Abstract
In the AAA+ ClpXP protease, ClpX uses repeated cycles of ATP hydrolysis to pull native proteins apart and to translocate the denatured polypeptide into ClpP for degradation. Here, we probe polypeptide features important for translocation. ClpXP degrades diverse synthetic peptide substrates despite major differences in side-chain chirality, size, and polarity. Moreover, translocation occurs without a peptide –NH and with 10 methylenes between successive peptide bonds. Pulling on homopolymeric tracts of glycine, proline, and lysine also allows efficient ClpXP degradation of a stably folded protein. Thus, minimal chemical features of a polypeptide chain are sufficient for translocation and protein unfolding by the ClpX machine. These results suggest that the translocation pore of ClpX is highly elastic, allowing interactions with a wide-range of chemical groups, a feature likely to be shared by many AAA+ unfoldases.
Introduction
Molecular machines of the AAA+ family (ATPases associated with various cellular activities) use ATP hydrolysis to drive repetitive conformational changes that perform mechanical work within cells (for review, see Hanson and Whiteheart, 2005). Many AAA+ enzymes function by translocating polypeptide or nucleic-acid polymers. Examples include ATP-dependent proteases, protein-secretion translocons, and DNA/RNA helicases, pumps, and viral packaging motors. For AAA+ proteases, ATP hydrolysis is coupled to conformational changes that are used to force unfolding of native protein substrates and then to drive polypeptide translocation into the degradation chambers of enzymes such as ClpXP, ClpAP, HslUV, Lon, FtsH, and the proteasome (for review, see Sauer et al., 2004).
The ClpXP protease of Escherichia coli, which consists of the hexameric ClpX ATPase and the tetradecameric ClpP peptidase, is an archetypal AAA+ protease. ClpP is formed by back-to-back stacking of two ClpP7 rings, placing the proteolytic active sites in an interior chamber accessible through a narrow axial portal in each ring (Wang et al., 1997). Six identical ClpX subunits, each containing a single AAA+ ATPase module, interact to form a hexameric ring with an axial pore. In ClpXP, the pores of one or two hexamers of ClpX align with the ClpP portals, creating channels for polypeptide translocation into the degradation chamber (Fig. 1; Grimaud et al., 1998; Ortega et al., 2000; Martin et al., 2007). Protein substrates are targeted to ClpXP by short peptide sequences (Flynn et al., 2003). For example, any protein bearing a C-terminal ssrA tag (AANDENYALAA) is a substrate for ClpXP degradation (Gottesman et al., 1998; Kim et al., 2000; Singh et al., 2000; Lee et al., 2001; Kenniston et al., 2003; 2004). The ssrA tag initially binds in the axial pore of ClpX (Siddiqui et al., 2004; Martin et al., 2008b; 2008c).
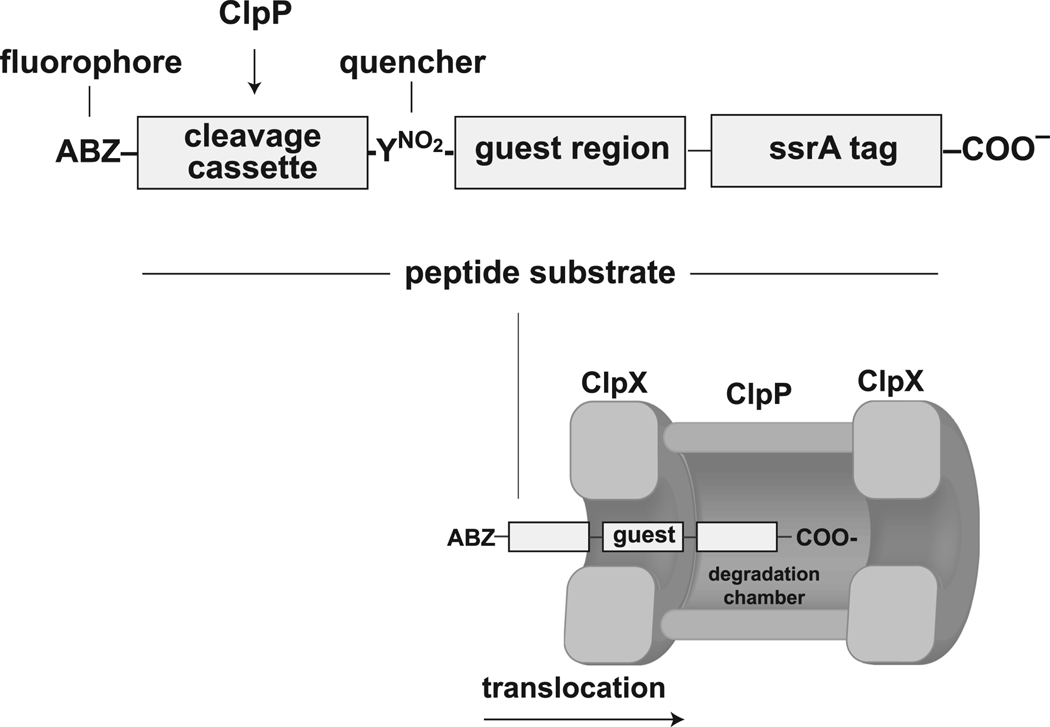
Figure 1.
Top — peptide substrates contained an N-terminal sequence (FAPHMALVP) that is cleaved efficiently by a ClpP, a central guest region of variable composition, and a C-terminal ssrA tag (AANDENYALAA). The cleavage cassette had an amino-benzoic acid fluorophore (ABZ) on the N-terminal side and a nitro-tyrosine quencher (YNO2) on the C-terminal side to allow detection of ClpP proteolysis. Bottom — ClpP cleavage between the ABZ and YNO2 groups of peptide substrates requires prior translocation of the guest region through the axial pore of ClpX.
Polypeptide translocation by ClpX is required for protein unfolding and for transporting denatured substrates into ClpP for degradation. Translocation of the ssrA tag of a native substrate appears to pull the attached protein structure against the entrance to the axial pore, thereby generating a denaturation force because the pore is smaller than the folded protein (for review, see Sauer et al., 2004). For a very stable native substrate, hundreds of cycles of ATP hydrolysis by ClpXP can be required before denaturation occurs, suggesting that enzymatic unfolding is a stochastic process with only a small probability of success per pulling event (Kenniston et al., 2003). A translocation-induced unfolding model is supported by the finding that mutations in the GYVG loops, which line the axial pore of ClpX, slow translocation of unfolded substrates, reduce the rate of unfolding of native substrates, and increase the ATP-hydrolysis cost of both processes (Martin et al., 2008c).
AAA+ enzymes use two kinds of “active” sites for ATP-dependent polypeptide translocation. One traditional “chemical” site mediates the binding and hydrolysis of ATP to generate conformational changes in the enzyme. The other “mechanical” site transmits force generated by these conformational movements to the substrate. What features of a polypeptide chain are recognized by the mechanical site of ClpX to allow the pulling events that lead to translocation and unfolding? The answer is unclear. There appears to be no obligatory directionality to translocation, as ClpXP can degrade substrates starting either from the N-terminus or from the C-terminus (Gottesman et al., 1998; Gonciarz-Swiatek et al., 1999; Lee et al., 2001; Flynn et al., 2003; Hoskins et al., 2002; Kenniston et al., 2005; Farrell et al., 2007). In principle, the pore loops of ClpX could bind to the peptide bonds, interact with certain types of side chains, or recognize the chiral branching of side chains in the unfolded polypeptide. Based on mutant studies in the related HslUV protease and the conservation of a critical aromatic side chain in the pore loops of all AAA+ unfoldases, it has been postulated that π -cation and π-π interactions between the unfoldase and aromatic side chains in a substrate may be important for translocation and unfolding (Park et al., 2005). Glycine/alanine-rich stretches and other low-complexity sequences appear to prevent unfolding of very stable domains by the 26S proteasome, suggesting that side-chain variety may be an important component in translocation-dependent denaturation of hyper-stable structures (Tian et al., 2005; Hoyt et al., 2006).
Here, we probe the chemical and structural features of a polypeptide chain that allow it to be translocated by ClpX and find that this process is remarkably promiscuous. Peptides can be translocated even when they contain homopolymeric tracts of amino acids that are chemically and structurally diverse, including D-amino acids, residues that lack a peptide –NH group, or amino acids bearing insertions of as many as nine methylene groups between successive peptide bonds. These results, which run counter to traditional “lock and key” notions of enzymatic specificity, have important implications for the mechanism of ClpX translocation and unfolding and may be a common feature of other ATP-dependent unfoldases.
Results
Design of substrates
All peptide substrates were prepared by solid-phase synthesis and contained three segments: an N-terminal module containing a ClpP cleavage site, a variable central guest region, and a C-terminal ssrA tag (Fig. 1). To detect peptide-bond cleavage, we used a peptide sequence (FAPHMALVP) that ClpP cleaves at a rate •104 min−1 ClpP14−1 (Thompson and Maurizi, 1994), flanked on one side by an aminobenzoic acid fluorophore (ABZ) and on the other side by a nitro-tyrosine quencher (YNO2). Cleavage within this segment results in an increase in fluorescence. The guest region was typically 10 residues in length, which exceeds the ClpX translocation step-size (Kenniston et al., 2005; Martin et al., 2008b). In an extended conformation, 10 residues are also sufficient to span the entire ClpX pore (S. Sundar, A. Martin, R.T.S., unpublished). Consequently, active translocation of the guest region of a peptide substrate is required for the cleavage module to enter ClpP for proteolysis (Fig. 1). To improve solubility, most substrates also had two lysines (KK) between the guest region and the ssrA tag. Table 1 lists the sequences of the peptides used for this study. We refer to peptides using the one letter code for the sequence of the guest region. For example, the [Q10] peptide contains ten glutamines in the guest region, and the [VG]5 peptide has a guest region with the sequence VGVGVGVGVG.
Table 1.
Steady-state kinetic parameters for ClpXP degradation of peptide substrates.
| name | Vmax min−1 ClpP−1 | KM µM | Vmax with SspB min−1 ClpP−1 | cost ATP/peptide with SspB | Sequence | length |
|---|---|---|---|---|---|---|
| [G7] | 13.7 ± 1.3 | 5.4 ± 1.9 | 13.5 ± 0.7 | 30 | ABZ-FAPHMALVPYNO2G7KKAANDENYALAA | 30 |
| [G10] | 14.3 ± 0.8 | 6.1 ± 0.2 | 11.5 ± 0.4 | 30 | ABZ-FAPHMALVPYNO2G10KKAANDENYALAA | 33 |
| [G5KKG5] | 14.5 ± 1.9 | 4.5 ± 2.2 | 12.1 ± 0.1 | 36 | ABZ-FAPHMALVPYNO2G5KKG5AANDENYALAA | 33 |
| [β]10 | 9.8 ± 0.9 | 4.1 ± 0.3 | 8.8 ± 1.1 | 43 | ABZ-FAPHMALVPYNO2β10KKAANDENYALAA ABZ- | 33 |
| [γ]10 | 6.4 ± 0.3 | 3.7 ± 0.6 | 6.5 ± 0.4 | 57 | FAPHMALVPYNO2γ10KKAANDENYALAA ABZ- | 33 |
| [ε]10 | 6.1 ± 0.6 | 4.7 ± 0.4 | 5.9 ± 0.3 | 61 | FAPHMALVPYNO2ε10KKAANDENYALAA | 33 |
| [O]5 | 7.0 ± 0.2 | 4.3 ± 0.4 | 6.8 ± 1.3 | 53 | ABZ-FAPHMALVPYNO2O5KKAANDENYALAA ABZ- | 33 |
| [U]4 | 6.2 ± 0.6 | 9.0 ± 2.0 | 7.8 ± 0.5 | 47 | FAPHMALVPYNO2U4KKAANDENYALAA | 27 |
| [P5] | 11.4 ± 0.2 | n.d. | 12.9 ± 1.1 | 45 | ABZ-FAPHMALVPYNO2P5KKAANDENYALAA ABZ- | 28 |
| [P10] | 6.4 ± 0.2 | 2.8 ± 0.2 | 6.5 ± 0.2 | 65 | FAPHMALVPYNO2P10KKAANDENYALAA ABZ- | 33 |
| [P15] | 3.4 ± 0.8 | 3.6 ± 0.7 | 3.3 ± 0.3 | 125 | FAPHMALVPYNO2P15KKAANDENYALAA | 38 |
| [VG]5 | 14.4 ± 1.7 | 3.4 ± 0.3 | 13.6 ± 0.4 | 30 | ABZFAPHMALVPYNO2 [VG]5KKAANDENYALAA | 33 |
| [DVG]5 | 14.0 ± 0.4 | 6.7 ± 1.6 | 14.2 ± 0.2 | 28 | ABZ-FAPHMALVPYNO2[dVG]5KKAANDENYALAA | 33 |
| [FG]5 | 12.1 ± 1.7 | 4.2 ± 1.0 | 14.1 ± 0.8 | 22 | ABZ-FAPHMALVPY [FG]5KKAANDENYALAA | 33 |
| [Q10] | 8.7 ± 0.5 | 9.1 ± 1.7 | 8.8 ± 0.8 | 48 | ABZ-FAPHMALVPYNO2Q10KKAANDENYALAA ABZ- | 33 |
| [E10] | 6.0 ± 0.1 | 2.0 ± 0.1 | 6.3 ± 0.2 | 62 | FAPHMALVPYNO2E10KKAANDENYALAA ABZ- | 33 |
| [K10] | 2.7 ± 0.1 | • 0.2 | 2.8 ± 0.1 | 160 | FAPHMALVPYNO2K10KKAANDENYALAA ABZ- | 33 |
| [R10] | 8.0 ± 0.4 | 0.2 ± 0.1 | 8.4 ± 0.1 | 53 | FAPHMALVPYNO2R10KKAANDENYALAA | 33 |
β (β-alanine); γ (γ-aminobutyric acid), ε (ε-amino caproic acid); O (8-aminooctanoic acid), U (11-aminoundecanoic acid).
Vmax and KM values are means of 2–3 independent determinations (n) with errors calculated as .
Degradation requires ATP-dependent translocation
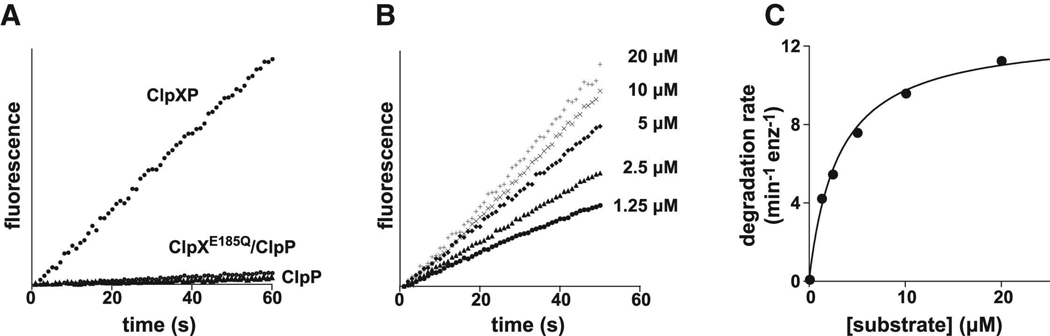
Using the fluorescence assay to monitor substrate cleavage, we assayed the rate of degradation of 10 µM [G10] peptide by ClpXP (Fig. 2A). Control experiments established that cleavage was almost entirely dependent on ATP-dependent translocation by ClpX. First, cleavage by ClpP alone occurred at a 40-fold slower rate than cleavage by ClpXP (Fig. 2A). Second, the ATPase and translocation defective ClpXE185Q mutant, which still binds ClpP and ssrA-tagged substrates in an ATP-dependent fashion (Hersch et al., 2005), did not markedly stimulate ClpP cleavage of this peptide substrate (Fig. 2A). Similar results were observed for all peptide substrates; peptide cleavage by ClpP alone was always at least 20-fold slower than that by ClpXP (not shown). We conclude that the vast majority of ClpXP peptide degradation in our assays occurs via active ATP-dependent translocation.
Figure 2.
(A) Efficient cleavage of the [G10] peptide by ClpP was observed in the presence of wild-type ClpX but not in the absence of ClpX or with ClpXE185Q, which cannot hydrolyze ATP. All reactions contained 10 µM of the [G10] substrate and 300 nM ClpP14. When present, the concentration of ClpX6 or the ATPase-defective mutant was 800 nM. (B) Degradation of different concentrations of the [VG]5 peptide by 800 nM ClpX6 and 300 nM ClpP14. (C) Steady-state rates of [VG]5 peptide degradation by ClpXP were calculated from the data in panel C and fit to the Michaelis-Menten equation (KM = 3.1 µM; Vmax = 12.7 min−1 ClpP−1).
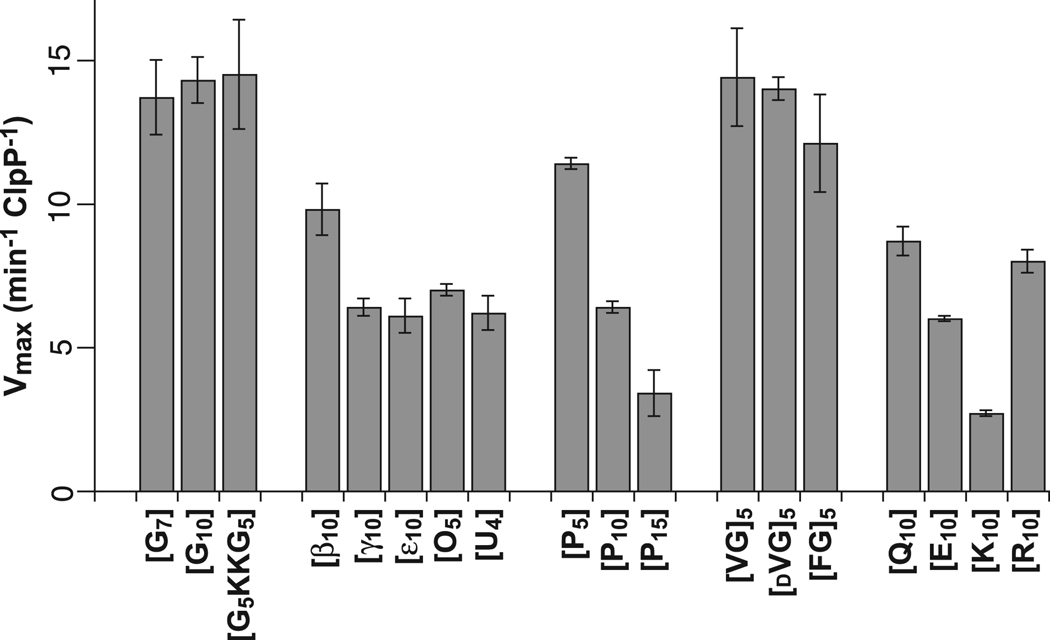
For each peptide, we measured steady-state rates of ClpXP degradation at different substrate concentrations and fit the data to obtain KM and Vmax values (Table 1). Fig. 2B and 2C show these experiments for the [VG]5 peptide. In all cases, we report maximal degradation rates normalized by the total concentration of ClpP. For example, the steady-state kinetic parameters obtained by fitting one set of [VG]5 degradation reactions were KM = 3.1 µM and Vmax = 12.7 min−1 ClpP−1. Although both KM and Vmax varied for different peptides, the latter parameter is more important for understanding effects on ClpX translocation. Indeed for different peptides, average Vmax values obtained from 2–3 experiments ranged from 2.7 to 14.5 min−1 ClpP−1 (Fig. 3; Table 1). These results show that the identity of residues in the peptide guest region influences the overall rate of ClpXP degradation.
Figure 3.
Maximum rates of ClpXP degradation of peptide substrates with different guest regions were determined from multiple experiments like those shown in Fig. 2B and Fig. 2C. See Table 1 for sequences of individual peptides and definition of error bars.
Polyglycine translocation
Glycine is the smallest amino acid, with only a hydrogen atom for a side chain. ClpXP degraded peptides with seven or ten glycines in the guest region with Vmax values of approximately 14 min−1 ClpP−1 (Table 1; Fig. 3), demonstrating successful translocation of polyglycine sequences. When we permuted the sequence of the [G10] peptide by moving the KK solubility sequence to the middle of the guest region in the [G5KKG5] substrate, Vmax for ClpXP degradation was unchanged (Table 1; Fig. 3).
Altered peptide-bond spacings
Successive peptide bonds in all natural proteins are separated by a single carbon atom and are related by two dihedral angles (Φ, Ψ), resulting in restrictions in possible backbone conformations. To probe the importance of this geometry, we synthesized peptides in which the guest region contained unnatural amino acids with additional carbon atoms between successive peptide bonds by inserting β-alanine (2-carbon spacing), γ-aminobutyric acid (3-carbon spacing), ε-aminocaproic acid (5-carbon spacing), 8-aminooctanoic acid (7-carbon spacing), or 11-aminoundecanoic acid (10-carbon spacing) in the guest region. Strikingly, peptides with 4–10 residues of these “stretched” amino acids in the guest region were degraded at 40–70% of the [G10] peptide degradation rate (Fig. 3). Because substrates with guest-region spacings of 2–10 methylene groups between successive peptide bonds were translocated and degraded at substantial rates compared to peptides with the normal single-carbon spacing, we conclude that the spacing of peptide bonds along the polypeptide backbone is not a major determinant of recognition during translocation of substrates by ClpXP.
Polyproline translocation
Proline lacks a peptide –NH group, and successive prolines severely constrain the polypeptide backbone and often adopt a left-handed polyproline-II helix (F = −75°, Y = 150°; Schulz and Schirmer, 1979; Adzhubei and Sternberg, 1993). The maximal rates of ClpXP degradation of the [P5], [P10], and [P15] peptides were 11.4, 6.4, and 3.4 min−1 ClpP−1, respectively (Table 1; Fig. 3). Hence, a peptide –NH group is not required for translocation nor is the ability to assume an α-helix, a β-strand, or other conformations incompatible with polyproline sequences. Because ClpX degradation slowed in proportion to the length of the polyproline segment, however, a polyproline helix might be difficult to translocate or may need to be disrupted to allow translocation.
Side-chain chirality and size
Natural amino acids, with the exception of glycine, are L-isomers. To assess the effect of side-chain chirality on ClpXP translocation, we measured degradation rates for substrates with five successive L-Val-Gly repeats in the guest region (Vmax = 14.4 min−1 ClpP−1) or five successive D-Val-Gly repeats (Vmax = 14.0 min−1 ClpP−1). Because these rates are essentially the same, we conclude that peptides containing D-isomers can be translocated as well as those containing L-isomers. Thus, ClpX appears to be indifferent to side-chain chirality.
The maximum degradation rates for the [G10], [VG]5, and [FG]5 peptides were 14.3 min−1 ClpP−1, 14.4 min−1 ClpP−1, and 12.1 min−1 ClpP−1, respectively. The similarities in these rates suggest that the presence of larger residues in a substrate, including β-branched and aromatic side chains, plays little role in ClpXP translocation.
Side-chain polarity and charge
Does the charge or polarity of amino-acid side chains affect ClpX translocation? To address this question, we determined Vmax values for ClpXP degradation of peptides with guest regions containing 10 lysines (Vmax = 2.7 min−1 ClpP−1), 10 arginines (Vmax = 8.0 min−1 ClpP−1), 10 glutamic acids (Vmax = 6.0 min−1 ClpP−1), or 10 glutamines (Vmax = 8.7 min−1 ClpP−1). These results show that ClpXP can translocate homopolymeric stretches of charged and polar side chains. Degradation of the [K10] peptide was slower than any of the other peptides tested in this study. However, the [R10] peptide was degraded about three-fold faster, showing that positive charge per se is not the sole cause of slow [K10] peptide degradation. The [Q10] peptide was degraded about 50% faster than the [E10] peptide. Thus, negatively charged glutamic-acid side chains are modestly more difficult for ClpX to translocate than uncharged but isosteric glutamine side chains.
ATP cost of translocation
Rates of substrate degradation by ClpXP need not be correlated with energetic efficiency, because the ATPase activity of ClpX can vary substantially for different substrates (Kenniston et al., 2003; 2004; Martin et al., 2008c). To assess energetic costs, we measured the rate of ATP hydrolysis and the maximum rate of peptide degradation during ClpXP proteolysis under the same conditions. To ensure saturation of the enzyme by substrate in these studies, we used 10–20 µM substrate in the presence of equimolar SspB adaptor, which reduces KM for ClpXP degradation of ssrA-tagged substrates to a value of 200 nM or less (Levchenko et al., 2000; Wah et al., 2003; Bolon et al., 2004). We then divided the ATPase rate by the degradation rate for the SspB-bound peptide to provide an estimate of the number of ATPs hydrolyzed during degradation of a single molecule of each peptide substrate. This value is an average. It includes energy consumed during productive and non-productive work (for example, substrate slipping or ATP hydrolyzed during engagement), much as the fuel economy of a vehicle traveling over rough muddy terrain might be reduced by occasional spinning of the wheels without net movement.
As shown in Table 1, the cost of degradation ranged from approximately 20 to 160 ATPs per substrate, with the highest costs associated with the slowest Vmax values. Peptides with non-polar amino acids in the guest region were degraded with the lowest costs, whereas peptides with “stretched” amino acids, prolines, or polar residues had higher costs. For the most efficient substrates, an average of about one ATP was hydrolyzed per amino acid translocated and degraded. For the least efficient substrate, an average of roughly five ATPs were hydrolyzed per amino acid translocated and degraded. For several substrates, we also performed experiments to calculate the ATP cost of peptide degradation in the absence of SspB and obtained values within 20% of those measured with the adaptor (not shown).
Translocation under load
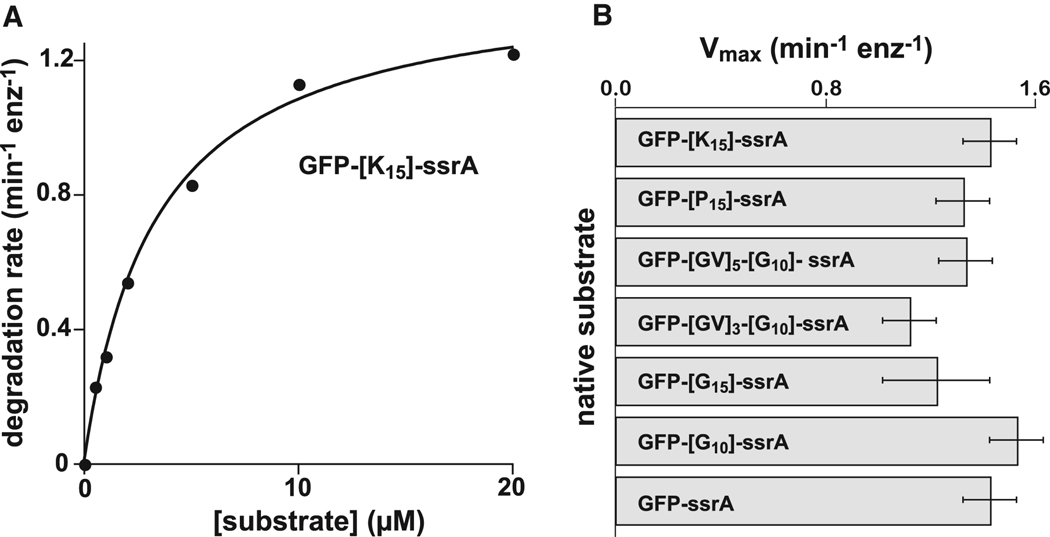
It might be argued that homopolymeric stretches of glycines, prolines, or other residues are easy to translocate in the absence of an opposing force, but may not allow ClpX to grasp a substrate firmly enough to allow it to unfold a stable native protein. To test this idea, we fused degradation tags containing stretches of glycine, proline, or lysine to green fluorescent protein (GFP). Denaturation is known to be the rate-limiting step in ClpXP degradation of ssrA-tagged variants of GFP (Kim et al., 2000). Michaelis-Menten experiments, like the one shown in Fig. 4A, revealed that ClpXP degraded all of these GFP substrates at comparable maximal rates (Fig. 4B). For polyglycine substrates, the maximal rate of degradation was similar regardless of whether the homopolymeric stretch was immediately adjacent to GFP, and thus would occupy the pore during unfolding, or was separated from GFP by several residues. Similar results for tags containing polyglycine have been obtained independently (P. Chien and T.A. Baker, unpublished). Hence, ClpX must grip polyglycine, polyproline, or polylysine sequences tightly enough to allow translocation-mediated unfolding of GFP.
Figure 4.
(A) Michaelis-Menten analysis of ClpXP degradation of GFP-[K15]-ssrA. The solid line is a non-linear least-squares fit (KM = 3.3 ± 0.3 µM; Vmax = 1.44 ± 0.05 min−1 ClpP−1 ). (B) Maximum rates of ClpXP degradation of native GFP substrates with homopolymeric sequences of lysine, proline, or glycine between the folded body of GFP and the ssrA degradation tag. Error bars represent the uncertainty of a non-linear least-squares fit of experimental data to the Michaelis-Menten equation. KM’s for the fits not shown in panel A were GFP-ssrA (3.3 ± 0.4 µM); GFP-[P15]-ssrA (7 ± 2 µM); GFP-[GV]5-[G10]-ssrA (2.4 ± 0.4 µM); GFP-[GV]3-[G10]-ssrA (2.0 ± 0.2 µM); GFP-[G15]-ssrA (2.1 ± 1 µM); GFP-[G10]-ssrA (4.1 ± 0.5 µM).
Discussion
Molecular translocation can be viewed as moving a biological polymer through a stationary machine or alternatively as tracking of a dynamic machine along a fixed polymer. For example, many DNA and RNA helicases track in a 3´ to 5´ direction along one strand of a nucleic-acid duplex, and simultaneously disrupt interactions with the complementary strand (Patel and Picha, 2000; Singleton et al., 2007; Pyle, 2008; Enemark and Joshua-Tor, 2008). For some helicases, including those belonging to the AAA+ family, the enzyme interacts with the sugar-phosphate backbone of the DNA/RNA strand and has a step size of one nucleotide per ATP hydrolyzed. This type of fixed step-size drive mechanism is analogous to the relationship between the teeth on a sprocket and the roller links on a bike chain, which allows forward pedal movement to be tightly coupled to the rotation of the bike wheel.
Although ClpX conceptually tracks along an unstructured polypeptide chain, our results suggest that this polypeptide-translocation machine operates by a rather different mechanism than related hexameric helicases. For example, there is no obligatory directionality to ClpX translocation in the sense that degradation can start near a degradation tag at either terminus of a protein substrate (Lee et al., 2001; Hoskins et al., 2002; Kenniston et al., 2005). Moreover, we find no evidence that ClpX translocation requires a fixed spacing between successive peptide bonds or side chains. These results appear to rule out drive-train mechanisms that rely on strict geometric coupling between the movement of ClpX machine parts and the properties of the polypeptide chain. Indeed, there may not be an invariant ClpX translocation step size. For unfolded proteins, the average step size of ClpXP translocation has been estimated to range from one to five amino acids per ATP hydrolyzed (Kenniston et al., 2005; Martin et al., 2008a), and we observed significant variations in the energetic cost of ClpXP degradation of the different peptides studied here, suggesting that they are also translocated with variable average step sizes. Another difference between ClpX and many hexameric helicases involves the order in which subunits around the hexameric ring hydrolyze ATP. A strictly sequential firing mechanism has been proposed for the T7 gp4 helicase, the Φ12 RNA packaging ATPase P4, and the pappilomavirus E1 helicase (Singleton et al., 2000; Mancini et al., 2004; Enemark and Joshua-Tor, 2006), whereas ClpX appears to employ a probabilistic mechanism in which the order of ATP hydrolysis in different subunits is not predetermined (Martin et al., 2005).
Our results show that ClpXP translocation is relatively indifferent to the chirality, size, polarity, or charge of protein side chains. The ClpXP enzyme from E. coli has hundreds of natural substrates (Flynn et al., 2003; Neher et al., 2006), and attaching an ssrA tag to numerous proteins makes them substrates for ClpXP degradation (Gottesman et al., 1998; Kim et al., 2000; Singh et al., 2000; Lee et al., 2001; Kenniston et al., 2003; 2004). During translocation of these substrates, the sequence of the polypeptide segment being actively moved through the ClpX pore changes continually. Thus, ClpX must be able to translocate an enormous number of different polypeptide sequences, each with distinct chemical properties and conformational preferences. Viewed from this perspective, our results make both functional and biological sense.
How has nature evolved a protein-degradation machine that is exquisitely specific in terms of substrate choice but cares little about the detailed chemical and structural properties of these substrates? The answer is that degradation, like many key cellular processes, is controlled at the level of initiation. Only proteins bearing degradation tags that bind specifically to the protease are engaged, translocated, and then degraded. For example, the ssrA tag of a substrate initially binds in the axial pore of ClpX, where it makes specific interactions with pore loops whose ATP-fueled movements subsequently drive translocation (Siddiqui et al., 2004; Martin et al., 2008b; 2008c). However, once translocation of the ssrA-tagged substrate commences, the chemical properties of the rest of the polypeptide chain seem to have only small influences on the rate of degradation. An analogy with a macroscopic machine is apt. Conveyor belts can move objects of vastly different sizes and shapes, but these objects must first be placed on the belt.
How does ClpX translocate polypeptide substrates without strict recognition of chemical or geometric features? One possibility is that the ClpX pore is relatively elastic and collapses around a polypeptide, allowing flexible pore loops to maintain atomic contact with the substrate. Then, during the power stroke of the ATPase cycle, conformational changes in ClpX could drag the substrate along by van der Waals forces that create friction between the enzyme and the unfolded polypeptide. Because van der Waals interactions occur between all types of atoms, they would be ideally suited for interactions with substrates like unfolded polypeptides, which have highly variable atomic compositions. Pore elasticity could also explain how ClpXP can simultaneously translocate multiple polypeptide chains during degradation of disulfide-bonded proteins (Burton et al., 2001; Bolon et al., 2004).
Variation in the average step size for ClpXP translocation (from one to five residues per ATP hydrolyzed Kenniston et al., 2005; Martin et al., 2008a) can be explained in several ways. First, different polypeptide sequences may adopt different conformations during translocation, and a power stroke of a fixed length might move more substrate residues in a compact conformation than in an extended conformation. Second, some substrates might not move during each power stroke, or might slip back afterwards, in a manner that depends upon the precise sequence and the elasticity of the pore. There is precedent for substrate slipping. For example, during attempts to unfold some native ssrA-tagged proteins, engaged substrates slip from of the grasp of ClpXP and are released without being translocated through the pore (Kenniston et al., 2005). In addition, mutating the GYVG pore loops of ClpX results in a smaller average translocation step size per power stroke. This result is expected if such mutations weaken substrate contacts and result in an increased number of mechanical cycles that fail to move the polypeptide substrate (Martin et al., 2008c).
Regardless of the exact mechanism, our results show that ClpXP can grip and translocate homopolymeric stretches of glycine, proline, and lysine forcefully enough to denature an attached GFP protein. Because spontaneous solution denaturation of GFP occurs with a half-life of years, enzymatic unfolding of this protein by ClpXP represents a major challenge (Kim et al., 2000). Taken together, these results indicate that minimal features of a polypeptide chain are adequate for ClpXP translocation, even when acting against a substantial resisting force. These findings are also consistent with experiments demonstrating that ClpXP can completely degrade some substrates containing several folded protein domains (Lee et al., 2001; Kenniston et al., 2005; Martin et al., 2008b; 2008c). In these instances, the primary degradation tag attached to the first domain is proteolyzed before the second domain is encountered, and translocation through the ClpX pore of a segment of the first domain or a linker drives unfolding of the second domain. Thus, there appears to be no requirement for translocation of specialized sequences to allow denaturation of attached native domains. It is possible, of course, that some polypeptide sequences are somewhat “slippery” during normal ClpX translocation and thus do not allow efficient force transfer and subsequent ClpXP denaturation of hyper-stable substrates, as has been demonstrated for the eukaryotic proteasome (Tian et al., 2005; Hoyt et al., 2006).
Given that our studies show that minimal sequence determinants are required for substrate translocation by ClpXP, it is reasonable to ask if this ability to translocate radically different natural and unnatural polypeptide sequences is a specialized adaptation or represents a general property of other ATP-dependent proteases as well. Because all AAA+ proteases share the ability to degrade a wide variety of protein substrates, we anticipate that translocation tolerance may be a common feature of this entire enzyme family.
Significance
Prior to this work, it would have been reasonable to assume that translocation by ClpX involves recognition either of regular chemical features of the polypeptide backbone or of specific side chains in a substrate. Strikingly, our experimental results fail to support either model. Instead, we find that translocation and subsequent degradation by ClpXP is remarkably tolerant to a wide range of natural and non-natural amino acids. We find no evidence that sequence diversity is necessary for normal translocation. For example, homopolymeric stretches of charged amino acids (Lys, Arg, Glu), polar residues (Gln), and small non-polar residues (Pro, Ala, Gly) all appeared to be translocated. Similarly, the presence of D-amino acids or residues with 2–10 methylenes between successive peptide bonds failed to halt translocation by ClpX. Moreover, previous studies had shown that there is no requisite directionality to ClpXP degradation and that more than one polypeptide chain can be translocated at the same time (Burton et al., 2001; Lee et al., 2001; Hoskins et al., 2002; Bolon et al., 2004; Kenniston et al., 2005). A new model is required to account for this collective information. The translocation pore of the ClpX hexamer must be elastic and highly adaptable, and general chemical features, such as van der Waals interactions, must allow ClpX to grip substrates tightly enough to couple nucleotide-dependent changes in hexamer structure to vectorial movement of the translocating polypeptide. It will be important to decipher the structural basis of this translocation mechanism and to test whether it also applies to other families of AAA+ proteases.
Experimental Procedures
Peptides and proteins
Peptides were synthesized by standard solid-phase techniques and were purified by reverse-phase HPLC chromatography on an LC-10AD-VP column (Shimadzu), using a gradient from 0 to 80% acetonitrile in 0.06% TFA. The expected masses of purified peptides were confirmed by MALDI-TOF mass spectrometry. Peptide concentrations were determined by nitro-tyrosine absorption at 381 nm (ε = 2200 M−1 cm−1; Means and Feeny, 1971).
Variants of E. coli ClpX with an N-terminal His6 tag and E. coli ClpP with a C-terminal His6 tag were purified as described (Kim et al., 2000; Hersch et al., 2004). PCR-mediated mutagenesis was used to construct GFP variants with an N-terminal His6 tag, a variable sequence, and a C-terminal ssrA tag after the GFP-coding sequence. These variants were expressed, under IPTG control, from a pACYC vector in E. coli BLR/lDE3 △clpX cells and were purified by Ni2+-NTA affinity after lysis under non-denaturing conditions. GFP concentrations were determined by absorbance at 280 nm (ε = 19770 M−1 cm−1). Purified GFP-ssrA and ClpXE185Q were gifts from Greg Hersch (MIT).
Assays
Degradation assays were performed at 30 °C in PD buffer (25 mM HEPES [pH 7.6], 200 mM KCl, 5 mM MgCl2, 0.032% NP-40, and 10% glycerol). For ClpXP degradation, assays included 300 nM Clp14, 800 nM wild-type or mutant ClpX6, and an ATP regeneration system consisting of 4 mM ATP, 16 mM creatine phosphate, and 0.32 mg/mL creatine phosphokinase. For ClpP degradation, ClpX and the ATP regeneration system were omitted. All reaction components except substrate were preincubated at 30 °C, and reactions were initiated by the addition of substrate and monitored by changes in fluorescence using a QM-2000-4SE spectrofluorimeter (Photon Technology International). For peptide degradation assays, samples were excited at 320 nm and fluorescence at 420 nm was monitored. In a control experiment, we found that complete ClpXP degradation of the [VG]5 peptide resulted in the same final fluorescence as degradation of this substrate by chymotrypsin, and that peptide-degradation rates calculated from changes in fluorescence corresponded well to rates determined by loss of the substrate peak following HPLC separation. For GFP degradation assays, samples were excited at 467 nm and fluorescence emission was monitored at 511 nm; assays monitored by SDS-PAGE showed similar rates of GFP degradation by ClpXP.
For measurement of rates of ATP hydrolysis during degradation, we used the SspB adaptor protein (a gift from Natalia Ivanova, MIT) to ensure that ClpX was saturated with the ssrA-tagged peptide substrate. For these experiments, reactions contained 800 nM ClpX6, alone or with 300 nM ClpP14, and equimolar SspB and peptide substrate (10–20 µM) in PD buffer. Rates of ATP hydrolysis at 30 °C were measured by changes in absorbance at 340 nm using a coupled assay system with 5 mM ATP, 1 mM NADH, 2 mM phosphoenolpyruvate, 3 U/ml lactate dehydrogenase, and 3 U/ml pyruvate kinase (Nørby, 1988). For the experiments containing ClpX6 and ClpP14, the ATPase activity of ClpX hexamers in the doubly capped ClpX6•ClpP14•ClpX6 complex was calculated by correcting for the activity of free ClpX hexamers (25% of total). In control experiments, we found that SspB enhanced the rate of ClpXP degradation of sub-saturating concentrations of peptide substrates but suppressed the very slow rate of ATP-independent proteolysis of these substrates by ClpXE185Q/ClpP and ClpP (not shown).
Acknowledgements
This research was supported by NIH grant AI-15706 and HHMI. We thank B.O. Cezairliyan, J. Sohn, N. Ivanova, E. Gur, J.H. Barkow, and J. Kalb for helpful discussions. T.A.B. and I.L. are employees of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adzhubei AA, Sternberg MJE. Left-handed polyproline II helices commonly occur in globular proteins. J. Mol. Biol. 1993;229:472–493. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell. 2004;16:343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr. Opin. Struct. Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell CM, Baker TA, Sauer RT. Altered specificity of a AAA+ protease. Mol. Cell. 2007;25:161–166. doi: 10.1016/j.molcel.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Gonciarz-Swiatek M, Wawrzynow A, Um SJ, Learn BA, McMacken R, Kelley WL, Georgopoulos C, Sliekers O, Zylicz M. Recognition, targeting, and hydrolysis of the lambda O replication protein by the ClpP/ClpX protease. J. Biol. Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with C-terminal peptide tails added by the SsrA tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud R, Kessel M, Beuron F, Steven AC, Maurizi MR. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell. Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hersch GL, Baker TA, Sauer RT. SspB delivery of substrates for ClpXP proteolysis probed by the design of improved degradation tags. Proc. Natl. Acad. Sci. USA. 2004;101:12136–12141. doi: 10.1073/pnas.0404733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Yanagihara K, Mizuuchi K, Wickner S. ClpAP and ClpXP degrade proteins with tags located in the interior of the primary sequence. Proc. Natl. Acad. Sci. USA. 2002;99:11037–11042. doi: 10.1073/pnas.172378899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Zich J, Takeuchi J, Zhang M, Govaerts C, Coffino P. Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 2006;25:1720–1729. doi: 10.1038/sj.emboj.7601058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Burton RE, Siddiqui SM, Baker TA, Sauer RT. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J. Struct. Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Sauer RT. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc. Natl. Acad. Sci. USA. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fueled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol. Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease: critical dependence on ATP-hydrolysis rates and energy landscapes. Nat. Struct. Mol. Biol. 2008a;15:139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell. 2008b;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 2008c;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means GE, Feeney RE. 1st edition. San Francisco: Holden-Day, Inc; 1971. Chemical Modification of Proteins. [Google Scholar]
- Neher SB, Villen J, Oakes E, Bakalarski C, Sauer RT, Gygi SP, Baker TA. Proteomic profiling of ClpXP substrates following DNA damage reveals extensive instability within SOS regulon. Mol. Cell. 2006;22:193–204. doi: 10.1016/j.molcel.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Nørby JG. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- Ortega J, Singh SK, Ishikawa T, Maurizi MR, Steven AC. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell. 2000;6:1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- Park E, Rho YM, Koh OJ, Ahn SW, Seong IS, Song JJ, Bang O, Seol JH, Wang J, Eom SH, Chung CH. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J. Biol. Chem. 2005;280:22892–22898. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ESC, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz GE, Schirmer RH. New York: Springer-Verlag; 1979. Principles of Protein Structure. [Google Scholar]
- Siddiqui SM, Sauer RT, Baker TA. Role of the protein-processing pore of ClpX, an AAA+ ATPase, in recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Rozycki J, Ortega J, Ishikawa T, Lo J, Steven AC, Maurizi MR. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J. Biol. Chem. 2001;276:29420–29429. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J. Biol. Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat. Struct. Mol. Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- Wah DA, Levchenko I, Rieckhof GE, Bolon DN, Baker TA, Sauer RT. Flexible linkers leash the substrate-binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol. Cell. 2003;12:355–363. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]