Abstract
Motivation: More and more genomes are being sequenced, and to keep up with the pace of sequencing projects, automated annotation techniques are required. One of the most challenging problems in genome annotation is the identification of the core promoter. Because the identification of the transcription initiation region is such a challenging problem, it is not yet a common practice to integrate transcription start site prediction in genome annotation projects. Nevertheless, better core promoter prediction can improve genome annotation and can be used to guide experimental work.
Results: Comparing the average structural profile based on base stacking energy of transcribed, promoter and intergenic sequences demonstrates that the core promoter has unique features that cannot be found in other sequences. We show that unsupervised clustering by using self-organizing maps can clearly distinguish between the structural profiles of promoter sequences and other genomic sequences. An implementation of this promoter prediction program, called ProSOM, is available and has been compared with the state-of-the-art. We propose an objective, accurate and biologically sound validation scheme for core promoter predictors. ProSOM performs at least as well as the software currently available, but our technique is more balanced in terms of the number of predicted sites and the number of false predictions, resulting in a better all-round performance. Additional tests on the ENCODE regions of the human genome show that 98% of all predictions made by ProSOM can be associated with transcriptionally active regions, which demonstrates the high precision.
Availability: Predictions for the human genome, the validation datasets and the program (ProSOM) are available upon request.
Contact: yves.vandepeer@psb.ugent.be
1 INTRODUCTION
Currently, the genomic sequence of over 50 eukaryotic organisms is available. Many more sequencing projects are to be finished in the next few years (Liolios et al., 2006), and so it becomes increasingly important to automate the identification of functional elements, such as genes and regulatory sequences. For protein coding sequences, there are many complementary approaches that can accurately identify the coding part of the gene (Brent, 2008; Guigó et al., 2006). For regulatory sequences, such as transcription factor binding sites, many methods have been developed with increasing success relying on motif searches and/or comparative techniques (Elnitski et al., 2006). However, the identification of the core promoter region and the localization of the transcription start site (TSS) remains a difficult problem (Bajic et al., 2006; Sonnenburg et al., 2006; Wang et al., 2007a; Xie et al., 2006). In light of the many genomes that become available, it is important to accurately identify these regions, as better core promoter predictions will improve the genome annotation and allow a better understanding of the transcription-initiation process.
The core promoter is the region immediately upstream of the TSS, where the transcription-initiation complex assembles (Pedersen et al., 1999; Smale and Kadonaga, 2003). It is located upstream of the coding part of the gene, sometimes up to several thousand base pairs, and is responsible for basal transcription as well as transcriptional regulation of the gene it is linked with (Choi et al., 2004). Genes encoding structural or regulatory RNAs instead of proteins also contain a core promoter, but have no coding part. This is often problematic for the current promoter prediction programs (PPPs) that are often trained to recognize the 5′ end of protein coding genes (Bajic et al., 2004).
Recently, it has been shown that genes usually do not have a single TSS. This finding is well documented in humans as an outcome of genome-wide investigations on gene expression (Carninci et al., 2006; The ENCODE Project Consortium, 2007). Most human genes have multiple promoters and each promoter has multiple TSSs (Frith et al., 2008; Sandelin et al., 2007). For most genes, transcription starts in a cluster of positions, with some positions favored over others. The choice of which alternative promoter to use depends on the conditions in the cell. The use of alternative promoters results in the complexity and diversity of the human transcriptome (Kawaji et al., 2006). All these studies imply that core promoter prediction techniques should not try to predict a single TSS but rather a cluster of TSSs. In addition, the use of alternative promoters depends on the promoter type: conserved tissue-specific promoters have a better defined TSS and are TATA-enriched, while more loosely defined CpG-rich promoters have a broader promoter region, often with multiple alternative promoters and many TSSs (Carninci et al., 2006).
Core promoters have distinct features that can be used to distinguish them from other sequences. On a nucleotide level, several motifs have been linked to the core promoter in eukaryotes. The best-known motif, the TATA box, is mainly present in tissue-specific genes. Estimations of the number of TATA-containing promoters in the human genome range from 5–30% (Deng and Roberts, 2005; Florquin et al., 2005; Smale and Kadonaga, 2003). Other motifs that are related to the core promoter are, among others, the initiator element and the TFIIB recognition element (Deng and Roberts, 2005). These motifs have specific consensus sequences that can be used to identify the motif. When scanning all promoters, only a limited number of promoters contain the motif as defined by the consensus sequences. Furthermore, when screening the whole genome for these sequences, many occurences of the consensus do not correspond to a functional site. Besides motifs, also more general sequence-dependent properties are promoter specific. One of the most prominent features is the distinct G+C content in the area around the TSS (Aerts et al., 2004).
Another way to represent the DNA sequence is by using physical properties of DNA. One such property, called base-stacking energy (Ornstein et al., 1978), models the local base-stacking energy. High values denote regions that destack or melt easily. This representation has been used before to characterize the promoter (Abeel et al., 2008; Baldi et al., 1998; Florquin et al., 2005; Kanhere and Bansal, 2005). When using this representation, the promoter has an interesting property: two regions that seem to melt easily are located around −30 from the TSS and on the TSS, and are embedded in a large-scale region that is significantly more stable (Abeel et al., 2008; Goni et al., 2007). The two high-value regions are the location where RNA polymerase II binds, and where transcription starts. The large-scale region is especially interesting because it points to the global variation of G+C around the TSS. We used this large-scale feature in earlier work to predict promoter regions in a wide range of species (Abeel et al., 2008). One may argue that the nucleotide information and the sequence-dependent physical properties are two sides of the same coin, but several studies have shown that there are indeed differences and that some properties are better suited for promoter prediction than others (Abeel et al., 2008; Baldi et al., 1998; Florquin et al., 2005; Liao et al., 2000). Future research may focus on aggregating the results of the different properties or on constructing a classifier that takes into account all the properties and selects the relevant ones (Saeys et al., 2007). However, this is not a trivial task and it is not further explored here. Techniques from the field of ensemble learning may prove useful in this context (Polikar, 2006).
In previous research, we have used a single average structural value over a window of 400 bp to predict core promoter regions (Abeel et al., 2008). This single property was based on a large-scale feature of the core promoter that is present in several eukaryotes (Abeel et al., 2008). Here, we use the small-scale properties of the core promoter that have been described, but then not used for promoter prediction. We will revisit these small-scale features below.
We present a novel promoter prediction technique, called ProSOM, that uses an unsupervised self-organizing map (SOM) to distinguish core promoter regions from the rest of the genome. Compared to other PPPs, ProSOM results in significantly fewer false predictions, a problem that has been inherent to promoter prediction since the very beginning (Fickett and Hatzigeorgiou, 1997). Furthermore, we propose a new validation strategy for promoter prediction, taking into account comparisons with experimentally verified TSS data. This novel validation strategy results in a more realistic evaluation of promoter predictors, and has the potential to become a new evaluation standard for future promoter predictors.
2 MATERIAL AND METHODS
2.1 Data
The sequences for the human genome assembly (hg17, May 2004) were retrieved from the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/) (Karolchik et al., 2008).
The cap analysis gene expression (CAGE) datasets have been compiled by Carninci et al. (2006) and retrieved from the Fantom3 project (http://fantom.gsc.riken.go.jp/). The dataset was compiled with the CAGE technique (Shiraki et al., 2003) and covers the entire human genome. This technique is based on the preparation and sequencing of concatamers of DNA tags derived from the initial 20 nucleotides of 5′ end mRNAs. It allows high-throughput analysis of gene expression and the identification of TSSs. The CAGE technique maps the TSS more accurately than other techniques. However, to remove any false hits from the CAGE technique we only retained tag clusters with at least two mapped tags, resulting in 123 400 unique TSSs for humans.
The Ensembl gene annotation was retrieved using the BioMart tool for Ensembl release 37 (Flicek et al., 2008).
The ENCODE regions and annotation were retrieved from the ENCODE home page (http://genome.ucsc.edu/ENCODE/) (The ENCODE Project Consortium, 2007).
For the training of the SOM we retrieved promoters, and transcribed and intergenic sequences. The sequences for promoters were extracted from the DBTSS database (Wakaguri et al., 2008). From the 1250 bp long sequences provided in this database, we extracted the region [−200, 50] around the TSS. Sequences containing ambiguous symbols were discarded. This resulted in 30 964 sequences. The transcribed and intergenic sequences were extracted from the genome assembly. We used the gene coordinates retrieved from Ensembl using the BioMart tool. From the edges of these sequences we stripped 5000 bp to remove border signals. Finally we selected 30 000 sequences of 250 bp on random locations from the set of transcribed and intergenic sequences. The large number of sequences lowers the influence that the few TSSs that may still be present in this set have on the outcome.
In total, we have three datasets, one with promoter sequences, one with intergenic sequences and one with transcribed sequences. Each of these sets contains 30 000 sequences, or slightly more in the case of the promoter set.
2.2 ProSOM implementation
ProSOM was implemented using the Java language version 1.5. The SOM implementation was taken from the Java Machine Learning Library (http://java-ml.sf.net/) and modified to suit our needs. This library is also required to run ProSOM. The program has no size constraints on the number of sequences that can be processed and is designed to run on multiple machines in parallel to process different sequences.
2.3 Structural profiles
The structural profile of a set of DNA sequences is calculated in two steps. First, the nucleotide sequence is converted into a sequence of numbers (i.e. a numerical profile) by replacing each dinucleotide with its energy value, which is obtained from experimentally validated conversion tables. We have used the conversion tables for base-stacking energy from Florquin et al. (2005). Thereafter, for each position, we take the average over all sequences for that position. The resulting numeric vector is called a structural profile.
While we take an average of several thousand sequences to calculate the profiles in the SOM, this is not possible for the sequences we extracted from the genome. The sequences for which we want to make predictions, are smoothed using a 3 bp sliding window. This was done to remove some of the noise in the profile of a single sequence.
2.4 Clustering and promoter prediction
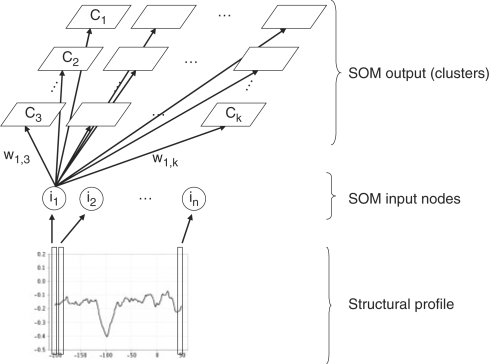
The clustering technique we used is the SOM (Kohonen, 2001), a special type of artificial neural network that can be used both for clustering and class prediction. An SOM consists of a rectangular grid of clusters, each of which has a weighted connection to every input node (Fig. 1). In our case, the input nodes represent the different values of a structural profile associated with a potential promoter region. The SOM provides a mapping from a higher-dimensional feature space (the structural profile) to a lower dimensional cluster space.
Fig. 1.
An SOM for clustering structural profiles. Every position in the structural profile is associated to an input node of the SOM. The output represents the different clusters organized into a grid structure.
During the learning phase, weights are updated by iteratively presenting samples to the network. Using the principle of competitive learning, the Euclidean distance to all weight vectors is calculated for each sample, and the cluster with the most similar weight vector is called the best matching unit (BMU). Subsequently, the weight vector of the BMU and its neighboring clusters in the grid are moved slightly towards this sample. The magnitude of the change decreases with distance of the weight vector from the BMU, resulting in a topological mapping of the input data. In a topological mapping, neighboring clusters represent more similar objects than distant clusters. After many cycles, the network ends up associating clusters with groups in the input data set.
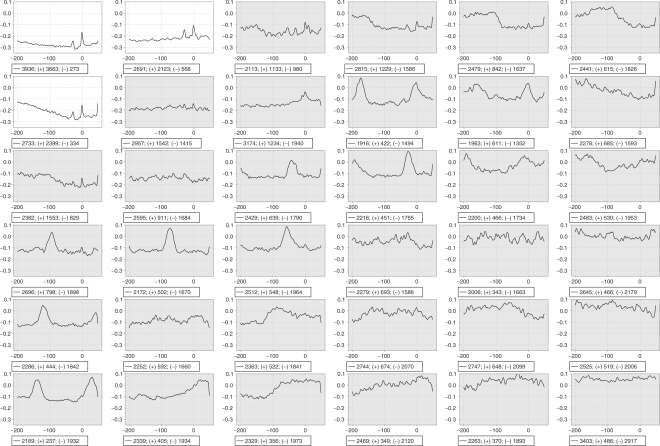
Once the cluster structure is learned, the SOM can be used to map new vectors to their corresponding clusters by determining the cluster whose weight vector is nearest to the new vector. To convert this procedure into a PPP, we determined which clusters are clearly associated with promoters (see Fig. 4 and further in the text) and predicted each sequence that is mapped to one of those clusters as a putative promoter.
Fig. 4.
Result of SOM clustering on a 6 × 6 grid. The profile in each graph is the average of all sequences that map to that cluster. The sequences are converted using the base-stacking energy. The X-axis shows the position relative to the putative TSS. The two left-most clusters in the top row and the left-most cluster in the second row are promoter-rich and show the typical core promoter profile. The promoter-rich clusters are displayed with a white background, the others with a gray one. The legend shows the total number, the number of promoter (+) and the number of other (−) sequences.
2.5 Validation
Our technique was validated on two sequence sets; first on the entire human genome assembly (hg17, May 2004) and secondly on the ENCODE regions. For both sets we retrieved a set of experimentally characterized TSSs and a gene annotation. While the training set and validation set overlap to a certain extent, this has little influence on our validation. The training was unsupervised, so the SOM clustering did not use the sequence type information that was provided with the training data. This means that the clustering was not trained to specifically recognize the sequences in the training set, but it clustered all sequences, and clusters with high promoter content emerged. Because the clustering separates promoters from other sequences in an unsupervised way, it can be used for promoter prediction.
An aggregate measure for the performance of a classifier that is often used in the machine learning field is the F-measure (Van Rijsbergen, 1979). This is the harmonic mean of the recall (sensitivity) and the precision (specificity). The higher this value, the better the classifier is able to separate promoter sequences from other sequences. The recall rate is the number of correctly predicted promoters divided by the total number of promoters. The precision rate is the number of correct predictions divided by the total number of predictions. To assess the number of correctly predicted promoters (true positives, TPs), false predictions (false positives, FPs) and unpredicted promoters (false negatives, FNs), we define the maximum distance that a prediction is allowed to be from the true site. Previous work typically set this number to rather high values of 1000 bp or even 2000 bp. Recently, a more stringent distance of 500 bp was used, and we will also use this more stringent validation for reference to earlier work (Abeel et al., 2008; Goni et al., 2007). For true core promoter prediction validation, predictions should not be further than 50 bp from the actual TSS. Here, we use a validation with a maximum distance of 50 bp to rank the PPPs.
There are two ways to evaluate a PPP. In the classic way to evaluate a PPP (Bajic et al., 2004), one starts from gene annotations, e.g. Ensembl or GENCODE annotation. A TP is then defined as the 5′ end of a gene that has a prediction within 500 bp. A FP is a prediction that lies inside the gene but not within the first 500 bp of the gene. A FN is a 5′ end of a gene that has no associated prediction within 500 bp. All predictions that fall within an intergenic region and are >500 bp from the 5′ end of a gene, are ignored. Unfortunately, this technique has some drawbacks and a new scheme has been proposed to evaluate PPPs (Abeel et al., 2008). A first major drawback of the classic scheme is that all intergenic predictions are ignored, while they may well be false predictions. The view of a single TSS at the 5′ end of a gene, on which this technique is based, is no longer viable in light of many recent studies (Carninci et al., 2006; Frith et al. 2008; Kawaji et al. 2006). We included the validation with the classic technique for reference purposes.
We have proposed a more objective way to assess the performance of a PPP based on the genomewide screening for TSSs (Abeel et al., 2008). This technique is based on the CAGE datasets that have been described above. The dataset contains locations where transcription starts. A TP is a known site that has a prediction within 500 bp of a true TSS, a FN is a TSS without a prediction and a FP is a prediction that has no associated TSS in the reference set within 500 bp. However, for core promoter prediction the 500 bp distance is still too wide. The core promoter is only a very small region, and therefore a much smaller distance should be used when validating predictions. Predictions should be within 50 bp of known sites to be counted as TP. Distances larger than 50 bp have little biological significance when talking about core promoters. For this stronger validation we can only use the CAGE dataset because it is the only one containing experimentally characterized TSSs.
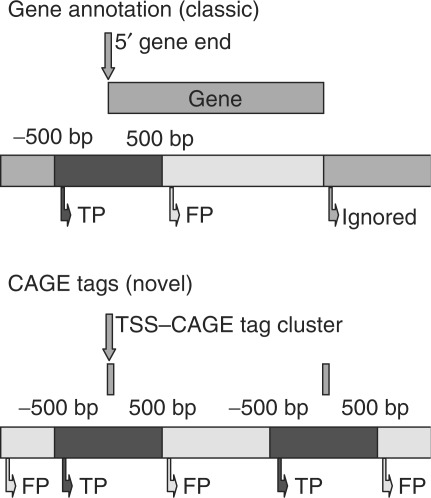
Figure 2 shows the two techniques to validate a PPP. The top panel shows the classic technique where the region around a known gene is depicted. The left side of the gene (suppose to be a forward strand gene) is the 5′ end. Genes with a prediction in the region [−500, +500] around the 5′ end are considered to be TPs, the predictions that fall in the white area [+500, 3′ end] are considered to be FPs. Predictions that fall in the gray area are all ignored in this technique. This will bias the evaluation towards protein-coding gene-oriented software that makes many intergenic predictions, because there is no penalty for false predictions in intergenic regions. The bottom panel of Figure 2 shows the new technique that uses CAGE tags as a reference. Two tags are depicted black. Predictions in the region [−500, +500] around each tag (black) are considered to be correct (TP). All other predictions are FPs. In both cases, genes or tags that have no associated prediction are FNs. For the validation to identify true core promoter predictions, all intervals are reduced to [−50, +50].
Fig. 2.
Validation techniques for PPPs. The top figure shows the classic technique, the bottom figure the new one. Genes or tags that have a prediction inside the black area are considered TPs. Predictions in the white area are FPs. All predictions inside the gray area are ignored. Notice that only the classic technique has ignored regions. For the strict validation the distance is 50 bp.
3 RESULTS
3.1 Sequence profiles
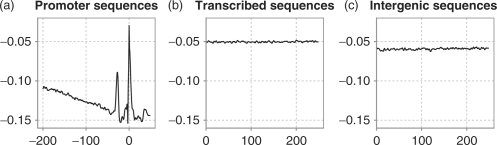
Figure 3 shows the average structural profile of base-stacking energy of the three datasets used for training the SOM. The promoter sequences show a very striking profile with overall lower values than the other two graphs. It has two clear peaks at position −30 (TATA-binding} protein location) and position 0 (TSS). The profile of the transcribed sequences is slightly higher than that of intergenic sequences, which may indicate that, on average, transcribed sequences require slightly less energy to melt than intergenic sequences. However, the difference is small, and no further investigation was done to prove this hypothesis. Our focus is on promoters and their remarkably low profile, which indicates a stable region.
Fig. 3.
Structural profile of promoter (a), transcribed (b) and intergenic (c) human sequences. The profiles are the averages over all sequences in the respective training sets. We used the base-stacking energy as physical property. (a) Region [−200, 50] around the TSS, while for (b) and (c) there is no reference point and the location are numbered from 0 to 250.
The low values in the profile of the core promoter indicate that the region requires a lot of additional energy to melt. This stable region around the TSS is typical for the core promoter (Abeel et al., 2008). However, two regions are less stable. The first region, where the TBP is known to bind, is located ∼30 bp upstream of the TSS. This region is crucial for the assembly of the transcription machinery. The second region is located around the TSS itself, and needs to denature to allow transcription to start. One possible explanation for this stable region can be that it provides a contrasting background for the highly unstable peaks, which are essential for transcription initiation. The overall rigid structure combined with the two peaks can be viewed as a guiding mechanism for the transcription apparatus to select the appropriate site to initiate transcription.
3.2 Unsupervised clustering identifies promoters
We clustered 30 964 promoters from the DBTSS database (Wakaguri et al., 2008), 30 000 intergenic and 30 000 transcript sequences retrieved from Ensembl (Flicek et al., 2008) using the SOM. Several other clustering techniques, such as k-means, ACO, AQBC, Cobweb, DB-scan, EM-clustering, farthest first, OPTICS and x-means, have been tested, but did not result in a proper separation of the different sequence types (results not shown). These techniques are thus not suitable for our promoter prediction task and have not been validated as promoter predictor.
Figure 4 shows the result of the SOM clustering of the 90 964 sequences. Each graph in the figure represents a cluster. On these graphs the X-axis gives the relative position from the TSS for the promoter set, or the position in the sequence for the other sets. On the Y-axis, we see the normalized base-stacking energy. The legend of each graph shows the total number of sequences, the number of promoters (+) and the number of other sequences (−). In the top row, the two left-most graphs contain significantly more promoter than non-promoter sequences. Also the left-most graph on the second row clearly contains more promoter sequences than other ones. Each of these three graphs shows the known core promoter profile with two peaks, one at position −30 associated with the TBP and one on the TSS. Other graphs contain profiles that do not correspond to known core promoter features, as expected, because these clusters contain few promoter sequences.
3.3 Parameter tuning
As stated above, two main parameters require tuning: first, the grid size used to do the clustering, and second, the prediction threshold. We have optimized both by applying the approach to the whole genome for each parameter combination. For the grid size we tested 4 × 4, 5 × 5, 6 × 6 and 7 × 7. Higher and lower values were not tested as the performance follows a normal distribution with a peak at 6 × 6. Each cluster has a different promoter probability. The promoter probability of a cluster is defined as the number of promoter sequences divided by the total number of sequences, e.g. for the top-left cluster in Figure 4 the probability is 0.93 (3663 divided by 3936). Each cluster has a different promoter probability and we tested each of these probabilities as threshold for promoter classification. All sequences that are mapped to a cluster with a promoter probability higher or equal to the threshold will be predicted to be a putative promoter. So when using the promoter probability from the cluster with the highest promoters rate, only sequences that map to that specific cluster will be predicted to be promoters. When using a lower threshold, all sequences that map to that cluster or to a cluster with a higher promoter probability will be predicted as putative promoters.
Table 1 shows the results for the different grid sizes and the different thresholds. The rows contain the different numbers of clusters to determine the threshold, and the columns the different grid sizes. Even though SOMs are not restricted to square grids, we have opted to check only those to limit the search space for the parameters. Further tuning may improve the promoter prediction slightly. From Table 1 it is clear that a 6 × 6 grid combined with the top three clusters performs best.
Table 1.
Evaluation of the different parameter combinations for ProSOM
| 4 × 4 | 5 × 5 | 6 × 6 | 7 × 7 | |
|---|---|---|---|---|
| 1 | 0.44 | 0.40 | 0.36 | 0.32 |
| 2 | 0.46 | 0.47 | 0.45 | 0.41 |
| 3 | 0.37 | 0.46 | 0.48 | 0.45 |
| 4 | 0.10 | 0.36 | 0.44 | 0.47 |
| 5 | 0.09 | 0.29 | 0.39 | 0.46 |
The columns denote the different grid sizes, the rows contain the different thresholds. The first line contains the results when using the threshold of the cluster with the highest promoter probability. The second line contains results when using the promoter probability of the cluster with second highest promoter probability as threshold, and so on. The values in the table are the F-measure calculated from the entire human genome when validating against the CAGE dataset. The maximum allowed distance is 500 bp. Bold value indicates highest value.
3.4 ProSOM validation
We used the trained SOM to predict promoter regions. To each cluster we attached a probability that a given sequence assigned to that cluster is a promoter. If the structural profile of a sequence maps to a cluster that has a probability equal to or above the threshold, we predict it as a promoter region.
When designing classification algorithms, researchers often use cross-validation to validate their technique. Cross-validation is usually employed when only a limited amount of data is available. In our case, we have plenty of real-world data that can be used for validation. Once we have trained our approach on the limited set of training samples that is available from DBTSS and Ensembl, we can apply the approach to the entire human genome and compare with experimental screening for TSSs. Since real-world validation is superior to cross-validation, no cross-validation was performed.
To compare programs that result in different recall and precision values different techniques can be utilized. We use the F-measure (harmonic mean of precision and recall) to calculate a final score for a program for several reasons. (i) The F-measure is a single measure that can compare different programs with different precision and recall scores. (ii) In the case of the validation in a genomewide context it is very hard to estimate the number of TNs correctly because most prediction programs do not classify each site (nucleotide) in the genome as being a promoter or not, but only make predictions of core promoter regions. Recall, precision and F-measure do not require knowledge about the number of TNs and are thus suitable to use in this context. (iii) Other comprehensive measures such as Receiver Operating Characteristic, Area Under the Curve and precision-recall curve have a major drawback: they require that the program is able to explore the whole range of precision and recall values. While this is possible for ProSOM, many of the other programs we tested do not support this, or take prohibitively long time to do this (several weeks on an 80 CPU cluster for one parameter setting).
To validate our predictions we use the dataset of CAGE tags from Carninci et al. (2006) and a set of genes from Ensembl. To compare with the state-of-the-art, we used a maximum allowed distance from the TSS of 50 bp. This value is more stringent, but larger values would mean that more distant predictions are also correct, which is not desirable. Previously, we used 500 bp (Abeel et al., 2008), which is still too loose to call it TSS prediction or core promoter prediction. To really validate whether a program can truly detect core promoters, the distance between a known TSS and the prediction should be no more than 50 bp. Table 2 shows the performance of ProSOM versus a number of other PPPs. The values for all programs (including ProSOM) are based on the predictions made by the program with default settings. We assume that authors provide the optimal set of defaults for their own program. In case no defaults were supplied, we optimized the parameters of the program on the CAGE dataset with a maximum allowed distance of 500 bp as was done for ProSOM. It should be noted that the results in the table only represent a single point on the precision–recall curve and may thus be an underrepresentation of the actual potential of a program. We compared all programs on the reference distance of 500 bp combined with Ensembl and CAGE data and on the strong validation of 50 bp with the CAGE data. Some of the more recent approaches are not considered, as neither the program nor the predictions for the entire human genome are available for academic use. Programs that are currently unavailable for this type of study include FProm (Solovyev et al., 2006), ProStar, AMOSA-based promoter prediction, EnsemPro, PSPA (Wang and Hannenhalli, 2006) and MetaProm. While the 500 bp columns in Table 2 are useful for reference with earlier work, the real strength of a PPP is best assessed on the 50 bp columns. The last column of Table 2 contains the number of predictions made by a program.
Table 2.
Evaluation of PPPs using the CAGE and Ensembl datasets with a maximum allowed distance of 500 bp and for the CAGE dataset with a maximum distance of 50 bp
| CAGE (500 bp) |
Ensembl (500 bp) |
CAGE (50 bp) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Program | Reference | Recall | Precision | F | Recall | Precision | F | Recall | Precision | F | No. of predictions |
| ProSOM | 0.38 | 0.66 | 0.48 | 0.48 | 0.43 | 0.45 | 0.17 | 0.30 | 0.22 | 62 804 | |
| Eponine | Down and Hubbard, 2002 | 0.28 | 0.75 | 0.41 | 0.36 | 0.51 | 0.42 | 0.14 | 0.35 | 0.20 | 60 247 |
| EP3 | Abeel et al., 2008 | 0.34 | 0.66 | 0.45 | 0.42 | 0.46 | 0.44 | 0.11 | 0.27 | 0.16 | 45 765 |
| ARTS | Sonnenburg et al., 2006 | 0.38 | 0.74 | 0.50 | 0.53 | 0.59 | 0.56 | 0.11 | 0.27 | 0.15 | 47 144 |
| FirstEF | Davuluri et al., 2001 | 0.41 | 0.42 | 0.42 | 0.58 | 0.34 | 0.43 | 0.13 | 0.15 | 0.14 | 101 985 |
| DragonGSF | Bajic and Brusic, 2003 | 0.31 | 0.75 | 0.44 | 0.45 | 0.63 | 0.53 | 0.09 | 0.28 | 0.13 | 35 410 |
| PromoterInspector | Scherf et al., 2000 | 0.29 | 0.81 | 0.43 | 0.38 | 0.70 | 0.49 | 0.07 | 0.39 | 0.13 | 21 576 |
| DragonPF | Bajic et al., 2002 | 0.51 | 0.11 | 0.18 | 0.65 | 0.11 | 0.19 | 0.32 | 0.07 | 0.11 | 603 389 |
| N-Scan | Gross and Brent, 2006 | 0.33 | 0.45 | 0.38 | 0.55 | 0.51 | 0.53 | 0.07 | 0.13 | 0.09 | 67 748 |
| CpGProD | Ponger and Mouchiroud, 2002 | 0.34 | 0.41 | 0.37 | 0.50 | 0.36 | 0.42 | 0.08 | 0.12 | 0.09 | 76 793 |
| PromoterExplorer | Xie et al., 2006 | 0.39 | 0.30 | 0.34 | 0.55 | 0.24 | 0.33 | 0.09 | 0.08 | 0.09 | 132 794 |
| McPromoter | Ohler et al., 2000 | 0.17 | 0.68 | 0.28 | 0.24 | 0.61 | 0.34 | 0.05 | 0.29 | 0.09 | 20 862 |
| PromFD | Chen et al., 1997 | 0.44 | 0.16 | 0.23 | 0.55 | 0.14 | 0.22 | 0.12 | 0.04 | 0.06 | 329 999 |
| PromoterScan | Prestridge, 1995 | 0.16 | 0.09 | 0.12 | 0.19 | 0.08 | 0.11 | 0.03 | 0.02 | 0.02 | 197 852 |
| Promoter2.0 | Knudsen, 1999 | 0.63 | 0.04 | 0.08 | 0.68 | 0.03 | 0.06 | 0.11 | 0.01 | 0.01 | 191 9363 |
| NNPP 2.2 | Reese, 2001 | 0.03 | 0.03 | 0.03 | 0.03 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 104 746 |
Bold values indicate ranks for the different programs.
The first three columns from Table 2 contain the results for the CAGE validation set with a maximum distance of 500 bp, the middle three columns the results for the Ensembl set with a maximum distance of 500 bp and the last three columns those for the CAGE set with a maximum distance of 50 bp. The first column for each set presents the recall rate, which is the percentage of reference sites that has been predicted; the second column for each set the precision, which is the percentage of correct predictions; finally, the third column the F-measure (Van Rijsbergen, 1979), which is the harmonic mean of the precision and recall. This measure provides a single number to quantify the quality of the predictions. The higher this value, the better. The rows in Table 2 are ranked according to the F-measure on the CAGE dataset with maximum distance of 50 bp. On the reference validation with maximum distance of 500 bp, ProSOM works slightly worse than the best program (ARTS). On the strong validation with only 50 bp allowed distance between the true TSS and the prediction, ProSOM performs slightly better than Eponine. Again, the performance of our approach is better balanced in terms of recall and precision.
Overall, the choice of the PPP depends on the task at hand. When predictions within 500 bp are good enough, ARTS or ProSOM are the way to go. They both provide a good F-measure. If the exact localization of the prediction is more important, ProSOM or Eponine are the best options, with Eponine having higher precision and ProSOM being more balanced. Thus, ProSOM has the best all-round performance independent of the task type.
While our new approach, ProSOM, performs well on the real TSS data, it has some problems with the Ensembl dataset. The reason is that exonic sequences often end up in the clusters that contain the promoters. The counting method used for the Ensembl dataset counts all predictions inside exons as false predictions, which explains the lower than expected precision for the Ensembl dataset. As we have previously shown, the technique to validate a promoter prediction technique with gene annotation is flawed (Abeel et al., 2008), especially since it only takes into account the 5′ TSS and will label any other recognized TSS as a false prediction.
We also analyzed the ENCODE regions of the human genome in more detail. The ENCODE project tries to annotate 1% of the human genome in great detail (The ENCODE Project Consortium 2007). The regions are partly chosen for their importance and partly random. In total, they cover ∼30 Mbp. For these regions, again are data available for real TSSs, compiled by the Riken Institute using the CAGE technique. There is also a high-quality gene annotation available from the E-GASP/GENCODE project (Guigó et al., 2006). Furthermore, there is a lot of experimental data available that may support TSS predictions. When we apply ProSOM to these regions and compare with the CAGE and GENCODE data and a maximum distance of 500 bp, we get an F-measure of 0.73 and 0.57, respectively. These values are nearly the same as for the EP3 program (0.72 and 0.56). For the maximum distance of 50 bp and the CAGE dataset we get an F-measure of 0.28. These values are higher than those for the validation on the whole genome, which indicates that indeed a lot of the so-called false predictions are, in fact, correct predictions that are missing in the validation set. To test this hypothesis, we used the Evidence For Transcriptional Activity dataset (Abeel et al., 2008). This dataset is a compendium of experimental indicators for transcription activity. Within this set 98% of our predictions has a hit with a maximum distance of 500 bp and 85% within 50 bp, again indicating that our approach is highly precise.
4 RELATED WORK
Currently, several core PPPs are available that are aimed at predicting TSSs on the whole human genome. Early programs were limited in the amount of data they could process and in their predictive power (Fickett and Hatzigeorgiou, 1997). Only in 2004, promoter prediction tools have been validated on the whole human genome (Bajic et al., 2004). Later, the same authors also reviewed some PPPs on the ENCODE regions of the human genome (Bajic et al., 2006). Gene annotation was used to validate the different PPPs, but in light of new insights gained from the ENCODE project, this is too conservative and even often wrong. The ENCODE project shows that transcription can also start at the 3′ end of a gene or inside exons. Nowadays, it makes more sense to compare real unbiased experimentally characterized TSSs for validation. Recently, we have reviewed several PPPs on a dataset of TSSs that have been determined using the CAGE technique over the whole human genome (Abeel et al., 2008). Wang and co-workers (2007a) focus on alternative promoters and present a novel PPP, MetaProm, based on artificial neural networks. Other recent PPPs include ProStar (Goni et al., 2007), AMOSA-based promoter prediction (Wang et al., 2007b) and EnsemPro (Won et al., 2008). Two of the recent PPPs (MetaProm and EnsemPro) are in fact no real promoter predictors but meta-predictors that aggregate the results of several other programs. These programs probably address the problem of merging the results of several PPPs to some extent, a definitely interesting way to advance the performance of PPPs. Unfortunately, none of these new programs were available for genomewide screening at the time this article was submitted.
5 DISCUSSION AND CONCLUSION
Self-organizing maps provide an intuitive way to cluster DNA sequences. They are unique among unsupervised clustering techniques in their ability to distinguish core promoters from other sequences. When applied to a set of promoter, transcribed and intergenic sequences, they create promoter-rich and promoter-poor clusters. The promoter-rich clusters show the same structural profile that has been observed for core promoter sequences (Abeel et al., 2008). Because the clustering technique is unsupervised we can conclude that the profile that emerges from this clustering technique is indeed one of the hallmarks of the core promoter. Furthermore, the physical description of the core promoter is more general than that based on core promoter motifs. The physical structure of this region is important for transcription initiation as it remains the same, regardless of the presence of motifs.
We packaged this technique as a full-fledged promoter prediction tool, called ProSOM. This technique is made available to academic researchers for free. We used the program to predict core promoters in the human genome and it performs as well as the best existing software packages. Furthermore, it is more balanced regarding the number of retrieved sites and false predictions. The technique is also very precise as 98% of all predictions in the ENCODE region have evidence of transcriptional activity within 500 bp.
ACKNOWLEDGEMENTS
We would like to thank A. De Rycke for the implementation of many of the clustering techniques we tried.
Funding: T.A. is indebted to Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) for a predoctoral fellowship. Y.S. is a postdoctoral fellow of the Research Foundation-Flanders.
Conflict of Interest: none declared.
REFERENCES
- Abeel T, et al. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 2008;18:310–323. doi: 10.1101/gr.6991408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts S, et al. Comprehensive analysis of the base composition around the transcription start site in Metazoa. BMC Genomics. 2004;5:34. doi: 10.1186/1471-2164-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic VB, Brusic V. Computational detection of vertebrate RNA polymerase II promoters. Methods Enzymol. 2003;370:237–250. doi: 10.1016/S0076-6879(03)70021-4. [DOI] [PubMed] [Google Scholar]
- Bajic VB, et al. Dragon Promoter Finder: recognition of vertebrate RNA polymerase II promoters. Bioinformatics. 2002;18:198–199. doi: 10.1093/bioinformatics/18.1.198. [DOI] [PubMed] [Google Scholar]
- Bajic VB, et al. Promoter prediction analysis on the whole human genome. Nat. Biotechnol. 2004;22:1467–1473. doi: 10.1038/nbt1032. [DOI] [PubMed] [Google Scholar]
- Bajic VB, et al. Performance assessment of promoter predictions on ENCODE regions in the EGASP experiment. Genome Biol. 2006;7(Suppl 1):S3.1–S3.13. doi: 10.1186/gb-2006-7-s1-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, et al. Computational applications of DNA structural scales. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:35–42. [PubMed] [Google Scholar]
- Brent MR. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat. Rev. Genet. 2008;9:62–73. doi: 10.1038/nrg2220. [DOI] [PubMed] [Google Scholar]
- Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Chen QK, et al. PromFD 1.0: a computer program that predicts eukaryotic pol II promoters using strings and IMD matrices. Comput. Appl. Biosci. 1997;13:29–35. doi: 10.1093/bioinformatics/13.1.29. [DOI] [PubMed] [Google Scholar]
- Choi CH, et al. DNA dynamically directs its own transcription initiation. Nucleic Acids Res. 2004;32:1584–1590. doi: 10.1093/nar/gkh335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, et al. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- Deng W, Roberts SGE. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Hubbard TJP. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, et al. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- Fickett JW, Hatzigeorgiou AG. Eukaryotic promoter recognition. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florquin K, et al. Large-scale structural analysis of the core promoter in mammalian and plant genomes. Nucleic Acids Res. 2005;33:4255–4264. doi: 10.1093/nar/gki737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, et al. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goni JR, et al. Determining promoter location based on DNA structure first-principles calculations. Genome Biol. 2007;8:R263. doi: 10.1186/gb-2007-8-12-r263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SS, Brent MR. Using multiple alignments to improve gene prediction. J. Comput. Biol. 2006;13:379–393. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- Guigö R, et al. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006;7(Suppl 1):S2.1–S2.31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Bansal M. Structural properties of promoters: similarities and differences between prokaryotes and eukaryotes. Nucleic Acids Res. 2005;33:3165–3175. doi: 10.1093/nar/gki627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC genome browser database: 2008 update. Nucleic Acids Res. 2008;36((Database issue)):D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, et al. Dynamic usage of transcription start sites within core promoters. Genome Biol. 2006;7:R118. doi: 10.1186/gb-2006-7-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- Kohonen T. Self-Organizing Maps. 3rd ed. Springer, Berlin: 2001. [Google Scholar]
- Liao GC, et al. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liolios K, et al. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res. 2006;34((Database issue)):D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, et al. Stochastic segment models of eukaryotic promoter regions. Pac. Symp. Biocomput. 2000;1:380–391. doi: 10.1142/9789814447331_0036. [DOI] [PubMed] [Google Scholar]
- Ornstein RL, et al. Optimized potential function for calculation of nucleic-acid interaction energies. 1. Base stacking. Biopolymers. 1978;17:2341–2360. doi: 10.1002/bip.1978.360171005. [DOI] [PubMed] [Google Scholar]
- Pedersen AG, et al. The biology of eukaryotic promoter prediction–a review. Comput. Chem. 1999;23:191–207. doi: 10.1016/s0097-8485(99)00015-7. [DOI] [PubMed] [Google Scholar]
- Polikar R. Ensemble based systems in decision making. IEEE Circuit Syst. Mag. 2006;6:21–45. [Google Scholar]
- Ponger L, Mouchiroud D. CpGProD: identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics. 2002;18:631–633. doi: 10.1093/bioinformatics/18.4.631. [DOI] [PubMed] [Google Scholar]
- Prestridge DS. Predicting Pol II promoter sequences using transcription factor binding sites. J. Mol. Biol. 1995;249:923–932. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- Saeys Y, et al. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- Sandelin A, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Scherf M, et al. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J. Mol. Biol. 2000;297:599–606. doi: 10.1006/jmbi.2000.3589. [DOI] [PubMed] [Google Scholar]
- Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Solovyev V, et al. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006;7 (Suppl 1):S10.1–S10. doi: 10.1186/gb-2006-7-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg S, et al. ARTS: accurate recognition of transcription starts in human. Bioinformatics. 2006;22:e472–e480. doi: 10.1093/bioinformatics/btl250. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rijsbergen CJ. Information Retrieval. 2nd edition. MA, USA: Butterworth-Heinemann Newton; 1979. [Google Scholar]
- Wakaguri H, et al. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36((Database issue)):D97–D101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hannenhalli S. A mammalian promoter model links cis elements to genetic networks. Biochem. Biophys. Res. Commun. 2006;347:166–177. doi: 10.1016/j.bbrc.2006.06.062. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. MetaProm: a neural network based meta-predictor for alternative human promoter prediction. BMC Genomics. 2007a;8:374. doi: 10.1186/1471-2164-8-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Prediction of transcription start sites based on feature selection using AMOSA. Comput. Syst. Bioinformatics Conf. 2007b;6:183–193. [PubMed] [Google Scholar]
- Won H-H, et al. Ensempro: an ensemble approach to predicting transcription start sites in human genomic DNA sequences. Genomics. 2008;91:259–266. doi: 10.1016/j.ygeno.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Xie X, et al. PromoterExplorer: an effective promoter identification method based on the AdaBoost algorithm. Bioinformatics. 2006;22:2722–2728. doi: 10.1093/bioinformatics/btl482. [DOI] [PubMed] [Google Scholar]