Abstract
Motivation: The prediction of biologically active compounds is of great importance for high-throughput screening (HTS) approaches in drug discovery and chemical genomics. Many computational methods in this area focus on measuring the structural similarities between chemical structures. However, traditional similarity measures are often too rigid or consider only global similarities between structures. The maximum common substructure (MCS) approach provides a more promising and flexible alternative for predicting bioactive compounds.
Results: In this article, a new backtracking algorithm for MCS is proposed and compared to global similarity measurements. Our algorithm provides high flexibility in the matching process, and it is very efficient in identifying local structural similarities. To predict and cluster biologically active compounds more efficiently, the concept of basis compounds is proposed that enables researchers to easily combine the MCS-based and traditional similarity measures with modern machine learning techniques. Support vector machines (SVMs) are used to test how the MCS-based similarity measure and the basis compound vectorization method perform on two empirically tested datasets. The test results show that MCS complements the well-known atom pair descriptor-based similarity measure. By combining these two measures, our SVM-based model predicts the biological activities of chemical compounds with higher specificity and sensitivity.
Contact:ycao@cs.ucr.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The discovery of novel biologically active compounds using high-throughput screening (HTS) technologies is a very costly and time consuming component in the discovery process of novel drugs and chemical genomics. With the recent advancements in compound synthesis, more and more compounds become available, but the time and effort required to screen these libraries has only been slightly reduced in the past years (Dobson, 2004). Because of this situation, there is a high demand for predictive computational methods that can enrich the number of potential drug candidates in new screening libraries. For instance, similarity search and clustering techniques are used to identify new drug-like compounds (Sheridan and Kearsley, 2002). In addition, computational filtering methods and models are used to predict druggability of small molecules (Cheng et al., 2007). As a result, HTS has developed into a combinatorial approach consisting of in silico and in vitro screening methods to jointly improve the turnaround time and success rate of the drug discovery process (Abt et al., 2001; Engels and Venkatarangan, 2001).
One of the basic principles behind ligand-based activity prediction models is the widely accepted similar property principle. This principle is based on the observation that chemicals of similar structures frequently share similar physicochemical properties and biological activities (Johnson and Maggiora, 1990). Because of the importance of this principle in drug discovery research, many similarity measures have been proposed to accurately quantify similarity between chemical structures and predict their bioactivity potential.
Substructure and superstructure relationships are among the most commonly used similarity measures. Given two chemical structures A and B, if structure A is fully contained in structure B, then A is a substructure of B, while B is a superstructure of A. According to the above similar property principle, A and B may share properties that are related to their common substructure. Therefore, a substructure that is putatively associated with certain properties of interest can be used as query to identify in databases all compounds that share this substructure (or superstructure) and possibly its activities. This type of similarity measures have several drawbacks. First, the utilized matching strategy is very rigid and has a high false-negative rate. Second, substructure searching is a knowledge-based approach, in which every utilized query substructure needs to be well defined. If this is not the case or a substructure model is not entirely correct, the search results will be of poor quality and of very limited predictive value. Most importantly, this search type does not generate any quantitative similarity measure, which makes it difficult to rank the search results in a meaningful manner.
Structural descriptor-based methods are also commonly used approaches for structural similarity searching and bioactivity predictions. Structural descriptors represent chemical structures in a way so that their similarity can be easily quantified. Usually, this is achieved by enumerating different structural subcomponents in chemical structures by a variety of methods. This type of search represents a much less stringent (in fact, far more flexible) approach than substructure search. Typically, the search uses a full structure or substructure as query, and identifies structures in the database that are globally ‘similar’ to the query structure. Structure similarity search does not require an exact match, and they can provide scores as a similarity measure for ranking the results. Since there is no clear definition of structural similarity, many divergent similarity measurement methods have been proposed and are now employed in various software applications in form of 2D structure fragment-based search methods, such as fingerprint, atom pair (AP), atom sequence (AS) and many others (Carhart et al., 1985; Chen and Reynolds, 2002; Dean, 1995; Girke et al., 2005; Johnson and Maggiora, 1990; Sheridan and Kearsley, 2002; Willett et al., 1998). Essentially these methods represent a chemical structure as a vector in a high-dimensional space. The generated similarity coefficients of these approaches provide a mathematical model for estimating structural similarities. Many model building methods have been applied for predicting biological activities based on structural similarities, such as neural networks (Burden, 1996), fuzzy adaptive least squares (Moriguchi et al., 1992), inductive logic programming (King and Srinivasan, 1997) and frequency-based fragment weighing schemes (Carhart et al., 1985). Although computationally simple and effective in practice, practically all of the structural descriptor-based methods share severe limitations. Most importantly, they are unable to identify local similarities between structures or those with large size differences, which is depicted in Figure 1. In addition, their false-negative rates increase vastly when it comes to the identification of weaker similarities.
Fig. 1.
Local similarity between compounds. The two structures share a common substructure (dashed boxes). The size difference will result in insignificant scores in 2D fragment-based similarity measures.
Maximum common substructure (MCS), as a metric for chemical similarity searching and activity predictions, is a promising alternative to the above approaches. The MCS of two compounds is the largest substructure that appears in both structures. Using MCS to measure similarity of chemical structures has several advantages. First, it is intuitive, as the largest common substructure of structurally related drugs is likely to be an important component of their activities. Second, the match can be visualized by highlighting the maximum common subgraph between two chemical structures. When comparing the MCS-based similarity measures to the sub/superstructure-based similarity measures, the latter can be seen as a special case of the former, and therefore MCS has all the advantages of the sub/superstructure-based methods, but lacks the drawback of requiring an exact match. Compared to structural descriptor methods, the MCS-based similarity measures can generate a similarity score for a pair of structures, which can be used to rank the search results and to supply them to machine learning approaches and other mathematical models. Unlike structural descriptor methods, the MCS-based approach will perform well in identifying local similarities where structural descriptor methods typically fail.
The MCS problem has been well studied as a general graph matching problem, and has found applications in many areas (Bunke, 2000). Garey and Johnson (1979) showed that the MCS problem is non-deterministic polynomial time (NP)-complete. However, many algorithms have been proposed to solve the problem either optimally (Cordella et al., 1998; McGregor, 1982), sub-optimally (Luo and Hancock, 2001; Wang et al., 1997; Wilson and Hancock, 1997) or with error tolerance (Berretti et al., 2001; Dumay et al., 1992; Tsai and Fu, 1979). A comprehensive review of different MCS algorithms is available in (Conte et al., 2004). However, most of these algorithms are focused on general graphs from the pattern recognition area. These methods do not meet the specific needs for efficient graph representations in the chemical compound area, which are sparse, small in size and with bounded degrees. Also, most of the algorithms convert input graphs to association graphs and convert the MCS problem to a clique detection problem. These conversions make it much harder to perform flexible matching effectively. For example, allowing a bromine atom to be matched to a chlorine atom when they are attached to an aromatic ring, but not elsewhere, is much harder to do with association graphs.
Surprisingly, after Cone et al. (1977) proposed to use MCS as a similarity measure, the approach has received much less attention than other similarity measure strategies. This is mainly due to the intractable computational complexity of the MCS problem. Hagadone (1992) has built an MCS-based chemical structure search program for 2D structure drug discovery databases. More recently, Raymond et al. (2002a) applied several heuristic strategies that are based on specific properties of chemical structures to improve the efficiency of the MCS-based similarity search algorithm (Raymond et al. 2002b). The most recent work on the MCS problem is from Yan et al. (2005). The result is restricted to the design of an efficient feature database and does not include a structure comparison step.
In this project, we propose and implement a new backtracking algorithm to compute MCS. New heuristics and strategies are introduced to reduce the computation time. The search space is properly ordered to further enhance the efficiency and to facilitate the application of the progressive optimization strategy to process large compound databases.
We test the MCS-based similarity measure by applying it to similarity searching in compound databases. The MCS-based method is compared with the AP-based method (Carhart et al., 1985; Chen and Reynolds, 2002) using a number of simulated searches with bioactive compounds as queries. Our results show that the MCS-based method outperforms the AP-based method in typical searches in chemical databases.
We also introduce a novel concept of basis compounds to combine the MCS-based similarity measure with modern machine learning methods. To illustrate this, models for predicting bioactive compounds from chemical databases are built by coupling the above similarity measures with support vector machines (or SVMs) (Christianini and Shawe-Taylor, 2000). Our experimental results show that the prediction model based on the MCS information outperforms and complements models based on traditional similarity measures. The basis compounds method also allows us to easily take advantage of multiple complementary similarity measures by integrating them into one prediction model. We have incorporated the MCS-based similarity measure and traditional AP similarity measure into a hybrid model. This model provides the most effective predictions. The prediction models proposed in this article can be applied to in silico pre-screening, which filters a large compound library for experimental screening, and to sequential screening that utilizes a process of alternating in silico and experimental screening steps (Blower et al., 2006).
2 COMPUTING MAXIMUM COMMON SUBGRAPHS
2.1 Overview
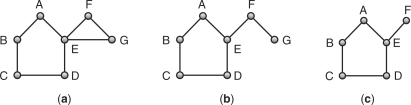
A pair of graphs are said to be isomorphic if there is a one-to-one correspondence between their vertices. Moreover, this correspondence has the property that an edge between two vertices exists in one graph if and only if an edge exists between the corresponding two vertices in the other graph. A graph Gs is said to be an induced subgraph of a graph G if all vertices of graph Gs belong to the set of vertices of graph G, and there is an edge between two vertices in graph Gs if and only if there is an edge between those two vertices in graph G. For example, in Figure 2, graph c is an induced subgraph of graph a. A common induced subgraph between the graphs G1 and G2 are two graphs Gs1 and Gs2, such that Gs1 is an induced subgraph of G1, Gs2 is an induced subgraph of G2 and Gs1 and Gs2 are isomorphic. In Figure 2, graph c is an induced subgraph of both graphs a and b, and is a common induced subgraph of graphs a and b. The largest common induced subgraph for a pair of graphs is referred to as the MCS between them.
Fig. 2.
Induced subgraph, common induced subgraph and MCS. Graph (c) is an induced subgraph of graphs (a) and (b). Therefore, it is common induced subgraph of the two graphs. It is also the MCS between them.
2.2 A new MCS algorithm
Many MCS algorithms convert the MCS problem into the maximum clique problem by introducing association graphs, also known as compatibility graphs (Barrow and Burstall, 1976; Cone et al., 1977; Levi, 1973). Using this approach, we can take advantage of the availability of different types of clique detection algorithms. However, due to the nature of chemical structures, the conversion will result in an association graph with a large and dense structure (Raymond et al., 2002a). In addition, such a conversion will complicate flexible matching approaches for finding common substructures. For instance, although it is possible to limit the solution to connected subgraphs (Koch, 2001), using association graphs it is not easy to further relax the constraints on their connectedness, such as limiting the solution to contain at most three connected components. Another example is matching different atoms under certain conditions, such as matching bromine and chlorine only when they are both attached to an aromatic ring. This level of flexibility in subgraph matching is also difficult to achieve with association graphs.
To avoid the difficulty in the association graph-based approaches, we propose a new backtracking algorithm for the MCS problem, which operates directly on the chemical structure graph. This algorithm is based on the VF algorithm, which is designed for the graph and subgraph isomorphism problems (Cordella et al., 2001). The pseudocode of our algorithm is included in the Supplementary Material Section.
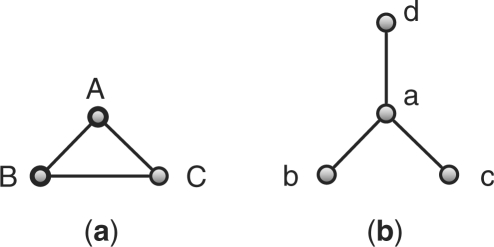
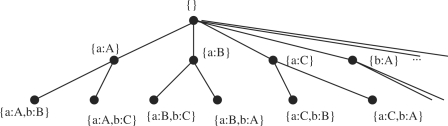
To illustrate the basic idea of our backtracking algorithm, one can consider the problem of finding the MCS of the graphs a and b in Figure 3. A common subgraph of two graphs can be represented by the vertex correspondences between the isomorphic subgraphs. For example, the subgraph of the graph 3a consisting of the vertices A and B and the edge (A,B) and the subgraph of the graph 3b consisting of the vertices a and b and the edge (a,b) are isomorphic. This common subgraph can be represented by matching A to a and B to b, and it is denoted by a set of vertex correspondences as {A : a,B : b}. The backtracking algorithm searches all possible combinations of vertex correspondences. It organizes these combinations in a search tree. Each node of the search tree is a set of correspondences. By moving down the tree, this set is expanded. Therefore, the set of correspondences at the parent node is always a subset of the set at any child node (Fig. 4). For example, the root of the tree is an empty set {}. The leftmost child of the root, {a : A}, has one correspondence, which matches vertex a to vertex A. The leaf nodes correspond to maximal common subgraphs, which cannot be further expanded. The final solution to the MCS problem corresponds to one of these leaf nodes. The backtracking algorithm searches this tree in a depth-first fashion, and will return the leaf node that contains the largest set of vertex correspondences as the solution to the MCS problem. In this example, the leftmost leaf node ({a : A,b : B}) may be returned as the MCS of the graphs 3a and 3b.
Fig. 3.
Two example graphs.
Fig. 4.
Search tree of a backtracking algorithm in search of the MCS between of the graphs a and b in Figure 3.
To speed up the computation, we introduce several strategies to reduce the search space. First, we limit the connectedness of the resultant MCS. Second, we use a heuristic based on the induced subgraph constraints to quickly remove branches with infeasible matches. Third, branch and bound strategies are employed to discard entire branches in the search tree. Finally, we order the search space such that the branch containing the solution can be searched as early as possible. This can improve the effectiveness of the branch and bound strategy, and it also helps when progressive optimization steps are used.
2.2.1 Connectedness of the resultant subgraphs
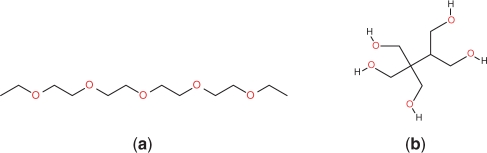
The MCS of two graphs may contain a number of disconnected structural fragments. This type of MCS is often not desirable when applied to similarity measure of chemical structures. For example, the MCS of two chemical structures in Figure 5 consists of five disconnected C–O fragments. When using the MCS as a similarity measure between chemical structures, it is often desired to only identify connected MCSs or MCSs consisting of only a limited number of disconnected fragments. Such a connectedness constraint can drastically reduce the search space in finding the MCS. To take advantage of this, our algorithm expands the current common subgraph by growing existing fragments if possible, and it keeps track of the connectedness of the current common subgraph. When it is required to start a new fragment in the common subgraph, the algorithm checks whether the number of disconnected fragments has reached its limit. If the limit has been reached, then the algorithm stops searching along the present branch and considers the common subgraph as the maximal one it can find along this branch of the search tree. In the pseudocode, this constraint is tested in the order subroutine. When the connectedness limit is reached and there is no way to further expand the current common subgraph without introducing a new disconnected fragment, the order subroutine will return a None value to stop searching the current branch. At this point the current common subgraph found will replace the global solution if it provides an improvement to the previous one.
Fig. 5.
Two example structures. The MCS of the two structures will be five disjoint C–O pairs.
2.2.2 Induced subgraph heuristic
A heuristic to quickly identify an infeasible set of correspondences can be derived from the definition of the induced subgraph. Consider the problem of finding the MCS between two graphs G1 and G2. Let u1 and v1 be unmatched in the vertices in G1 and G2, respectively. To check whether adding a correspondence, u1 : v1, to the current set of correspondences will lead to a feasible set of correspondences, the heuristic retrieves a set S1 of matched neighbors of u1 in G1, and a set S2 of matched neighbors of v1 in G2. If the elements in these two sets do not have a one-to-one correspondence in the current set of correspondences, then the correspondence u1 : v1 cannot be added. This heuristic is implemented in the compatible routine in the pseudocode.
For example, for finding the MCS between the graph in Figure 3a and the one in Figure 3b , when the current set of correspondences is {b : A}, it is not feasible to add the correspondence c : B, because c is a neighbor of b in graph 3b while A and B are not neighbors in graph 3a.
2.2.3 Branch and bound
Our algorithm employs the branch and bound strategy to discard branches of the search tree that cannot lead to an improvement of the candidate solution. An upper bound on the sizes of the common subgraphs at the leaf nodes of the present branch can be estimated. If this upper bound is worse than the candidate solution, then the branch can be discarded immediately and the algorithm backtracks to another branch.
The upper bound is estimated by using the above induced subgraph heuristic. At some point in traversing the search tree, let m be the size of the correspondence set, U be the set of the unmatched vertices in G1 and V be the set of the unmatched vertices in G2. For each vertex v in U, if v has not been marked as infeasible by previous search, we apply the above heuristic to test whether it is allowed to match to some vertex in V. If n vertices of U find potential matches in V, then an upper bound on the sizes of the common subgraphs at the leaf nodes of the present branch is m+n.
A tighter bound can be achieved by building a bipartite graph from these unmatched vertices. A bipartite G=(U+V,E) is built. Initially E is an empty set. For u1 from U and v1 from V, if the correspondence u1 : v1 passes the induced subgraph heuristic test, then edge (u1,v1) is added to E. To obtain a tighter upper bound, the size of the maximum matching in this bipartite is computed, and then added to m.
2.2.4 Ordering the search space
When using branch and bound, a proper order of the search space can help save a lot of computation time. We would like to search the branches that most likely contain the optimal or suboptimal solutions first, to potentially allow the candidate solution to be more quickly improved at an earlier stage. In this way, more branches can be discarded during the branch and bound steps.
A proper order of the search space also benefits progressive optimization. Progress optimization is a strategy that returns a suboptimal solution as early as possible to the user, and later progressively improves it over time. For example, progressive optimization can be applied to similarity searches that involve time-consuming computation of the similarity values between the query compound and compounds in the databases. To perform this search, a suboptimal solution is built using the approximate similarity values that can be computed faster. If the user is not satisfied with the result, the algorithm can improve it over time by progressively increasing the accuracy of similarity values. Our MCS algorithm can easily be adopted in progressive optimization-based similarity searches. This is achieved by suspending the MCS computation at any time, and using the candidate solution as an approximate result. To further refine the result, the MCS computation can be resumed on demand. Since ordering of search space in our MCS algorithm aims at more quickly improving the candidate solution at an earlier age, the result of progress optimization will be close to the optimal result at an earlier stage and it will stabilize sooner to an acceptable result.
At any point in traversing the search tree for computing the MCS between graphs G1 and G2, our algorithm chooses the next node to visit based on the following strategy. It finds the vertex (or vertices) in graph G1 with the most neighbors in the current common subgraph. Among all the unvisited nodes in the search tree relevant to this vertex (or these vertices), it chooses the one that can improve the aforementioned upper bound the most. Experiments have shown that this ordering strategy can reduce the average computation time.
The above algorithm has been implemented in C. The corresponding software will be made available for public download in the near future.
3 VECTOR REPRESENTATION OF COMPOUNDS FOR BIOACTIVITY PREDICTION
Traditional similarity measures, such as structural descriptors, represent a chemical structure as a vector in a high-dimensional space. Such vector representations may not be effective in statistical analyses and many machine learning techniques. This is because the dimensionality of the vectors is typically very high. For example, the widely used Daylight fingerprints use a 1024-bit string to represent a chemical structure (James et al., 1995). Fragment-based descriptors, such as AP descriptors, use a list of descriptors to represent chemical structures. These descriptors can easily be converted to a bit string, where 1 indicates the existence of a feature and 0 indicates the absence of a feature. Such a string may have an arbitrarily large dimensionality. In addition to this high dimensionality, the data is not numerical in nature, and is not suitable for most machine learning techniques.
These vector representations can be applied to bioactivity prediction models that are based on k nearest neighbors (KNN). Such approach requires only a suitable distance measure. Although the vector representations are not appropriate for statistical analyses or most machine learning procedures, they are suitable for measuring distances between chemical structures, and therefore they are useful for KNN-based modeling and prediction methods.
The MCS approach provides a useful distance measure between chemical structures, and therefore it can also be used for KNN. However, KNN-based modeling that employs MCS as a distance measure is computationally very expensive, because KNN requires all-against-all comparisons. To avoid all pairwise MCS calculation, and to vectorize chemical structures using the MCS approach, the concept of basis compounds is introduced. Basis compounds are a designed set of n diverse compounds, C1,C2,…,Cn, such that each compound Di from the compound database can be represented by an n-dimensional vector
In other words, each compound is represented by a vector, where each element is the size of an MCS between the compound and a basis compound. The set of basis compounds can be carefully designed or randomly chosen from a large set of diverse compounds. It is important to note that vectorization of a chemical structure using basis compounds requires only a numerical similarity measure and is not specific to MCS. Therefore, the MCS-based similarity measure can be replaced by any other similarity measure scheme, such as the AP-based similarity measure. When using basis compounds, each chemical structure can be represented as a multi-dimensional vector. Since the dimensionality is controllable and each vector contains real numerical values, these vectors can be used in statistical modeling and machine learning techniques. In this article, we use SVMs as an example modeling technique to demonstrate the utility of the above vectorization method in predicting bioactive compounds.
4 EVALUATION
4.1 Datasets
To test the performance of the proposed methods and models, experiments on similarity search and bioactivity prediction have been performed on two publicly available compound datasets, the NCI AIDS Antiviral Screen dataset and a subset of the NCI Human Tumor Cell Line Screen dataset. Both datasets have been published in the PubChem BioAssay database (Wheeler et al., 2007), and can be retrieved by BioAssay IDs 179 and 85, respectively. The NCI AIDS Antiviral Assay tested 40 000 compounds for evidence of anti-HIV activity. The NCI Human Tumor Cell Line Screen checked interesting compounds for anti-cancer activity by measuring growth inhibition of the MDA-MB-435 human breast tumor cell line. Both datasets record confirmatory activity information as well as concentration–response relationships, but only the confirmatory activity information was used in our experiments.
The NCI AIDS Antiviral Screen dataset contains structure and activity information for 44 150 compounds, among which 1812 are classified as having antiviral activity. After removing compounds with missing structure information, the dataset contains 42 689 compounds, 1504 of which are classified as active. The subset of the NCI Human Tumor Cell Line Screen dataset covers 27 706 compounds, including 1822 compounds that show anti-cancer activity. After removal of compounds without structure information, 26 366 compounds remain in the dataset, 1647 of which are active compounds.
4.2 Evaluation methods
4.2.1 Comparison of similarity measures by similarity searches
We first evaluated the performance of the two similarity measures by applying them in similarity searches. As similarity metrics we used for the MCS approach the size of the MCS between the query and each compound in the database, and for the AP method the Tanimoto coefficient. The AP method was chosen for benchmarking, because Chen and Reynolds (2002) showed that it outperforms other 2D structural descriptor-based similarity measures, such as the approach based on Molecular ACCess System (MACCS) keys proposed by Molecular Design Limited (MDL).
The performances of the two similarity search methods were compared by calculating their average positive predictive values (PPV; Altman and Bland, 1994) on the search results concerning the two chosen compound datasets. A series of simulated similarity searches were performed using a random set of active compounds as queries. Each simulated similarity search ranked all compounds from the most similar one to the least similar one relative to the query. Considering the set of the k top-ranked compounds, the ratio of active compounds in this set was defined as the PPV. PPV expresses the capability of a similarity search method to enrich active compounds at the top of the search results. The higher the PPV, with the same k-value, the better a similarity search performs.
4.2.2 Bioactivity prediction
The MCS- and AP-based similarity measures were also compared with regard to their performances in building SVM models for predicting bioactive candidates in compound databases. The proposed basis compound concept was used in converting chemical structures into vectors, which were then used in the training and testing of SVM-based prediction models. Between 20 to 140 randomly selected compounds from the whole dataset were used as basis compounds in the tests.
Three models were built and tested independently. The three models varied in their representations of chemical compounds. The first model uses the MCS similarity measure to build a vector for each compound (see Section 3). The second one uses the AP similarity measure for vector building. The third one—the hybrid model—concatenates the vectors from both previous models. The three models were tested in a series of experimental settings which varied in the size of the training set, the number of basis compounds, and the coefficients used in defining similarity between compounds. The output of each model was a ranking of the compounds, in which the compounds were sorted by how likely they were bioactive based on the prediction. From these ranked results, an ROC curve was generated by plotting the true positive rates against the false-positive rates (Provost and Fawcett, 2001). The performance of the prediction models for different experimental settings were then evaluated by measuring the corresponding areas under the ROC curves (AUC). Each experimental setting was tested in multiple runs, and the average AUC was used for comparisons, in which a larger AUC value indicates a better performance.
4.3 Experimental Results
4.3.1 MCS calculation speed
To evaluate the speed performance of our MCS algorithm, we performed the following test. We calculated the MCS between 1000 randomly selected pairs of compounds from the NCI anticancer data which had on average 24 non-hydrogen atoms and 27 bonds. The tests were performed in single thread mode on a workstation with a 2.0 GHz Intel Xeon processor and 2 GB of RAM. The average time required to finish an MCS calculation was 27.4 ms, with a SD of 35.6 ms. We also tested flexible matching on the same dataset by allowing one atom mismatch and disjoint MCSs. As expected, the flexible matching required more computation time. For example, when allowing one mismatch, the average time for one MCS calculation was about 2.2 s with a SD of 1.6 s. Unfortunately, we were not able to perform direct speed and accuracy comparisons with other methods because their implementations are currently not freely available in the public domain. In addition, the provided speed estimates are not comparable to those recorded in previous publications, because of major differences in their implementation, such as heuristic filtering and preprocessing steps before the MCS computation (e.g. Raymond et al., 2002a; Yan et al., 2005).
4.3.2 Similarity search
Using the method described in the previous section, we have plotted the average PPV across the simulated similarity searches with various k-values representing the number of predicted positives. The plot (see Supplement Fig. A1) shows that PPVs of both search methods decrease when k increases. This means that if one moves down the ranked search result list, the cumulative ratio of active compounds decreases. Interestingly, the MCS method performs better for larger k-values (>29) and the AP method for smaller k-values. This result is expected, because the AP method performs well in detecting compounds with high global similarities which are more likely to share the same bioactivity than compounds with local similarities. On the other hand, the MCS method is able to rank global and more sophisticated local similarities equally well. Therefore, the MCS method identifies a larger set of bioactive candidates, but it does not necessarily rank compounds with global similarities at the top of the list. The following example illustrates this performance benefit of the MCS approach. For instance, if the user is interested in more than 29 of the top compounds, the MCS searches exhibit a better performance than the AP searches. If the top 100 compounds are of interest, which is a reasonable choice for similarity searches, the MCS method has a PPV of 0.30, and the AP method has a PPV of 0.27.
4.3.3 Bioactivity prediction
To compare the predictive power of the MCS method against the AP method and the hybrid approach, each method was coupled with SVMs to build a model for predicting bioactive compounds. The input of the SVMs was the vector representation of compounds generated by each method. The output was either 0 for inactive compounds, or 1 for active compounds. SVM-based models were used to predict the bioactive compounds. The AUC values were recorded and used for evaluating the qualities of the predictions.
The SVM implementation provided in the R package e1071 (Dimitriadou et al., 2005) was used for the tests. This package is based on the widely used C++-based LibSVM from Chang and Lin (2001). The default regression parameters defined in the package were used in the tests. In one of our tests we also used the ROC Curve for Binary SVM tool distributed with LibSVM, to achieve a fair comparison against data reported elsewhere. In this test, cross-validation was used to select the parameters (C and γ) for the radial kernel function-based classifiers.
4.3.4 Varying the size of the training set
One of the key factors impacting the prediction performance is the size of the training set. For real applications, it is desirable to achieve an acceptable performance with small training set sizes, because the prediction model is often used to select compound subsets from a large compound library to be used in the experimental screening process. The prediction models are only practically useful if the training set, obtained from an experimental screening process, is much smaller than the whole screening library. This is especially important for sequential screening, in which only a small initial set of compounds are experimentally tested and used to train the initial models. For both datasets, we performed experiments with training sets consisting of 1, 5, 10 and 25% of the whole dataset. 20 basis compounds were selected randomly from the whole dataset for each test. The average AUC values and SD are listed in Table 1.
Table 1.
Average AUC values using different prediction models and different training set sizes
| Models | Training set size |
|||
|---|---|---|---|---|
| 1% | 5% | 10% | 25% | |
| NCI antiviral dataset | ||||
| MCS-based | 57.9 (3.0) | 64.0 (2.4) | 67.0 (1.3) | 70.0 (0.9) |
| AP-based | 58.2 (3.1) | 63.7 (1.8) | 65.8 (1.8) | 68.9 (1.5) |
| Hybrid | 61.3 (3.4) | 66.7 (1.9) | 69.2 (1.3) | 71.6 (1.2) |
| NCI anticancer dataset | ||||
| MCS-based | 60.3 (2.8) | 65.4 (1.8) | 68.0 (1.7) | 70.9 (1.3) |
| AP-based | 59.3 (3.3) | 65.2 (1.8) | 67.8 (1.7) | 70.9 (1.8) |
| Hybrid | 62.7 (3.2) | 69.2 (1.8) | 71.8 (1.4) | 74.8 (1.2) |
The MCS-based model uses the absolute MCS sizes to represent a chemical structure as a vector. The AP-based model uses the AP-based similarity, and the hybrid model concatenates the vectors from both previous models. SDs are given in parentheses.
Table 1 shows that the performance of all three models improves with the size of the training dataset. The hybrid model achieves in all cases the best performance, while the MCS model performs almost consistently better than AP model. Moreover, the hybrid model outperforms the AP model with much smaller training sets. For instance, in both datasets, the hybrid model performs with a training set of 10% of the dataset better than the AP model with a training set that is 2.5 times as large.
4.3.5 Overlap of active compounds identified by the models
The superior performance of the hybrid approach can be explained by the performance differences of the individual models in identifying similarities with different structural features (e.g. global or local similarities). For instance, in one of the tests with the NCI antiviral dataset, the active compound NSC 79521 was ranked by the AP model at position 1, while the MCS model ranked it at position 10 568. On the other hand, the active compound NSC 683278 was ranked at position 23 by the MCS model, and at position 2 673 by the AP-based model. The hybrid model ranked both of these actives at the very top of the list, at positions 33 and 4, respectively.
To evaluate these performance differences more carefully, we analyzed the overlaps between the active compounds identified by each pair of the three models. For a pair of models A and B, if the set of active compounds identified by A is S(A), and the set of active compounds identified by B is S(B), we define A's coverage of B, or CA(B) as
| (1) |
The larger the coverage is, the better model A can cover the chemical space covered by B. If both CA(B) and CB(A) are small, the two models cover different subspaces of the chemical space and they complement each other. We studied the cross coverage between each pair of models by plotting the corresponding coverage values with different numbers of predicted active compounds, n. The plot (shown in Supplementary Fig. A2) shows that with a typical choice of n, one's coverage of the other is relatively small between the AP model and the MCS model. On the other hand, the hybrid model's coverages of the other two models are relatively high. For example, if 5% of the compounds are predicted to be active, 62% of the active compounds identified by the AP model are also identified by the MCS model, and 59% vice versa. On the other hand, the hybrid model identifies 85% of the active compounds of the AP model and 73% of the MCS model. These data show that the AP model and the MCS model cover different subspaces of the chemical space and they complement each other. The hybrid model effectively utilizes both the MCS and the AP similarity information, and covers a wider chemical space than any one of the other two models alone.
4.3.6 Varying the MCS coefficients
In addition to the absolute size of the MCS between structures, a normalized MCS size has been considered in previous studies to measure similarity between a pair of structures (Bunke and Shearer, 1998). We performed tests using different MCS-based similarity measures, or MCS coefficients. The following coefficients are included in these comparisons:
| (2) |
| (3) |
| (4) |
Here, G1 is the chemical structure to be vectorized, and G2 is one of the basis compounds. Each coefficient is also used to derive a hybrid model, which is included in the comparisons. All the models have been tested using training sets of different sizes and multiple sets of 20 randomly selected basis compounds on both compound sets. Part of the results is listed in Table 2.
Table 2.
Average AUC values using the prediction models based on different MCS coefficients
| Models | Databases | |
|---|---|---|
| NCI antiviral | NCI anticancer | |
| MCS | 69.8 (0.9) | 69.9 (1.3) |
| MCS c1 | 70.0 (1.9) | 71.1 (1.3) |
| MCS c2 | 71.0 (0.9) | 71.0 (1.2) |
| MCS c3 | 70.5 (1.9) | 71.4 (0.9) |
| Hybrid | 71.5 (1.2) | 73.8 (1.2) |
| Hybrid c1 | 71.8 (1.7) | 73.8 (1.2) |
| Hybrid c2 | 72.3 (0.9) | 74.4 (1.1) |
| Hybrid c3 | 72.3 (1.2) | 74.2 (1.3) |
SDs are given in parentheses. The NCI antiviral dataset wastested with a training set of 10 000 compounds and the NCI anticancer dataset was tested with a training set of 5000 compounds. The MCS model uses the absolute MCS sizes. The models MCS c1, MCS c2 and MCS c3 use the MCS coefficients listed in Equations (2), (3) and (4), respectively. The hybrid model uses the absolute MCS sizes and the AP information. The models hybrid c1, hybrid c2 and hybrid c3 use MCS coefficients listed in Equation (2), (3) and (4), respectively, and the AP information. More data corresponding to different training set sizes are listed in Supplementary Table A1.
The coefficient from Equation (3) exhibits in general the best performance (Table 2), although the advantage is marginal. When using only MCS information, MCS c2 generates the best results in the NCI antiviral dataset, and close to the best results in the NCI anticancer dataset. When both the MCS and the AP information are used, the hybrid c2 model [Equation (3)] shows the best performance in both datasets.
4.3.7 Varying the number of basis compounds
Another factor that might affect the performance of prediction models is the choice of the basis compounds. In principle, the set of basis compounds should be as diverse as possible and should cover the chemical space as thoroughly as possible. To evaluate the sensitivity of our models regarding the choice of basis compounds, we performed a series of tests on both compound datasets. We varied the number of basis compounds from 20 to 140 in increments of 20 compounds. The obtained AUC values are listed in Table 3.
Table 3.
Average AUC values using the prediction models with different numbers of basis compounds
| Models | Number of basis compounds | ||||||
|---|---|---|---|---|---|---|---|
| 20 | 40 | 60 | 80 | 100 | 120 | 140 | |
| NCI antiviral dataset | |||||||
| AP | 70.7 | 72.4 | 73.3 | 73.9 | 74.0 | 72.9 | 72.9 |
| MCS c2 | 73.0 | 74.6 | 74.6 | 75.8 | 75.5 | 75.4 | 75.2 |
| Hybrid c2 | 74.4 | 75.2 | 75.4 | 76.2 | 76.1 | 75.4 | 75.6 |
| NCI anticancer dataset | |||||||
| AP | 69.5 | 72.4 | 72.6 | 73.2 | 73.9 | 74.7 | 73.7 |
| MCS c2 | 71.0 | 74.2 | 75.2 | 75.5 | 75.9 | 76.6 | 76.4 |
| Hybrid c2 | 74.4 | 75.9 | 75.9 | 76.1 | 76.5 | 77.2 | 76.9 |
For the NCI antiviral dataset, 25 000 randomly selected compounds were used as the training set. For the NCI anticancer dataset, 5000 randomly selected compounds were used as the training set.
The results in Table 3 indicate that for all the listed models, the AUC values increase with the number of basis compounds. This trend peaks at around 80 to 120 basis compounds. After this, no significant improvement can be achieved. It is important to point out, that the peak performance of each method in Table 3 should be used for comparisons between the three methods, and not their performance within the same number of basis compounds.
4.3.8 Comparisons with other prediction approaches
We compared the performance of our prediction models to results reported by (Deshpande et al. (2005). In this study, the authors listed the prediction performance measured by the AUC values of several representative approaches. The authors applied five-way cross-validation to all models using several datasets, among which was the NCI antiviral dataset. To compare our proposed approach with these models, we used the ROC Curve for Binary SVM tool to calculate the AUC value of our proposed hybrid model by applying five-way cross-validation on the NCI antiviral dataset. The result for our hybrid model is listed in Table 4 along with the data reported by Deshpande et al. (2005).
Table 4.
AUC values for different prediction models applied to the NCI antiviral dataset
| Models | hybrid c2 | physicochemical-based | descriptor-based | SUBDUE | SubdueCL | FSG |
|---|---|---|---|---|---|---|
| AUC | 82.3 | 47.3 | 72.1 | 58.5 | 65.2 | 79.4 |
Hybrid c2 is our proposed hybrid model using the coefficient from Equation (3) and 80 randomly selected compounds as basis compounds. Radial kernel function-based classifier was used, with C set to 64 and γ set to 0.0625. The physicochemical-based method is described in Deshpande et al. (2005). The descriptor-based method combines 166 MACCS keys from the MDL and Daylight fingerprints. SUBDUE (Holder et al., 1994) and SubdueCL (Gonzalez et al., 2001) are methods based on heuristic substructure discovery. FSG is the method proposed by (Deshpande et al. (2005) using topological subgraphs but not geometrical subgraphs.
According to these comparisons, our method outperforms all the methods listed in Table 4. In addition to its improved performance, our method uses a feature space with a much lower dimensionality. Therefore, it is computationally less complex to train the SVM model and make predictions. For example, to achieve the performance exhibited in the table, the Frequent SubGraph (FSG) model used 18 542 features to represent a compound. The authors had to apply feature selection to reduce this number to 2460. However, this feature selection step also reduced the prediction quality and cut the AUC value to 78.5. In comparison, our method used only 160 features to achieve the better performance without requiring any feature selection step. Furthermore, it is easy to incorporate other similarity measures into our hybrid model to further advance its performance. For example, one can easily incorporate the similarity measure used in FSG into our hybrid model.
5 CONCLUSION AND FUTURE WORK
This article describes the development of a novel MCS algorithm and the concept of basis compounds for measuring the similarity between chemical structures. Both methods are applied to similarity searching of chemical databases and are incorporated into SVM models to efficiently predict biologically active compounds.
More specifically, a new backtracking algorithm for finding the MCS between a pair of graphs is designed and its performance is tested. This algorithm can be applied to chemical structure graphs directly (instead of association graphs), and hence supports several matching contraints as well as relaxations that can be useful when applied to comparison of chemical compounds. Several strategies are introduced to increase the efficiency and flexibility of the algorithm. This algorithm can be effectively used with a progressive optimization strategy, which is very beneficial for ranking a large number of chemical structures by their similarities to query structures.
In order to combine the MCS-based similarity measure and other similarity measures with modern machine learning techniques to form effective bioactivity prediction models, the concept of basis compounds is proposed. This concept allows for a vector representation of chemical structures, similar to traditional fingerprint approaches, while avoiding many drawbacks of the traditional fingerprint approaches. Finally, the derived vectors are incorporated into SVMs to build prediction models of biological activities of compounds.
Our experimental results show that the MCS-based similarity measure is more effective in searching chemical databases than the well-known AP-based method. They also show that the SVM models based on vectors derived from basis compounds are effective for identifying bioactive compounds. Moreover, the MCS-based similarity measure complements the AP-based measure effectively. Our proposed hybrid model, which combines the MCS and the AP similarity measures, provides the most effective predictions in comparison with the existing approaches.
In the future we will work on several improvements of our approach. First, chemical structures have certain properties that can be used to achieve more effective heuristics in computing the MCS. Second, the MCS-based similarity can be easily adapted to cluster chemical structures. Another interesting application is the identification of the MCS among more than two structures. Finally, it will be interesting to combine the MCS-based similarity measure and the hybrid vectorization method with similarity measures based on physicochemical property descriptors.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the support from the Bioinformatics Core Facility, the Center for Plant Cell Biology (CEPCEB) and the Institute for Integrative Genome Biology (IIGB) at UC Riverside.
Funding: This work was supported by grants from the National Science Foundation (IOB-0420033, IOB-0420152, IGERT-0504249 and IIS-0711129).
Conflict of Interest: none declared.
REFERENCES
- Abt M, et al. Sequential approach for identifying lead compounds in large chemical databases. Statist. Sci. 2001;16:154–168. [Google Scholar]
- Altman D, Bland J. Diagnostic tests 2: predictive values. Br. Med. J. 1994;309:102. doi: 10.1136/bmj.309.6947.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow H, Burstall R. Subgraph isomorphism, matching relational structures and maximal cliques. Inf. Process. Lett. 1976;4:83–84. [Google Scholar]
- Berretti S, et al. Efficient matching and indexing of graph models in content-based retrieval. IEEE Trans. Pattern Anal. Mach. Intell. 2001;23:1089–1105. [Google Scholar]
- Blower P, et al. Comparison of methods for sequential screening of large compound sets. Comb. Chem. High Throughput Screen. 2006;9:115–122. doi: 10.2174/138620706775541882. [DOI] [PubMed] [Google Scholar]
- Bunke H. Graph matching: theoretical foundations, algorithms and applications. Proceedings of the International Conference on Vision Interface; Montreal, Quebec, Canada. 2000. pp. 82–88. [Google Scholar]
- Bunke H, Shearer K. A graph distance metric based on the maximal common subgraph. Pattern Recognit. Lett. 1998;19:255–259. [Google Scholar]
- Burden F. Using artificial neural networks to predict biological activity from simple molecular structural considerations. Quant. Struct.-Act. Rel. 1996;15:11. [Google Scholar]
- Carhart R, et al. Atom pairs as molecular features in structure-activity studies: definition and applications. J. Chem. Inf. Comput. Sci. 1985;25:64–73. [Google Scholar]
- Chang C, Lin C. LIBSVM: a library for support vector machines. 2001 Software available at http://www.csie.ntu.edu.tw/cjlin/libsvm.
- Chen X, Reynolds C. Performance of similarity measures in 2D fragment-based similarity searching: comparison of structural descriptors and similarity coefficients. J. Chem. Inf. Comput. Sci. 2002;42:1407–1414. doi: 10.1021/ci025531g. [DOI] [PubMed] [Google Scholar]
- Cheng AC, et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- Christianini N, Shawe-Taylor J. An Introduction to Support Vector Machines and Other Kernel-based Learning Methods. New York: Cambridge University Press; 2000. [Google Scholar]
- Cone M, et al. Molecular structure comparison program for the identification of maximal common substructures. J. Am. Chem. Soc. 1977;99:7668–7671. [Google Scholar]
- Conte D, et al. Thirty years of graph matching in pattern recognition. Inter. J. Pattern Recognit. Artif. Intell. 2004;18:265–298. [Google Scholar]
- Cordella L, et al. Proceedings of the 14th International Conference Pattern Recognition. Vol. 2. Brisbane, Australia: 1998. Graph matching: a fast algorithm and its evaluation; pp. 1582–1584. [Google Scholar]
- Cordella L, et al. Proceedings of the 3rd IAPR TC-15 Workshop on Graphbased Representations in Pattern Recognition. 2001. An improved algorithm for matching large graphs; pp. 149–159. [Google Scholar]
- Dean P. Molecular Similarity in Drug Design. London: Blackie Academic & Professional; 1995. [Google Scholar]
- Deshpande M, et al. Frequent Sub-structure-based approaches for classifying chemical compounds. IEEE Trans. Knowled. Data Eng. 2005;17:1036–1050. [Google Scholar]
- Dimitriadou E, et al. Wien: TU Wien; 2005. e1071: misc functions of the department of Statistics (e1071) [Google Scholar]
- Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- Dumay A, et al. Proceedings of the 11th Image, Speech and Signal Analysis (IAPR) International Conference on Patten Recognition. Vol.III. Netherlands: The Hague; 1992. Consistent inexact graph matching applied to labelling coronarysegments in arteriograms. Pattern Recognition; pp. 439–442. [Google Scholar]
- Engels M, Venkatarangan P. Smart screening: approaches to efficient HTS. Curr. Opin. Drug Discov. Devel. 2001;4:275–283. [PubMed] [Google Scholar]
- Garey M, Johnson D. Computers and Intractability: a Guide to the Theory of NP-Completeness. New York, NY, USA: WH Freeman & Co; 1979. [Google Scholar]
- Girke T, et al. ChemMine. A compound mining database for chemical genomics. Plant Physiol. 2005;138:573–577. doi: 10.1104/pp.105.062687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, et al. Proceedings of the Predictive Toxicology Challenge Workshop. Freiburg, Germany: 2001. Application of graph-based concept learning to the predictive toxicology domain. [Google Scholar]
- Hagadone T. Molecular substructure similarity searching:efficient retrieval in two-dimensional structure databases. J. Chem. Inf. Comput. Sci. 1992;32:515–521. [Google Scholar]
- Holder L, et al. Substructure discovery in the subdue system. Proc. AAAI. 1994;94:169–180. [Google Scholar]
- James C, et al. Daylight Theory Manual. Aliso Viejo, California, USA: Daylight Chemical Information Systems Inc; 1995. [Google Scholar]
- Johnson M, Maggiora G. Concepts and Applications of Molecular Similarity. New York: Wiley; 1990. [Google Scholar]
- King R, Srinivasan A. The discovery of indicator variables for QSAR using inductive logic programming. J. Comput. Aided Mol. Des. 1997;11:571–580. doi: 10.1023/a:1007967728701. [DOI] [PubMed] [Google Scholar]
- Koch I. Enumerating all connected maximal common subgraphs in two graphs. Theor. Comput. Sci. 2001;250:1–30. [Google Scholar]
- Levi G. A note on the derivation of maximal common subgraphs of two directed or undirected graphs. Calcolo. 1973;9:341–352. [Google Scholar]
- Luo B, Hancock E. Structural graph matching using the EM algorithm and singular value decomposition. IEEE Trans. Pattern Anal. Mach. Intell. 2001;23:1120–1136. [Google Scholar]
- McGregor J. Backtrack search algorithms and the maximal common subgraph problem. Software-Pract. Exper. 1982;12:23–34. [Google Scholar]
- Moriguchi I, et al. Fuzzy adaptive least squares and its application to structure-activity studies. Quant. struct. activ. relation. 1992;11:325–331. [Google Scholar]
- Provost F, Fawcett T. Robust classification for imprecise environments. Mach. Learn. 2001;42:203–231. [Google Scholar]
- Raymond J, et al. Heuristics for similarity searching of chemical graphs using a maximum common edge subgraph algorithm. J. Chem. Inf. Comput. Sci. 2002a;42:305–316. doi: 10.1021/ci010381f. [DOI] [PubMed] [Google Scholar]
- Raymond J, et al. RASCAL:calculation of graph similarity using maximum common edge subgraphs. Comput. J. 2002b;45:631. [Google Scholar]
- Sheridan RP, Kearsley SK. Why do we need so many chemical similarity search methods? Drug Discov. Today. 2002;7:903–911. doi: 10.1016/s1359-6446(02)02411-x. [DOI] [PubMed] [Google Scholar]
- Tsai W, Fu K. Error-correcting isomorphisms of attributed relational graphs for pattern analysis. IEEE Trans. Syst. Man Cybern. 1979;9:757–768. [Google Scholar]
- Wang Y, et al. Genetic-based search for error-correcting graph isomorphism. IEEE Trans. Syst. Man Cybern. Part B. 1997;27:588–597. doi: 10.1109/3477.604100. [DOI] [PubMed] [Google Scholar]
- Wheeler D, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007;35((Database issue)):D5. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett P, et al. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998;38:983–996. [Google Scholar]
- Wilson R, Hancock E. Structural matching by discrete relaxation. IEEE Trans. Pattern Anal. Mach. Intell. 1997;19:634–648. [Google Scholar]
- Yan X, et al. SIGMOD'05: Proceedings of the 2005 ACM SIGMOD International Conference on Management of Data. New York, NY, USA: ACM Press; 2005. Substructure similarity search in graph databases; pp. 766–777. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.