Abstract
Motivation: Understanding the role of genetics in diseases is one of the most important aims of the biological sciences. The completion of the Human Genome Project has led to a rapid increase in the number of publications in this area. However, the coverage of curated databases that provide information manually extracted from the literature is limited. Another challenge is that determining disease-related genes requires laborious experiments. Therefore, predicting good candidate genes before experimental analysis will save time and effort. We introduce an automatic approach based on text mining and network analysis to predict gene-disease associations. We collected an initial set of known disease-related genes and built an interaction network by automatic literature mining based on dependency parsing and support vector machines. Our hypothesis is that the central genes in this disease-specific network are likely to be related to the disease. We used the degree, eigenvector, betweenness and closeness centrality metrics to rank the genes in the network.
Results: The proposed approach can be used to extract known and to infer unknown gene-disease associations. We evaluated the approach for prostate cancer. Eigenvector and degree centrality achieved high accuracy. A total of 95% of the top 20 genes ranked by these methods are confirmed to be related to prostate cancer. On the other hand, betweenness and closeness centrality predicted more genes whose relation to the disease is currently unknown and are candidates for experimental study.
Availability: A web-based system for browsing the disease-specific gene-interaction networks is available at: http://gin.ncibi.org
Contact: radev@umich.edu
1 INTRODUCTION
The completion of the Human Genome Project has opened the door to new research opportunities and challenges. One of the major goals of the post-genome era is to understand the role of genetics in human health and diseases International Human Genome Sequencing Consortium, 2001; Venter et al., 2001. While fewer than 100 gene-disease associations were known before the project started in 1990, currently more than 1400 have been identified.1 Determining gene-disease associations will enhance the development of new techniques for prevention, diagnosis and treatment of the diseases.
One of the most well-known databases that stores gene-disease associations is Online Mendelian Inheritance in Man (OMIM, 2007), which provides summaries of publications about gene-disease relationships. However, it usually takes time before new discoveries are included in the curated databases. Given that the amount of biomedical literature regarding the identification of disease genes is increasing rapidly, one of the challenges that scientists in this domain face is that most of the relevant information remains hidden in the unstructured text of the published papers.
Another challenge is that the identification of new disease genes requires laborious experiments. For example, the genetic linkage analysis method is successfully used to determine the genomic regions that are associated with a disease. However, these regions often contain hundreds of genes and experimentally identifying the actual disease genes out of the large amount of candidate genes require considerable effort and time.
To address these challenges, we propose an approach based on integrating automatic text mining and network analysis methods to extract known disease genes and to predict unknown disease genes, which can be good candidates for experimental study. We started by collecting an initial set of genes (seed genes) known to be related to a disease from curated databases such as OMIM. We then used an information extraction approach based on dependency parsing (de Marneffe et al., 2006) and support vector machines (SVM) (Joachims, 1999) to build a disease-specific gene-interaction network. A syntactic parse tree represents the syntactic constituent structure of a sentence. On the other hand, a dependency parse tree captures the semantic predicate-argument dependencies among the words of a sentence. The nodes of a dependency parse tree represent the words of a sentence and the edges represent the types of the dependencies among the words such as subject, object and modifier. We generated the dependency parses of the sentences that contain at least two seed or neighbor genes (genes that interact with seed genes), and extracted the paths between all pairs of genes from the dependency parse trees. The motivating assumption is that the path between a pair of gene names in the dependency parse tree of a sentence captures the semantic relationship between them. We defined an edit distance-based kernel function among these dependency paths and used SVM to classify the sentences as describing an interaction between a gene pair or not. We have introduced this interaction extraction approach in (Erkan et al., 2007) and have achieved significant improvement (55.61% F-score performance for the AIMED data set2) compared to previous results in the literature.
Our main hypothesis is that the most central genes in an interaction network for a disease are likely to be related to the disease. Therefore, after extracting the interactions from the literature, we constructed a disease-specific gene-interaction network, where the nodes are the seed genes and their neighbors, and two genes are linked, if we have extracted an interaction between them. Next, we ranked the genes in the network by degree, eigenvector, betweenness and closeness network centrality metrics. To our knowledge, this is the first effort of building a gene-interaction network by automatic literature mining and applying network centrality to predict gene-disease associations on that network.
2 RELATED WORK
The number of biomedical publications is increasing rapidly. Currently, there are over 14 million articles indexed in PubMed.3 It is difficult for curators to detect and curate the information available in the biomedical literature. Therefore, the curated databases can cover only a small portion of the available information. Thus, extracting the available knowledge from the huge amount of biomedical literature has become a major challenge. Most of the previous studies that use text mining to extract gene-disease associations from the biomedical literature are based on the co-occurrence frequencies of genes and diseases. For example, Adamic et al. (2002) presented a method based on determining whether the frequency of occurrence of a gene in articles that mention a certain disease is statistically significantly higher than the expected frequency of occurrence computed by the Binomial distribution. They evaluated their approach for breast cancer and confirmed the relevance of 7 out of 10 highest ranked genes to breast cancer by using a human edited breast cancer gene database.4 Another relevant study is conducted by Al-Mubaid and Singh (2005). Given a disease name, a set of documents that contain the disease name (positive-document set) and a randomly-selected document set (negative-document set) are extracted. Co-occurrence and term frequency-based concepts from information theory are used to determine the genes that are significantly associated with the disease. The authors found six genes significantly associated with Alzheimer's disease and confirmed the correctness of their results through articles from PubMed.
Determining the genes that cause a disease usually requires laborious experiments over a large number of candidate genes. Therefore, another challenge in the domain is predicting and prioritizing candidate disease genes, which can further be validated by detailed experiments. Most proposed data mining approaches make use of available curated databases and predict gene-disease associations by using keyword similarity to known disease genes and phenotypes. For example, GeneSeeker (van Driel et al., 2002) is a web-based system that integrates positional and expression/phenotypic data from nine different human and mouse databases and provides a quick overview of interesting candidate genes. The authors evaluated their approach for ten syndromes. On average, the system reduced a list of 163 candidate genes to a list of 22 genes, which still contained the correct disease gene. Freudenberg and Propping (2002 proposed a method based on clustering diseases based on their phenotypic similarity, which is computed by considering the similarity of the disease index terms in the OMIM database. Candidate genes for a disease in a cluster are predicted by selecting functionally similar genes to the genes associated with the other diseases in the cluster. The authors performed a leave-one-out cross-validation of 878 diseases using 10 672, genes. They reported that in roughly one-third of the diseases, the correct disease gene was within the top scoring 321 genes, and in the two-third of the diseases, the correct disease gene was within the top scoring 1 600 genes. The G2D system (Perez-Iratxeta et al., 2002, 2005) uses a method based on fuzzy logic and co-occurrence of relevant keywords in biomedical abstracts to associate pathological conditions with gene ontology (GO) terms (Ashburner et al., 2000). Prediction of candidate genes is performed by searching for genes homologous to the GO-annotated and disease-associated genes. The authors evaluated their system with 100 known disease-associated genes and found that the correct disease gene was among the 8 top-scoring genes with 25% chance, and among the 30 top-scoring genes with 50% chance.
Protein interactions play important roles in vital biological processes such as cell cycle control, metabolic and signaling pathways and disease pathways. These interactions can be represented as complex networks, where the nodes are the proteins and the edges represent the interactions between the pairs of proteins they connect. This representation makes it possible to analyze protein-interaction networks from a graph theory and complex networks perspective. Most graph-theoretic studies of protein-interaction networks extract the interactions from curated databases (Jeong et al., 2001; Schwikowski et al. 2000; Spirin and Mirny 2003; Wuchty et al. 2003). There are also recent studies that analyze protein-interaction networks constructed by mining the literature (Chen and Sharp 2004; Hoffmann and Valencia 2005). It has been shown that the interaction networks constructed in either way, share similar topological properties such as being small-world and scale-free, with each other and with various non-biological complex systems such as the WWW, the Internet, and social networks (Chen and Sharp 2004; Hoffmann and Valencia 2005; Jeong et al., 2001.
Graph-theoretic analysis of protein-interaction networks has been successfully applied in many biological domains. For example, protein-interaction networks have been used for evolutionary comparisons among organisms (Wuchty et al., 2003), for identifying functional modules and network motifs (Spirin and Mirny 2003) and for predicting functional annotations based on network connectivity (Schwikowski et al., 2000). Schwikowski et al. (2000) used a majority-rule method that assigns to a protein the function that occurs most commonly among its neighbors and reported an accuracy of 70% for the yeast protein-interaction network.
Recently, protein-interaction networks have also been used to predict gene-disease associations (Chen et al., 2006; Gonzalez et al., 2007). Chen et al. (2006) used an initial gene list (seed genes) for Alzheimer's from the OMIM database, and built an interaction network by extracting the interactions of the corresponding proteins from the Online Predicted Human Interaction Database (OPHID) (Brown and Jurisica 2005). They defined a heuristic scoring function for the genes based on their connectedness in the graph. When building the network, only the interactions among the seed genes and the interactions of seed genes with their neighbors were considered. The interactions among the neighbors were not taken into account. Thus, this approach is biased in favor of the seed genes. A total of 19 of the top scoring genes are seed and only one is a non-seed (inferred) gene. Gonzalez et al. (2007) started with a list of seed genes obtained from the automatically mined CBioC database and created an interaction network by extracting the interactions of the seed genes from the CBioC database (Baral et al., 2005) and curated databases such as BIND (Bader et al., 2003) and MINT (Zanzoni et al., 2002). Like (Chen et al., 2006), they did not take into account the interactions among the non-seed genes. To eliminate the bias in favor of the seed genes, they refined the scoring function by considering just the interactions with seed genes and including a measure for the impact of each gene on the connectivity of the network. A total of 45% of their top scoring 20 genes are non-seed and 66.67% of these non-seed genes are correctly inferred genes, i.e. reported in OMIM or in the literature as being related to the disease.
Our approach is different from previous approaches in two aspects. First, we create a gene-interaction network by automatic literature mining. Second, we use degree, eigenvector, betweenness and closeness centrality to rank the gene-disease associations. Centrality measures have successfully been applied in other biological domains. For example, Jeong et al. (2001) studied the protein–protein interaction network of yeast in order to predict lethal mutations. They showed that the network is tolerant to random errors, whereas errors related to the most central proteins (in terms of degree) cause lethality. Similarly, Joy et al. (2005) and Hahn and Kern (2005) found that there is an association between the betweenness centrality and the essentiality of a gene (a gene is essential if the organism dies when the gene malfunctions). Goh et al. (2007) showed that central genes based on degree are also essential. Centrality measures have originally been developed and used in non-biological domains. For example, the Pagerank algorithm underlying the popular search engine Google is based on eigenvector centrality to rank the web pages (Page et al., 1998). Recently, eigenvector centrality has also been used in document summarization to identify the most important sentences (Erkan and Radev 2004) as well as to identify the most influential members of the US Senate (Fader et al., 2007).
We built a disease-specific interaction network around a list of seed genes that are known to be related to a disease. Besides the interactions involving seed genes, we also considered the interactions among non-seed genes (genes that interact with at least one seed gene). We used centrality measures to infer gene-disease associations. Our hypothesis is that, the genes that are central in the created disease-specific network are likely to be related to the disease. Our results confirmed this hypothesis. We achieved a 75% non-seed gene proportion among the top 20 central genes and 93.33% accuracy in relatedness of these non-seed genes to the specified disease.
3 METHODS
3.1 Corpus
To construct the literature-mined gene-interaction network we used 48 245 articles from PubMed Central (PMC) Open Access,5 which is an open access digital archive of biomedical and life science journals. Unlike PubMed, articles in PMC Open Access are full-text.
We pre-processed the corpus by segmenting the articles into sentences with MxTerminator (Reynar and Ratnaparkhi 1997). Gene names are annotated with the Genia Tagger (Tsuruoka et al., 2005), whose developers report an F-score performance of 71.37% for biological named entity recognition.6
3.2 Initial list of seed genes
To build an interaction network for a disease and to infer gene-disease associations from the network properties, we started with an initial list of seed genes known to be related to the disease.
We evaluated our system for prostate cancer. We compiled 15 prostate cancer seed genes from the Morbid Map component of the OMIM database. OMIM Morbid Map shows the cytogenetic map location of disease-associated genes described in OMIM. Table 1 lists the seed genes for prostate cancer.
Table 1.
The prostate cancer seed genes retrieved from OMIM Morbid Map
| Gene | Description |
|---|---|
| AR | Androgen Receptor |
| BRCA2 | Breast cancer 2, early onset |
| MSR1 | Macrophage scavenger receptor 1 |
| EPHB2 | EPH receptor B2 |
| KLF6 | Kruppel-like factor 6 |
| MAD1L1 | MAD1 mitotic arrest deficient-like 1 (yeast) |
| HIP1 | Huntingtin interacting protein 1 |
| CD82 | CD82 molecule |
| ELAC2 | ElaC homolog 2 (Escherichia coli) |
| MXI1 | MAX interactor 1 |
| PTEN | Phosphatase and tensin homolog |
| RNASEL | Ribonuclease L (2′, 5′-oligoisoadenylate synthetase-dependent) |
| HPC1 | Hereditary prostate cancer 1 |
| CHEK2 | CHK2 checkpoint homolog (Schizosaccharomyces pombe) |
| PCAP | Predisposing for prostate cancer |
3.3 Gene name normalization
A gene name might have several different synonyms. For instance, AR which stands for the androgen receptor gene, might appear as AIS, NR3C4, SMAX1, HUMARA, DHTR or SBMA in biological text. To normalize the gene names tagged by Genia Tagger and the seed gene names so that each gene is represented by a single node in the interaction network, we used the HUGO Gene Nomenclature Committee (HGNC) database7 (Wain et al. 2004), which contains 24 680 records. We matched the tagged gene names against the approved symbol, approved name, previous symbols, previous names, aliases and name aliases fields of the database. We unified each tagged gene name with its corresponding approved gene symbol.
3.4 Extracting the gene-interaction network from the literature
Although there are public databases that store the interactions among proteins, they only cover a small portion of the information available in the rapidly increasing biomedical literature. Therefore, the development and application of text mining approaches to automatically extract protein interactions from text is crucial to utilize the information hidden in the unstructured text of biomedical articles.
3.4.1 Sentence filtering
We used the initial list of seed genes to build a disease-specific gene-interaction network mined automatically from the literature. Before applying our text mining approach to extract gene-interactions, we selected the potential interaction sentences from the PMC Open Access corpus. A list of interaction words, which consists of 45 noun and 53 verb roots, was compiled from the literature. We extended the list to contain all the inflected forms of the words and spelling variations such as coactivate/co-activate and localize/localise. Our assumption is that a sentence that describes an interaction between a pair of genes should contain at least two genes and an interaction word. We created an expanded gene list, by including the seed genes and all the genes that appear in the same sentence with a seed gene. We filtered out the sentences that do not contain an interaction word and at least two genes from the expanded gene list.
3.4.2 Sentence classification based on dependency parsing and SVM
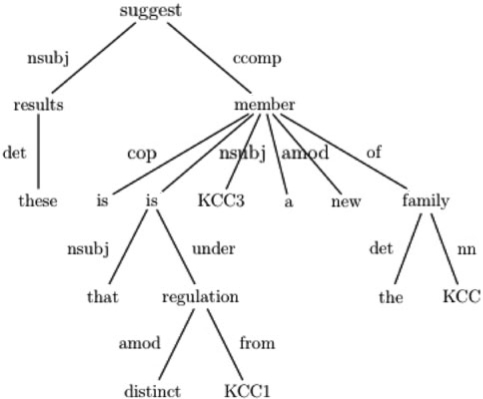
To extract the gene interactions from text, we generated the dependency parses of the sentences that we analyze, making use of the dependency relationships among the words. We parsed the sentences with the Stanford Parser8 (de Marneffe et al., 2006). From the dependency parse tree of each sentence we extracted the shortest paths between all gene pairs. There may be multiple paths between a gene pair, if either of the genes appears multiple times in the sentence. Figure 1 shows the dependency tree we obtained for the sentence ‘These results suggest KCC3 is a new member of the KCC family that is under distinct regulation from KCC1’. The shortest path (in this case the only path) between the genes KCC3 and KCC1 is ‘KCC3-nsubj-member-is-under-regulation-from-KCC1’.
Fig. 1.
The dependency tree of the sentence ‘These results suggest KCC3 is a new member of the KCC family that is under distinct regulation from KCC1’.
Next, we defined the similarity between two dependency paths based on word-based edit distance. Edit distance between two strings is the minimum number of edit operations that have to be performed to transform the first string into the second. The operations are defined as insertion, deletion or substitution of a single word. We normalized edit distance by dividing it by the length (number of words) of the longer path, so that it takes values in the range [0,1]. We converted the distance measure into a similarity measure as follows.
| (1) |
A well-defined kernel function should be symmetric and positive definite. Cortes et al. (2004) proved that the edit kernel is not always positive definite. However, it is possible to make the kernel matrix positive definite by adjusting the γ parameter, which is a positive real number. We tuned the γ parameter with cross-validation experiments to 4.5.
We integrated this similarity measure as a kernel function to SVM by plugging it in the SVMlight package (Joachims 1999). We trained the system by combining the AIMED9 and CB10 data sets, which were pre-processed by replicating each sentence for each different gene pair.11 The summary of the pre-processed training data sets is provided in Table 2. The trained system is used to classify the new sentences as describing an interaction between a gene pair or not. We have introduced this interaction extraction approach in a recent study (Erkan et al., 2007), and shown that it achieves an F-score performance of 84.96% for the CB data set and 55.61% for the AIMED data set, which is to our knowledge higher than the performances reported for the AIMED data set so far. The reader can refer to (Erkan et al., 2007) for details of the gene interaction extraction method. We have also used this method to provide annotations for the BioCreative Meta-Server by classifying abstracts as describing a protein interaction or not (Leitner et al., 2008).
Table 2.
Training data sets
| Data set | Sentences | +Sentences | − Sentences |
|---|---|---|---|
| AIMED | 4026 | 951 | 3075 |
| CB | 4056 | 2202 | 1854 |
3.5 Network centrality for inferring gene-disease associations
Centrality of a node in a graph defines how important a node in the graph is.
3.5.1 Degree centrality
A graph can be represented by an adjacency matrix A, where Aij=1 if there is an edge between nodes i and j; and Aij=0 if there does not exist an edge between nodes i and j. Degree centrality is the simplest network centrality measure. It only takes into account the degree of a node, which is the number of nodes that a given node is connected to (Freeman 1979). The degree ki of node i is calculated as follows.
| (2) |
Degree centrality measures the extent of influence that a node has on the network. The more neighbors a node has, the more important it is.
3.5.2 Eigenvector centrality
In degree centrality each neighbor contributes equally to the centrality of a node. However, in many real-world situations not all the relationships (connections) between nodes in a network are equally important in determining the centrality of a node. This notion is defined as ‘prestige’ in social networks. Intuitively, the prestige of a person does not only depend on the number of acquaintances he has, but also how prestigious his acquaintances are. A node in a network is more central if it is connected to many central nodes. The centrality xi of node i is proportional to the sum of the centralities of its neighbors (Newman 2003):
| (3) |
Let's represent the centralities of the nodes as a vector x=(x1,x2, …, xn) and rewrite Equation 3 in matrix form.
| (4) |
Here, x is an eigenvector of the adjacency matrix A with eigenvalue λ. By Perron–Frobenius theorem, there is only one eigenvector x with all centrality values non-negative and this is the unique eigenvector that corresponds to the largest eigenvalue λ (Newman 2003). Eigenvector centrality assigns each node a centrality that not only depends on the quantity of its connections, but also on their qualities.
3.5.3 Closeness centrality
The closeness centrality of a node measures the centrality of a node based on how close it is to other nodes in the network. The smaller the total distance of a node to other nodes, the higher its closeness is. The distance between two nodes is defined as the length of the shortest path between them. We calculate closeness centrality measure for a node by inverting the sum of the distances from it to other nodes in the network (Freeman, 1979).
3.5.4 Betweenness centrality
The betweenness centrality of a node is the number of shortest paths between other nodes that run through the node in interest (Freeman 1977). For a node x, this measure is computed by taking the sum of the number of shortest paths between pairs of nodes that pass through node x divided by the total number of shortest paths between pairs of nodes. Betweenness centrality characterizes the control of a node over the information flow of the network. A node is considered central if it appears on many paths that connect pairs of nodes (i.e. it acts as a bridge between pairs of nodes in the network).
4 RESULTS AND DISCUSSION
4.1 Properties of the prostate cancer network
The prostate cancer-related gene-interaction network consists of 226 nodes (distinct genes) and 1 187 edges (interactions among these genes). The resulting graph is a small world network with diameter 6 and average shortest path 2.57. The clustering coefficient (Watts and Strogatz 1998) is 0.4497, which is significantly higher than the clustering coefficient of a random graph with the same number of vertices (0.0487). The degree distribution of the network is a power law with exponent 2.24. The power-law degree distribution and small-world characteristics of the network confirm the results of previous studies (Chen and Sharp 2004; Hoffmann and Valencia 2005; Jeong et al., 2001).
4.2 Centrality and gene-disease associations
We used the Prostate Gene DataBase (PGDB) (Li et al., 2003), which is a curated database of genes related to prostate cancer, for the initial evaluation of the methods. In the next sub-section we analyze the most central 20 genes in more detail.
Table 3 shows the precisions of the methods for the top ranked n genes, i.e. the percentage of the top ranked ‘n’ genes that are marked by PGDB as being related to prostate cancer. The entire network (226 genes) is the neighborhood of the seed genes and 17.70% of the 226 genes are related to prostate cancer. As the centrality score of the genes decreases (i.e. as ‘n’ increases), the percentage of the genes related to prostate cancer decreases, and the performances of the four methods converge to each other. For genes with high centrality, eigenvector, degree and betweenness metrics achieve similar performances, whereas closeness centrality performs worse than them.
Table 3.
Percentage of top n genes associated with prostate cancer based on the PGDB
| Top n | Degree | Eigenvector | Betweenness | Closeness | Baseline |
|---|---|---|---|---|---|
| 10 | 80.00 | 80.00 | 90.00 | 70.00 | 50.00 |
| 20 | 75.00 | 80.00 | 70.00 | 55.00 | 45.00 |
| 30 | 60.00 | 63.33 | 63.33 | 56.67 | 43.33 |
| 40 | 55.00 | 57.50 | 52.50 | 47.50 | 32.50 |
| 50 | 46.00 | 50.00 | 48.00 | 42.00 | 28.00 |
| 75 | 33.33 | 36.00 | 34.67 | 33.33 | 34.67 |
| 100 | 26.00 | 28.00 | 26.00 | 27.00 | 27.00 |
| 125 | 23.20 | 25.60 | 23.20 | 23.30 | 22.40 |
| 150 | 20.67 | 22.00 | 20.00 | 20.00 | 18.67 |
| 175 | 18.29 | 20.57 | 18.29 | 18.29 | 17.14 |
| 200 | 17.50 | 19.00 | 18.50 | 17.00 | 15.00 |
| 226 | 17.70 | 17.70 | 17.70 | 17.70 | 13.27 |
For baseline evaluation, we created a co-occurrence network by linking two genes if they appear in the same sentence and at least one of them is a seed gene. We ranked the genes by the number of connections they make with the seed genes.
Betweenness centrality achieves the highest precision (90%) for the top 10 genes. The precision of degree and eigenvector centrality measures is 80%, and the precision of closeness centrality is 70%. The baseline approach performs considerably worse (50% precision).
When we consider the top 20 genes, the highest precision is achieved by eigenvector centrality (80%). Degree centrality follows eigenvector centrality with 75% precision, whereas the precision of betweenness centrality drops to 70% and the precision of closeness centrality drops to 55%. Degree, eigenvector and betweenness centrality perform significantly better than the baseline method (P-value <0.05, Fisher's Exact Test (Fisher, 1970).
To analyze the error tolerance of the gene-disease identification approach, we performed experiments by randomly removing edges from the gene-interaction network. When up to 25% of the edges were removed randomly from the network, there was no decrease in the precisions of the centrality metrics for the top 20 genes. An insignificant decrease in the precisions of the metrics was observed when 40% of the edges were removed. The precision of degree centrality dropped by 13.3% (from 75 to 65%), eigenvector centrality by 6.25%, betweenness centrality by 7.14% and closeness centrality by 9.1%. This shows that the proposed approach is robust against random errors.
4.3 Detailed analysis of the most central genes
For each centrality method, we performed a detailed evaluation for the top 20 ranked genes by finding evidence of their association to the disease from various resources as presented in Table 4. The descriptions of the genes are presented in Table 5. Seed genes are known to be related to the disease. To verify the newly found (inferred) genes, we first used the PGDB database. If a gene is not marked by PGDB as being related to prostate cancer, we manually searched for articles indexed in PubMed that state that the gene is related to prostate cancer and also checked whether the gene appears in the KEGG pathway for prostate cancer,12 which is a manually drawn pathway map of the currently known molecular interaction and reaction network for prostate cancer.
Table 4.
Genes inferred by degree, eigenvector, closeness and betweenness centralities
| Gene | Degree | Eigenvector | Closeness | Betweenness | Evidence | |
|---|---|---|---|---|---|---|
| TP53 | + | + | + | + | PGDB | |
| BRCA1 | + | + | + | + | PGDB | |
| EREG | + | + | + | + | None | |
| AKT1 | + | + | + | + | PGDB | |
| MAPK1 | + | + | + | + | Literature | (Hao et al.2007; Sarfaraz et al.2006) |
| TNF | + | + | + | + | PGDB | |
| CCND1 | + | + | + | + | PGDB | |
| MYC | + | + | + | + | PGDB | |
| APC | + | + | − | − | PGDB | |
| CDKN1B | + | + | + | − | PGDB | |
| MAPK8 | + | + | + | + | PGDB | |
| NR3C1 | − | + | + | − | Literature | (Wei et al.2007) |
| VEGFA | + | + | + | − | PGDB | |
| MDM2 | + | + | + | − | KEGG and Literature | (Wang et al.2003; Zhang et al.2003) |
| POLD1 | − | − | + | + | None | |
| SNORA62 | − | − | + | + | None | |
| CNTN2 | − | − | − | + | None | |
| PPA1 | − | − | − | + | None | |
| TMEM37 | − | − | + | − | None | |
| FZR1 | − | − | + | − | PGDB | |
| SSSCA1 | − | − | + | − | None | |
| BCL2 | + | − | − | − | PGDB | |
| INS | + | − | − | − | KEGG and Literature | (Ho et al.2003) |
‘+’indicates that the given gene is found by the centrality method with score ranking within the top 20 and ‘−’ indicates that the gene is not among the top 20 genes inferred by the method. Evidences for each gene-disease relationship are confirmed by using PGDB database, KEGG pathway for prostate cancer and articles indexed in PubMed.
Table 5.
Gene names normalized by Hugo and their description
| Gene | Description |
|---|---|
| TP53 | Tumor protein p53 (Li-Fraumeni syndrome) |
| BRCA1 | Breast cancer 1, early onset |
| EREG | Epiregulin |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| MAPK1 | Mitogen-activated protein kinase 1 |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) |
| CCND1 | Cyclin D1 |
| MYC | V-myc myelocytomatosis viral oncogene homolog (avian) |
| APC | Adenomatosis polyposis coli |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| MAPK8 | Mitogen-activated protein kinase 8 |
| NR3C1 | Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| VEGFA | Vascular endothelial growth factor A |
| MDM2 | Mouse double minute 2, human homolog of; p53-binding protein |
| POLD1 | Polymerase (DNA directed), delta 1, catalytic subunit 125kDa |
| SNORA62 | Small nucleolar RNA, H/ACA box 62 |
| CNTN2 | Contactin 2 (axonal) |
| PPA1 | Pyrophosphatase (inorganic) 1 |
| TMEM37 | Transmembrane protein 37 |
| FZR1 | Fizzy/cell division cycle 20 related 1 (Drosophila) |
| SSSCA1 | Sjogren's syndrome/scleroderma autoantigen 1 |
| BCL2 | B-cell CLL/lymphoma |
| INS | Insulin |
Twelve of the genes in Table 4 are confirmed to be related to prostate cancer by using the PGDB database. The centrality methods were able to find four genes, which are not included in PGDB, but were confirmed to be related to prostate cancer by manually searching for evidence in the literature (articles indexed in PubMed) and in the KEGG pathway for prostate cancer. Two genes (MDM2 and INS) are part of the KEGG pathway for prostate cancer. For these genes, we also found articles in the literature that support their association to prostate cancer. For example, (Wang et al. (2003) and Zhang et al. (2003)) state that ‘MDM2 has a role in prostate cancer growth via p53-dependent and p53-independent mechanisms’. For the INS (insulin) gene, Ho et al. (2003) state that ‘Polymorphism of the insulin gene is associated with increased prostate cancer risk’. Supportive evidence for the association of NR3C1 to prostate cancer is presented by Wei et al. (2007), who show that it is differentially expressed in androgen-independent prostate cancer. For the gene MAPK1, Sarfaraz et al. (2006) state that ‘apoptosis induced by cannabinoid receptor CB1 and CB2 agonists leads to activation of ERK1/2 leading to G1 cell cycle arrest in prostate cancer cells’. Here, ERK2 is a synonym of MAPK1. Another article that provides supportive evidence for the MAPK1-prostate cancer association includes the statement ‘lysophosphatidic acid (LPA), the receptor LPA(1), ERK2 and p38alpha are important regulators for prostate cancer cell invasion and thus could play a significant role in the development of metastasis’ (Hao et al., 2007). For the remaining seven genes in the table, we found neither positive nor negative evidence for their association to prostate cancer.
Using degree centrality, among its top 20 ranking genes, 5 genes of the original 15 seed genes are found (AR, BRCA2, CD82, PTEN and CHEK2). The remaining 15 genes (75% of the top 20 genes) are inferred genes in which we were able to confirm the association of 14 genes (93.33% of the inferred genes) to prostate cancer, except for 1 gene: EREG. For this exceptional gene, we did not find negative nor positive evidence, which implies that the gene may still potentially be a prostate cancer gene.
The result of eigenvector centrality is as successful as degree centrality method with 95% of the top ranked 20 genes having supportive evidence. Eigenvector centrality found 6 seed genes (AR, BRCA2, CD82, MXI1, PTEN and CHEK2) and 14 inferred genes. Out of the 14 inferred genes, 13 are confirmed (92.86% of the inferred genes) and the same gene EREG is not.
Using closeness centrality, we found 2 seed genes (AR and BRCA2) and inferred 18 new genes. A total of 13 of the inferred genes (72.22% of the inferred genes) have evidence, which indicate that they are related to prostate cancer and 5 inferred genes (EREG, POLD1, SNORA62, TMEM37 and SSSCA1) do not have such affirmative evidence.
Betweenness centrality found the most seed genes among the four centrality methods. In its result, we have 7 seed genes (AR, BRCA2, CD82, MXI1, PTEN, CHEK2 and KLF6) and 13 inferred genes, of which 8 inferred genes (61.54% of the inferred genes) are verified to have relation to the disease. The five inferred genes that we were not able to confirm are EREG, POLD1, SNORA62, CNTN2 and PPA1.
Table 6 lists the definitions used in Table 7, which shows the summary of the results for the top 20 genes.
Table 6.
Definitions used in the evaluation of the top 20 genes
| term | definition |
|---|---|
| Seed gene: | A gene, which is one of the prostate cancer genes retrieved from OMIM Morbid Map (i.e. one of the genes in Table 1) |
| Inferred gene: | A non-seed gene |
| Percentage of inferred genes: | (Number of inferred genes / 20) × 100 |
| Confirmed inferred gene: | An inferred gene found to be related to prostate cancer based on PGDB, KEGG pathway for prostate cancer and published articles |
| Percentage of confirmed inferred genes: | (Number of confirmed inferred genes / Number of inferred genes) × 100 |
| Percentage of confirmed genes: | ((Number of confirmed inferred genes + Number of seed genes) / 20) × 100 |
Table 7.
Summary of the results for the top 20 genes
| Degree | Eigenvector | Betweenness | Closeness | Baseline | |
|---|---|---|---|---|---|
| Number of seed genes | 5 | 6 | 7 | 2 | 3 |
| Number of inferred genes | 15 | 14 | 13 | 18 | 17 |
| Percentage of inferred genes | 75 | 70 | 65 | 90 | 85 |
| Number of confirmed inferred genes | 14 | 13 | 8 | 13 | 10 |
| Percentage of confirmed inferred genes | 93.33 | 92.86 | 61.54 | 72.22 | 58.82 |
| Percentage of confirmed genes | 95 | 95 | 75 | 75 | 65 |
We observed that degree and eigenvector centrality methods generate highly accurate results; 95% of the top ranked 20 genes are actually related to prostate cancer. They are significantly better than the baseline method in which only 65% of the top 20 genes are prostate cancer genes. We used Fisher's Exact Test (Fisher 1970) to measure the significance level of the differences in performances between the centrality methods and the baseline method. Degree and eigenvector centrality perform significantly better (P-value <0.05) than the baseline approach in terms of the percentage of the confirmed genes and confirmed inferred genes. These methods are good candidates for use in practice for mining existing genes related to a particular disease. On the other hand, although closeness and betweenness centrality methods are not statistically significantly better than the baseline method in finding known prostate cancer genes, compared to degree and eigenvector centrality they introduce more genes that are not currently identified as related to the disease of interest. These methods can be used to generate new hypothesis on gene-disease research, which are candidates for experimental validation. In our experiments, even though we were not able to find evidence of whether gene EREG is related to prostate cancer or not; the fact that all four centrality methods suggest that this gene gives more confidence to EREG-prostate cancer relation. We believe that EREG is a strong candidate for prostate cancer gene research.
As discussed in Section 2, the scoring function proposed by Chen et al. (2006) is based on the connectedness of the genes. However, the interactions among the non-seed genes are not considered. Thus, their approach is biased toward seed genes. Out of the top 20 genes, 19 are seed and only 1 gene is an inferred (non-seed) gene. Gonzalez et al. (2007) alleviate this bias by computing connectedness by considering only the interactions with the seed genes. However, they do not consider the interactions among the non-seed genes either. A total of 45% of their top scoring 20 genes are non-seed and 66.67% of these non-seed genes are correctly inferred genes. Our approach of building the network by literature mining, including the interactions among the non-seed genes, and applying network centrality measures achieved a higher proportion of non-seed (inferred) genes and a higher accuracy of the inferred genes. For example, with closeness centrality the proportion of inferred genes is 90 and 72.22% of these inferred genes are correct; with degree centrality the proportion of inferred genes is 75 and 93.33% of these genes are correct.
5 CONCLUSION
We have presented a new approach to predict gene-disease associations based on text mining and network analysis. We collected an initial list of seed genes known to be related to a disease and constructed a disease-specific gene-interaction network by extracting the interactions among the seed genes and their neighbors automatically from the biomedical literature by using SVM with dependency path edit kernel. Next, we used degree, eigenvector, closeness and betweenness centrality metrics to rank the genes in the network according to their relevance to the disease. We hypothesized that the genes that are central in the constructed disease-specific network are likely to be associated with the disease.
We evaluated our approach for prostate cancer and showed that degree and eigenvector centrality metrics achieve highly accurate results (95% of the top 20 genes are actually related to the disease), whereas closeness and betweenness centrality metrics introduce genes that are currently unknown to be related to the disease. We were able to extract genes, which are not marked as being related to prostate cancer by the curated PGDB even though there are recent articles that confirm the association of these genes with the disease. The proposed approach can be used to extract known gene-disease associations from the literature, as well as to infer unknown gene-disease associations which are good candidates for experimental analysis.
ACKNOWLEDGEMENTS
We would like to thank David J. States for his helpful comments, Alex Ade for his help with the PMC Open Access corpus and the members of the CLAIR (Computational Linguistics And Information Retrieval) group at the University of Michigan, in particular Anthony Fader and Joshua Gerrish, for their assistance with this project.
Funding: This work was supported in part by the NIH Grant U54 DA021519 to the National Center for Integrative Biomedical Informatics, NIH Grant R01 LM008106 and NSF Grant IIS 0534323.
Conflict of Interest: none declared.
Footnotes
11The pre-processed data sets are available at:http://belobog.si.umich.edu/ clair/biocreative
REFERENCES
- Adamic LA, et al. A literature based method for identifying gene-disease connections. Proceedings of the IEEE Computer Society Conference on Bioinformatics; Stanford, CA. 2002. pp. 109–117. [PubMed] [Google Scholar]
- Al-Mubaid H, Singh RK. A new text mining approach for finding protein-to-disease associations. Am J Biochem Biotechnol. 2005;1:145–152. [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G, et al. Bind – the biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral C, et al. The Genetics Society of America 1st International Biocurator Meeting. CA: Pacific Grove; 2005. Cbioc: web-based collaborative curation of molecular interaction data from biomedical literature. [Google Scholar]
- Brown K, Jurisica I. Online predicted human interaction database ophid. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- Chen H, Sharp BM. Content-rich biological network constructed by mining pubmed abstracts. BMC Bioinformatics. 2004;5:147–159. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, et al. Mining Alzheimer disease relevant proteins from integrated protein interactome data. Pac. Symp. Biocomput. 2006;11:367–378. [PubMed] [Google Scholar]
- Cortes C, et al. Rational kernels: theory and algorithms. J. Mach. Learn. Res. 2004;5:1035–1062. [Google Scholar]
- de Marneffe M-C, et al. Generating typed dependency parses from phrase Structure Parses. Proceedings of 5th International Conference on Language Resources and Evaluation (LREC2006); Genoa, Italy. 2006. [Google Scholar]
- Erkan G, Radev DR. Lexrank: graph-based lexical centrality as salience in text summarization. J. Artif. Intell. Res. (JAIR) 2004;22:457–479. [Google Scholar]
- Erkan G, et al. Semi-supervised classification for extracting protein interaction sentences using dependency parsing. Proceedings of the Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL); Prague, Czech Republic. 2007. pp. 228–237. [Google Scholar]
- Fader A, et al. MavenRank: identifying influential members of the US senate using lexical centrality. Proceedings of the Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL); Prague, Czech Republic. 2007. pp. 658–666. [Google Scholar]
- Fisher RA. Statistical Methods for Research Workers. 14th edn. London: Collier-Macmillian; 1970. [Google Scholar]
- Freeman LC. A set of measures of centrality based on betweenness. Sociometry. 1977;40:35–41. [Google Scholar]
- Freeman LC. Centrality in social networks: conceptual clarification. Soc. Networks. 1979;1:215–239. [Google Scholar]
- Freudenberg J, Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18(Suppl. 2):S110–S115. doi: 10.1093/bioinformatics/18.suppl_2.s110. [DOI] [PubMed] [Google Scholar]
- Goh K-I, et al. The human disease network. Proc. Natl Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez G, et al. Mining gene-disease relationships from biomedical literature: weighting protein-protein interactions and connectivity measures. Pac. Symp. iocomput. 2007;12:28–39. [PubMed] [Google Scholar]
- Hahn MW, Kern AD. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 2005;22:803–806. doi: 10.1093/molbev/msi072. [DOI] [PubMed] [Google Scholar]
- Hao F, et al. Lysophosphatidic acid induces prostate cancer pc3 cell migration via activation of lpa(1), p42 and p38alpha. Biochim. Biophys. Acta. 2007;1771:883–892. doi: 10.1016/j.bbalip.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho G, et al. Polymorphism of the insulin gene is associated with increased prostate cancer risk. Br. J. Cancer. 2003;88:263–269. doi: 10.1038/sj.bjc.6600747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R, Valencia A. Implementing the ihop concept for navigation of biomedical literature. Bioinformatics. 2005;21(Suppl. 2):ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jeong H, et al. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Joachims T. Advances in Kernel Methods-Support Vector Learning. Cambridge, MA, USA: MIT-Press; 1999. Making Large-Scale SVM Learning Practical. [Google Scholar]
- Joy M, et al. High-betweenness proteins in the yeast protein interaction network. J. Biomed. Biotechnol. 2005;2:96–103. doi: 10.1155/JBB.2005.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner F, et al. Introducing meta-services for biomedical information extraction. Genome Biol. 2008 doi: 10.1186/gb-2008-9-s2-s6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, et al. Pgdb: a curated and integrated database of genes related to the prostate. Nucleic Acids Res. 2003;31:291–293. doi: 10.1093/nar/gkg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167. [Google Scholar]
- OMIM. Online Mendelian inheritance in man, OMIM (TM). McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD) 2007 Available at http://www.ncbi.nlm.nih.gov/omim/last accessed November 19, 2007.
- Page L, et al. Technical report, Stanford Digital Library Technologies Project. Stanford University; 1998. The pagerank citation ranking: bringing order to the web. Technical Report. [Google Scholar]
- Perez-Iratxeta C, et al. Association of genes to genetically inherited diseases using data mining. Nat. Genet. 2002;31:316–319. doi: 10.1038/ng895. [DOI] [PubMed] [Google Scholar]
- Perez-Iratxeta C, et al. G2d: a tool for mining genes associated with disease. BMC Genet. 2005;6:45. doi: 10.1186/1471-2156-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynar JC, Ratnaparkhi A. A maximum entropy approach to identifying sentence boundaries. Proceedings of the Fifth Conference on Applied Natural Language Processing; Washington D.C. 1997. pp. 16–19. [Google Scholar]
- Sarfaraz S, et al. Cannabinoid receptor agonist-induced apoptosis of human prostate cancer cells lncap proceeds through sustained activation of erk1/2 leading to g1 cell cycle arrest. J. Biol. Chem. 2006;281:39480–39491. doi: 10.1074/jbc.M603495200. [DOI] [PubMed] [Google Scholar]
- Schwikowski B, et al. A network of protein-protein interactions in yeast. Nat. Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc. Natl Acad. Sci. USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruoka Y, et al. Developing a robust part-of-speech tagger for biomedical text. Proceedings of the 10th Panhellenic Conference on Informatics; Volos, Greece, LNCS 3746. 2005. pp. 382–392. [Google Scholar]
- van Driel MA, et al. A new web-based data mining tool for the identification of candidate genes for human genetic disorders. Eur. J. Hum. Genet. 2002;11:57–63. doi: 10.1038/sj.ejhg.5200918. [DOI] [PubMed] [Google Scholar]
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wain HM, et al. Genew: the human gene nomenclature database, 2004 updates. Nucleic Acids Res. 2004;32((D255-7)):1257–1261. doi: 10.1093/nar/gkh072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. Experimental therapy of human prostate cancer by inhibiting mdm2 expression with novel mixed-backbone antisense oligonucleotides: in vitro and in vivo activities and mechanisms. Prostate. 2003;54:194–205. doi: 10.1002/pros.10187. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of small-world networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Wei Q, et al. Global analysis of differentially expressed genes in androgen-independent prostate cancer. Prostate Cancer Prostatic Dis. 2007;10:167–174. doi: 10.1038/sj.pcan.4500933. [DOI] [PubMed] [Google Scholar]
- Wuchty S, et al. Evolutionary conservation of motif constituents in the yeast protein interaction network. Nat. Genet. 2003;35:176–179. doi: 10.1038/ng1242. [DOI] [PubMed] [Google Scholar]
- Zanzoni A, et al. Mint: a molecular interaction database. FEBS Lett. 513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. Antisense therapy targeting mdm2 oncogene in prostate cancer: effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc. Natl Acad. Sci. 2003;100:11636–11641. doi: 10.1073/pnas.1934692100. [DOI] [PMC free article] [PubMed] [Google Scholar]