Abstract
Motivation: Chemical compounds like small signal molecules or other biological active chemical substances are an important entity class in life science publications and patents. Several representations and nomenclatures for chemicals like SMILES, InChI, IUPAC or trivial names exist. Only SMILES and InChI names allow a direct structure search, but in biomedical texts trivial names and Iupac like names are used more frequent. While trivial names can be found with a dictionary-based approach and in such a way mapped to their corresponding structures, it is not possible to enumerate all IUPAC names. In this work, we present a new machine learning approach based on conditional random fields (CRF) to find mentions of IUPAC and IUPAC-like names in scientific text as well as its evaluation and the conversion rate with available name-to-structure tools.
Results: We present an IUPAC name recognizer with an F1 measure of 85.6% on a MEDLINE corpus. The evaluation of different CRF orders and offset conjunction orders demonstrates the importance of these parameters. An evaluation of hand-selected patent sections containing large enumerations and terms with mixed nomenclature shows a good performance on these cases (F1 measure 81.5%). Remaining recognition problems are to detect correct borders of the typically long terms, especially when occurring in parentheses or enumerations. We demonstrate the scalability of our implementation by providing results from a full MEDLINE run.
Availability: We plan to publish the corpora, annotation guideline as well as the conditional random field model as a UIMA component.
Contact:roman.klinger@scai.fraunhofer.de
1 INTRODUCTION AND RELATED WORK
Finding relevant information is one of the most important challenges in our time. In particular in life science and chemical research a huge amount of new publications, research reports and patents is produced every year. For users of huge text corpora like MEDLINE, document categorization, ranking and finding entity-related information is an important help in their daily research and work life. The automated identification of entities of interest in text in a domain and their mapping to database entries strongly improves information retrieval, information extraction and information aggregation. For such tasks tools have been successfully developed in the last decades especially for finding mentions of proteins and genes. Those provide basic methods to extract e.g. protein–protein relations. For that field of interest, the BioCreative competition (Hirschman et al., 2007) provides an evaluation of state-of-the-art techniques on publicly available data sources. Dictionary-based systems allow a direct mapping of the recognized entities to reference objects (e.g. EntrezGene identifiers for genes). An inherent drawback of such approaches, however, is the dependence on the quality and completeness of the dictionary and the methods of the underlying algorithm to recognize spelling variants and to resolve ambiguous names.
Dictionary independent methods (rule-based systems as well as machine learning-based system) are well suited to find names where no comprehensive dictionary is available. An example for a rule-based approach is to find mentions matching a given set of regular expressions. Machine learning approaches are based on an annotated training set from which statistical information can be obtained about the inherent dependencies in the data. This extracted information can then be applied on unseen data to classify word tokens, i.e. to label parts of text with different classes. The best approaches in the BioCreative sub task of gene mention recognition have an F1 measure between 86% and 87% (Wilbur et al., 2007). From 21 submissions, 11 use conditional random fields (CRF), a machine learning method based on undirected graphical models (Bishop, 2006) which leads to competitive results, in our hands 86.33% F1 measure (Klinger et al., 2007b).
Finding mentions of chemical compounds in text is of interest for several reasons. An annotation of the entities enables a search engine to return documents containing elements of this entity class (semantic search), e.g. together with a disease. This can be helpful to find relations e.g. to adverse reactions or diseases. Mapping the found entities to corresponding structures leads to the possibility to search for relations between different chemicals. This enables a chemist to search for similar structures or substructures and combine the knowledge in the text with knowledge from databases or to integrate other tools handling chemical information (e.g. solubility or mass calculation).
Chemical names can be distinguished into different classes: to deal with complex structures, different methods of nomenclature are used, e.g. mentions of the sum formula or names according to the Simplified Molecular Input Line Entry Specification (SMILES; Weininger, 1988) or the successor of SMILES, the IUPAC International Chemical Identifier (InChI). Because of a limited readability of such specifications for humans, trivial names and the nomenclature published by the International Union of Pure and Applied Chemistry (IUPAC; McNaught and Wilkinson, 1997) is commonly applied (Eller, 2006) in text. Also combinations of the different types of names as well as abbreviations, especially of often used substances, are in use.
Trivial names can be searched for with a dictionary-based approach and directly mapped to the corresponding structure at the same time. For example, the dictionary-based named entity recognition system ProMiner (Hanisch et al., 2005) uses a DrugBank1 (Wishart et al., 2006) dictionary for the recognition of drug names in MEDLINE abstracts (Kolářik et al., 2007). Other systems use the drug dictionary from MedlinePlus2 (e.g. EbiMed; (Rebholz-Schuhmann et al., 2007) for the recognition of drug names.
For other representations of chemical structures like SMILES, InChI or IUPAC names such an enumeration is only possible for the most common substances. The full chemical space cannot be enumerated. Several systems address that problem regarding chemical entities with a variety of approaches.
Narayanaswamy et al. (2003) describe a manually developed set of rules relying upon lexical information, linguistic constraints of the English language and contextual information for the detection of several entity classes. The reason for choosing this approach is stated as the lack of an annotated corpus. The evaluation was done on a small hand-selected corpus containing 55 MEDLINE abstracts selected by searching for acetylates, acetylated and acetylation. They found 158 chemical names from which 22 were ambiguous and classified into different classes and 13 chemical parts with two ambiguous ones. The F1 measure for the first is 90.86% (93.15% precision, 86.08% recall). The latter has an F1 measure of 91.67% (100% precision, 84.62% recall).
Similarly, (Kemp and Lynch, 1998) identify chemical names in patent texts with handcrafted rules using dictionaries with chemical name fragments. They claim to identify 97.4% from 14 855 specific chemical names in 70 patent descriptions taken from documents from the IPC class CO 7D. The false positive rate is reported to be 4.2%.
The concept described by Anstein et al. (2006) for which the preconditions are described by Reyle (2006) uses a grammar for the analysis of fully specified (e.g. 7-hydroxyheptan- 2-one), trivial (e.g. benzene) and semi-systematic (e.g. benzene-1,3,5-triacetic acid) as well as underspecified (e.g. deoxysugar) compound names. The advantage of that approach is that the grammatical analysis can be used as a basis for a conversion to the chemical structure. A possible problem is the difficulty to recognize names not following the specification to a certain degree as well as the completeness and maintenance of a changing standard.
A molecular similarity search is used by (Rhodes et al., 2007) to enable a user to ‘search for related Intellectual Property’ in US patents based on a specified drawn molecule. They report to find 3 623 248 unique chemical structures from 4 375 036 US patents. The absolute numbers of found patents for the top 25 drugs listed by Humana (2005) are given.
The program developed by (Sun et al., 2007) focuses on finding sum formulas like CH3(CH2)2OH in text using support vector machines (Schölkopf and Smola, 2002) and CRFs (Lafferty et al., 2001).
In the work of Corbett et al. (2007), first-order Hidden Markov Models (Rabiner, 1989) implemented in the toolkit LingPipe3 are combined with other methods for the identification of chemical entities. The program finds e.g. structural classes, atoms and elements, fragments, trivial names, SMILES and InChI as well as IUPAC names. They give an inter-annotator F1 measure of 93% for chemical names on their annotated corpus. The performance is evaluated on different corpora, recall rates are between 69.1% and 80.8% and precision rates beween 64.1% and 75.3% (Corbett and Murray-Rust, 2006). A seperate evaluation of the included named entity recognition modules from the toolkit LingPipe results in an F1 measure of 74% (Corbett et al., 2007). To our knowledge, their implementation—the open source program OSCAR34 (Open Source Chemistry Analysis Routines; Corbett, 2007)—is the only software available to the academic community.
We prefer to have a method identifying IUPAC and IUPAC-like names only and to have additional approaches to recognize other chemical name classes (e.g. brand names or elements): IUPAC and IUPAC-like names can be recognized based on their morphological structure with higher performance than with methods based on dictionaries (Kolářik et al., 2008). We therefore introduce a system for the recognition of IUPAC and IUPAC-like names while trivial names are found with a dictionary approach not described here. These IUPAC-like terms do not only include correct IUPAC names but also names not following the nomenclature strictly. This enables a higher recall regarding mentioned chemicals, which is important for document retrieval purposes.
In addition to the correct recognition of IUPAC and IUPAC-like names, the aim is to transform these names using name-to-structure converters to allow the usage of chemical tools on the extracted data. Therefore enumerations have to be detected with all parts while modifying tokens (e.g. substitutes, analogs) have to be tagged separately (cf. Fig. 1). We use a CRF approach and present our development of a training corpus as well as our experiences regarding inter-annotator agreement in comparison to the work of Corbett et al. (2007). Next to the training corpus we describe test corpora especially on MEDLINE and on patents.
Fig. 1.
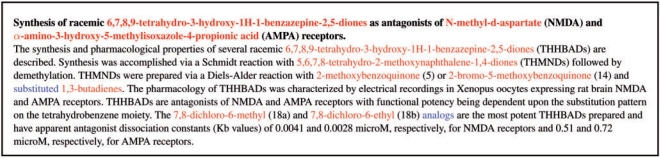
Example abstract with tagged entities (PMID 9240357; Guzikowski et al., 1997). IUPAC entities are depicted in red while MODIFIER entities are shown in blue.
Additionally, to the presentation of the results an exhaustive analysis of the influences of different CRF orders and offset conjunctions is shown and the impact of the different feature sets on the results are evaluated and discussed.
2 METHODS
2.1 Overview
We apply a CRF to build a model for finding IUPAC and IUPAC-related MODIFIER entities. A training corpus has been annotated by two independent annotators and the inter-annotator agreement is discussed in Section 2.3. The model selection is performed by bootstrapping (Efron and Tibshirani, 1993) and evaluated on two independent test corpora, one consisting of sampled abstracts from MEDLINE, the other one on hand selected paragraphs from bio-chemical patents. We analyze the use of name-to-structure converters as a basis of a possible normalization, a mapping of the found entities to a unique structure.
2.2 Entity types
The entities in which we focus are IUPAC and MODIFIER mentions. As described, chemical entities in general are named following different nomenclatures which are also combined by the authors of biomedical texts. Only concentrating on correct IUPAC terms is not sufficient, so we define a IUPAC entity to be a chemical substance mentioned in a IUPAC-like manner. Additionally to correct IUPAC names, it includes IUPAC names in which a part is abbreviated, fragments and group names. In Figure 1 an example abstract from MEDLINE with annotations of the two entity classes is shown. Next to full names like ‘1,2,3,4-tetrahydronaphthalene-1-carboxylic acid’ or ‘4-[[(3-chlorophenyl)amino]methyl]-6,7-dihydroxychromen-2-one’, fragments, e.g. in enumerations, are tagged separately like ‘acridine-4-’ and ‘phenazine-1-carboxamide’ in ‘…both the acridine-4- and ‘phenazine-1-carboxamide series…’ or ‘3α-[bis(4-fluoro-’ and ‘4-chlorophenyl)methoxy] tropane’ in ‘…N- and 2-substituted-3α-[bis(4-fluoro- or 4-chlorophenyl)methoxy]tropane…’.5
The alternative to the separate way of annotating parts in enumerations would have been an annotation including the connecting word (in that example ‘or’). This is not meaningful because parts of names are sometimes divided by long text passages. With our kind of annotation, a possible enumeration resolution of the found parts in the text is prepared.
The MODIFIER entity describes similarities to a mentioned substance like in ‘[IUPAC-entity] analogues’ or ‘[IUPAC-entity] modifier’ or ‘3-substituted-IUPAC-entity’.
2.3 Corpus generation and inter-annotator agreement
Three main corpora are generated for building the model and evaluating our approach following a developed annotation guideline. A training corpus consisting of MEDLINE abstracts (abbreviated as TrainM), a test corpus containing MEDLINE abstracts (TestM) and a test corpus made up of parts of patents (TestP).
The training corpus is built in two steps. First, a preliminary corpus (abbreviated as Trainpr) is built in the same manner as described by Friedrich et al. (2006). For that, in the BioCreative training corpus (Hirschman et al., 2007), the gene and protein names are replaced by randomly selected correct IUPAC names from PubChem (NCBI, 2007). This leads to an artificial corpus with 15 000 sentences with 1 216 341 tokens. It includes 24 325 entities. On that corpus a CRF is trained and used for tagging 10 000 sampled abstracts from MEDLINE. From these, 463 abstracts are selected which include 161 591 tokens in 3700 sentences6 with 3712 IUPAC and 1039 MODIFIER entities.
For evaluation of the system, 1000 MEDLINE records with 124 122 tokens in 5305 sentences are sampled equally distributed from full MEDLINE containing 151 IUPAC and 14 MODIFIER entities resulting in the corpus TestM.
Passages from 26 patents dealing with chemical processes were hand selected according to occurring enumerations of chemicals, especially using different and mixed nomenclatures to detect possible problems. These paragraphs consist of 4309 words in 152 sentences with 411 IUPAC entities forming the corpus TestP.
The training corpus is annotated by two independent annotators. An assessed inter-annotator F1 measure for the IUPAC entity is relatively low with 78% [in contrast to 93% claimed by Corbett et al. (2007)]. One reason for the difference in comparison to Corbett and his colleagues is our differentiation of the IUPAC entity to other chemical mentions, which is not always easy to decide while all chemical mentions in the corpus generated by Corbett are combined in one entity. Another reason are the different experience levels of our annotators: while the first annotator collaborated on the development of the annotation guideline and annotated several corpora, the second annotator based his annotations directly on the provided guideline.
For building the conclusive training corpus both annotations are combined by an independent person. The F1 measure between the resulting training corpus (TrainM) and the first-annotated corpus is 94%.
2.4 Conditional random fields
CRFs (Lafferty et al., 2001; McDonald and Pereira, 2005) are a family of probabilistic, undirected graphical models for computing the probability 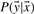 of a possible label sequence
of a possible label sequence 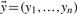 given the input sequence
given the input sequence  . In the context of named entity recognition this observation sequence
. In the context of named entity recognition this observation sequence  corresponds to the tokenized text. This is the sequence of tokens which is defined by a process called tokenization—splitting the text at white space, punctuation marks and parentheses. A straightforward idea for the tokenization of IUPAC names is to keep the whole name together to use the sheer length for their identification. That is not possible because of leading and successive brackets and other symbols as well as often used wrong white spaces (based on e.g. converted line breaks). So we use a very fine tokenization, also splitting at all number-letter changes in the text.
corresponds to the tokenized text. This is the sequence of tokens which is defined by a process called tokenization—splitting the text at white space, punctuation marks and parentheses. A straightforward idea for the tokenization of IUPAC names is to keep the whole name together to use the sheer length for their identification. That is not possible because of leading and successive brackets and other symbols as well as often used wrong white spaces (based on e.g. converted line breaks). So we use a very fine tokenization, also splitting at all number-letter changes in the text.
The label sequence is encoded in a label alphabet similar to ℒ={I-<entity>,O,B-<entity>} where yi=O means that xi is not an entity, yi=B-<entity> means that xi is the beginning of an entity and yi=I-<entity> means that xi is the continuation of an entity. In our case we use the alphabet
as described in Section 2.2. An example for an observation sequence with a label sequence is depicted in Figure 2.
Fig. 2.
Example for observation and label sequence for the text snippet: ‘…of cyclohepta-1,3-diene …’ after tokenization.
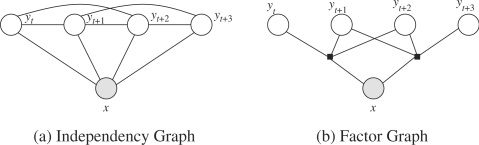
A CRF in general is an undirected probabilistic graphical model
| (1) |
where Ψj are the different factors given through an independency graph like in Figure 3 (Kschischang et al., 2001). These factor functions combine different features fi of the considered part of the text and the label sequence. We mainly use morphological features of the text tokens for every possible label transition.7 A subset of the used features is depicted in Table 1. They usually have a form similar to
The feature set used in our approach is described in Section 2.4.1.
Fig. 3.
First-order linear-chain CRF.
Table 1.
Features used in the CRF
| Name | Explanation |
|---|---|
| Static morphol. features | Reg.Ex. |
| All Caps | [A-Z]+ |
| Real Number | [-0-9]+[.,]+[0-9.,]+ |
| Is Dash | [- – — −] |
| Is Quote | [,, “ ” ” ‘ ’] |
| Is Slash | [\ /] |
| Autom. generated features | |
| Autom. Prefixes/Suffixes | Automatic generation of a feature for every token: match that prefix or suffix (length 2) |
| Bag-Of-Words | Automatic generation of a feature for every token: match that token |
| Spaces | |
| Spaces_left | white space preceding token |
| Spaces_right | white space following token |
| Lists | |
| Prefix/Suffix lists | Prefixes and suffixes (length 4) of intermediate or last words generated from IUPAC names |
A special case of the general CRF, in fact the one shown in Figure 3, is the linear-chain CRF where the factors are given in the form
| (2) |
so that the CRF can be written as
| (3) |
The normalization to [0,1] is given by
| (4) |
Here 𝒴 is the set of all possible label sequences.
To compute the normalization factor, the forward-backward algorithm known from Hidden Markov Models (Rabiner, 1989) can be incorporated. Optimization of the parameters (training) can be done by applying the limited-memory Broyden–Fletcher–Goldfarb–Shanno algorithm (L-BFGS; Nocedal, 1980) on the convex function ℒ(𝒯) with the training data 𝒯:
These algorithms can also be used in CRF with a higher order, given by
| (5) |
where q is the order of the CRF (cf. Fig. 4).
Fig. 4.
Second-order linear-chain CRF.
Our own implementation of the named entity recognizer for IUPAC terms is based on MALLET (McCallum, 2002), a widely used and successfully applied system for linear-chain CRF. A more detailed description of these models and their relation to other graphical models is e.g. given by Klinger and Tomanek (2007).
2.4.1 Feature set
Many of the evaluated features are extracted by standard methods, especially the morphological ones. Some of them are shown in Table 1. Next to these commonly used features we incorporate special IUPAC-related features. These are the membership of a token to a list of often used prefixes and suffixes of length four in IUPAC names or a list of typical last tokens of the names. These lists are extracted from all IUPAC names mentioned in the data available from PubChem (NCBI, 2007).
The list of prefixes of length 4 has 714 members, the list of suffixes of the same length has 661 members. Another list includes 300 suffixes of the last tokens of IUPAC names to improve the detection of the end of a IUPAC name. The general idea of these lists is to provide the system with a possibility to generalize in excess of the training data. Another feature usually not used in the context of other entities is the specification of a token being preceded or succeeded by white space. This is important especially in enumerations or abbreviations of IUPAC names or trivial names to separate them from each other, in particular with reference numbers like shown in Figure 1. This feature is necessary due to the need of a fine tokenization.
Additionally, we use the so-called offset conjunction (OC) that adds features of the preceding and succeeding tokens for every token, incorporating contextual information to the token to be labeled.
2.5 Conversion of IUPAC names to structures
To normalize the found names, one solution is to convert them to a structure representation. Several tools have been developed for that task.
Eigner-Pitto et al. (2007) show a short evaluation of three commercial tools. One is LexiChem8 (OpenEye, 2007) by OpenEye, a product capable of conversions from IUPAC names as well as other names to structures and vice versa. Another program is ACDName by ACDLabs, which generates chemical structures from systematic names, derivatives, semi-systematic and trivial names as well as incorrect names, not strictly following the nomenclature (ACDLabs, 2007), but it focuses more on correct names than the program Name=Struct by CambridgeSoft (CambridgeSoft, 2007; Eigner-Pitto et al., 2007).
We use the open source converter included in OSCAR3, called OPSIN,9 the only software to our knowledge, which is freely available for academic evaluation purposes. It converts names to the Chemical Markup Language (CML, Murray-Rust (1997)) which we translate to SMILES using the Chemistry Development Kit10 (CDK; Steinbeck et al., 2003).
3 RESULTS
In a first step a CRF is trained on the preliminary, tweaked corpus Trainpr mentioned in Section 2.3 and evaluated by 50-fold bootstrapping. The result is an F1 measure of 97.92% (98.08% precision, 97.76% recall) with a first-order CRF with first-order offset conjunction and the same parameter set as described in Section 3.1. These results are comparable to those published by Friedrich et al. (2006). Evaluating this model on the annotated MEDLINE training corpus TrainM shows a low F1 measure (with 19.5%) and 38.4% recall. The performance on the sampled MEDLINE test corpus TestM is even worse with 1.1% F1 measure and a recall of 29.1%. These results show that there is a fundamental difference in tagging the tweaked corpus Trainpr (which seems to be simple, considering the F1 measure) and real world texts (as TrainM and TestM). The analysis of the different corpora shows two main problems: On the one hand, only correct IUPAC names are included in Trainpr, but fragments occur frequently in real text. On the other hand, a big problem are missing negative examples in the tweaked training data representing what is not a IUPAC name: nearly all isolated numbers, single letters, expressions in or around brackets are found wrong.
Based on the experiences on the tweaked training corpus Trainpr, a CRF is trained on the annotated training corpus based on MEDLINE abstracts TrainM using a selected parameter set. The evaluation of the different parameters is given.
3.1 Parameter selection
For model selection, the impact of the following parameters of the CRF are evaluated by applying 30-fold bootstrapping on the training set TrainM:
features representing the text like Bag-Of-Words or morphological features (cf. Table 1),
the order of the CRF and
the order of the offset conjunction.
The feature set of the system with the best performance consists of automatically added features based on Bag-Of-Words as well as Autom. Prefixes/Suffixes of length two. Additionally, the membership to Prefix/Suffix lists containing prefixes or suffixes of length four of last or intermediate tokens from IUPAC names is considered. From the set of static morphological features, All Caps, Real Number, Is Dash, Is Slash and Is Quote are used. The Spaces features to determine if the token is preceded or succeeded by white space is also included. Many other features, mainly from the field of gene and protein recognition were also tested, e.g. mapping the token to regular expressions representing greek letters, combinations of alpha-numerical symbols, natural numbers, etc. For lists of different features see McDonald et al. (2004), McDonald and Pereira (2005), Settles (2005) and Klinger et al. (2007a,b).
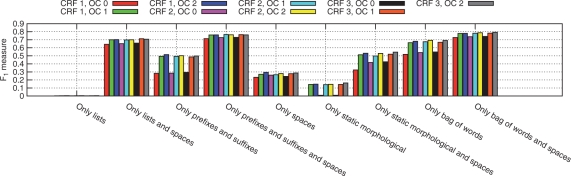
To evaluate the impact of the different features we omit one from the best feature set in several experiments (Fig. 5) and train models only with small feature sets (depicted in Fig. 6). The automatically generated features Bag-Of-Words and Autom. Prefixes/Suffixes have the highest impact on the performance together with the Spaces feature. Especially the last one is essential to obtain good results with impacts between 6.5% (CRF 2, OC 2) and 13.64% (CRF 1, OC 0). In contrast, the static morphological features and the Prefix/Suffix lists bring nearly no loss omitting them and low results when used as the only feature. Nevertheless, together with the feature Spaces, the results are surprisingly high (70% F1 measure). Interesting is that using only Autom. Prefixes/suffixes or Bag-Of-Words together with the Spaces feature and CRF order 3 and offset conjunction order 2 results in an F1 measure of 76.03% or 79.31%, respectively.
Fig. 5.
Results on the training data TrainM with 30-fold bootstrapping with different feature sets, different orders q of the CRF (given as CRF q above) and different orders p of the offset conjunction (given as OC p). The best results were obtained with the feature set presented in Table 1. For more details see Sections 2.4.1 and 3.1.
Fig. 6.
Results on the training data TrainM with some very small feature sets on different orders of the CRF and offset conjunctions. In contrast to Figure 5, where variations of a larger feature set are shown, the importance of features is presented in the context of very small feature sets. (Note the different scales between Figs 5 and 6.)
We evaluate different configurations of the features with different orders of offset conjunction (adding context in the form of features of the last p and next p tokens, where p is the order of the offset conjunction) as well as the order of the CRF, which includes information from the last q labels (q is the order of the CRF). The results of some of the features for different orders of offset conjunction and CRF can also be seen in Figure 5. The importance of the different features is nearly the same for all the different orders. The divergence in the results is high for different feature sets, but it is also very important to have the context information provided by the offset conjunction. The best F1 measure can be obtained with an offset conjunction of order 2 and a CRF order of 2 or 3. The difference between a CRF without an offset conjunction (i.e. order 0) to a CRF with order 1 offset conjunction are much higher than between order 1 and order 2 offset conjunctions. The increase of the order comes along with a high increase in the number of weights λi [compare to Equation (2) and (3)]. We have (for the CRF with order 3) 119 884 weights without an offset conjunction, 315 377 with an offset conjunction order of 1 and 521 179 weights with an offset conjunction order of 2. This corresponds to the training and tagging durations depicted in Figure 7.
Fig. 7.
Results on the sampled MEDLINE corpus TestM with different orders q of the CRF (given as order q) and orders p of the offset conjunction (given as OCp). The upper chart shows the F1 measures for the different configurations, the lower one the tagging and training durations.
Inspecting the tagging errors, we find that especially boundary errors at the end or at the beginning of the name are more frequent for a lower order of the offset conjunction. Other taggings that can be correctly identified with an offset conjunction order of 2 are formulations like ‘… through the 7- or 12-methylene carbon with …’ where the high context information is necessary to classify ‘7-’ correctly. A similar example is ‘… 2,3-substituted …’ with a tagging of ‘2,3’ as IUPAC with an offset conjunction of 1 but a correct result with an offset conjunction of 2.
3.2 Evaluation of the named entity recognition
Using the best configuration identified in the previous section, the resulting model is evaluated on the sampled MEDLINE test corpus TestM. In Figure 7 different orders of the CRF and the offset conjunction together with tagging and training durations are depicted. Similar to the results estimated with bootstrapping on the training corpus TrainM, highest performance is obtained with the most context information included by a CRF order of 3 and an offset conjunction order of 2. The F1 measure for IUPAC entities is 85.6% with a precision of 86.5% and a recall of 84.8%. The MODIFIER entities are found with an F1 measure of 84.6% (91.7% precision and 78.6% recall). Higher orders have not been applied because of prohibitive training durations. However, it can be seen that our best result is obtained at the expense of a high training time and, what is more important, on a higher tagging time of 307 s then other configurations of the CRF. For tagging a higher amount of data like the full MEDLINE database one could prefer to use a faster configuration like the one with order 2 and offset conjunction of 1 which only takes 215 s for tagging the test corpus. The F1 measure for IUPAC entities is lower with 77.7%, but the recall is nearly on the same level with 82.1% (MODIFIER: 55% precision, 78.6% recall). It can be concluded, that there is a trade-off between tagging time and performance, so it depends on the application which configuration should be preferred. The analysis of the errors show frequent problems in the recognition of short chemical names. On the one hand, chemical names are recognized by the system which are not specified as IUPAC-like. On the other hand, short names, similar to trivial names, specified as IUPAC-like by the annotators are most frequently unrecognized by the system. Nearly 50% of the other false positive errors are boundary errors. In addition, names morphological similar to IUPAC names like enzymes (e.g. ‘2-phospho-D-glyceratehydro-lyase’ or ‘pyruvate O2-phosphotransferase’) are detected as false positive matches.
Applying the best system trained on the MEDLINE training corpus, TrainM, for tagging the patent test corpus, TestP, shows a decrease in F1 measure in comparison to the MEDLINE test corpus TestM due to the bias of hand selecting difficult paragraphs instead of sampling from a set of sentences or text snippets. We get an F1 measure of 81.5% with a precision of 77.2% and a recall of 86.4%.
3.3 Annotation of full MEDLINE
We performed a run of the best CRF model on the full MEDLINE with 16 848 632 MEDLINE article entries (version as of July 13, 2007). In these entries, we have 8 975 073 abstracts. We tag titles and abstracts, altogether 2.2×109 tokens, in which there are 1 715 263 IUPAC entities in 875 102 MEDLINE database entries. The tagging is performed on a computer cluster using 48 machines with two Opteron AMD double core processors with 2.6 GHz and 8 GB main memory on each machine in 76.65 h (3.19 days). The operating system is Suse Linux Enterprise Server 9 (x86 64) with the Sun N1 Grid Engine 6.
From the found IUPAC entities, only 142 181 could be transformed to a structure (16.24%). The top 15 found terms from MEDLINE are shown in Table 2, the top 5 of the converted structures in Table 3 together with the most often used terms which lead to the normalization. To get an upper bound of convertible IUPAC names, we sample 100 000 correct names from data provided by NCBI (2007). From these, 30 028 (30%) are converted to structure information by OPSIN.
Table 2.
Top 15 found terms with their number of occurrences
| Frequency | Name |
|---|---|
| 16811 | N-methyl-D-aspartate |
| 15275 | 5-hydroxytryptamine |
| 11690 | 5-fluorouracil |
| 9001 | 6-hydroxydopamine |
| 7023 | glucose-6-phosphate |
| 6685 | N-ethylmaleimide |
| 5932 | N-acetylcysteine |
| 5178 | 12-O-tetradecanoylphorbol-13-acetate |
| 5032 | methyl |
| 4742 | N-acetylglucosamine |
| 4311 | benzo[a]pyrene |
| 4164 | 3-methylcholanthrene |
| 3991 | 4-aminopyridine |
| 3931 | 2,3,7,8-tetrachlorodibenzo-p-dioxin |
| 3979 | 5-hydroxyindoleacetic acid |
Table 3.
Top 5 found converted structures [applying OPSIN and CDK, drawn with Marvin (ChemAxon, 2007)] with their frequency and the frequency of occurrences of the top 3 terms which lead to the SMILES string
 |
4 SUMMARY AND DISCUSSION
In this article, we present our approach of finding IUPAC-like terms in text with CRF. We demonstrate that our IUPAC recognizer identifies entities with an F1 measure of 86.5% on a sampled independent test corpus built from MEDLINE. This corpus gives an estimation for all available abstracts from that database. These results show that restricting the recognition to the special class of IUPAC-like terms instead of all mentions of chemical names as focused on in Oscar3 (Corbett et al., 2007) increases the performance.
Using a tweaked corpus with correct IUPAC names shows that incorporating only complete IUPAC names in the training corpus is not sufficient. Obviously, the challenge is to recognize fragments and parts of IUPAC names. An error analysis of the final system on MEDLINE shows that boundary problems and the recognition of shorter chemical names lead to the main performance loss. This may be founded in ambiguities in the training data regarding these names and should be considered in a further extension of the training corpus. Preliminary results on an extended annotation of short names show an increase of precision to 91.4% (Kolářik et al., 2008).
When the IUPAC recognizer is applied to a hand-sampled patent corpus containing long enumerations and mixtures of different chemical nomenclatures the drop in performance is unexpectedly low with an F1 measure of 81.5%. Apparently, the loss of F1 measure in comparison to the MEDLINE corpus is due to a loss of precision rather then recall. Typical problems are finding the right borders of the chemical names in enumerations. From these results we cannot generalize that it is harder to find IUPAC names in patents than in abstracts.
In the feature evaluation we show that automatically generated features like Bag-of-Words and Autom. Prefixes/Suffixes together with Space information are the most important features influencing the performance of the system. The usage of combinations of these features alone e.g. Space together with prefixes and suffixes result in an F1 measure of 76.03%. In contrast, the static morphological features which are usually very important for a good generalization (together with other morphological features), in particular on the entity class of genes and proteins (Klinger et al., 2007b) do not have such a high impact here. Remarkably, the Prefix/Suffix lists (used for generalization purposes) appear to be of very low importance indicated by nearly no loss when left out. When used as the only feature, no positive result can be obtained. However, when combined with the feature Spaces, the results are surprisingly high (70.71% F1 measure).
Higher orders of the CRF in combination with high order offset conjunctions lead to the best results observed (F1 measure 85.6%) on the MEDLINE test corpus with an CRF order 3 and an offset conjunction of 2. On this corpus also the direct dependency of training and labeling durations to the orders of CRF and offset conjunction are shown (cf. Fig. 7).
Despite of the analysis given here for IUPAC entities, the open question remains, in which cases a representation of context information on the labels should be preferred in comparison to a representation of context information in the text, in form of features, used here by incorporating offset conjunction. To our knowledge, no deeper analysis is published about that topic so far. Our own experiments with different orders of CRF and offset conjunction in the field of gene and protein names showed that with higher orders the results tend to get worse, probably because of more needed training data when more complex dependencies are modeled (data not shown here).
In a final test, the full MEDLINE was labeled showing the scalability of the implementation. The highest frequency (without normalization) is almost 17 000 mentions of one term (Table 2). A conversion of the names to its corresponding structure show that only a minor part (below 20%) can be processed (without evaluating the correctness of the conversion). From the 15 most frequent chemical names only one can be converted (4-aminopyridine; cf. Table 3). Even from correct names provided by the NCBI in the database PubChem, only 30% can be converted. Unfortunately, it is not allowed to evaluate the conversion rate of the commercial tools for academic applications. Difficulties in the name to structure conversion are mixed nomenclatures and formally incorrect IUPAC and chemical names instead of correct nomenclature. We conclude that name-to-structure conversion in its current form seems to be a persistent scientific challenge.
In the future it is necessary to combine different existing tools and programs to be developed which find mentions of trivial names, formulas, IUPAC names, InChI, SMILES, group names, etc. and determine the intersection in their results and enable them also to find combined terms (e.g. in which part of the names follows the nomenclature of SMILES and another part follows IUPAC nomenclature). For that purpose, a representative, comprehensive test corpus including all these entities has to be developed.
Another goal is to combine the knowledge in drawn structures with the information in text, using the results provided by tools like chemOCR (Algorri et al., 2007; Zimmermann et al., 2005) which converts drawn structures into computer interpretable representations.
ACKNOWLEDGEMENTS
We thank all our co-workers, especially Erfan Younesi, Harsha Gurulingappa for annotation and Katrin Tomanek (University of Jena) and Theo Mevissen for fruitful discussions.
Funding: This work has been partially funded by the MPG-FhG Machine Learning Collaboration (http://lip.fml.tuebingen.mpg.de/).
Conflict of Interest: none declared.
Footnotes
2U.S. National Library of Medicine (2007), http://mplus.nlm.nih.gov/medlineplus/druginformation.html
5The colors here show the entity: red for IUPAC entities, blue for MODIFIER entities
6Number of sentences is detected with the JULIELab sentence splitter (http://www.julielab.de/,Tomanek et al., 2007).
7In the I,O,B-format like mentioned above for the existence of one entity there are eight possible transitions: B→B, O→O, I→I, B→I, I→B, I→O, B→O and O→B.
9Version of October 11, 2006, http://oscar3-chem.sourceforge.net
10Version 1.0.1 of June 26, 2007, cdk.sourceforge.net/
REFERENCES
- ACDLabs. ACDName. Software. 2007 Available at http://www.acdlabs.com/products/name_lab/name/(last accessed date December 18, 2007)
- Algorri M-E, et al. Reconstruction of chemical molecules from images. Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society EMBC 2007; IEEE Engineering in Medicine and Biology Society (EMBS), Lyon, France. 2007. pp. 4609–4612. [DOI] [PubMed] [Google Scholar]
- Anstein S, et al. Identifying and classifying terms in the life sciences: the case of chemical terminology. In: Calzolari N, et al., editors. Proceedings of the Fifth Language Resources and Evaluation Conference. Genoa, Italy: ELRA-ELDA; 2006. pp. 1095–1098. [Google Scholar]
- Bishop CM, et al. Pattern Recognition and Machine Learning. Berlin: Springer; 2006. [Google Scholar]
- CambridgeSoft. Name=struct. Software. 2007 Available at http://www.cambridgesoft.com/databases/details/?db=16(last accessed date December 18, 2007)
- ChemAxon. Marvin. Software. 2007 Available at http://www.chemaxon.com/marvin/(last accessed on January 11, 2008)
- Corbett P, Murray-Rust P. High-throughput identification of chemistry in life science texts. In: Berthold MR, et al., editors. 2nd International Symposium on Computational Life Science (CompLife 2006, LNBI 4216) Berlin, Heidelberg: Springer-Verlag; 2006. pp. 107–118. [Google Scholar]
- Corbett P, et al. BioNLP 2007: Biological, Translational, and Clinical Language Processing. Prague: Association for Computational Linguistics; 2007. Annotation of chemical named entities; pp. 57–64. [Google Scholar]
- Corbett P. Oscar3. Software. 2007 Available at http://oscar3-chem.sourceforge.net, (last accessed date December 13, 2007)
- Efron B, Tibshirani RJ. An Introduction to the Bootstrap. New York: Chapman & Hall/CRC; 1993. [Google Scholar]
- Eigner-Pitto V, et al. Mining, storage, retrieval: the challenge of integrating chemoinformatics with chemical structure recognition in text and images. Talk on 5th Fraunhofer Symposium on Text Mining in the Life Sciences. 2007 Available at http://www.scai.fhg.de/tms07.html(last accessed date December 18, 2007)
- Eller GA. Improving the quality of published chemical names with nomenclature software. Molecules. 2006;11:915–928. doi: 10.3390/11110915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich CM, et al. Biomedical and chemical named entity recognition with conditional random fields: the advantage of dictionary features. In: Ananiadou S, Fluck J, editors. Proceedings of the Second International Symposium on Semantic Mining in Biomedicine (SMBM 2006) Vol. 7. UK: BioMed Central Ltd, London; 2006. pp. 85–89. BMC Bioinformatics 2006. [Google Scholar]
- Guzikowski AP, et al. Synthesis of racemic 6,7,8,9-tetrahydro-1h-1-benzazepine-2,5-diones as antagonists of n-methyl-d-aspartate (nmda) and α-amino-3-hydroxy-5-methylisoxazole-4- propionic acid (ampa) receptors. J. Med. Chem. 1997;40:2424–2429. doi: 10.1021/jm960724e. [DOI] [PubMed] [Google Scholar]
- Hanisch D, et al. ProMiner: rule-based protein and gene entity recognition. BMC Bioinformatics. 2005;6(Suppl. 1)((S14)) doi: 10.1186/1471-2105-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman L, et al., editors. Proceedings of the Second BioCreative. Madrid, Spain: Challenge Evaluation Workshop. Centro Nacional de Investigaciones Oncologicas, CNIO; 2007. [Google Scholar]
- Humana I. Top 50 drugs brand-name prescribed. 2005 Available at http://apps.humana.com/prescription_benefits_and_services/includes/Top50BrandDrugs.pdf(last accessed date December 14, 2007.
- Kemp N, Lynch M. The extraction of information from the text of chemical patents. 1. identification of specific chemical names. J. Chem. Inf. Comput. Sci. 1998;38:544–551. [Google Scholar]
- Klinger R, Tomanek K. Technical Report TR07-2-013. Department of Computer Science, Dortmund University of Technology; 2007. Classical Probabilistic Models and Conditional Random Fields. ISSN 1864-4503. [Google Scholar]
- Klinger R, et al. Identifying gene specific variations in biomedical text. J. Bioinform. Computat. Biol. 2007a;5:1277–1296. doi: 10.1142/s0219720007003156. [DOI] [PubMed] [Google Scholar]
- Klinger R, et al. Proceedings of the Second BioCreative Challenge Evaluation Workshop. CNIO, Madrid, Spain: Centro Nacional de Investigaciones Oncologicas; 2007b. Named entity recognition with combinations of conditional random fields; pp. 89–91. [Google Scholar]
- Kolářik C, et al. Identification of new drug classification terms in textual resources. Bioinformatics. 2007;23:i264–i272. doi: 10.1093/bioinformatics/btm196. [DOI] [PubMed] [Google Scholar]
- Kolářik C, et al. Workshop on Building and evaluating resources for biomedical text mining (6th edition of the Language Resources and Evaluation Conference) 2008. Chemical names: terminological resources and corpora annotation. [Google Scholar]
- Kschischang F, et al. Factor graphs and the sum-product algorithm. IEEE T. Inform. Theory. 2001;47:498–519. [Google Scholar]
- Lafferty JD, et al. Proceedings of the Eighteenth International Conference on Machine Learning (ICML 2001) San Francisco, CA, USA: Morgan Kaufmann Publishers; 2001. Conditional random fields: probabilistic models for segmenting and labeling sequence data; pp. 282–289. [Google Scholar]
- McCallum AK. MALLET: a machine learning for language toolkit. 2002 Available at http://mallet.cs.umass.edu(last accessed May 5, 2008)
- McDonald RT, et al. An entity tagger for recognizing acquired genomic variations in cancer literature. Bioinformatics. 2004;20:3249–3251. doi: 10.1093/bioinformatics/bth350. [DOI] [PubMed] [Google Scholar]
- McDonald R, Pereira F . Identifying gene and protein mentions in text using conditional random fields. BMC Bioinformatics. 2005;6(Suppl. 1):S6. doi: 10.1186/1471-2105-6-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaught AD, Wilkinson A. Compendium of Chemical Terminology – the Gold Book. Oxford, UK: Blackwell Science; 1997. [Google Scholar]
- Murray-Rust P. Chemical markup language: a simple introduction to structured documents. World Wide Web J. 1997;2:135–147. [Google Scholar]
- Narayanaswamy M, et al. Proceedings of the Pacific Symposium on Biocomputing. 2003. A biological named entity recognizer; pp. 427–438. [DOI] [PubMed] [Google Scholar]
- NCBI. Pubchem data. Online. 2007 Available at ftp://ftp.ncbi.nlm.nih.gov/pubchem/Compound/CURRENT-Full/XML/(last accessed date September 5, 2007)
- Nocedal J. Updating Quasi-Newton matrices with limited storage. Math. Comput. 1980;35:773–782. [Google Scholar]
- OpenEye. Lexichem. Software. 2007 Available at http://www.eyesopen.com/products/toolkits/lexichem.html(last accessed date December 18, 2007)
- Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- Rebholz-Schuhmann D, et al. Ebimed – text crunching to gather facts for proteins from medline. newblock. Bioinformatics. 2007;23:237–244. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- Reyle U. Understanding chemical terminology. Terminology. 2006;12:111–136. [Google Scholar]
- Rhodes J, et al. Mining patents using molecular similarity search. Proceedings of the Pacific Symposium on Biocomputing. 2007;Vol. 12:304–315. [PubMed] [Google Scholar]
- Schölkopf B, Smola AJ. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond: Support Vector Machines, Regularization, Optimization and Beyond (Adaptive Computation and Machine Learning) Cambridge, MA: The MIT Press; 2002. [Google Scholar]
- Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics. 2005;21:3191–3192. doi: 10.1093/bioinformatics/bti475. [DOI] [PubMed] [Google Scholar]
- Steinbeck C, et al. The chemistry development kit (cdk): an open-source java library for chemo- and bioinformatics. J. Chem. Inf. Comput. Sci. 2003;43:493–500. doi: 10.1021/ci025584y. Available at cdk.sourceforge.net/(last accessed date December 18, 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, et al. Proceedings of the International World Wide Web Conference. Canada: Banff, Alberta; 2007. Extraction and search of chemical formulae in text documents on the web; pp. 251–260. [Google Scholar]
- Tomanek K, et al. A reappriasal of sentence and token splitting for life science documents. Proceedings of the 12th World Congress on Medical Informatics. 2007 Available at http://www.julielab.de/(last accessed date December 16, 2007) [PubMed]
- U.S. National Library of Medicine. Medlineplus. 2007 Available at http://www.nlm.nih.gov/medlineplus/druginformation.html(last accessed date September 1, 2008)
- Weininger D. Smiles, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- Wilbur J, et al. Proceedings of the Second BioCreative Challenge Evaluation Workshop. CNIO, Madrid, Spain: Centro Nacional de Investigaciones Oncologicas; 2007. Biocreative 2. gene mention task; pp. 7–9. [Google Scholar]
- Wishart DS, et al. Drugbank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. Available at http://redpoll.pharmacy.ualberta.ca/drugbank/(last accessed date July 16, 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, et al. Proceedings of the 1st German Conference on Chemoinformatics. Germany: Goslar; 2005. Combating illiteracy in chemistry: towards computer-based chemical structure reconstruction. [Google Scholar]