Development of FuGO: an ontology for functional genomics investigations. OMICS. 2006a;10:199–204. doi: 10.1089/omi.2006.10.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetzel PL, et al. The MGED ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006b;7:866–873. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]