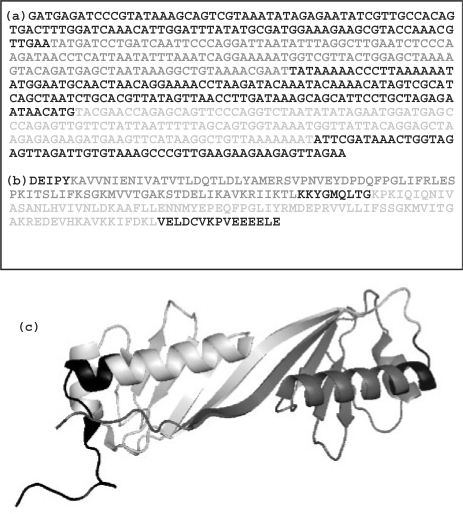
Fig. 1.
Example of repeat found at the three levels in the Tata-box Binding Protein (TBP) of Sulfolobus acidocaldarius (1MP9). (a) DNA (137 nt of repeat length), (b) amino acid sequence (82 aa), (c) 3D structure (83 aa). Repeats are shown in light gray and non-repeated regions are shown in black. Amino acid and 3D repeats are almost perfectly coincident, but the DNA repeat is smaller and within the region of the other repeats. Among homologous elements, similarity decreases with divergence time at different rates. It decreases quicker at the DNA and slower at the protein structural levels (Chothia and Lesk, 1986). This frequently results in smaller repeats in DNA than at the other levels. Edges of very degenerated repeats may also not precisely coincide at the different levels due to terminal mismatches at some but not at all levels. This is a typical feature of methods aiming at optimizing local alignments.