Abstract
Epidermal growth factor (EGF) receptor (EGFR)/MAPK signaling can induce a switch in MCF-7 breast cancer cells, from an estrogen receptor (ER)α-positive, Luminal-A phenotype, to an ERα-negative, Basal-like phenotype. Although mechanisms for this switch remain obscure, Basal-like cancers are typically high grade and confer a poorer clinical prognosis. We previously reported that miR-206 and ERα repress each other’s expression in MCF-7 cells in a double-negative feedback loop. We show herein that miR-206 coordinately targets mRNAs encoding the coactivator proteins steroid receptor coactivator (SRC)-1 and SRC-3, and the transcription factor GATA-3, all of which contribute to estrogenic signaling and a Luminal-A phenotype. Overexpression of miR-206 repressed estrogen-mediated responses in MCF-7 cells, even in the presence of ERα encoded by an mRNA lacking a 3′-untranslated region, suggesting miR-206 affects estrogen signaling by targeting mRNAs encoding ERα-associated coregulatory proteins. Furthermore, EGF treatments enhanced miR-206 levels in MCF-7 cells and ERα-negative, EGFR-positive MDA-MB-231 cells, whereas EGFR small interfering RNA, or PD153035, an EGFR inhibitor, or U0126, a MAPK kinase inhibitor, significantly reduced miR-206 levels in MDA-MB-231 cells. Blocking EGF-induced enhancement of miR-206 with antagomiR-206 abrogated the EGF-inhibitory effect on ERα, SRC-1, and SRC-3 levels, and on estrogen response element-luciferase activity, indicating that EGFR signaling represses estrogenic responses in MCF-7 cells by enhancing miR-206 activity. Elevated miR-206 levels in MCF-7 cells ultimately resulted in reduced cell proliferation, enhanced apoptosis, and reduced expression of multiple estrogen-responsive genes. In conclusion, miR-206 contributes to EGFR-mediated abrogation of estrogenic responses in MCF-7 cells, contributes to a Luminal-A- to Basal-like phenotypic switch, and may be a measure of EGFR response within Basal-like breast tumors.
MicroRNA-206 targets mRNAs encoding ERα, SRC-1, SRC-3 and GATA-3, and contributes to the EGF-induced transition from a “Luminal” to a “Basal-like” subtype in breast cancer.
Several subtypes of breast cancers have emerged through the use of expression profiling (1,2,3). Two of these subtypes include Luminal-A and Basal-like. Luminal-A cancers express estrogen receptor-α (ERα) and progesterone receptor (PGR) but do not display overexpression or amplification of epidermal growth factor receptor (EGFR)/ErbB1 or HER2/ErbB2. Basal-like tumors express basal-specific cytokeratins and may display myoepithelial-like characteristics (e.g. smooth muscle α-actin). Basal-like cancers do not express ERα, PGR, or HER2 but frequently overexpress the EGFR (4,5,6,7). Newly diagnosed Basal-like breast cancer is of a higher grade, more difficult to treat, and confers a poorer prognosis (i.e. higher recurrence score, lower predicted overall survival) than the Luminal-A subtype. The ontogeny of Basal-like breast cancer is unclear. Some evidence suggests that this tumor type might originate from ERα-negative progenitor cells (6,8,9,10), whereas other studies indicate that ERα-positive tumors or preneoplastic legions acquiring additional oncogenic signaling (e.g. EGFR overexpression) promotes the selection of a more transformed, estrogen-independent, ERα-negative cell type (11,12).
The interaction between EGFR signaling and ERα is complex, involving both positive and negative cross talk, depending on the cellular context. For example, an emerging model of tamoxifen resistance in breast cancer involves the binding of tamoxifen to ERα at the cell membrane, and subsequent activation of a pathway that results in the release of membrane heparin-bound EGF, and activation of the EGFR (13,14). Consequently, tamoxifen (and estrogen) becomes engaged in the stimulation of both proliferative (i.e. MAPK) pathways, and anti-apoptotic/pro-survival (i.e. phosphoinositol 3-kinase/Akt) pathways. In contrast to this cooperative interaction between ERα and growth factor signaling during the emergence of resistance to hormonal therapy in breast cancer, the expression of ERα and EGFR in newly diagnosed breast cancer is frequently inversely correlated. El-Ashry and colleagues have demonstrated that augmentation of MAPK signaling leads to the loss of ERα expression at the mRNA and protein levels in MCF-7 cells (15,16,17). This group has also used mRNA profiling from MCF-7 cells in which stable overexpression of EGFR, or of several downstream effectors within the MAPK pathway, was achieved to establish a MAPK signature of gene expression (12). Notably, the mRNA profiles from EGFR-overexpressing cells were similar to those of ERα-negative, Basal-like breast tumors.
We have recently examined whether ERα might be regulated by microRNA, which are a subclass of noncoding regulatory RNAs (18,19). MicroRNAs represent an important component in the process of posttranscriptional gene silencing, in that microRNA-mRNA interactions primarily result in mRNA degradation and/or translational inhibition of the targeted transcript. The targeting of a specific mRNA by one or more microRNAs may completely eliminate the expression of the corresponding protein, or maintain the level of the protein below a threshold level or tipping point (20). Numerous microRNAs have been found to be differentially expressed in breast cancer (21,22,23,24,25), and specific microRNAs have been implicated in the regulation of ERα and ErbB-related receptors, cell cycle traverse, survival, and metastasis in breast cancer (26,27,28,29,30,31,32). In particular, we have shown that microRNA-206 (miR-206) targets and represses ERα expression in MCF-7 and T47D breast cancer cell lines (33). Given that miR-206 levels are elevated in ERα/HER2-negative vs. ERα-positive human breast tumor specimens (22), miR-206 up-regulation may play a role in ERα-positive tumors transitioning to an ERα-negative, Basal-like lesion.
We also reported that 17β-estradiol (E2) and the ERα-selective agonist, propyl pyrazole triol (PPT), repressed miR-206 expression in a double-negative feedback loop (33). This relationship between ERα and miR-206 conforms to a recently described network motif (i.e. an interaction that occurs significantly more frequently than expected by chance) in Caenorhabditis elegans (34). This network motif is composed of a feedback loop in which a transcription factor and a microRNA regulate each other’s expression. Steady-state or oscillatory network motifs involved a single-negative feedback loop in which a transcription factor stimulates the expression of a specific microRNA, and that microRNA inhibits the expression of the transcription factor (34). Bistable systems (i.e. stable expression of one of two possible phenotypes) utilize double-negative feedback loops in which a transcription factor and a microRNA repress each other’s expression (34). We propose that the double-negative feedback loop between ERα and miR-206 contributes to the bistable ERα-positive vs. ERα-negative phenotype observed in breast cancer.
In the current study, we extend our work on miR-206 in three general ways. First, we hypothesized that coregulatory proteins should be included in transcription factor-microRNA feedback network motifs. We show that miR-206 coordinately targets the mRNAs of two ERα coactivator proteins, SRC-1 and SRC-3, along with GATA-3, a transcription factor that cooperates with ERα in the regulation of gene expression, normal ductal growth, and differentiation of a luminal phenotype (35,36,37,38). Moreover, miR-206 sufficiently suppresses estrogenic responses in the presence of elevated ERα that is not targeted by miR-206, strongly indicating that miR-206 targets and represses the expression of multiple proteins involved in mediating estrogenic responses. Second, we demonstrate that EGFR signaling tips the balance of the bistable ERα-positive vs. ERα-negative phenotype in breast cancer cells, by showing that EGF represses ERα, SRC-1, and SRC-3 expression and activity through a mechanism that involves the up-regulation of miR-206. Finally, we show that forced overexpression of miR-206 in ERα-positive MCF-7 cells regulates numerous genes involved in breast cancer, and as expected in 17β-estradiol/ERα-addicted cells, causes an overall decrease in cell proliferation and survival rates in the absence of an ancillary oncogenic signaling pathway. These findings demonstrate how a single microRNA, in the presence of active oncogenic EGFR signaling, can coherently regulate several signaling networks involved in the coordinate repression of an ERα-positive, Luminal-A phenotype in breast cancer cells.
Results
Overexpression of miR-206 dysregulates the ERα-signaling regulatory network in MCF-7 cells
As a member of the nuclear hormone receptor family, ERα exerts transcriptional regulation of genes through the interaction with coregulatory proteins (39,40,41,42,43). Whereas coactivator proteins are recruited by ERα to genes that are stimulated by estrogen, corepressor proteins are recruited by ERα to genes that are repressed by estrogen. Because active ERα signaling represses miR-206 expression as part of a double-negative feedback loop, we initially hypothesized that mRNAs encoding corepressor proteins would be targeted by miR-206 and included within the ERα/miR-206 double-negative feedback network motif. However, in silico screens using TargetScan, PicTar, and RNA Hybrid databases indicated that the 3′-untranslated region (UTR) of mRNAs encoding two ERα-interacting corepressors, nuclear receptor corepressor 1 (NCOR1) and prohibitin 2 (PHB2), as well as ERβ, are not predicted to harbor potential miR-206 target sites. In contrast, mRNAs encoding two ERα-interacting coactivators, steroid receptor coactivator-1 (SRC-1; also called NCOA1) and steroid receptor coactivator-3 [SRC-3; also called NCOA3 or amplified in breast cancer-1 (AIB-1)] each contained two putative miR-206 target sites within their respective 3′-UTRs (Table 1). Given the results from in silico analysis, and the fact that ERα, SRC-1, and SRC-3 expression predominates over miR-206 levels in MCF-7 cells, we focused instead on the empirical confirmation of the miR-206 sites within SRC-1 and SRC-3 mRNAs.
Table 1.
In silico analysis of putative miR-206 target sites in various coregulatory molecules
| Gene name (official full name) | Gene symbol (human Refseq Id) | Gene function (specific to ERα) | Predicted target sites (nucleotides from start of the 3′-UTR) | TargetScan (context score) | PicTar (probability) | RNAHybrid (ΔG in kcal/mol) |
|---|---|---|---|---|---|---|
| Nuclear receptor | NCOA1 | AF-2 coactivator for ERα | Site 1 = 1903–1924 | Site 1 = −0.17 | Site 1 = 0.93 | Site 1 = −24.2a |
| Coactivator 1 | NM_147223 | Histone acetyltransferase | Site 2 = 2129–2150 | Site 2 = −0.14 | Site 2 = 0.88 | Site 2 = −21.4 |
| Nuclear receptor | NCOA3 | AF-2 coactivator for ERα | Site 1 = 232–253 | Site 1 = −0.10 | Site 1 = 0.88 | Site 1 = −19.3 |
| Coactivator 3 | NM_181659 | Histone acetyltransferase | Site 2 = 763–784 | Site 2 = −0.07 | Site 2 = 0.87 | Site 2 = −15.9 |
| Estrogen receptor α | ESR1 | Homodimerizes with ERα | Site 1 = 87–108a | Site 1 = N/A | Site 1 = N/A | Site 1 = −25.7a |
| NM_000125 | Transcription factor | Site 2 = 1137–1158 | Site 2 = −0.07 | Site 2 = 0.89 | Site 2 = −25.7 | |
| Estrogen receptor β | ESR2 | Heterodimerizes with ERα | No sites detected | N/A | N/A | N/A |
| NM_001437 | Inhibition of ERα activity | |||||
| Nuclear receptor | NCOR1 | AF-2 corepressor of ERα | No sites detected | N/A | N/A | N/A |
| Corepressor 1 | NM_006311 | Histone deacetylase | ||||
| Prohibitin 2 | PHB2 | Indirect corepressor | No sites detected | N/A | N/A | N/A |
| NM_007273 | Blocks SRC-1/ERα binding |
Table depicts the multiple putative miR-206 target sites within the coactivators NCOA1 (SRC-1) and NCOA3 (SRC-3), along with ESR1 (ERα), using the TargetScan, PicTar, and RNAHybrid prediction databases. The analysis gives a description of gene function with respect to ERα activity, the locations of putative miR-206 target sites from the start of the 3′-UTR, and the respective scoring from each of the databases mentioned above, where a lower context score, higher probability, and lower ΔG indicates a stronger miR-206 target site. No target sites were found in ESR2 (ERβ), or the corepressors NCOR1 and PHB2.
Valid miR-206 site from our previous study (33), not found by the TargetScan and PicTar databases.
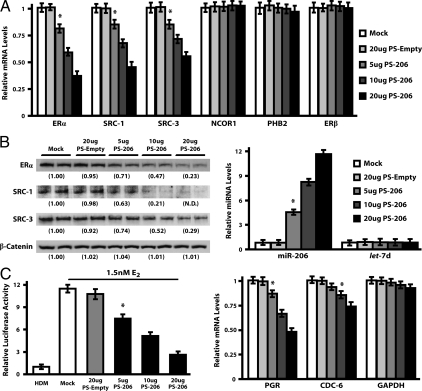
The regulation of SRC-1 and SRC-3 expression by miR-206 was first examined by overexpression of miR-206 using an simian virus 40 pSilencer expression plasmid containing the 85-nucleotide (nt) pre-miR-206 sequence [PS-206 (33)]. In MCF-7 cells, transfection of 5–20 μg PS-206 resulted in a subsequent increase in mature miR-206 levels, up to 11.5 ± 1.21-fold with 20 μg PS-206, whereas let-7d levels remained unchanged (Fig. 1B, right panel). Real-time PCR analysis determined that the overexpression of miR-206 caused a dose-dependent repression of both SRC-1 and SRC-3 mRNA levels, whereas NCOR1, PHB2, and ERβ levels remained unchanged, as compared with mock or PS-empty transfected MCF-7 cells (Fig. 1A). Western blot analyses of protein lysates generated from the same experimental conditions noted above revealed that after 48 h of transfection, 20 μg PS-206 significantly reduced SRC-3 expression 3.5 ± 0.27-fold and was also able to ablate SRC-1 expression to nondetectable levels (Fig. 1B, left panel). These values were normalized to total cellular β-catenin expression, which did not change among treatment conditions. Because SRC-1 and SRC-3 are involved in the transcriptional activation of genes by ERα, we assessed whether PS-206 abrogated luciferase activity generated from an estrogen response element (ERE)-luciferase reporter construct in MCF-7 cells (Fig. 1C, left panel). MCF-7 cells cultured in 1.5 nm E2 for 24 h generated an 11.8 ± 0.79-fold increase in ERE-luciferase activity when compared with hormone-depleted cells. However, transfection with 20 μg PS-206 reduced this enhancement of ERE-luciferase activity to only 3.1 ± 0.36-fold. Real time PCR analyses also indicated that 20 μg PS-206, in the presence of 1.5 nm E2, reduced expression of endogenous estrogen-responsive genes such as PGR and cell division cycle 6 (CDC-6) by 2.1 ± 0.11-fold and 1.4 ± 0.07-fold, respectively, whereas no significant change in glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels was observed (Fig. 1C, right panel). Collectively, these data show that miR-206 represses the expression of proteins involved in estrogen signaling, and therefore abrogates estrogen-mediated responses in MCF-7 cells.
Figure 1.
miR-206 Represses SRC-1/SRC-3 Expression and estrogen response in MCF-7 cells. A, MCF-7 cells transfected with 0–20 μg PS-206 or 20 μg PS-Empty for 48 h were harvested for subsequent real-time PCR analysis of ERα, SRC-1, SRC-3, NCOR1, PHB2, or ERβ transcript levels. B, Western blot analysis of ERα, SRC-1, SRC-3, and β-catenin protein expression in MCF-7 whole-cell lysates generated from the same transfection conditions noted above (left panel). Values below each blot represent the mean densitometric measurements from three independent experiments with duplicate samples per experiment and are reported as relative to the mock transfected control. The levels of miR-206 and let-7d under the current transfection paradigm were assessed by microRNA real-time PCR assays (right panel). C, Hormone-depleted MCF-7 cells treated with 0–1.5 nm E2 for 48 h were transfected with either 0–20 μg PS-206 or 20 μg PS-Empty for 24 h and assessed for ERE-luciferase activity (left panel). Values reported were normalized to Renilla, set relative to the hormone-depleted control, and depicted as the mean ± sem (see Materials and Methods). Real-time PCR assays were performed to measure levels of estrogen-responsive genes PGR, CDC-6, and GAPDH in 0–20 μg PS-206 or PS-empty transfected MCF-7 cells cultured in HDM with 1.5 nm E2 (right panel). All real-time PCR results were obtained from three independent experiments with four replicates per experiment, normalized to RPL-19 (mRNA analysis) or 5s (microRNA analysis), and depicted as the mean ± sem, relative to mock-transfected samples. N.D., Nondetectable densitometric reading; *, P < 0.05, as compared with the mock control.
Validation of the putative miR-206 target sites within the coactivators SRC-1 and SRC-3
To determine whether miR-206 directly targets the 3′-UTR of the SRC-1 and SRC-3 coactivators, we performed heterologous luciferase assays on each of the potential miR-206 target sites derived from the in silico analysis as performed in our previous study (33). Using the same pIS-0 vector, 60-nt regions of the endogenous target mRNA, which included the presumed miR-206 target sequence, were inserted into the 3′-UTR of the firefly luciferase gene [wild type (WT)]. For each site (denoted as A or B) a 4-nt mutation was also made to disrupt part of the target site that was predicted to hybridize to the 5′-seed sequence of miR-206 (denoted as 5′-mut). MCF-7 cells were then transfected with each construct plus a Renilla luciferase control, with or without various amounts of pre-miR-206, pre-let-7d, or pre-miR-neg, for 24 h. We found that in MCF-7 cells endogenous levels of miR-206 reduced SRC-1-A-WT, SRC-1-B-WT, SRC-3-A-WT, and SRC-3-B-WT luciferase activity by 1.4 ± 0.11-fold, 2.0 ± 0.13-fold, 1.3 ± 0.08-fold, and 1.6 ± 0.09-fold, respectively, when compared with each of the 5′-mut luciferase constructs (Fig. 2A–D, white bars in left panels). After addition of 200 nm pre-miR-206, suppression increased to 2.8 ± 0.19-fold, 5.1 ± 0.31-fold, 2.6 ± 0.21-fold, and 3.7 ± 0.28-fold, respectively (Fig. 2, A–D, black bars in left panels). The enhanced suppression is solely due to miR-206, because addition of either pre-miR-neg or pre-let-7d had no effect on the activity generated by pIS-SRC-WT or pIS-SRC-5′-mut constructs (Fig. 2, A–D, left panels).
Figure 2.
Luciferase reporter assays indicate miR-206 directly targets both SRC-1 and SRC-3. Various SRC-1 and SRC-3 constructs (500 ng) were transfected into MCF-7 cells, additionally transfected with either 200 nm pre-miR-neg, 200 nm pre-let-7d, or 0–20 nm pre-miR-206 (A–D, left panels), or with 50 nm antagomiR-neg, 50 nm antagomiR-let-7d, or 0–50 nm antagomiR-206 (A–D, right panels), and assayed for firefly luciferase activity 24 h after transfection. Specifically, activity assays were performed on two predicted miR-206 target sites within SRC-1 [pIS-SRC-1-A and pIS-SRC-1-B, (panels A and B) and two predicted sites within SRC-3 [pIS-SRC-3-A and pIS-SRC-3-B (panels C and D). Luciferase activity levels generated by the various pIS-SRC-wild-type (WT) constructs, containing the endogenous miR-206 target sites, were set relative to the activity of the pIS-SRC-5′-mutant (5′mut) constructs, containing a 4- to 6-nt mismatch mutation in the respective target sites, under the mock-transfected, untreated condition, where (*) represents a P value of <0.05. All values were normalized to Renilla luciferase activity, generated by 200 ng pRL-TK, and are reported as the mean activity ± sem from three independent experiments with each treatment condition performed in duplicate. Ant, AntagomiR; neg, scrambled RNA sequence.
We further analyzed the putative miR-206 target sites within SRC-1 and SRC-3 by reducing endogenous miR-206 levels using a synthetic 2′-O-methyl antisense RNA to miR-206 (antagomiR-206). Despite the various levels of luciferase activity generated from each of the pIS-SRC-WT constructs, 50 nm antagomiR-206 was sufficient to enhance the luciferase activity to that of the respective pIS-SRC-5′-mut constructs (Fig. 2, A–D, black bars in right panels). Moreover, 50 nm antagomiR-let-7d, and antagomiR-neg (a scrambled antisense RNA) resulted in no enhancement in the suppression of luciferase activity conferred by the pIS-SRC-WT constructs (Fig. 2, A–D, right panels). These data demonstrate that the two miR-206 target sites identified within the 3′-UTR of SRC-1 and SRC-3 specifically interact with miR-206, allowing for the direct repression of the corresponding mRNA and protein levels.
GATA-3, a luminal marker in breast cancer, is also targeted and repressed by miR-206
In addition to coregulatory proteins, ERα also interacts with several other transcription factors to promote estrogen-mediated responses. Expression profiling using the small, targeted Breast Cancer and Estrogen Receptor Signaling quantitative PCR (qPCR) array from SuperArray Biosciences (see Materials and Methods) indicated that the transcription factor GATA-3 was suppressed by miR-206 (see Fig. 9A). GATA-3 is required for normal ductal morphogenesis of terminal end bulb development in mouse mammary glands, as well as for luminal differentiation of epithelial cells (37,44). GATA-3 and ERα expression are also highly correlated in breast cancer, and comparative meta-analyses of coexpressed genes in ERα or GATA-3 expressing breast tumors indicate a significant overlap in their respective gene expression patterns (35). Furthermore, GATA-3 and ERα mutually stimulate each other’s expression by direct transcriptional regulation (45). These known interactions between GATA-3 and ERα, as well as our finding of GATA-3 down-regulation in miR-206 overexpressing cells, prompted us to determine whether miR-206 directly targeted and suppressed the expression of GATA-3 in MCF-7 cells. Real-time PCR assays performed on 0–20 μg PS-206 or 20 μg PS-Empty transfected MCF-7 cells for 48 h, indicated that miR-206 decreased GATA-3 expression in a dose-dependent manner, in which 20 μg PS-206 induced a 4.1 ± 0.12-fold reduction in GATA-3 mRNA levels (Fig. 3A). Additionally, in silico examination of microRNA binding sites within the 3′-UTR of GATA-3 mRNA revealed one predicted miR-206 target site, which raised the possibility of direct, posttranscriptional regulation of GATA-3 by miR-206. Direct analysis of this site using a pIS-GATA-WT luciferase reporter with 200 nm pre-miR-206 revealed that it is a relatively weak, 2.1 ± 0.11-fold as compared with the pIS-GATA-3–5′-mut construct, but a bona fide miR-206 target site (Fig. 3B, top panel). Given that GATA-3 expression in MCF-7 cells requires ERα, miR-206 could also decrease GATA-3 mRNA levels indirectly, by repressing ERα and ERα-related coactivators. Collectively, these data show that miR-206 suppresses ERα-dependent and luminal-specific gene expression through the coordinate repression of two transcription factors and two coregulators.
Figure 9.
Overexpression of miR-206 in MCF-7 cells confers an antitumorigenic phenotype. A, ER signaling qPCR arrays were performed on MCF-7 cells transfected with either 0 μg or 10 μg PS-206 (see Materials and Methods). Values were normalized to a pool of housekeeping genes on the array by ΔΔCt and are reported as the average fold change in gene expression relative to the mock control and are sorted into GOTERMs. B, Trypan blue viability assays were performed on MCF-7 cells transfected with 15 μg PS-Empty or 15 μg PS-206 for 0–144 h (left panel). For each time point, values were reported as the number of viable cells × 105. MCF-7 cells transfected with 20 μg PS-Empty or 0–20 μg PS-206 for 48 h were assessed for BrdU incorporation (right panel). Values were reported as the percentage of cells positive for nuclear staining relative to the mock control, which was set to 100%. C, TUNEL labeling assays of MCF-7 cells under the same transfection paradigm as noted above (left panel). Results are reported as a relative fold increase in the number of TUNEL positive cells compared with the mock-transfected condition. MCF-7 cells transfected with 20 μg PS-empty or 0–10 μg PS-206 for 48 h were analyzed for BCL-2, cleaved PARP, cyclin D1, and β-catenin protein expression via Western blot analysis (right panel). Numbers below the band represent average densitometric measurements from three independent experiments, relative to the mock control. *, P < 0.05, compared with the mock control.
Figure 3.
The GATA-3 transcription factor is directly targeted and repressed by miR-206. A, Real-time PCR analysis of GATA-3 and ERβ mRNA levels was performed on MCF-7 cells treated with 0–20 μg PS-206 or 20 μg PS-empty for 48 h. Results were obtained from three independent experiments with four replicates per experiment, normalized to RPL-19, and depicted as the mean ± sem, relative to mock-transfected samples, where (*) denotes a P value of <0.05 as compared with the mock control. B, MCF-7 cells were transfected with 500 ng of either pIS-GATA-3-WT or pIS-GATA-3-5′-mut reporter constructs for 24 h and assessed for firefly luciferase activity. Cells were additionally transfected with either 200 nm pre-miR-neg, 200 nm pre-let-7d, or 0–200 nm pre-miR-206 (top panel), or with 50 nm antagomiR-neg, 50 nm antagomiR-let-7d, or 0–50 nm antagomiR-206 (bottom panel), and the resultant luciferase activity levels were normalized to Renilla luciferase activity and set as relative to the pIS-GATA-3–5′-mut activity under the mock-transfected, untreated condition, where (*) represents a P value of <0.05. Values are reported as the mean activity ± sem from three independent experiments. Ant, AntagomiR; neg, scrambled RNA sequence.
The miR-206-induced blunting of estrogenic response is not solely due to the loss of ERα levels
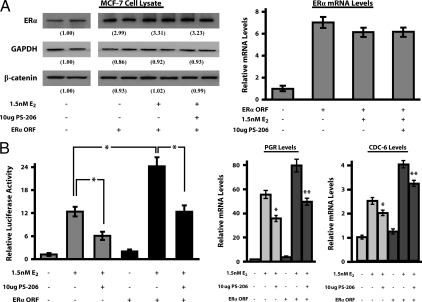
To examine whether the ability of miR-206 to alter the estrogenic response in MCF-7 cells was due solely to its targeting of ERα, or due to the targeting of other coactivator proteins such as SRC-1, and SRC-3, we transfected MCF-7 cells with an ERα open-reading frame-only cytomegalovirus (CMV) expression construct (pCMV6-ERα-ORF) that encoded an mRNA devoid of the 3′-UTR, and thus, lacking miR-206 target sites. In hormone-depleted MCF-7 cells, a 24-h transfection of 500 ng pCMV6-ERα-ORF led to a 2.9 ± 0.31-fold and 6.7 ± 0.79-fold increase in ERα protein expression and mRNA levels, respectively, as compared with the hormone-depleted, mock-transfected condition (Fig. 4A). As expected, Western blot and real-time PCR analyses indicated that cotransfection of pCMV6-ERα-ORF with 10 μg PS-206 had no significant effect on the level of ERα overexpression in MCF-7 cells.
Figure 4.
miR-206 Blunts estrogenic responses in MCF-7 cells independently of ERα levels. A, Western blot analysis of ERα, GAPDH, and β-catenin expression in hormone-depleted MCF-7 cells treated with 0–1.5 nm E2 after either mock, pCMV6-ERα-ORF (500 ng), and/or PS-206 (10 μg) transfection for 24 h (left panel). The values below each set of bands represent the mean densitometric intensities of each condition from three independent experiments and are reported as relative to the mock-transfected, hormone-depleted control. Results from real-time PCR assays monitoring the ERα mRNA levels in MCF-7 cells under the treatment/transfection paradigm noted above (right panel). B, ERE-luciferase activity assays on hormone-depleted MCF-7 cells ± 1.5 nm E2 after mock, and/or pCMV6-ERα-ORF (500 ng), and/or PS-206 (10 μg) transfection for 24 h (left panel). Values reported were normalized to Renilla activity, set relative to hormone-depleted control, and depicted as the mean of three independent experiments ± sem, where (*) represents P < 0.05. PGR and CDC-6 mRNA levels from the treatment paradigm above, as determined by real-time PCR analysis (right panel). *, P < 0.05, as compared with 1.5 nm E2; **, P < 0.05, as compared with 1.5 nm E2 + ERα-ORF. All real-time PCR results were obtained from three independent experiments with four replicates per experiment, normalized to RPL-19, and depicted as the mean ± sem, relative to mock-transfected, hormone-depleted samples.
To measure the effect of pCMV6-ERα-ORF on the transcriptional estrogenic response, ERE-luciferase activity assays (Fig. 4B, left panel) and real-time PCR analysis of PGR and CDC-6 mRNA levels (Fig. 4B, right panel) were performed on hormone-depleted MCF-7 cells treated with 0–1.5 nm E2, and transfected with either 0–10 μg PS-206 and/or 500 ng pCMV6-ERα-ORF for 24 h. The pCMV6-ERα-ORF plus 1.5 nm E2 condition generated 2.1 ± 0.61-fold higher ERE-luciferase activity, as well as the 1.5 ± 0.21-fold, and 1.6 ± 0.19-fold higher PGR and CDC-6 mRNA levels, respectively, when compared with 1.5 nm E2 alone. Thus, it is clear that pCMV6-ERα-ORF generates functionally active ERα. Importantly, the approximate 2.2-fold repression of ERE-luciferase activity, as well as the 1.6- and 1.3-fold repression in PGR and CDC-6 mRNA levels, respectively, mediated by 10 μg PS-206 occurs even when the pCMV6-ERα-ORF is generating constant levels of ERα. These data indicate that the targeting of proteins other than ERα contributes to the repression of this ERα-mediated transcriptional response. These data also suggest, but do not prove, that the targeting of SRC-1, SRC-3, and/or GATA-3 by miR-206 contributes to the overall effect of this microRNA on the estrogenic response in MCF-7 cells.
The addition of EGF blunts E2/ERα-mediated responses in hormone-depleted MCF-7 cells
ERα and EGFR expression are frequently inversely correlated in breast cancer (29), and EGFR overexpression has been shown to repress ERα expression in MCF-7 cells (15). To determine whether EGF reduced the transcript levels of genes involved in mediating estrogen response (i.e. ERα, SRC-1, and SRC-3), total RNA was harvested from MCF-7 cells cultured in hormone-depleted media (HDM) for 24 h and treated with 0–200 ng/ml EGF for an additional 48 h. Real-time PCR analysis revealed that EGF reduced the mRNA levels of ERα, as well as the coactivators SRC-1 and SRC-3, in both a dose- and time-dependent manner (Fig. 5, A and B). In contrast, NCOR1, PHB2, and ERβ mRNA levels remained constant across all treatment conditions. Specifically, 200 ng/ml EGF repressed ERα, SRC-1, and SRC-3 gene expression by 2.2 ± 0.12-fold, 1.8 ± 0.14-fold, and 1.6 ± 0.12-fold, respectively (Fig. 5A). Moreover, the effect of EGF displayed relatively slow kinetics, requiring approximately 16 h of treatment before significant reductions in ERα, SRC-1, and SRC-3 mRNA levels were observed (Fig. 5B). Western blot analyses indicated that 48 h of the 200 ng/ml EGF treatment also resulted in an approximate 2- to 2.5-fold decrease in ERα, SRC-1, and SRC-3 protein expression (Fig. 5C, left panel). Levels of GATA-3 mRNA were also assessed and found to be down-regulated by EGF in both a dose- and time-dependent manner (supplemental Fig. S1 published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org).
Figure 5.
EGF inhibits SRC-1/SRC-3 expression and global estrogen response in MCF-7 cells. Hormone-depleted MCF-7 cells were treated with 0–200 ng/ml EGF for 48 h (A), or 200 ng/ml EGF for 0–32 h (B), and total RNA was harvested for subsequent real-time PCR analysis of ERα, SRC-1, SRC-3, NCOR1, PHB2, and ERβ levels. C, Western blot analysis of ERα, SRC-1, SRC-3, and β-catenin protein expression in hormone-depleted MCF-7 cells treated with 0–200 ng/ml EGF for 48 h (left panel). The numbers below the bands represent the average densitometric measurements from three independent experiments and are reported as relative to the hormone-depleted control. ERE-luciferase activity assays were performed on hormone-depleted MCF-7 cells treated with 0–200 ng/ml EGF ± 1.5 nm E2 for 48 h (right panel). The mean activity levels ± sem were normalized to Renilla and set relative to the hormone-depleted control. D, PGR and CDC-6 mRNA levels were assessed via real-time PCR assays in hormone-depleted MCF-7 cells ± 1.5 nm E2, and/or 200 ng/ml EGF for 48 h. Values from real-time PCR experiments were normalized to RPL-19 and reported as the mean ± sem, relative to hormone-depleted or untreated conditions. *, P < 0.05, as compared with untreated (A and B) or E2 treated (C and D) conditions.
We then used a luciferase reporter construct harboring a validated estrogen response element [ERE-Luc; (46)] to test the effect of EGF on the global estrogen response in MCF-7 cells. Whereas 1.5 nm E2 stimulated luciferase activity 11.4 ± 0.42-fold when compared with hormone-depleted cells, this response was severely blunted (4.5 ± 0.21-fold increase in activity) by the addition of 200 ng/ml EGF (Fig. 5C, right panel). Furthermore, 200 ng/ml EGF reduced the ability of 1.5 nm E2 to enhance the expression of PGR and CDC-6 (1.9 ± 0.14-fold and 1.3 ± 0.09-fold, respectively), when compared with the 1.5 nm E2 treatment alone (Fig. 5D). Taken together, these data clearly indicate that EGF initiates a global down-regulation of the estrogen response in MCF-7 cells by reducing the expression of ERα, the steroid hormone receptor coactivators SRC-1 and SRC-3, as well as the transcription factor GATA-3.
EGF and EGFR signaling selectively enhances mature miR-206 levels in breast cancer cell lines
We previously reported that miR-206 was down-regulated by E2 as part of a feedback mechanism to maintain ERα positivity in breast cancer cell lines (33). Given the data above, this estrogen-dependent repression of miR-206 would presumably be antagonized by the addition of EGF. Indeed, in MCF-7 cells cotreatment studies showed that E2 or PPT, an ERα-selective agonist, plus EGF, blocked the ability of E2 to repress miR-206 levels (supplemental Fig. S2A). As a result, we examined whether EGF up-regulated miR-206, as part of the mechanism by which EGF represses ERα and estrogen responsiveness in MCF-7 cells. In this series of experiments the Basal-like MDA-MB-231 cells were used as a positive control, because this particular cell line has approximately 8.3-fold enhanced expression of EGFR compared with the MCF-7 cell line (supplemental Fig. S2B). Both cell types were cultured in HDM for 24 h and treated with 0–200 ng/ml EGF for an additional 48 h, as noted above. Real-time PCR experiments indicated that 200 ng/ml EGF increased mature miR-206 levels 1.9 ± 0.13-fold and 3.8 ± 0.29-fold in MCF-7 and MDA-MB-231 cells, respectively (Fig. 6A). Additionally, EGF significantly increased miR-206 levels 16 h after treatment in hormone-depleted MCF-7 and MDA-MB-231 cells (1.3 ± 0.08-fold and 3.2 ± 0.18-fold, respectively), as compared with the 0 h time point (Fig. 6B). Throughout these experiments EGF did not enhance the levels of mature let-7d, another microRNA presumably able to target ERα as determined by the miRanda database/algorithm (47).
Figure 6.
Active EGF/EGFR signaling stimulates miR-206 levels in breast cancer cell lines. Hormone-depleted MCF-7 (A and B, left panel) and MDA-MB-231 (A and B, right panel) cells were treated with either 0–200 ng/ml EGF for 48 h (A), or 200 ng/ml EGF for 0–32 h (B), and total RNA was harvested for subsequent real-time PCR analysis of miR-206 and let-7d microRNA levels. C, MCF-7 (left panel) and MDA-MB-231 (right panel) cells were hormone depleted, treated with 0–200 ng/ml EGF for 48 h, and analyzed for miR-206 activity using pIS-ERα-1-WT, pIS-ERα-1-SNP, and pIS-ERα-1-5′-mut luciferase constructs. Reported values represent average luciferase activity ± sem and were generated from three independent experiments after normalization to Renilla luciferase, relative to miR-206 activity in the respective untreated conditions. Values from real-time PCR experiments were obtained from three independent experiments performed in triplicate, normalized to 5s RNA, and reported as the mean ± sem, relative to hormone-depleted controls. *, P < 0.05, as compared with the hormone-depleted control.
To determine whether EGF enhances miR-206 activity, as opposed to level, MCF-7 and MDA-MB-231 cells were hormone depleted for 24 h and then transfected with various pIS-ERα-1 luciferase constructs, previously shown to be indicators of functional miR-206 activity (33), for 48 h in the presence of 0–200 ng/ml EGF. These experiments confirmed that EGF increased miR-206 activity in both cell lines, as determined by enhanced suppression of luciferase activity generated by the pIS-ERα-1-WT and pIS-ERα-1-SNP constructs, which was not observed with the pIS-ERα-1–5′-mut construct (Fig. 6C).
To test whether EGF enhancement of miR-206 levels and activity depends upon intact EGFR/MAPK signaling, MDA-MB-231 cells, which highly express the EGFR, were transfected with 0–50 nm siEGFR [smart pool of four small interfering RNAs (siRNAs) directed toward EGFR] for 24 h and monitored for subsequent miR-206 levels. Western blot analysis confirmed 25 nm siEGFR induced a 5.6 ± 1.1-fold knockdown in EGFR expression, as compared with the mock transfected condition, while β-actin and β-catenin levels remained unchanged (Fig. 7A, left panel). Importantly, real-time PCR analysis indicated that 25 nm siEGFR effectively repressed miR-206 levels by 2.2 ± 0.95-fold, whereas let-7d levels did not change under any treatment condition (Fig. 7A, right panel). These findings were validated by treating the MDA-MB-231 cells with 0–10 μm PD153035 (EGFR inhibitor) or 0–10 μm U0126 (MAPK kinase inhibitor) for 48 h (Fig. 7B). Real-time PCR analyses indicated that 10 μm PD153035 or U0126 reduced miR-206 levels by 3.2 ± 0.15-fold or 2.3 ± 0.21-fold, respectively, as compared with vehicle-treated cells. In summary, these findings demonstrate that EGF enhances miR-206 levels and activity via the EGFR/MAPK signaling pathway in both MCF-7 and MDA-MB-231 cell lines, probably through similar cellular mechanisms.
Figure 7.
Inhibition of EGFR/MAPK Signaling in MDA-MB-231 cells reduces miR-206 levels. A, MDA-MB-231 cells transfected with 0–50 nm siEGFR (smart pool of four siRNAs directed toward EGFR) for 24 h, analyzed for EGFR, β-actin, and β-catenin expression via Western blot analysis (left panel) and subsequent miR-206 and let-7d levels by real-time PCR assays (right panel). Numbers below representative blots are average densitometric measurements generated from each condition, from three independent experiments, and are reported as relative to the mock-transfected control. B, MDA-MB-231 cells were treated with either 0–10 μm PD153035 (left panel) or 0–10 μm U0126 (right panel) for 48 h, and total RNA was harvested for subsequent real-time PCR analysis of miR-206 and let-7d microRNA levels. All values from real-time PCR experiments were obtained from three independent experiments performed in triplicate, normalized to 5s RNA, and reported as the mean ± sem, relative to mock-transfected or untreated controls. *, P < 0.05, as compared with the mock-transfected or vehicle control.
EGF-mediated repression of the estrogen response in MCF-7 cells requires miR-206 activity
EGF represses the expression of ERα-related genes in MCF-7 cells as early as 16 h after treatment, which is approximately the time when EGF begins to significantly elevate miR-206 levels and activity. To test whether the EGF-mediated repression of ERα response required miR-206, MCF-7 cells were hormone depleted for 24 h and transfected with 50 nm of either antagomiR-206, antagomiR-neg, or no antagomiR (mock), in the presence or absence of 200 ng/ml EGF for 48 h (Fig. 8, A and B). Real-time PCR analysis of total RNA harvested from MCF-7 cells under this treatment paradigm indicated that despite the presence of EGF, levels of ERα, SRC-1, and SRC-3 were enhanced 1.5 ± 0.09-fold, 1.3 ± 0.06-fold, and 1.4 ± 0.07-fold, respectively, when treated with 50 nm antagomiR-206 vs. either mock or antagomiR-neg controls (Fig. 8A). This effect was selective because NCOR1, PHB1, and ERβ transcript levels remained unchanged in the presence or absence of either EGF and/or antagomiR-206.
Figure 8.
EGF-mediated repression of estrogen response in MCF-7 cells requires miR-206. A, Hormone-depleted MCF-7 cells treated with either 0 ng/ml or 200 ng/ml EGF for 48 h, and/or 50 nm antagomiR-neg or 50 nm antagomiR-206 for 24 h, were harvested for subsequent real-time PCR analysis of ERα, SRC-1, SRC-3, NCOR1, PHB2, or ERβ levels. To analyze levels of the estrogen-responsive genes PGR, CDC-6, and GAPDH, all cells under the treatment conditions mentioned above, were also given 1.5 nm E2 for 48 h (B, left panel). Real-time PCR results were obtained from three independent experiments with four replicates per experiment, normalized to RPL-19, and depicted as the mean ± sem relative to mock-transfected hormone-depleted samples. In these experiments * denotes a P value of <0.05 when compared with the mock-transfected, EGF treatment condition. ERE-luciferase activity assays were performed on hormone-depleted MCF-7 cells treated with either 1.5 nm E2 and/or 200 ng/ml EGF for 48 h, and/or 50 nm antagomiR-neg or 25–50 nm antagomiR-206 for 24 h (B, right panel). Values are depicted as the mean ± sem of three independent experiments, and set relative to the hormone-depleted samples. * denotes P < 0.05 when compared with the mock-transfected, E2 + EGF treatment condition. Ant, AntagomiR; neg, scrambled RNA sequence.
To determine whether the same treatment paradigm resulted in enhanced estrogen response, real-time PCR assays were performed to measure levels of transcripts downstream of ERα (Fig. 8B, left panel). In the presence of 1.5 nm E2 and 200 ng/ml EGF, 50 nm antagomiR-206 enhanced PGR and CDC-6 mRNA levels 1.7 ± 0.13-fold and 1.3 ± 0.11-fold, respectively, as compared with the antagomiR-neg and mock-transfected controls. However, the transcript levels of another estrogen-responsive gene, GAPDH, was not elevated, indicating that miR-206 only partially augments EGF-mediated repression of estrogen response in MCF-7 cells. To evaluate the extent of miR-206 involvement in the EGF-induced repression of ERα signaling, MCF-7 cells were hormone depleted for 24 h, transfected with the various antagomiRs, as well as the ERE-firefly luciferase reporter and a Renilla luciferase control construct, all in the presence or absence of EGF and/or E2 (Fig. 8B, right panel). In the cultures that received both 200 ng/ml EGF and 1.5 nm E2, 50 nm antagomiR-206 enhanced ERE-luciferase activity 1.9 ± 0.12-fold when compared with either 50 nm antagomiR-neg or mock-transfected MCF-7 cells. Given that the 50 nm antagomiR-206 treatment subsequently led to a significant increase in ERE-luciferase activity, as well as PGR and CDC-6 mRNA levels while in the presence of EGF, strongly indicates that miR-206 is partially required for effective EGF-mediated repression of estrogenic responses within MCF-7 cells.
The overexpression of miR-206 promotes an antitumorigenic phenotype in MCF-7 cells
To assess the consequences of forced miR-206 expression in the MCF-7 cell line, a real-time qPCR array (see Materials and Methods) was used to examine the changes in expression of genes known to be involved in breast cancer and estrogen-mediated signaling in PS-206 vs. mock-transfected MCF-7 cells (Fig. 9A). To determine the major biological process associated with each gene, we performed a categorical analysis using GOTERMS and the DAVID/EASE PROGRAM (48). Of the 76 genes analyzed, 10 μg of PS-206 resulted in the dysregulation of 48 genes, a majority of which were involved in the regulation of programmed cell death, cell cycle, cell adhesion, and cellular metabolism, whereas 28 genes remained unchanged. Further in silico analysis of these genes using the TargetScan and miRanda databases indicated that some genes were potentially direct targets of miR-206 (i.e. BCL2, CCND1, MKI67, HMGB1, and, as confirmed above, GATA3), whereas the reduced expression of the other genes, as well as the enhanced expression of Fas ligand and IL-6, may be due to indirect effects of PS-206. Given the dependence MCF-7 cells have on estrogen and ERα signaling, it is not surprising to observe that overexpression of miR-206 in general down-regulates both antiapoptotic and proproliferative genes. Thus, in the absence of ancillary oncogenic signaling, miR-206 overexpression and the consequent repression of ERα signaling appear to impede proliferation and survival in MCF-7 cells.
The effect of miR-206 overexpression on proliferation was assessed directly by performing Trypan Blue growth curve assays over the course of 96 h post PS-206 transfection. MCF-7 cells transfected with 15 μg of PS-206 had a 1.8 ± 0.09-fold reduction in the number of viable cells as compared with 15 μg PS-Empty or mock-transfected conditions (Fig. 9B, left panel). To confirm this finding, cell cycle traverse was measured using bromodeoxyuridine (BrdU) incorporation assays (Fig. 9B, right panel). We found that 48 h after transfection, 20 μg of PS-206 reduced the relative percentage of BrdU-labeled cells by 3.6 ± 0.32-fold as compared with the mock or 20 μg PS-Empty-transfected MCF-7 cells. All values were normalized to the total number of cells within the assay as determined by hematoxylin counterstaining. Furthermore, Western blot analysis indicated that at 48 h cyclin D1 expression was reduced 4.3 ± 0.32-fold in 20 μg PS-206 vs. mock or 20 μg PS-Empty-transfected cells (Fig. 9C, right panel).
Overexpression of miR-206 also enhanced the frequency of apoptosis in MCF-7 cells, which was assessed directly by a terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) assay (Fig. 9C, left panel). Transfection of 20 μg PS-206 for 48 h resulted in a 2.1 ± 0.12-fold increase in the number of cells undergoing DNA fragmentation when compared with mock or PS-Empty-transfected cells. Western blot analyses were performed on lysates generated from MCF-7 cells 48 h after PS-206 transfection to measure the expression of particular apoptotic markers. These analyses indicated that although whole-cell β-catenin levels remained unchanged, the antiapoptotic protein BCL-2 was reduced 2.3 ± 0.15-fold, whereas levels of cleaved-poly ADP ribose polymerase (PARP), a marker of terminal cell death, was enhanced 7.8 ± 0.39-fold in cells transfected with 20 μg PS-206 as compared with mock-transfected cells (Fig. 9C, right panel). In summary, miR-206 exerts an overall tumor-suppressive action in E2/ERα-addicted MCF-7 cells, by stalling cell-cycle traverse and repressing cell survival cues.
Discussion
Breast cancer represents a heterogeneous disease for which several subtypes have been characterized, including the Luminal-A and Basal-like subtypes (1,2,3,4,49). Luminal-A cancers represent the majority (i.e. ∼65%) of newly diagnosed breast cancers, express ERα and PGR, but display little or no expression of EGFR and HER2. Luminal-A cancers tend to be of a lower grade, confer a positive prognosis, and usually respond, at least initially, to adjuvant hormonal therapy (e.g. antiestrogens and aromatase inhibitors). Basal-like tumors are relatively rare, do not express ERα, PGR, or HER2, but frequently overexpress EGFR. Basal-like cancers tend to be of a higher grade, confer a poor prognosis, and cannot be treated with antihormone therapy, although EGFR inhibitors may be useful in some cases.
Due to the different behavior and treatment options of these two subtypes, it is important to understand their origins and underlying genetic and epigenetic lesions. El-Ashry and colleagues (15,16,17) have shown that forced expression of EGFR or downstream MAPK signaling components converts MCF-7 cells, which fall into the Luminal-A subtype, into ERα-negative cells. In fact, expression profiling of EGFR (plus EGF)-overexpressing MCF-7 cells closely resembled that of Basal-like breast cancer (12). These findings support the possibility that some forms of ERα-negative, Basal-like cancers arise from ERα-positive, Luminal A cancers through the oncogenic gain of EGFR overexpression. Our findings now identify miR-206 as one component of the EGFR signaling pathway in breast cancer that directly suppresses aspects of the luminal phenotype.
In the current study, we hypothesized that the network motif, as defined by a negative feedback loop between a transcription factor and a microRNA, would also include the coregulatory proteins that are recruited by the transcription factor (34). As part of a double-negative feedback loop between ERα and miR-206, we expected to find miR-206 target sites in mRNAs encoding ERα corepressor proteins, such as NCOR1, which would be involved in maintaining the low basal levels of miR-206 found within these cells. However, no predicted miR-206 sites were found in the direct corepressor, NCOR1, or the indirect corepressor, PHB2. In contrast to our expectations, in silico analyses revealed two putative miR-206 target sites each in the 3′-UTRs of the mRNAs encoding the two coactivator proteins, SRC-1 and SRC-3. Although miR-206 sites may reside in other corepressors of ERα, we decided instead to focus on the miR-206 target sites predicted to be within the 3′-UTR of SRC-1 and SRC-3 mRNAs.
SRC-1 and SRC-3 are both coactivators of several members of the nuclear receptor family, including ERα, as well as other transcription factors, such as E2F1 (39). Both SRC-1 and SRC-3 have been implicated in normal breast development (39), and SRC-3 [also called amplified in breast cancer (AIB-1)] is amplified or overexpressed in breast cancer, usually in the ERα-negative, HER2 subtype (50). SRC-3 was previously shown to be a target of miR-17-5p, which is expressed at low levels in breast cancer as opposed to normal breast tissue (51). In this study, we found that expression of miR-206 reduced endogenous SRC-1 and SRC-3 levels in MCF-7 cells, and that all four miR-206 target sites conferred miR-206-specific repression in heterologous luciferase reporter assays. Therefore, it is likely that the targeting of these two coactivators by miR-206 contributes to the dampening of estrogen-dependent responses on which this particular cell type is reliant for growth and survival.
Another ERα-interacting protein down-regulated by miR-206 overexpression was GATA-3, as determined by our qPCR array. GATA-3 is one of six members of the GATA zinc-finger transcription factor family (52) and is required for normal ductal development and luminal differentiation of mammary epithelia (37,38). GATA-3 and ERα expression are highly correlated in breast cancer (36), and comparative meta-analysis found a significant overlap in the coexpression signature of genes expressed in either ERα- or GATA-3-positive tumors (35). Because ERα and GATA-3 mutually stimulate each other’s expression (45), it is likely that miR-206 indirectly represses GATA-3 through its effects of ERα, SRC-1, and SRC-3. Additionally, we found one predicted miR-206 target site in the 3′-UTR of GATA-3 mRNA, and this site conferred a modest repression when assayed in a heterologous reporter system.
The finding that miR-206 targets a number of mRNAs encoding several ERα-interacting proteins raises the possibility that miR-206 suppresses estrogenic signaling/responses in MCF-7 cells through the coherent down-regulation of multiple components within this system, as opposed to ERα alone. To address this possibility, we used a DNA construct that encodes an ERα open reading frame devoid of a 3′-UTR (pCMV6-ERα-ORF), and as a result were able to overexpress a form of ERα that was not targeted by miR-206. The fact that miR-206 blunted estrogen-mediated responses, even in the face of a constant level of ERα (pCMV6-ERα-ORF), supports a model that involves the targeting of ERα, along with coactivators and other ERα-associated proteins, by miR-206. Although further studies are required to fully sort out the contributions of each of these miR-206 targets to the overall reduction of estrogen signaling by this microRNA, our studies demonstrate that miR-206 suppresses ERα expression and ERα-mediated signaling at several levels.
The second general finding of this study is that EGF and EGFR represent one oncogenic signaling pathway that tips the equilibrium in favor of miR-206. EGF stimulates miR-206 expression, and miR-206 is required for a significant degree of the repressive effect EGF has on the expression of ERα, SRC-1, and SRC-3. Thus, in Luminal-A breast cancer, ERα and coactivator proteins are robustly expressed, whereas EGFR and miR-206 expression is low (Fig. 10). However, the onset of EGFR overexpression, which occurs in more than half of Basal-like breast cancers (7), would stimulate miR-206 expression. Consequently, ERα, SRC-1, SRC-3, and GATA-3 expression would be repressed. Because EGFR also stimulates other proliferative and antiapoptotic pathways (53), the transformed cells could then dispense with ERα-dependent signaling. Furthermore, the loss of GATA-3 would not only amplify ERα silencing, but would also lead to a loss of luminal differentiation (37,38). Eventually other changes, such as hypermethylation of the ERα promoter (54), could occur to stabilize this loss of ERα expression,. These findings lead one to predict that overexpression of EGFR, along with miR-206, would generate a Basal-like breast cancer cell type from MCF-7 cells, a prediction supported by the findings of Creighton et al. (12). Conversely, it will be important to determine whether knockdown of miR-206 blocks the ability of EGFR overexpression, plus EGF, to induce this Basal-like transcriptional program and phenotypic switch.
Figure 10.
A model of EGF/EGFR-induced inhibition of a Luminal-A phenotype in MCF-7 cells. MCF-7 breast cancer cells are of a Luminal-A phenotype, express ERα and PGR, and are negative for HER2 amplification. The activity of miR-206 can influence this phenotype through the direct targeting and repression of not only ERα, but of coactivators (SRC-1 and SRC-3) and transcription factors (GATA-3) that are required to mediate estrogenic responses within this cell type. The expression and activity of miR-206 are enhanced by active EGF/EGFR signaling, a hyperactive signaling pathway found in a majority of ERα-negative Basal-like breast tumors. This supports the notion that ERα-positive tumors acquiring ancillary EGFR signaling would have enhanced miR-206 levels, which would push the bi-stable system within the tumor from an ERα-positive to an ERα-negative phenotype. Also, given that EGFR expression induces a Basal-like gene expression pattern in MCF-7 cells (12), miR-206 may also actively promote a Basal-like phenotype.
The mechanism by which EGFR becomes overexpressed in Basal-like cancers is presently unknown. One possible approach is through the oncogenic loss of microRNAs that target EGFR mRNA in Luminal-A cancers. Webster et al. (55) recently reported the targeting of EGFR mRNA at two sites by miR-7 in several cancer cell lines, including the MDA-MB-468 breast cancer cell line. Interestingly, we recently observed that miR-7 was increased 2.4-fold in PPT vs. ICI 182780 (also called Faslodex; an antiestrogen)-treated MCF-7 cells, using small RNA library construction and deep sequencing (our unpublished data). Thus, estrogen/ERα signaling in Luminal-A breast cancers may keep EGFR levels low by stimulating the expression of miR-7 (and possibly other microRNAs). Conversely, microRNAs that directly regulate EGFR expression, such as miR-128b, frequently show a loss of heterozygosity in human tumor samples (56). Thus, it is likely that microRNA-dependent mechanisms contribute to the low and high expression of EGFR in Luminal-A and Basal-like breast cancer, respectively.
The mechanism by which active ERα or EGFR signaling regulates the expression of miR-206 has not yet been determined. Although it is known that the miR-206 gene is a monocistronic, discrete gene at chromosomal location 6p12.2 (http://www.ncbi. nlm.nih.gov), almost no work has been performed on the transcriptional regulation of this gene in humans. In silico analysis of the human genomic sequence near the predicted transcriptional start site of miR-206 revealed a putative serum response element, which might confer transcriptional regulation by the EGFR pathway through c-Fos/c-Jun activation and the subsequent recruitment to this serum-response element site. However, further work is still required to examine whether estrogen and/or EGFR directly regulates miR-206 expression at the transcriptional level, and if so, specifically which signaling pathways and DNA response elements are involved in this regulation.
The third general finding of this study is that, in the absence of compensatory EGFR signaling, or any other oncogenic signaling, miR-206 overexpression has a tumor-suppressive effect on ERα-dependent MCF-7 cells. Whereas overexpression of miR-206 decreased proliferation and increased apoptosis in MCF-7 cells after 48 h, miR-206 also altered the gene expression of these cells, as detected by a small, focused qPCR array. As discussed above, this array initially revealed the repression of GATA-3 by miR-206, which was confirmed by real-time PCR analysis. It is also interesting to note that miR-206 repressed ErbB2/HER2 expression without having an effect on ErbB1/EGFR. Thus, EGFR signaling would be expected to suppress HER2 expression, which is consistent with low expression of HER2 in Basal-like breast cancers (7). The repression of ERα and the progesterone receptor (PGR; a target gene of ERα) was expected, whereas the repression of the androgen receptor (AR) was not. Some studies have reported a correlation of AR expression with ERα and PGR positivity in tumor samples (57), although a clear role for AR expression in either ERα-positive and ERα-negative breast cancer has not emerged.
Overexpression of miR-206 also enhanced the expression of six mRNAs, presumably through the indirect repression of a transcriptional inhibitor of these genes. One of these genes, IL-6, is the founding member of the IL-6 cytokine subfamily and regulates inflammatory and immune responses (58). However, IL-6 has also been implicated in tumor progression in several cancers, and a high serum level of IL-6 confers a poor prognosis in breast cancer patients (59). Gao et al. (60) recently provided evidence that certain forms of EGFRs containing somatic activating mutations stimulate IL-6 gene expression, which, in turn, stably activates oncogenic STAT-3 activity through autocrine/paracrine signaling in lung cancer. Also, Sansone et al. (61) recently reported that autocrine/paracrine signaling by IL-6 activates oncogenic Notch/Jagged signaling in breast cancer progression. Interestingly, IL-6 was only detected in Basal-like cancers, although a functional link between EGFR and IL-6 was not examined in this study. Further work is needed to examine whether miR-206 targets a transcription factor that negatively regulates the IL-6 gene. It is worth noting that 17β-estradiol has been shown to inhibit IL-6 production (62), whereas a shift in the ERβ/ERα might increase IL-6 expression (63). Clearly, more work is needed to examine the mechanism by which miR-206 promotes IL-6 production in breast cancer.
Our previous findings also indicated that ERα and miR-206 exist within a double-negative feedback loop. Double-negative feedback loops between transcription factors and miRNA are a feature of bistable systems (20,64). An example of such a regulatory loop is the role of EGFR signaling during Drosophila eye development (65). The transcription factor, Yan, and miR-7 mutually repress each other. Before photoreceptor differentiation, Yan maintains low expression of miR-7. In response to EGFR signaling, Yan is transiently degraded, allowing cells to express miR-7 and stably repress Yan expression, contributing to photoreceptor differentiation. The photoreceptor phenotype is stabilized by the ability of another transcription factor Pointed P1, to outcompete Yan for overlapping binding sites in the miR-7 promoter and stimulate miR-7 expression. This regulatory loop has several parallels to the findings of the current study, in that EGFR stimulates the expression of miR-206, which is repressed by, but also targets, the transcription factor ERα. Further work is needed to examine the extent to which miR-206 contributes to the established ability of EGFR to cause a switch from an ERα-positive, Luminal-A phenotype to an ERα-negative, Basal-like phenotype (12,15). In this respect, it is interesting to note that miR-206 is a myogenic microRNA (66), and overexpression of miR-206 induces smooth muscle α-actin levels, while repressing nonmuscle β-actin in MCF-7 cells (our unpublished findings). Thus, miR-206 may not only suppress a luminal phenotype, but may actively promote a basal one with myoepithelial differentiation.
In summary, we have demonstrated that the double-negative feedback loop between miR-206 and ERα signaling is a coherent, comprehensive one involving the targeting of at least four mRNAs encoding functionally interactive proteins. Furthermore, we have demonstrated how the EGFR, acting through one microRNA, can shift an ERα-positive, luminal phenotype to an ERα-negative, basal phenotype in breast cancer cells. Given this, silencing miR-206 in ERα-negative, Basal-like breast cancers may provide a therapeutic approach to this aggressive type of cancer. Alternatively, enhancing miR-206 levels in ERα-positive, Luminal-A breast cancers may induce cell quiescence and death.
Materials and Methods
Cell line maintenance
All tissue culture reagents were purchased from Invitrogen (Carlsbad, CA) and Sigma (St. Louis, MO). MDA-MB-231 and MCF-7 cell lines were obtained through American Type Culture Collection (Manassas, VA). Cell lines were maintained in DMEM/F12 supplemented with 10% fetal bovine serum and 1% Pen/Strep for standard culturing or without antibiotics (transfection media). Cells cultured in HDM were grown in phenol red-free DMEM/F12 media with 10% charcoal-treated dextran-stripped fetal bovine serum from Hyclone Laboratories (Logan, UT).
Cell treatments and transfections
MCF-7 cells were transfected with either 0–20 μg PS-206, a miR-206 expression construct, or with 20 μg PS-empty, for various periods of time using Lipofectamine 2000 from Invitrogen, and assessed for subsequent changes in mRNA levels, protein expression, ERE-luciferase activity, and other functional assays described below. For siRNA experiments, MDA-MB-231 cells were transfected with 0–50 nm siEGFR (smart pool of four siRNAs directed toward EGFR) from Dharmacon (Lafayette, CO) for 24 h, and subsequent protein and RNA lysates were analyzed for EGFR and miR-206 expression by Western blot and microRNA real-time PCR assays, respectively.
MCF-7 cells were also treated with various hormones and selective inhibitors, purchased from Tocris Bioscience (Ellisville, MO). Cells at 70% confluence were washed and switched to HDM every 24 h. Cells were treated 48 h later with either 1–1.5 nm E2, 10 nm propyl pyrazole triol (PPT), 10 nm diarylpropionitrile, 1 nm progesterone, and/or 50–200 ng/ml EGF (Sigma-Aldrich) for 24–48 h. Resultant changes in mRNA/microRNA levels, protein expression, and luciferase activity were then measured. In instances where transfection of synthetic RNAs or various DNA constructs were required, the 24-h transfection occurred before hormone/growth factor treatments. For selective inhibitor experiments, MDA-MB-231 cells were treated with either 0–10 μm PD153035 or 0–10 μm U0126 for 48 h, and RNA lysates were analyzed for microRNA levels by real-time PCR.
Quantitative real-time PCR detection assay
For mRNA and microRNA detection, real-time PCR assays were performed on 100 ng/μl total RNA isolated from cell lines under various treatment conditions using Trizol reagent from Invitrogen as previously described (33). A list of the specific human primers generated is reported in supplemental Table S1. Analysis of the data by ΔΔCt was done using GraphPad Prism 4.0 and Microsoft Excel. All experiments were performed in triplicate, normalized to either RPL-19 or 5s RNA, and values were reported as relative levels compared with the mock or untreated controls.
Western blot analysis
Western blot analyses were performed as previously described (33). Antibodies to ERα (1:1000) and SRC-1 (1:1000) from Millipore Corp. (Billerica, MA), SRC-3 (1:1000), and cleaved-PARP (1:500) from BD Biosciences (San Jose, CA), BCL-2 (1:250) from Cell Signaling Technology (Danvers, MA), β-catenin (1:1000) from Transduction Laboratories (Lexington, KY), β-actin (1:1000) from Abcam, Inc. (Cambridge, MA), EGFR (1:1000) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and cyclin D1 (1:1000) from Thermo Scientific (Fremont, CA) were used in conjunction with either an antimouse, antirat, or antirabbit IgG horseradish peroxidase-conjugated secondary antibody, from Santa Cruz Biotechnology. Expression was detected via ECL Plus from GE Healthcare Life Sciences (Piscataway, NJ) and chemiluminescent film from Eastman Kodak (Rochester, NY). Band intensities were quantified using the NIH Image J software.
Luciferase reporter assays
To determine whether miR-206 activity in both MCF-7 and MDA-MB-231 cell lines was altered under various treatment conditions, firefly luciferase assays using 500 ng of the various pIS-ERα-1 constructs were performed (33). For these assays the values generated from each construct were normalized to 200 ng of Renilla luciferase (pRL-TK) and reported as mean luciferase activity ± the sem, relative to either the untreated or mock-transfected conditions.
To determine whether miR-206 targeted SRC-1, SRC-3, and GATA-3, pIS-0 luciferase constructs were modified to contained 60-nt regions of the respective endogenous sequence that harbored the putative miR-206 target site as described previously (33). Sequences used to generate these constructs are depicted in supplemental Table S2. Experiments using these luciferase constructs were performed as mentioned above, except that values generated from each construct were set relative to the 5′-mutant control under the mock transfection condition. For these assays, MCF-7 cells were additionally transfected with 0–200 nm pre-let-7d, pre-miR-206, or pre-miR-neg synthetic RNAs from Ambion, Inc. (Austin, TX), or, 0–50 nm antagomiR-206, antagomiR-let-7d, or antagomiR-neg from Dharmacon, Inc. (Lafayette, CO).
Luciferase reporter assays were also performed to determine estrogen responsiveness in the MCF-7 transfectants via an ERE-Luc reporter construct (46). After hormone depletion for 24 h, cells were treated with 1.5 nm E2 and/or various amounts of EGF and transfected with 1.0 μg of ERE-Luc and 200 ng of pRL-TK for an additional 24 h. In some experiments, 0–20 μg PS-206 was transfected into the cells after hormone depletion and addition of 1.5 nm E2. Luciferase activity was measured and reported as indicated above, and the values depicted were set relative to the hormone-depleted condition. All luciferase assays were performed in triplicate and are from three independent experiments.
ERα-open reading frame (ORF) transfection experiments
A myc-DDK-tagged ESR1 TrueORF cDNA (RefSeq: NM_000125.2) clone (pCMV6-ERα-ORF) was purchased from Origene (Rockville, MD). MCF-7 cells cultured in six-well plates were washed once in 1× PBS, given HDM for 8 h, washed again in 1× PBS, and cultured in HDM for an additional 16 h. Cells were then transfected with 500 ng of pCMV6-ERα-ORF, and/or 10 μg PS-206, and/or 1.5 nm E2. When performing ERE-luciferase activity assays, all conditions were additionally cotransfected with 1.0 μg of ERE-Luc and 200 ng of pRL-TK. RNA and protein lysates were subsequently isolated from each condition 24 h after transfection and analyzed for ERα expression, ERE-luciferase activity, as well as PGR and CDC-6 mRNA levels as described above.
ER signaling qPCR array
MCF-7 cells were cultured in two 100-mm dishes until approximately 70% confluency, mock transfected or given 10 μg PS-206 for 24 h, and cultured with fresh transfection media for an additional 24 h. Total RNA was isolated from each condition via Trizol from Invitrogen (Carlsbad, CA) and converted to cDNA using the RT2 First Strand Kit and loaded onto the Breast Cancer and Estrogen Receptor Signaling RT2 Profiler PCR Array as per manufacturer’s instructions (both products from SuperArray Biosciences (Frederick, MD). The qPCR array was performed three times on the isolated RNA described above, and values are depicted as the mean relative fold change in expression over the mock-treated condition. The resultant gene pool was then categorized using GOTERMS via the DAVID/EASE database (48). The genes within each category were ranked based on the fold change in expression compared with the mock condition, where ≥2.5:1 is the highest and ≤ 1:2.5 is the lowest change.
Trypan blue proliferation assays
The Trypan blue viability assays were performed on MCF-7 cells over the course of 144 h after initial seeding. After seeding 1 × 105 cells in each well of a six-well culture plate, the cells were transfected at the 48-h time point with either 15 μg PS-206 or PS-empty, or mock treated. The number of viable cells were determined every 24 h by trypsinization of cells, resuspension in growth medium, 1:4 dilution with 0.4% Trypan blue stain, and hemocytometer counts. Values generated are representative of three independent experiments with four replicates per experiment and are reported as the average viable number of cells ± sem.
BrdU incorporation assays
MCF-7 cells were cultured in transfection media in six-well plates until 70% confluency was obtained. Twenty four hours after mock, PS-empty, or PS-206 transfection, cells were washed and incubated in transfection media containing 10 μm BrdU. After an additional 24 h MCF-7 cells were washed with PBS, fixed in 3.7% formaldehyde, incubated with 2 n HCl for 1 h, rinsed with 0.1m borate followed by PBS, and then incubated with a monoclonal BrdU antibody from Sigma-Aldrich (St. Louis, MO) for 1 h. Cells were then rinsed with PBS, stained with an antimouse horseradish peroxidase/DAB UltraVision Detection system from ThermoScientific (Fremont, CA), and counterstained with hematoxylin. The resultant BrdU-positive cells were counted under ×20 magnification on a Zeiss IM-35 Inverted Microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY), and values were normalized to the total number of hematoxylin-positive cells within a given field. Five random counts were performed within each well by two independent observers, where each condition was performed in triplicate per experiment. The values reported are depicted as the mean percentage of stained cells relative to the mock-transfected control from three independent experiments ± sem.
In situ TUNEL stain
MCF-7 cells were cultured in transfection media in six-well plates until 70% confluency was obtained. Forty eight hours after mock, PS-empty, or PS-206 transfection, cells were washed and prepared for TUNEL. The TUNEL stain was performed using the TACS™ TdT In Situ Apoptosis Detection Kit, from R&D Systems, Inc. (Minneapolis, MN), according to manufacturer’s instructions. The resultant TUNEL-positive cells were counted under ×20 magnification on a Zeiss IM-35 Inverted Microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY), and values were normalized to the total number of cells within a given field, as determined by Nuclear Fast Red counterstaining. Four random counts were performed within each well by two independent observers, where each condition was performed in triplicate. The values reported are depicted as the fold increase in TUNEL+ cells relative to the mock-transfected control from three independent experiments ± sem.
Statistical analysis
Unless noted otherwise, values reported in all analyses were expressed as the mean ± sem, whereas densitometric readings of Western blot analyses were reported as averages only. Changes between treatment and control groups were analyzed by ANOVA with a Bonferroni multiple comparison or Dunnett posttest using Graphpad Instat 3.0, and statistical significance was accepted at P < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Carolyn Smith (Baylor College of Medicine) for providing us with the ERE-Luciferase reporter construct and Dr. Louise McCullough (University of Connecticut Health Center, Farmington, CT) for providing the necessary reagents including BCL-2 and cleaved-PARP antibodies. We also thank Dr. H. Furneaux, Department of Molecular, Microbial, and Structural Biology, Drs. T. Hla, L. Shapiro, and K. Claffey, Department of Cell Biology, and B. Graveley, Translational Genomics Core (University of Connecticut Health Center) for providing equipment, reagents, technical advice, and data analysis for this study. A special thanks to Ms. Emina Begic for performing the numerous blinded cell counts required for the PS-206 functional studies reported here.
Footnotes
This research was funded by Grant 1-R21-DK073456 from the National Institutes of Health; the communication and publication costs associated with this study were supported by the Carole and Ray Neag Comprehensive Cancer Center at the University of Connecticut Health Center.
Disclosure Summary: B.D.A., D.M.C., and B.A.W. have no conflicts of interest to declare.
First Published Online May 7, 2009
Abbreviations: AR, Androgen receptor; BrdU, bromodeoxyuridine; CDC-6, cell division cycle 6; CMV, cytomegalovirus; E2, 17β-estradiol; EGF, epidermal growth factor; EGFR, EGF receptor; ERα, estrogen receptor-α; ERE, estrogen response element; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDM, hormone-depleted media; miR-206, microRNA-206; NCOR1, nuclear receptor corepressor 1; nt, nucleotide; ORF, open reading frame; PARP, poly ADP ribose polymerase; PGR, progesterone receptor; PPT, propyl pyrazole triol; PS-206; psilencer pre-miR-206; qPCR, quantitative PCR; siEGFR, smart pool of four siRNAs directed toward EGFR; siRNA, small interfering RNA; SRC, steroid receptor coactivator; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling; UTR, untranslated region; WT, wild type.
References
- Culhane AC, Howlin J 2007 Molecular profiling of breast cancer: transcriptomic studies and beyond. Cell Mol Life Sci 64:3185–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, et al. 2006 The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7:96–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan PB, Millikan RC 2007 Molecular subtyping of breast cancer: opportunities for new therapeutic approaches. Cell Mol Life Sci 64:3219–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM 2006 Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol 19:264–271 [DOI] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Ellis IO 2008 Impact of basal-like breast carcinoma determination for a more specific therapy. Pathobiology 75:95–103 [DOI] [PubMed] [Google Scholar]
- Korsching E, Jeffrey SS, Meinerz W, Decker T, Boecker W, Buerger H 2008 Basal carcinoma of the breast revisited: an old entity with new interpretations. J Clin Pathol 61:553–560 [DOI] [PubMed] [Google Scholar]
- Kobayashi S 2008 Basal-like subtype of breast cancer: a review of its unique characteristics and their clinical significance. Breast Cancer 15:153–158 [DOI] [PubMed] [Google Scholar]
- LaMarca HL, Rosen JM 2008 Minireview: hormones and mammary cell fate—what will I become when I grow up? Endocrinology 149:4317–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarala M, Wicha MS 2008 Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol 26:2813–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, El-Ashry D, Wicha MS 2004 Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab 15:193–197 [DOI] [PubMed] [Google Scholar]
- Allred DC, Brown P, Medina D 2004 The origins of estrogen receptor α-positive and estrogen receptor α-negative human breast cancer. Breast Cancer Res 6:240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D 2006 Activation of mitogen-activated protein kinase in estrogen receptor α-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor α-negative human breast tumors. Cancer Res 66:3903–3911 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Jordan VC 2008 Selective estrogen modulators as an anticancer tool: mechanisms of efficiency and resistance. Adv Exp Med Biol 630:206–219 [DOI] [PubMed] [Google Scholar]
- Schiff R, Massarweh SA, Shou J, Bharwani L, Arpino G, Rimawi M, Osborne CK 2005 Advanced concepts in estrogen receptor biology and breast cancer endocrine resistance: implicated role of growth factor signaling and estrogen receptor coregulators. Cancer Chemother Pharmacol 56(Suppl 1):10–20 [DOI] [PubMed] [Google Scholar]
- Oh AS, Lorant LA, Holloway JN, Miller DL, Kern FG, El-Ashry D 2001 Hyperactivation of MAPK induces loss of ERα expression in breast cancer cells. Mol Endocrinol 15:1344–1359 [DOI] [PubMed] [Google Scholar]
- El-Ashry D, Miller DL, Kharbanda S, Lippman ME, Kern FG 1997 Constitutive Raf-1 kinase activity in breast cancer cells induces both estrogen-independent growth and apoptosis. Oncogene 15:423–435 [DOI] [PubMed] [Google Scholar]
- Holloway JN, Murthy S, El-Ashry D 2004 A cytoplasmic substrate of mitogen-activated protein kinase is responsible for estrogen receptor-α down-regulation in breast cancer cells: the role of nuclear factor-κB. Mol Endocrinol 18:1396–1410 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2004 MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM 2007 microRNA functions. Annu Rev Cell Dev Biol 23:175–205 [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC 2008 Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 9:831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM 2005 MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070 [DOI] [PubMed] [Google Scholar]
- Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H 2008 miR-206 Expression is down-regulated in estrogen receptor α-positive human breast cancer. Cancer Res 68:5004–5008 [DOI] [PubMed] [Google Scholar]
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavaré S, Caldas C, Miska EA 2007 MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 8:R214–R229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C 2006 Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 5:24–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN 2007 Altered microRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 67:11612–11620 [DOI] [PubMed] [Google Scholar]
- Iorio MV, Casalini P, Tagliabue E, Ménard S, Croce CM 2008 MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur J Cancer 44:2753–2759 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM 2006 MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 66:7390–7394 [DOI] [PubMed] [Google Scholar]
- Barbarotto E, Schmittgen TD, Calin GA 2008 MicroRNAs and cancer: profile, profile, profile. Int J Cancer 122:969–977 [DOI] [PubMed] [Google Scholar]
- Adams BD, Guttilla IK, White BA 2008 Involvement of microRNA in breast cancer. Semin Reprod Med 26:522–536 [DOI] [PubMed] [Google Scholar]
- Sassen S, Miska EA, Caldas C 2008 MicroRNA: implications for cancer. Virchows Arch 452:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Weinberg RA 2008 Micromanagers of malignancy: role of microRNA in regulating metastasis. Trends Genet 24:448–456 [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA 2007 microRNA as oncogenes and tumor suppressors. Dev Biol 302:1–12 [DOI] [PubMed] [Google Scholar]
- Adams BD, Furneaux H, White BA 2007 The micro-ribonucleic acid (microRNA) miR-206 targets the human estrogen receptor-α (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol 21:1132–1147 [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ 2008 A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev 22:2535–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BJ, Giguère V 2008 Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor α pathway. Mol Cancer 7:49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voduc D, Cheang M, Nielsen T 2008 GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol Biomarkers Prev 17:365–373 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE 2007 Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9:201–209 [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z 2006 GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127:1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li Q 2003 Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol 17:1681–1692 [DOI] [PubMed] [Google Scholar]
- Smith CL, O'Malley BW 2004 Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev 25:45–71 [DOI] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW 2007 Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell 27:691–700 [DOI] [PubMed] [Google Scholar]
- Duong V, Augereau P, Badia E, Jalaguier S, Cavailles V 2008 Regulation of hormone signaling by nuclear receptor interacting proteins. Adv Exp Med Biol 617:121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf IM, Heitzer MD, Grubisha M, DeFranco DB 2008 Coactivators and nuclear receptor transactivation. J Cell Biochem 104:1580–1586 [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Kim JW, Bechis SK, Werb Z 2008 GATA-3 and the regulation of the mammary luminal cell fate. Curr Opin Cell Biol 20:164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M 2007 Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res 67:6477–6483 [DOI] [PubMed] [Google Scholar]
- Nawaz Z, Lonard DM, Smith CL, Lev-Lehman E, Tsai SY, Tsai MJ, O'Malley BW 1999 The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol 19:1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004 Human microRNA targets. Plos Biol 2:363–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R 2003 DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:R60–R70 [PubMed] [Google Scholar]
- Peppercorn J, Perou CM, Carey LA 2008 Molecular subtypes in breast cancer evaluation and management: divide and conquer. Cancer Invest 26:1–10 [DOI] [PubMed] [Google Scholar]
- Yan J, Tsai SY, Tsai MJ 2006 SRC-3/AIB1: transcriptional coactivator in oncogenesis. Acta Pharmacol Sin 27:387–394 [DOI] [PubMed] [Google Scholar]
- Hossain A, Kuo MT, Saunders GF 2006 Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol 26:8191–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viger RS, Guittot SM, Anttonen M, Wilson DB, Heikinheimo M 2008 Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol Endocrinol 22:781–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess AW 2008 EGFR family: structure physiology signalling and therapeutic targets. Growth Factors 26:263–274 [DOI] [PubMed] [Google Scholar]
- Yang X, Phillips DL, Ferguson AT, Nelson WG, Herman JG, Davidson NE 2001 Synergistic activation of functional estrogen receptor (ER)-α by DNA methyltransferase and histone deacetylase inhibition in human ER-α-negative breast cancer cells. Cancer Res 61:7025–7029 [PubMed] [Google Scholar]
- Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ 2009 Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284:5731–5741 [DOI] [PubMed] [Google Scholar]
- Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA, Varella-Garcia M, Bunn Jr PA, Haney J, Helfrich BA, Kato H, Hirsch FR, Franklin WA 2008 EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol 19:1053–1059 [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Hai E, Matsumoto K, Ikeda K, Tokunaga S, Nagahara H, Sakurai K, Inoue T, Nishiguchi Y 2008 Androgen receptor expression in breast cancer: relationship with clinicopathological factors and biomarkers. Int J Clin Oncol 13:431–435 [DOI] [PubMed] [Google Scholar]
- Kishimoto T 2005 Interleukin-6: from basic science to medicine–40 years in immunology. Annu Rev Immunol 23:1–21 [DOI] [PubMed] [Google Scholar]
- Bachelot T, Ray-Coquard I, Menetrier-Caux C, Rastkha M, Duc A, Blay JY 2003 Prognostic value of serum levels of interleukin 6 and of serum and plasma levels of vascular endothelial growth factor in hormone-refractory metastatic breast cancer patients. Br J Cancer 88:1721–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, Bromberg JF 2007 Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest 117:3846–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, Chieco P, Bonafè M 2007 IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest 117:3988–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J, Mak YT, Papaioannou S, Evans BA, Fogelman I, Hampson G 2003 Interleukin-6 (IL-6), IL-1, receptor activator of nuclear factor κB ligand (RANKL) and osteoprotegerin production by human osteoblastic cells: comparison of the effects of 17-β oestradiol and raloxifene. J Endocrinol 177:423–433 [DOI] [PubMed] [Google Scholar]
- Ogura E, Kageyama K, Hanada K, Kasckow J, Suda T 2008 Effects of estradiol on regulation of corticotropin-releasing factor gene and interleukin-6 production via estrogen receptor type β in hypothalamic 4B cells. Peptides 29:456–464 [DOI] [PubMed] [Google Scholar]
- Hobert O 2006 Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb Symp Quant Biol 71:181–188 [DOI] [PubMed] [Google Scholar]
- Li X, Carthew RW 2005 A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123:1267–1277 [DOI] [PubMed] [Google Scholar]
- McCarthy JJ 2008 MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta 1779:682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.