Abstract
The transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator (PGC)-1α is involved in the coordinate induction of changes in gene expression in the liver that enable a homeostatic response to alterations in metabolic state, environmental cues, and nutrient availability. In exploring the specific pathways under PGC-1α regulation in the liver, we have made the surprising observation that this coactivator can induce the expression of CYP11A1 and CYP17A1, key rate-limiting enzymes involved in the initial steps of steroidogenesis. Both of these enzymes function to produce C19-steroids, converting cholesterol into pregnenolone, and then to dehydroepiandrosterone (DHEA). Estrogen-related receptor (ERR)-α mediates PGC-1α’s induction of CYP11A1 and binds within the first intron of the CYP11A1 gene. Both ERR-α and hepatocyte nuclear factor-4α are required for PGC-1α-mediated induction of CYP17A1, and specific binding sites for these receptors have been identified in the regulatory regions of this gene. The potential physiological significance of these observations was highlighted in rats where fasting induced hepatic expression of PGC-1α and CYP17A1 and was associated with an increase in hepatic levels of DHEA. These data suggest that DHEA could be playing a role as an intracellular signaling molecule involved in modulating hepatic activity in response to fasting conditions.
PGC-1α regulates hepatic synthesis of DHEA by inducing expression of CYP11A1 and CYP17A1 through HNF4α and ERRα. This study highlights hepatic DHEA production as a novel fasting response.
Homeostatic control of nutrient levels in the body is a primary function of the liver, which modulates circulating levels of sugars, lipids, and proteins in response to hormones and other internal and external cues. Fasting causes a metabolic switch in liver that includes activation of glycogenolysis and gluconeogenesis and increases in fatty acid oxidation to accommodate increased mobilization of free fatty acids from adipose stores. In the fed state, these processes are usually repressed; however, in pathological conditions such as type 2 diabetes, elements of this control are lost, and hepatic glucose production persists despite elevated glucose levels (1). Sustained imbalances in glucose levels as well as distinct metabolic defects that influence lipid handling can cause fatty acids and triglycerides to accumulate in the liver, leading to hepatic steatosis (2).
The transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator (PGC)-1α is a key integrator of many of the signaling pathways that are induced in the liver and muscle upon fasting (3). Notably, glucagon and glucocorticoids induce the expression of PGC-1α in the liver through cAMP response element-binding protein (4), and the expressed coactivator serves to integrate signals from afferent signaling pathways that influence its ability to regulate transcription. Insulin suppresses expression of gluconeogenic enzymes partly through AKT/protein kinase B-mediated phosphorylation and inactivation of PGC-1α (5). In addition, acetylation of PGC-1α by the acetyltransferase, GCN5 reduces its activity (6), whereas the sirtuin, SirT1, an NAD+ sensor, deacetylates and increases its activity (7).
When expressed and active, PGC-1α acts as a potent transcriptional activator and can interact with a number of transcription factors to provide metabolic control at the transcriptional level. It interacts with the orphan nuclear receptor hepatocyte nuclear factor (HNF)-4α to induce expression of glucose-6-phosphatase and phosphoenolpyruvate carboxykinase, key enzymes involved in gluconeogenesis (3,8). It also interacts with HNF4α to increase hepatic synthesis of apolipoproteins A-IV, CII, and CIII (9). Estrogen-related receptor (ERR)-α is another key partner for PGC-1α, providing direct and indirect control of numerous genes involved in oxidative phosphorylation (10,11), fatty acid oxidation (12), and the tricarboxylic acid cycle (13). Finally, PGC-1α can also interact with and coactivate several other nuclear receptors, most notably the peroxisome proliferator-activated receptors, as well as other transcription factors including nuclear respiratory factor 1 and 2 (NRF-1 and -2), myocyte enhancer factor 2C (MEF2C), and for khead box O1 (FoxO1) (reviewed in Ref. 14).
We have previously performed an extensive analysis of the changes in gene expression that occur in response to PGC-1α expression in hepatic cells (13) and discovered that the mRNAs corresponding to several of the rate-limiting enzymes in steroidogenesis are dramatically and unexpectedly induced. Under normal circumstances, steroidogenesis takes place in the adrenals and gonads, where cytochrome p450 hydroxylases and steroid dehydrogenases modify cholesterol to form the different steroid hormones. The initial enzymatic step is catalyzed by cytochrome p450 cholesterol side-chain cleavage enzyme, cytochrome p450 (CYP)-11A1, which oxidizes cholesterol at the 20 and 22 carbon positions and cleaves the side chain between these carbons to produce pregnenolone. The enzyme CYP17A1 catalyzes the next steps, converting pregnenolone to dehydroepiandrosterone (DHEA) through its 17α-hydroxylase and 17,20-lyase activities. Additional enzymes can then modify DHEA to form sex steroids or pregnenolone and 17α-OH pregnenolone to generate glucocorticoids and mineralocorticoids (reviewed in Refs. 15 and 16).
Although the adrenal and gonads secrete the majority of hormones in circulation, a number of tissues are capable of locally synthesizing steroids. In postmenopausal women, for instance, the concentration of estrogen in breast tumors has been reported to be significantly higher than circulating plasma levels (17). This elevated concentration is thought to be due to local conversion of androgens to estrogens by the ectopically expressed aromatase enzyme (CYP19) in tumor stromal cells (reviewed in Ref. 18). In the brain, most steroids (neurosteroids) are thought to be synthesized locally, with both neurons and glia expressing many of the enzymes and transporters necessary for steroidogenesis from cholesterol (reviewed in Refs. 19 and 20). Finally, and of direct relevance to the studies described below, it has been determined that human fetal liver expresses most of the steroidogenic enzymes but at levels considerably lower than those found in primary steroidogenic organs (21). Fetal rat liver expresses CYP17A1 mRNA, and at birth, expression in liver is comparable to that in testis. These levels rise and peak after 8 d and then steadily decline to undetectable levels after puberty (22). Additionally, expression analysis of the genes induced in the liver of fasted mice found that CYP17A1 was robustly induced by a 24- and 48-h fast (23). However, it has not been determined whether or not these enzymes are expressed and functional in human liver or how their expression fits within the metabolic functions of the liver.
In this study, we report that CYP11A1 and CYP17A1, rate-limiting enzymes in steroidogenesis, are functionally induced in human hepatic cells by PGC-1α. Although these genes have previously been shown to be regulated by cAMP and the orphan nuclear receptor steroidogenic factor 1 (SF-1) (reviewed in Ref. 24), we show in this study that ERRα and HNF4α are novel regulators of these genes, mediating their induction in hepatic cells by PGC-1α. Interestingly, steroidogenesis in PGC-1α-expressing hepatic cells does not progress beyond DHEA to classical sex steroids, suggesting that this steroid or an unidentified metabolite could be involved in a novel hepatic signaling pathway in response to fasting conditions.
Results
Induction of CYP11A1 and CYP17A1 expression in hepatic cells is regulated by PGC-1α
The transcription coactivator PGC-1α has been shown to interact with and modulate the transcriptional activity of several nuclear receptors and unrelated transcription factors. This has made it difficult to link specific aspects of PGC-1α biology to its activity on an individual transcription factor. To circumvent this specificity problem, we previously developed a modified PGC-1α construct, PGC-1α 2x9, which interacts selectively with ERRα and HNF4α. Expression of this modified coactivator in HepG2 cells enabled us to use microarray-based approaches to define the target genes that are regulated by the PGC-1α-(ERRα/HNF4α) axis (13). A gene ontology analysis of the target genes regulated by both PGC-1α and PGC-1α 2x9 led to the identification of a number of genes involved in steroid metabolism, including CYP11A1 and CYP17A1.
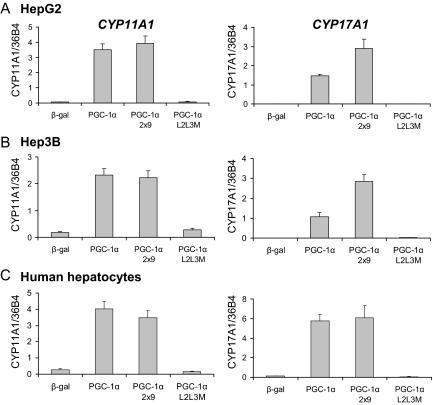
To confirm that CYP11A1 and CYP17A1 are regulated by PGC-1α, HepG2 cells were treated with adenoviruses expressing β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M, a mutant form of PGC-1α in which the nuclear receptor-interacting domain has been disrupted (11,13). Expression of the mRNAs encoding these enzymes was analyzed using quantitative PCR (qPCR), revealing that both CYP11A1 and CYP17A1 are robustly induced by either PGC-1α or PGC-1α 2x9 but not β-galactosidase (β-gal) or PGC-1α L2L3M (Fig. 1A). A similar response was also observed in other human hepatic cells, including Hep3B cells (Fig. 1B), and in primary human hepatocytes (Fig. 1C). Although CYP17A1 was induced in primary rat hepatocytes (supplemental Fig. 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend. endojournals.org), CYP11A1 was not found to be expressed in rat livers in any of the conditions we tested (data not shown). We completed this survey of PGC-1α responsiveness by analyzing CYP11A1 and CYP17A1 expression in a series of cells that were not of hepatic origin. In this manner, we observed that PGC-1α did not induce the expression of either gene in H295R adrenal cells (supplemental Fig. 1) or MCF7 breast cancer cells (data not shown). However, PGC-1α did induce expression of CYP11A1 in U251 cells and CYP17A1 in AGS stomach carcinoma cells (supplemental Fig. 1). These data suggest that PGC-1α induces elements of the steroidogenic pathway in a limited set of cell types.
Figure 1.
PGC-1α and PGC-1α 2x9 induce gene expression of CYP11A1 and CYP17A1 in hepatic cells. Gene expression of CYP11A1 and CYP17A1 was measured by quantitative PCR in HepG2 cells (A), Hep3B cells (B), or primary human hepatocytes (C) infected with adenoviruses expressing β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M. Gene expression was normalized to 36B4 expression. Error bars represent sem of three replicates, and each graph is representative of at least three independent experiments.
PGC-1α increases the functional activity of CYP11A1 and CYP17A1
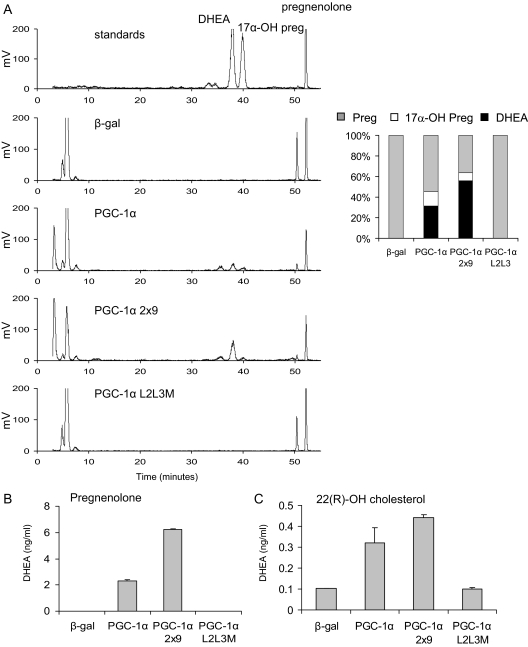
Both CYP11A1 and CYP17A1 require additional proteins and redox partners for their enzymatic activity. Specifically, CYP11A1 requires ferridoxin reductase (FDXR) and ferridoxin, whereas the p450 oxidoreductase (POR) and cytochrome b5 are obligate partners of CYP17A1 (reviewed in Ref. 25). Interestingly, we found that p450 oxidoreductase is induced by 3- to 3.5-fold in the PGC-1α-expressing cells (P < 0.001) in the original array (13). Levels of ferridoxin reductase were unaffected by PGC-1α expression. Therefore, we considered whether the observed increases in the mRNA expression of these enzymes results in increased functional activity when PGC-1α is expressed. To this end, [3H]pregnenolone was incubated with HepG2 cells for 6 h, expressing β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M. The steroids in the cells at the end of the incubation period were extracted and separated using HPLC. Cells expressing either PGC-1α or PGC-1α 2x9 displayed increased synthesis of a product that comigrates with standards for 17α-OH pregnenolone and DHEA (Fig. 2A). Notably absent on the chromatograph were peaks corresponding to other sex steroids including androstenedione, which migrates on this column at 23.2 min, estrogen at 27.3 min, testosterone at 31.3 min, or progesterone at 50.2 min (data not shown). This observation suggests that the steroidogenic pathway induced by PGC-1α does not appear to progress beyond DHEA. Additionally, these data reflect our inability to detect significant mRNA expression of 3β-hydroxysteroid dehydrogenases 1 and 2, CYP21, and CYP11B1 in HepG2 cells, the encoded proteins of which are the enzymes required to convert DHEA to either sex steroids or glucocorticoids (supplemental Fig. 2). Some additional differences between the chromatograms derived from the PGC-1α-expressing cells and the control cells were apparent, particularly in the rapidly migrating polar products that are likely to be metabolites of DHEA. The percent conversion of pregnenolone to DHEA and 17α-OH pregnenolone was calculated by determining the area under the curves in each sample. As illustrated in the bar graph (Fig. 2A), 14 and 8% of the pregnenolone was converted to 17α-OH pregnenolone in PGC-1α- and PGC-1α 2x9-expressing cells, respectively, and 31 and 56% of the pregnenolone was converted to DHEA.
Figure 2.
PGC-1α and PGC-1α 2x9 increase the functional activity of CYP11A1 and CYP17A1. A, HepG2 cells were infected with adenoviruses expressing β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M and then were incubated with 200 nm [3H]pregnenolone (Preg) for 6 h. Steroids were extracted and separated by HPLC. Bar graph represents area under the curve. B and C, HepG2 cells were infected as above and incubated with 200 nm pregnenolone (B) or 10 μm 22(R)-OH cholesterol (C), and DHEA was measured using a RIA. Error bars represent sem of three replicates, and each RIA is representative of three independent experiments.
Additional evidence that the product identified was DHEA was obtained by a RIA to specifically measure this steroid in the media derived from HepG2 cells incubated with pregnenolone for 6 h. This analysis confirmed that DHEA is synthesized in HepG2 cells expressing either PGC-1α or PGC-1α 2x9 (Fig. 2B). Finally, because we observed that CYP11A1 was also expressed in HepG2 cells, we examined whether PGC-1α expression would permit DHEA synthesis de novo from a CYP11A1 substrate. To test this idea, the cell-permeable cholesterol metabolite 22(R)-OH cholesterol was used as a substrate, and DHEA synthesis was measured by RIA. HepG2 cells expressing either PGC-1α or PGC-1α 2x9 showed increased synthesis of DHEA (Fig. 2C), indicating that CYP11A1 was also functionally active after the expression of PGC-1α in these cells.
PGC-1α-dependent induction of CYP11A1 and CYP17A1 requires ERRα
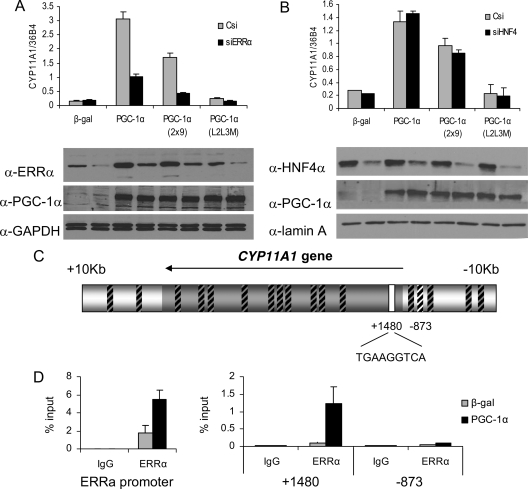
We have determined that CYP11A1 is induced by both PGC-1α and PGC-1α 2x9, but not a mutant PGC-1α L2L3M that is incapable of interacting with nuclear receptors. This result implicates ERRα and/or HNF4α, in the regulation of this gene. Thus, we next examined the impact of small interfering RNA (siRNA)-mediated knockdown of each of these receptors on CYP11A1 expression in HepG2 cells. The results of this analysis, shown in Fig. 3A, indicate that reducing the expression of ERRα substantially abrogated the induction of CYP11A1 by PGC-1α and PGC-1α 2x9.
Figure 3.
ERRα mediates PGC-1α induction of CYP11A1. A and B, HepG2 cells were transduced with lentiviruses expressing siRNA to either a nonspecific control si (Csi), or to ERRα (A) or HNF4α (B), followed by adenoviral delivery of β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M. Gene expression of CYP11A1 was measured by qPCR and normalized to 36B4 expression, and relevant protein expression is shown by Western blot. C, Schematic of the putative ERRE sites around the CYP11A1 gene tested by ChIP. Regions not bound by ERRα are indicated by boxes with black hash marks. The white box indicates the site identified by ChIP scanning in D. Numbering is relative to the start site. D, ChIP of ERRα and amplification of putative ERREs by qPCR, where the ERRα promoter was used as a positive control.
To examine regulation of CYP11A1 by HNF4α, we designed an siRNA to knock down HNF4α expression. This construct was able to substantially reduce protein expression of HNF4α (Fig. 3B) as well as reduce expression of known HNF4α target genes (supplemental Fig. 3). However, reduced expression of HNF4α had little or no effect (Fig. 3B) on the expression of CYP11A1.
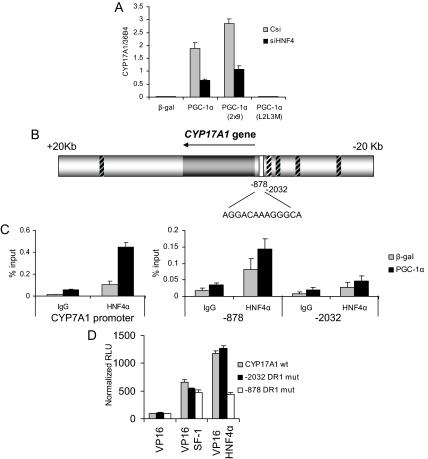
Using the consensus site and matrix previously described for ERRα (26), the sequence surrounding the CYP11A1 gene was scanned for putative ERRα response elements (ERREs) using Transcription Element Search Software (TESS). This analysis identified 18 putative sites within the gene and the 10-kb surrounding region (Fig. 3C). Chromatin immunoprecipitation (ChIP) was used to determine whether an ERRα-binding site could be identified in the region surrounding CYP11A1. ERRα has previously been described to be subject to autoregulation by a defined binding site in its own promoter (27), and this site was used as a positive control for ERRα binding. PGC-1α expression further increases ERRα binding to its own promoter, as also described. We tested ERRα binding to each of the putative response elements around the CYP11A1 gene by designing primers to amplify each region and measuring the response by qPCR. ERRα was found to associate most strongly with a site in the first intron of CYP11A1 (Fig. 3D and supplemental Fig. 4). PGC-1α is required for ERRα to bind to this site, because almost no binding is observed in cells expressing β-gal. ERRα exhibited very minimal binding to all other putative sites, including a site at −873 bp (Fig. 3D), and three additional sites that are perfect matches to the ERRα consensus (supplemental Fig. 4). We conclude from these studies both that ERRα is required for PGC-1α-mediated induction of CYP11A1 expression and that this likely results from its interaction with a specific ERRE within the gene.
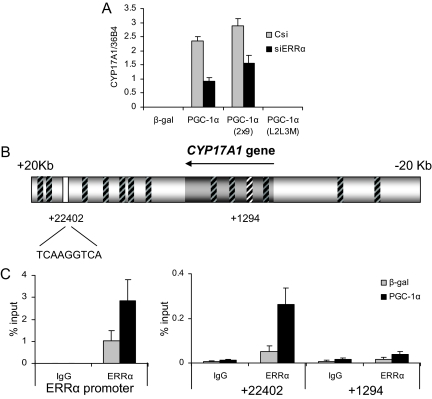
We next evaluated the role of ERRα in PGC-1α-mediated induction of CYP17A1 gene expression. Again using the siRNA-mediated knockdown approach, we observed reduced expression of CYP17A1 by PGC-1α and PGC-1α 2x9 in cells in which ERRα expression was reduced (Fig. 4A). Interestingly, however, we routinely found that even with substantial knockdown of ERRα, we achieved only approximately a 50% decrease in CYP17A1 expression, a result that implicated either HNF4α or a non-nuclear receptor target of PGC-1α in the regulation of this gene (see below). Scanning the region 20 kb around the CYP17A1 gene with the same method that we used for CYP11A1, 14 putative ERRE sites were identified (Fig. 4B). ERRα association with each of these putative ERREs was tested using ChIP (supplemental Fig. 5). ERRα was found to be associated most strongly with an element 22 kb downstream of the transcriptional start of CYP17A1 (Fig. 4C), and as in the case of the ERRE in CYP11A, PGC-1α expression was required for this interaction, because no binding was observed in cells expressing β-gal alone. This site is contained within the gene region of the hypothetical protein C10orf26; however, expression of PGC-1α or siERR has no effect on the expression of this transcript (data not shown). Notably, ERRα does not bind to any significant degree to any of the other sites identified by this method, including the site 1294 bp downstream from the transcription start site (Fig. 4C). Thus, we have identified a potential site through which the ERRα/PGC-1α complex could regulate expression of CYP17A1.
Figure 4.
CYP17A1 is regulated by PGC-1α through ERRα. A, HepG2 cells were transduced with lentiviruses expressing either a nonspecific control si (Csi), or siERRα, followed by adenoviral transduction of β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M. Gene expression of CYP17A1 was measured by qPCR and normalized to 36B4 expression. B, Schematic of the putative ERRE sites found in and around the CYP17A1 gene region. Regions not bound by ERRα are indicated by boxes with black hash marks. White boxes indicate the position of sites identified by ChIP scanning in C. Numbering is relative to the start site. C, ChIP of ERRα and amplification of putative ERREs by qPCR, where the ERRα promoter serves as a positive control.
HNF4α is required for maximal induction of CYP17A1 by PGC-1α
Given the incomplete inhibition of PGC-1α-mediated CYP17A1 expression in ERRα knockdown cells, we next queried the role of HNF4α in the regulation of this enzyme. The results of these analyses revealed that knockdown of HNF4α protein (Fig. 3B) was able to significantly impair PGC-1α-dependent induction of CYP17A1 (Fig. 5A) (∼50%), indicating that HNF4α is also involved in regulating the expression of CYP17A1. Because HNF4α binds a direct repeat with 1-bp spacing (DR-1 element) (28), nuclear hormone receptor (NHR) scan (29) was used to identify DR-1 elements in the genome within CYP17A1 or in its surrounding 20-kb region. This scan identified six putative sites (Fig. 5B), and we tested the association of HNF4α with each site using ChIP. Previously, CYP7A1 has been shown to be a direct transcriptional target of HNF4α (30) and therefore was used as a positive control for the ChIP analysis in this study. HNF4α was found most strongly associated with the DR-1 site most proximal to the transcriptional start of CYP17A1 (Fig. 5C) and may additionally bind the site at −6837 (supplemental Fig. 6). At both sites, PGC-1α expression appears to increase HNF4α binding.
Figure 5.
CYP17A1 is regulated by PGC-1α through HNF4α. A, HepG2 cells were transduced with lentiviruses expressing either a nonspecific control si (Csi), or siHNF4α, followed by adenoviral transduction of β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M. Gene expression of CYP17A1 was measured by qPCR and normalized to 36B4 expression. B, Schematic of the DR-1 sites found in and around the CYP17A1 gene region. Regions not bound by HNF4α are indicated by boxes with black hash marks. White box indicates the position of site identified by ChIP scanning shown in C. C, ChIP of HNF4α and amplification of regions by qPCR, where HNF4α binding to the CYP7A1 promoter served as a positive control. D, Activity of a reporter gene fused to 3.2-kb promoter region upstream of CYP17A1 tested in combination with HNF4α fused to a VP16 activation domain. VP16-SF-1 is a positive control. Each DR-1 site was mutated to examine binding activity. mut, Mutant; wt, wild type.
The functionality of the −878 site was next examined. To this end, we used a reporter construct containing approximately 3.2 kb of the promoter region immediately upstream of the start of the CYP17A1 gene (31). Two DR-1 elements are within this region of the promoter, and both half-sites of each element were mutated to examine binding. A VP16 activation domain was fused to HNF4α and SF-1, a well characterized regulator of CYP17A1. Although mutation of either DR-1 site did not affect activation of the reporter by VP16-SF-1, mutation of the most proximal DR-1 site at −878 bp significantly reduced the activity of VP16-HNF4α (Fig. 5D). Mutation of the more distal DR-1 site at −2032 bp had no effect on the reporter activity, indicating that HNF4α binds the proximal DR-1 site in the CYP17A1 promoter.
Fasting induces CYP17A1 and increases hepatic DHEA concentration
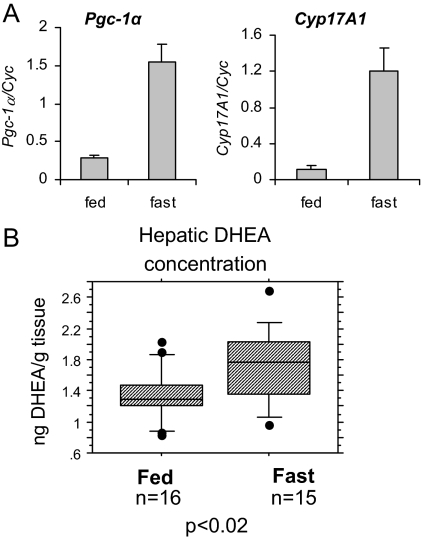
All the studies presented thus far have involved the transient ectopic expression of PGC-1α or its derivatives in cells, with a subsequent analysis of CYP11A1 and CYP17A1 expression. We felt therefore that it was necessary to recapitulate this regulation in a system where the physiological significance was more apparent. Previously, PGC-1α expression has been shown to be induced in hepatocytes during fasting (3). We confirmed that the expression of PGC-1α mRNA is significantly induced in rat livers after a 14- to 16-h fast (Fig. 6A). More importantly, however, we observed that CYP17A1 gene expression is also elevated in rat liver under the same conditions (Fig. 6A). These data highlight the physiological relevance of the regulation studies that we previously performed in cultured cells.
Figure 6.
Fasting induces Cyp17A1 expression and function. A and B, Rats were fasted for 14–16 h. RNA was collected for gene expression analysis by qPCR, gene expression was normalized to cyclophilin (A), and liver samples were collected for steroid extraction and analysis of DHEA concentration by RIA (B). Boxes represent the interquartile range (25–75th percentile), with median value in the center. Whiskers mark the 10th and 90th percentiles, and dots represent measurements less than the 10th percentile or more than the 90th percentile. Statistical significance was calculated by a Student’s t test.
Because DHEA is a product of CYP17A1 activity and is a predominant steroid resulting from overexpression of PGC-1α in HepG2 cells (Fig. 2A), hepatic DHEA levels were examined in fasted and fed rats. This analysis revealed a small but significant increase in DHEA in the livers of the fasted rats (Fig. 6B). Preliminary data indicate that serum levels of DHEA do not appear to be altered by fasting conditions (data not shown), suggesting that increased hepatic DHEA concentration results from local synthesis. However, application of more sensitive methods is required to fully investigate this issue, and these are being developed. Thus, as was observed in cultured cells, fasting-induced expression of CYP17A1 leads to an increase in hepatic production of DHEA.
DHEA reduces the concentration of free amino acids in hepatic cells
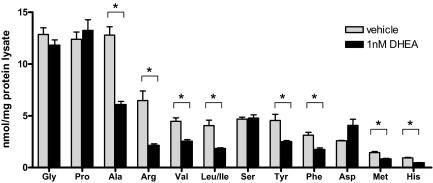
DHEA’s role in metabolism has been controversial, but several studies have shown that DHEA can reduce abdominal visceral fat and insulin levels in humans and animal models (32,33). However, for the most part, these studies have used concentrations of the steroid that we would not expect to achieve under the conditions we have studied to date. Thus, we were faced with defining a biological process in hepatic cells that could occur after the administration of DHEA at the levels we have determined to result from fasting-induced expression of steroidogenic enzymes. To this end, we asked whether low concentrations of DHEA might have an impact on any relevant metabolic markers in hepatic cells. Specifically, HepG2 cells were treated with 1 nm DHEA, and tandem mass spectrometry was used to measure various metabolic parameters of the cells, including acylcarnitines and amino acids. No changes in acylcarnitines were evident (data not shown). However, although glycine, serine, proline, and aspartate levels did not change in treated cells, we reproducibly observed that the levels of alanine, arginine, valine, leucine, isoleucine, tyrosine, phenylalanine, methionine, and histidine levels were reduced about 50% by 1 nm DHEA (Fig. 7).
Figure 7.
DHEA reduces the concentration of free amino acids in HepG2 cells. HepG2 cells were treated with 1 nm DHEA. Cell lysates were collected for analysis of free amino acid concentrations and normalized to total protein concentration. Error bars represent the se of three replicates, and the graph is representative of three independent experiments. Asterisks indicate significant difference (P < 0.05) by independent Student’s t test.
We have not yet been able to account for the loss in amino acids and do not know whether they are incorporated into new proteins, exported, or subject to catabolism. In addition, because a specific receptor or target for DHEA has not yet been characterized, we are unable to directly link PGC-1α-mediated induction of DHEA synthesis to the changes in amino acid biology. Regardless, the robust effects of nanomolar concentrations of DHEA puts the PGC-1α-mediated regulation of the synthesis of this hormone in liver into some physiological context and sets up future studies on this enigmatic steroid.
Discussion
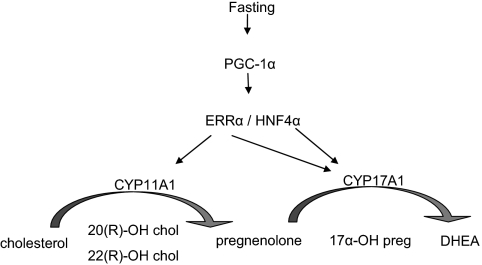
PGC-1α has emerged as a key integrator of signaling pathways that regulate metabolism in a variety of tissues. In this study, we present evidence that PGC-1α regulates production of steroids in the liver, most notably DHEA. Our studies provide definitive evidence to show that PGC-1α can induce gene expression of CYP11A1 and CYP17A1 in both hepatic cell lines and in primary cultures of hepatocytes. Importantly, the necessary auxiliary proteins required for the functionality of these enzymes are expressed at a sufficient level or are coinduced by PGC-1α, because we have observed conversion of either pregnenolone or 22(R)-OH cholesterol to DHEA in human hepatic cell models. The physiological significance of these observations is underscored by the observation that hepatic levels of DHEA are increased in fasted rats. A model describing our view of the relationship between fasting, PGC-1α, and steroidogenesis is presented in Fig. 8.
Figure 8.
Model for fasting-induced expression of steroidogenic enzymes and synthesis of hepatic steroids. chol, Cholesterol; preg, pregnenelone.
Previously, Bauer et al. (23) have reported that CYP17A1 is induced in livers of fasted mice. Our studies extend this previous work by providing a firm understanding of the mechanisms by which fasting leads to the increased synthesis of this enzyme as well as CYP11A1. Although these enzymes are expressed predominantly in the adrenals and gonads where they are involved in mediating the initial steps in steroidogenesis, these studies show that they are also induced and functional in hepatic cells. Although the increase in hepatic DHEA we have observed in whole livers is relatively small, we have demonstrated in culture that this is sufficient to affect a wholesale change in the intracellular concentrations of amino acids. This latter activity highlights the potential physiological significance of PGC-1α-induced increases in DHEA synthesis, and we are currently investigating its role in the fasting response.
Regulation of the expression of steroidogenic genes by PGC-1α, ERRα, and HNF4α
In the classical steroidogenic tissues (adrenals and gonads), gene expression of steroidogenic enzymes is regulated by the hypothalamic-pituitary axis through cAMP-mediated pathways. The precise mechanisms regulating the coordinated expression of these genes have not been fully delineated; however, the nuclear receptor SF-1 has been shown to bind multiple regions of each promoter and regulate their expression (reviewed in Ref. 16). Interestingly, the ERRE within the ERRα gene responsible for its auto-induction is almost identical to the canonical SF-1 response element. Furthermore, it has been shown that ERRα can activate reporters containing SF-1 response elements in transient transfection assays (34). We found that ERREs are overrepresented in the region around the CYP11A1 and CYP17A1, genes. However, we noted that the binding of ERRα is very selective, interacting in a PGC-1α-dependent manner with only one of 18 and one of 14 putative ERREs located with the CYP11A1 and CYP17A1 genes, respectively. This highly selective response suggests that other factors or mechanisms are also involved in regulating ERRα’s binding and transcriptional activation of these genes in hepatic cells.
Although these studies identify putative ERRα regulatory sites, ERRα could bind additional sites that contain a more degenerate consensus site, or ERRα could regulate expression through other transcription factors.
HNF4α is a key transcriptional regulator of many hepatic processes, being reported to associate with the promoter regions of up to 12% of the genes expressed in hepatic cells that are represented on a Hu133K array (35). It was not surprising, therefore, that we were able to show that HNF4α is involved in regulating expression of CYP17A1 in hepatic cells. However, whether or not the activities of these transcription factors can be regulated by other coactivators, permitting an uncoupling of the response observed in the presence of PGC-1α remains to be determined.
A possible role for DHEA in regulating metabolic function in hepatocytes
Our results indicate that DHEA is synthesized in HepG2 cells when PGC-1α is expressed. However, downstream conversion of this steroid to androgens, such as androstenedione or testosterone, was not observed. This was in agreement with our inability to detect significant expression of 3β-hydroxysteroid dehydrogenase 1 or 2 in unmanipulated cells or those expressing PGC-1α. It is therefore tempting to speculate that the primary purpose of these enzymes during fasting is to produce DHEA and that it has an important role in the regulation of liver metabolism.
DHEA administration has been shown to have dramatic effects on metabolism in animal models, reducing hyperglycemia and/or hyperinsulinemia of diabetic db/db mice (36), streptozotocin-induced diabetes (33), Zucker rats (37), and obese ob/ob mice (36). Administration of DHEA to rats increases absolute and relative liver weight, protein, DNA, RNA, and lipid and mitochondrial content within 7 d (38), indicating that DHEA can impact liver metabolism in rodents.
DHEA, or one of its yet to be identified metabolites, could have a role as an intracellular signaling molecule in hepatic cells during fasting. However, this hypothesis is difficult to test because its biological role is still unclear beyond that of an androgen precursor. A specific receptor for DHEA has not been identified, although this steroid has been reported to be able to modulate the activity of other nuclear receptors, such as the estrogen receptor (39,40) and an unidentified membrane-bound receptor (41).
Intriguing data also suggest that DHEA can protect endothelial cells from apoptosis via a Gαi receptor-mediated induction of the phosphatidylinositol 3-kinase/Akt-mediated pathway (42). In fact, these protective effects were also observed using nanomolar concentrations of DHEA (0.1–10 nm) and could be functioning in hepatic cells to protect them from an elevated oxidative state brought on by fasting. Alternatively, induction of the Akt pathway in hepatic cells has been shown to inhibit the activity of PGC-1α (5). Therefore, PGC-1α’s induction of DHEA may constitute a component of a negative feedback loop that controls the gluconeogenic activity of PGC-1α in extended periods of fasting. Further work to explore these possibilities is currently underway.
Regulation of cholesterol homeostasis: generation of other ligands
Whereas we have observed a significant increase in the production of DHEA in cells expressing activated PGC-1α, it is possible that CYP11A1 and CYP17A1 may induce the synthesis of other molecules that could be important in regulating the fasting response. For instance, it has been shown that CYP11A1 catalyzes three different modifications on cholesterol, generating cholesterol metabolites hydroxylated at the 20 and 22 carbons (25). These oxysterols are known ligands for liver X receptors (LXRs), which regulate cholesterol homeostasis and lipid metabolism (43). Additionally, CYP17A1 has been described to have monooxygenase activity on squalene, a cholesterol precursor, producing squalene epoxide (44). Intriguingly, this squalene epoxide can be shunted into an alternative pathway to produce 24(S),25-epoxicholesterol, which is also a potent LXR ligand (45). Therefore, induction of both of these enzymes leads to the production of LXR ligands by two independent pathways, potentially providing another mechanism by which fasting and PGC-1α can regulate LXR activity.
Regulation of steroidogenesis in other tissues
Although these studies highlight the ability of PGC-1α, ERRα, and/or HNF4α to regulate the steroidogenic pathway in hepatic cells, it raises the possibility that they could also regulate this process in other tissues. Classical steroidogenic tissues such as the adrenals and gonads should be considered because cAMP induces expression of both PGC-1α and most of the steroidogenic enzymes.
Another possibility is that PGC-1α could be regulating a steroidogenic program in the brain. Neurosteroids are thought to be primarily synthesized de novo in both neurons and glia where they have a role in signaling and neuroprotection (20). PGC-1α has also been shown to have a neuroprotective role in the brain, resulting in part from its induction of radical oxygen scavengers (46). We have observed that PGC-1α is able to induce expression of CYP11A1 in the U251 glioma cell line (supplemental Fig. 1), suggesting that synthesis of neurosteroids could contribute to PGC-1α’s neuroprotective role in the brain.
Conclusion
Although the functional significance of PGC-1α-mediated induction of functional CYP11A1 and CYP17A1 enzymes in the liver is still under investigation, these results have revealed a potential role for DHEA in regulating hepatic metabolism under conditions of fasting.
Materials and Methods
Plasmids
Lentiviral vectors pLL5.0, VSV-G, Rev, and Gag/Pol were a gift from Dr. Jim Bear (University of North Carolina Chapel Hill, Chapel Hill, NC). Hairpins were designed as described in Cai et al. (47) to include a 19-mer siRNA sequence. siRNA for ERRα was constructed using the sequence described in Schreiber et al. (48), and the oligo used for cloning is listed in supplemental Table 1. siRNA for HNF4α was designed using Oligo engine, and the oligos used for cloning are listed in supplemental Table 1. Oligos were inserted into Hpa1 and Xho1 sites in pLL5.0.
The −3.2-kb-uas-cyp17 reporter construct (31) was a gift from Dr. Walter Miller (University of California, San Francisco, CA). Mutations of the DR-1 elements of the CYP17 promoter were made by transferring the promoter region to pENTRT7 using Xho1 and HindIII sites and using excite mutagenesis with the primers listed in supplemental Table 1. The mutated promoter regions were cloned back into the original vector using Xma1 and NcoI sites surrounding the DR-1 at −2032 and using AgeI and Xho1 sites surrounding the DR-1 site at −878. The accuracy of these mutated clones was verified by sequencing. HNF4α was PCR amplified from HepG2 cell RNA using primers with restriction sites for EcoRI and XbaI on the 5′ and 3′ ends, respectively, and then cloned into sites in pENTR3C (Invitrogen, Carlsbad, CA). HNF4α was recombined into pVP16-GW, a Gateway (Invitrogen) compatible destination vector modified from pVP16 (Clontech, Palo Alto, CA). VP16-SF1 was subcloned from pcDNA3.1-Zeo-SF1 (kind gift from Keith Parker) into pVP16 (Clontech) at EcoR1 and BamH1 sites.
Cell culture
Culture, transfection, infection, and reporter assays using HepG2 (hepatoma), Hep3B (hepatoma), and HeLa (human cervical carcinoma) cells were maintained at 37 C with 5% CO2 in minimal essential media (Invitrogen) supplemented with 8% fetal bovine serum, 0.1 mm nonessential amino acids, and 1 mm sodium pyruvate. HepG2 cells were cultured on dishes coated with 0.1% gelatin. Primary human hepatocytes (Lonza, Basel, Switzerland) were cultured in hepatocyte basal medium supplemented with HCM SingleQuots (Lonza).
For transient transfections, cells were plated on 24-well plates 24 h before transfection with Lipofectin. A total of 3 μg plasmid DNA was used per 24-well triplicate in transfections, and cytomegalovirus β-gal was used for normalization. Luciferase and β-gal readings were measured as described previously (49).
Adenoviruses expressing β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M were constructed as described previously (13) and were amplified and purified by CsCl2 centrifugation (a kind gift from John Bisi at GlaxoSmith Kline). Cells were infected at a multiplicity of infection of 5–50 for 48–72 h.
siRNA was delivered using lentiviruses that were grown in 293FT cells as described in Cai et al. (47). Media was filtered through a 0.45-μm syringe filter and applied to cells with 4 μg/ml polybrene. Fresh culture medium was applied after 16 h, and cells were cultured for 2–3 d and then plated on six-well plates at 400,000 cells per well. Adenoviral infection followed immediately as described above.
qPCR
Total RNA was collected using either QIAGEN (Valencia, CA), Sigma Chemical Co. (St. Louis, MO), or Bio-Rad (Hercules, CA) RNA purification columns and using deoxyribonuclease from either Ambion (Austin, TX), Sigma, or Bio-Rad. One microgram of RNA was used to generate cDNA using iScript (Bio-Rad). cDNA was diluted 1:50, using 5 μl/well in a 13-μl PCR with iQ SYBRGreen supermix (Bio-Rad) and 0.3 μm of each primer. The sequences of the primers used for these studies are included in supplemental Table 1. Data were analyzed by the standard curve method (50), normalizing expression to 36B4.
Western blots
Cells were lysed in 20 mm Tris (pH 8.0), 137 mm NaCl, 10% glycerol, 1% Nonidet P-40, 2 mm EDTA, and 30 μg protein was loaded on 8% SDS-PAGE gels, transferred onto nitrocellulose membranes, and blocked with 5% milk in PBS with 0.1% Tween. PGC-1α antibody (H-300) (Santa Cruz Biotechnology, Santa Cruz, CA), ERRα antibody (51), HNF4α antibody (C-19) (Santa Cruz; sc-6556), or GapDH (V-18) (Santa Cruz; sc-20357) were used to detect protein expression.
Steroid analysis
HepG2 cells were plated on six-well dishes coated with 0.1% gelatin at 250,000 cells per well and infected at a multiplicity of infection of 20–30 with adenoviruses expressing β-gal, PGC-1α, PGC-1α 2x9, or PGC-1α L2L3M. After 24 h, the medium was changed to white MEM with 8% charcoal-filtered serum, 0.1 mm nonessential amino acids, and 1 mm sodium pyruvate for another 24 h. For HPLC analysis, cells were incubated with 200 nm [7-3H]pregnenolone (0.5 μCi/ml) (PerkinElmer, Norwalk, CT) for 6 h. Medium was collected, any cell debris was pelleted by centrifugation, and supernatant was transferred to fresh tubes and snap frozen. Steroids were extracted from the medium with 3:2 ethyl acetate/hexane. This solvent was evaporated, and samples were reconstituted in 100 μl methanol and vortexed, and 40 μl was injected onto a Breeze model 1525 HPLC binary pump system equipped with model 717 plus autoinjector (Waters Corp., Milford, MA) and a Waters Symmetry C18 4.6- × 150-mm, 5-μm reverse-phase column. A methanol gradient was used starting at 50% methanol and holding this concentration for 20 min. This was followed by a gradient to 60% methanol for 22 min and then a gradient to 87% for 7 min. At 49 min, the concentration of methanol was returned to the initial 50% for 10 min before the next injection. The column effluent was analyzed with a model 2487 dual-wavelength UV detector set to 280 nm and a β-RAM model 3 in-line radioactivity detector (IN/US Systems, Inc., Tampa, FL). [3H]DHEA, [3H]17OH-pregnenolone, and [3H]pregnenolone (PerkinElmer, Waltham, MA) were used as standards.
For RIAs measuring DHEA, cells were plated and infected as described for HPLC experiments but were incubated with 200 nm pregnenolone (Sigma) or 50 μm 22(R)-OH cholesterol (Sigma) in 2 ml medium for 6 h. Medium was collected and centrifuged to remove any cell debris. DHEA was measured directly using a RIA kit (MP Biomedicals, Irvine, CA; or Diagnostic Systems Laboratories, Webster, TX). For measurements of DHEA in rodents, liver tissues were harvested, sectioned into approximately 400-mg pieces, weighed, andthen homogenized in equal volumes of solvent (3:2 mixture of ethyl acetate to hexane). Mixture was left at room temperature for 16 h and then centrifuged. The organic layer was removed and evaporated under nitrogen gas and residue was resuspended in 110 μl PBS containing 5 mg/ml BSA, and 100 μl was used in RIA for DHEA. Values were normalized to starting tissue mass.
ChIP
HepG2 cells (4 × 106) were plated on 15-cm plates coated with 1% gelatin and infected the next day with adenoviruses expressing β-gal or PGC-1α for 48 h. Cells were washed with PBS with 1 mm MgCl2 and fixed with PBS plus 1 mm MgCl2 plus 1% formaldehyde for 10–15 min. Glycine was added to 125 mm, and cells were washed three times with PBS, scraped off plate, and snap-frozen. Pellet was resuspended in sonication buffer [50 mm HEPES (pH 7.8), 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% sodium dodecyl sulfate (SDS)] and sonicated at 50% power using a Misonic XL-2000 sonicator for 10 sec followed by 10 sec on ice, repeated 10 times. Cell debris was removed by centrifugation, and 100 μl of 50% protein A/G beads containing 200 μg/ml salmon sperm DNA and 500 μg/ml BSA was used to preclear lysate. One percent of lysate was reserved for input samples, and 2.5 μl ERRα polyclonal antibody (a kind gift from Vincent Giguere) or 5 μg normal mouse IgG (sc-2025, 200 μg/0.5 ml) was used to test for ERRα binding. Five micrograms of HNF4α (either sc-6556 or sc-8987 from Santa Cruz) or IgG control (goat IgG sc-2028 or rabbit IgG sc-2027, respectively) was used to test for HNF4α binding. Antibodies were incubated with 1 ml lysate for 4 h at 4 C, followed by addition of 100 μl 50% protein A/G beads overnight. Beads were washed twice with sonication buffer, twice with wash A [50 mm HEPES (pH 7.8), 500 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS], twice with wash B [20 mm Tris (pH 8.0), 1 mm EDTA, 250 mm LiCl, 0.5% Nonidet P-40, 0.5% Na-deoxycholate], and twice with TE [10 mm Tris (pH 8.0), 1 mm EDTA]. Samples were eluted in 50 mm Tris (pH 8.0), 1 mm EDTA, 1% SDS at 65 C for 20 min, and cross-link was reversed using 0.2 m NaCl and incubating at 65 C overnight. Samples and reserved input were incubated with EDTA (4.2 mm) and proteinase K at 42 C for 1 h, purified on PCR purification columns (QIAGEN), eluted in 40 μl 10 mm Tris, and diluted 1:7 with water for analysis by qPCR. Data was analyzed as percentage of input. DNA shearing quality was monitored to ensure that fragment sizes were around 500 bp. Putative ERREs were identified using the matrix described by Sladek et al. (26), used in TESS at 90% stringency. Putative HNF4 sites were identified using NHR scan (http://www.cisreg.ca/cgi-bin/NHR-scan/nhr_scan.cgi?rm=advanced) (29). Positive control primers for ERRα binding to its own promoter were described by Laganière et al. (27). Primers were designed using Genscript’s real-time PCR primer design tool (https://www.genscript.com/ssl-bin/app/primer) to amplify a region within 300 bp of each putative response element. List of primers are provided in supplemental Table 1.
Animal studies
Animal studies were conducted in accordance with Institutional Animal Care and Use Committee standards according to protocol A-212-05-07 at Duke University. Male Wistar rats weighing 225–250 g were fed standard chow and housed in a controlled environment with 12-h light, 12-h dark cycles. Fasted rats had their food removed in the evening before the experiment and remained without food for 14–16 h before being killed. Animals were sedated with Nembutal (∼0.1 ml/100 g), serum was collected from the portal vein, and livers were harvested and flash frozen in liquid nitrogen.
Amino acid profiling
HepG2 cells were plated at a density of 500,000 cells per well in six-well dishes coated with 0.1% gelatin. After 24 h, cells were dosed with DHEA or vehicle (ethanol) at the indicated concentration. DHEA medium was replaced each day for 2–3 d. Cells were washed and then lysed in 300 μl water, and sonicated, and debris was pelleted by centrifugation. Supernatant was transferred to fresh tubes and flash frozen in liquid nitrogen. Amino acids and acylcarnitines were analyzed using stable isotope dilution techniques. Measurements were made by flow injection tandem mass spectrometry using sample preparation methods described previously (52). The data were acquired using a Micromass Quattro MicroTM system equipped with a model 2777 autosampler, a model 1525 HPLC solvent delivery system and a data system controlled by MassLynx 4.1 operating system (Waters).
Supplementary Material
Acknowledgments
We thank David Vance and Richard Auchus at University of Texas Southwestern for their help and expertise with HPLC.
Footnotes
This work was supported by DoD W81XWH-06-1-0444 (to L.L.G.), National Institutes of Health (NIH) Grant DK074652 (to D.P.M.), and NIH Grant DK59913 (S.R.H.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online April 23, 2009
Abbreviations: ChIP, Chromatin immunoprecipitation; CYP, cytochrome P450; DHEA, dehydroepiandrosterone; ERR, estrogen-related receptor; ERRE, ERRα response element; β-gal, β-galactosidase; HNF, hepatocyte nuclear factor; LXR, liver X receptor; NHR, nuclear hormone receptor; PGC, peroxisome proliferator-activated receptor-γ coactivator; qPCR, quantitative PCR; SDS, sodium dodecyl sulfate; SF-1, steroidogenic factor 1; siRNA, small interfering RNA.
References
- Muoio DM, Newgard CB 2006 Obesity-related derangements in metabolic regulation. Annu Rev Biochem 75:367–401 [DOI] [PubMed] [Google Scholar]
- Browning JD, Horton JD 2004 Molecular mediators of hepatic steatosis and liver injury. J Clin Invest 114:147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM 2001 Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131–138 [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M 2001 CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179–183 [DOI] [PubMed] [Google Scholar]
- Li X, Monks B, Ge Q, Birnbaum MJ 2007 Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature 447:1012–1016 [DOI] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P 2006 GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab 3:429–438 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P 2005 Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM 2003 Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc Natl Acad Sci USA 100:4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J, Ge H, Yang W, Fan M, Handschin C, Cooper M, Lin J, Li C, Spiegelman BM 2006 Partnership of PGC-1α and HNF4α in the regulation of lipoprotein metabolism. J Biol Chem 281:14683–14690 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, Schulman IG, Heyman RA, Lander ES, Spiegelman BM 2004 Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA 101:6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A 2004 The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA 27:6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Torra IP, Staels B, Giguère V, Kelly DP 2004 Estrogen-related receptor α directs peroxisome proliferator-activated receptor α signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol 24:9079–9091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard S, Grasfeder LL, Haeffele CL, Lobenhofer EK, Chu TM, Wolfinger R, Kazmin D, Koves TR, Muoio DM, Chang CY, McDonnell DP 2006 Receptor-selective coactivators as tools to define the biology of specific receptor-coactivator pairs. Mol Cell 24:797–803 [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM 2005 Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1:361–370 [DOI] [PubMed] [Google Scholar]
- Miller WL 2008 Steroidogenic enzymes. Endocr Dev 13:1–18 [DOI] [PubMed] [Google Scholar]
- Payne AH, Hales DB 2004 Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr Rev 25:947–970 [DOI] [PubMed] [Google Scholar]
- Pasqualini JR, Chetrite G, Blacker C, Feinstein MC, Delalonde L, Talbi M, Maloche C 1996 Concentrations of estrone, estradiol, and estrone sulfate and evaluation of sulfatase and aromatase activities in pre- and postmenopausal breast cancer patients. J Clin Endocrinol Metab 81:1460–1464 [DOI] [PubMed] [Google Scholar]
- Simpson ER 2003 Sources of estrogen and their importance. J Steroid Biochem Mol Biol 86:225–230 [DOI] [PubMed] [Google Scholar]
- Garcia-Ovejero D, Azcoitia I, Doncarlos LL, Melcangi RC, Garcia-Segura LM 2005 Glia-neuron crosstalk in the neuroprotective mechanisms of sex steroid hormones. Brain Res Brain Res Rev 48:273–286 [DOI] [PubMed] [Google Scholar]
- Mellon SH, Griffin LD, Compagnone NA 2001 Biosynthesis and action of neurosteroids. Brain Res Brain Res Rev 37:3–12 [DOI] [PubMed] [Google Scholar]
- Pezzi V, Mathis JM, Rainey WE, Carr BR 2003 Profiling transcript levels for steroidogenic enzymes in fetal tissues. J Steroid Biochem Mol Biol 87:181–189 [DOI] [PubMed] [Google Scholar]
- Vianello S, Waterman MR, Dalla Valle L, Colombo L 1997 Developmentally regulated expression and activity of 17α-hydroxylase/C-17,20-lyase cytochrome P450 in rat liver. Endocrinology 138:3166–3174 [DOI] [PubMed] [Google Scholar]
- Bauer M, Hamm AC, Bonaus M, Jacob A, Jaekel J, Schorle H, Pankratz MJ, Katzenberger JD 2004 Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol Genomics 17:230–244 [DOI] [PubMed] [Google Scholar]
- Lala DS, Ikeda Y, Luo X, Baity LA, Meade JC, Parker KL 1995 A cell-specific nuclear receptor regulates the steroid hydroxylases. Steroids 60:10–14 [DOI] [PubMed] [Google Scholar]
- Miller WL 2005 Regulation of steroidogenesis by electron transfer. Endocrinology 146:2544–2550 [DOI] [PubMed] [Google Scholar]
- Sladek R, Bader JA, Giguère V 1997 The orphan nuclear receptor estrogen-related receptor α is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol 17:5400–5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganière J, Tremblay GB, Dufour CR, Giroux S, Rousseau F, Giguère V 2004 A polymorphic autoregulatory hormone response element in the human estrogen-related receptor α (ERRα) promoter dictates peroxisome proliferator-activated receptor γ coactivator-1α control of ERRα expression. J Biol Chem 279:18504–18510 [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ 2008 Regulation of hepatocyte nuclear factor 4 α-mediated transcription. Drug Metab Pharmacokinet 23:2–7 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW 2005 Prediction of nuclear hormone receptor response elements. Mol Endocrinol 19:595–606 [DOI] [PubMed] [Google Scholar]
- Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY 1998 Transcriptional activation of the cholesterol 7α-hydroxylase gene (CYP7A) by nuclear hormone receptors. J Lipid Res 39:2192–2200 [PubMed] [Google Scholar]
- Rodriguez H, Hum DW, Staels B, Miller WL 1997 Transcription of the human genes for cytochrome P450scc and P450c17 is regulated differently in human adrenal NCI-H295 cells than in mouse adrenal Y1 cells. J Clin Endocrinol Metab 82:365–371 [DOI] [PubMed] [Google Scholar]
- Villareal DT, Holloszy JO 2004 Effect of DHEA on abdominal fat and insulin action in elderly women and men: a randomized controlled trial. JAMA 292:2243–2248 [DOI] [PubMed] [Google Scholar]
- Coleman DL, Leiter EH, Schwizer RW 1982 Therapeutic effects of dehydroepiandrosterone (DHEA) in diabetic mice. Diabetes 31:830–833 [DOI] [PubMed] [Google Scholar]
- Bonnelye E, Vanacker JM, Dittmar T, Begue A, Desbiens X, Denhardt DT, Aubin JE, Laudet V, Fournier B 1997 The ERR-1 orphan receptor is a transcriptional activator expressed during bone development. Mol Endocrinol 11:905–916 [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA 2004 Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman DL, Leiter EH, Applezweig N 1984 Therapeutic effects of dehydroepiandrosterone metabolites in diabetes mutant mice (C57BL/KsJ-db/db). Endocrinology 115:239–243 [DOI] [PubMed] [Google Scholar]
- Cleary MP, Shepherd A, Jenks B 1984 Effect of dehydroepiandrosterone on growth in lean and obese Zucker rats. J Nutr 114:1242–1251 [DOI] [PubMed] [Google Scholar]
- Mohan PF, Cleary MP 1988 Effect of short-term DHEA administration on liver metabolism of lean and obese rats. Am J Physiol 255:E1–E8 [DOI] [PubMed] [Google Scholar]
- Bruder JM, Sobek L, Oettel M 1997 Dehydroepiandrosterone stimulates the estrogen response element. J Steroid Biochem Mol Biol 62:461–466 [DOI] [PubMed] [Google Scholar]
- Maggiolini M, Donzé O, Jeannin E, Andò S, Picard D 1999 Adrenal androgens stimulate the proliferation of breast cancer cells as direct activators of estrogen receptor α. Cancer Res 59:4864–4869 [PubMed] [Google Scholar]
- Liu D, Ren M, Bing X, Stotts C, Deorah S, Love-Homan L, Dillon JS 2006 Dehydroepiandrosterone inhibits intracellular calcium release in β-cells by a plasma membrane-dependent mechanism. Steroids 71:691–699 [DOI] [PubMed] [Google Scholar]
- Liu D, Si H, Reynolds KA, Zhen W, Jia Z, Dillon JS 2007 Dehydroepiandrosterone protects vascular endothelial cells against apoptosis through a Gαi protein-dependent activation of phosphatidylinositol 3-kinase/Akt and regulation of antiapoptotic Bcl-2 expression. Endocrinology 148:3068-3076 [DOI] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ 1996 An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature 383:728–731 [DOI] [PubMed] [Google Scholar]
- Liu Y, Yao ZX, Papadopoulos V 2005 Cytochrome P450 17α-hydroxylase/17,20 lyase (CYP17) function in cholesterol biosynthesis: identification of squalene monooxygenase (epoxidase) activity associated with CYP17 in Leydig cells. Mol Endocrinol 19:1918–1931 [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, Willson TM 1997 Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem 272:3137–3140 [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM 2006 Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127:397–408 [DOI] [PubMed] [Google Scholar]
- Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE 2007 Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 128:915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A 2003 The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα). J Biol Chem 278:9013–9018 [DOI] [PubMed] [Google Scholar]
- Norris J, Fan D, Aleman C, Marks JR, Futreal PA, Wiseman RW, Iglehart JD, Deininger PL, McDonnell DP 1995 Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J Biol Chem 270:22777–22782 [DOI] [PubMed] [Google Scholar]
- Bookout AL, Mangelsdorf DJ 2003 Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal 1:e012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard S, Dwyer MA, McDonnell DP 2007 Definition of the molecular basis for estrogen receptor-related receptor-α-cofactor interactions. Mol Endocrinol 21:62–76 [DOI] [PubMed] [Google Scholar]
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB 2004 Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med 10:268–274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.