Summary
Regulation of eukaryotic gene expression is far more complex than one might have imagined thirty years ago. However, progress towards understanding gene regulatory mechanisms has been rapid and comprehensive, which has made the integration of detailed observations into broadly connected concepts a challenge. This review attempts to integrate the following concepts: 1) a well-defined organization of nucleosomes and modification states at most genes, 2) regulatory networks of sequence-specific transcription factors, 3) chromatin remodeling coupled to promoter assembly of the general transcription factors and RNA polymerase II, and 4) phosphorylation states of RNA polymerase II coupled to chromatin modification states during transcription. The wealth of new insights arising from the tools of biochemistry, genomics, cell biology, and genetics is providing a remarkable view into the mechanics of gene regulation.
Keywords: gene regulation, chromatin, genome-wide, pre-initiation complex, histone modification, transcription cycle, noncoding RNA
Optional Keywords: SAGA, TFIID, ubiquitylation, histone methylation, histone acetylation, RNA polymerase II CTD, nucleosomes
Introduction
Eukaryotic transcription is regulated by a large number of proteins, ranging from sequence-specific DNA binding factors to chromatin regulators to the general transcription machinery and their regulators (reviewed by Berger, 2000; Li et al., 2007a; Orphanides and Reinberg, 2002; Pugh, 2000; Struhl et al., 1998). Their collective function is to express a subset of genes as dictated by a complex interplay of environmental signals that is only partly understood. Classical biochemistry and cleverly devised genetic screens have led to discoveries of important components of the transcription machinery, and have provided insight into mechanisms involved in transcription by RNA Polymerase II (Pol II). Recent genome-wide expression profiling and factor location profiling have imbued our understanding of the organization of the transcription machinery and nucleosomes throughout the genome. The prevailing view of transcriptional activation is that many sequence-specific regulators interact with their cognate DNA motifs in response to cellular signals. They recruit transcriptional coactivators to alter the local chromatin environment and facilitate assembly of the pre-initiation complex (PIC), which is composed of the general transcription factors (GTFs) and Pol II.
The breadth of information about how genes are regulated has become sufficiently vast that it is becoming increasingly difficult to comprehend how the different aspects of transcriptional regulation fit together. This review attempts to integrate some of the major stages in gene regulation including those involving activators, chromatin remodeling and modifications, PIC assembly, and transcription elongation, which necessitates a limited depth of coverage on any one topic. To avoid “overload”, a representative name of the many involved complexes is used, rather than indicating all family members and other functionally related complexes. This review draws from a number of model eukaryotic systems, but places particularly emphasis on lessons learned from the budding yeast Saccharomyces cerevisiae. Much of the basic mechanisms of gene control remain highly conserved in eukaryotes, and yeast has provided the most simplified route towards a basic understanding of this control. The integration present here, in the context of genome-scale inquiry, is intended to provoke new questions about how the various stages are coordinated in a genome-wide response to environmental signals.
ChIP-chip and ChIP-seq identify the location and level of a protein binding anywhere in any genome
Chromatin immunoprecipitation (ChIP) has become an invaluable tool for mapping protein interactions along genomic DNA in vivo (Figure 1), and thus has been the single most informative assay in assessing the assembly of proteins on DNA in vivo. A key feature of the ChIP assay is that it preserves physiologically relevant interactions in the cell through formaldehyde crosslinking. Formaldehyde is an ideal crosslinking agent (Orlando et al., 1997; Solomon and Varshavsky, 1985; Toth and Biggin, 2000) because: i) it quickly permeates the cell and traps native interactions before the cell mounts a physiological response, ii) its single-carbon crosslinker length efficiently generates protein-DNA crosslinks in vivo (protein-protein and protein-RNA crosslinks are also formed), and iii) its readily reversible crosslink is important for subsequent DNA detection methods.
Figure 1. ChIP assays to measure protein binding across a genome.
Two DNA detection methods that make use of ChIP are illustrated. 1) In both methods, formaldehyde is used to crosslink transcription factors (TF) to the genome in vivo. 2) The DNA is then fragmented. 3) The protein is immuno-purified to remove DNA that is not bound to the TF.
In ChIP-chip, the DNA crosslinked to the protein is then attached to a red dye and detected by hybridization to an array of immobilized DNA probes (microarray chip), whose sequence matches specific genomic locations. Often a reference DNA sample (illustrated in green) is co-hybridized so that probe-to-probe variation can be controlled. In ChIP-chip, the low-resolution first generation microarrays contained probes spanning all genic and/or intergenic regions. Second generation microarrays provided higher detection resolution by tiling probes across the genome at 5 base pair spacing in yeast, and ~40 bp spacing in the larger genomes of fly and human.
In Chip-seq, each DNA molecule is directly sequenced. ChIP-seq achieves single base-pair detection resolution through sequencing, although the median DNA length of the ChIP sample preparation limits the spatial resolution that can be achieved by ChIP-seq. The sequencing read counts at each genomic coordinate are shown as a bar graph in a searchable genome browser screenshot (http://atlas.bx.psu.edu/).
Since its inception (Ren et al., 2000), ChIP coupled to microarray detection (ChIP-chip, Figure 1) has proven to be a powerful tool in understanding the interplay of the transcription machinery and chromatin (Kim and Ren, 2006; Pugh and Gilmour, 2001). It can determine the occupancy level of essentially any crosslinkable and immuno-purifiable protein across an entire genome. Early ChIP-chip microarrays have had two important limitations. First, the fabrication of such microarrays has required a sequenced genome. Second, spatial resolution of binding along a genome was limited by probe length and spacing. Today, this has been largely alleviated in the highly tiled (probes every 5–40 bp) second-generation microarrays (Figure 1). Recent break-throughs in cost-effective whole-genome shotgun sequencing has also eliminated the first limitation.
Detection of genomic segments bound by a protein has recently been taken to another level of resolution by coupling the ChIP assay with massively parallel DNA sequencing, called ChIP-seq (Figure 1) (Margulies et al., 2005; Schuster, 2008). The mapping of nucleosome positions across genomes was one of the first applications of ChIP-seq. In the past few years, ChIP-seq has produced whole-genome nucleosome maps for yeast (Albert et al., 2007; Mavrich et al., 2008a; Shivaswamy et al., 2008), fly (Mavrich et al., 2008b), worm (Johnson et al., 2006; Valouev et al., 2008), and human (Barski et al., 2007; Boyle et al., 2008; Schones et al., 2008). Others have used ChIP-seq to map the locations of transcription factors (Jothi et al., 2008; Nielsen et al., 2008; Robertson et al., 2007).
Some genes are packaged into repressive chromatin structures
Since genes within the eukaryotic genome are compacted within the nucleus in the form of chromatin, the transcription machinery must overcome a formidable structural barrier to access the underlying cis-regulatory elements and coding regions. Early work on the nuclear packaging of genomic DNA found that chromosomal DNA is composed of a beads-on-a-string configuration (Hewish and Burgoyne, 1973; Olins and Olins, 1974), which is thought to be the predominant form of transcriptionally-competent chromatin. Each bead is a nucleosome. The nucleosome core particle contains 147 base pairs of DNA wrapped 1.7 times around a histone octamer containing two copies of histones H2A, H2B, H3 and H4 (Luger et al., 1997). In contrast to the active state, genes may be repressed through compaction of chromatin into a 30 nanometer fiber (Figure 2A).
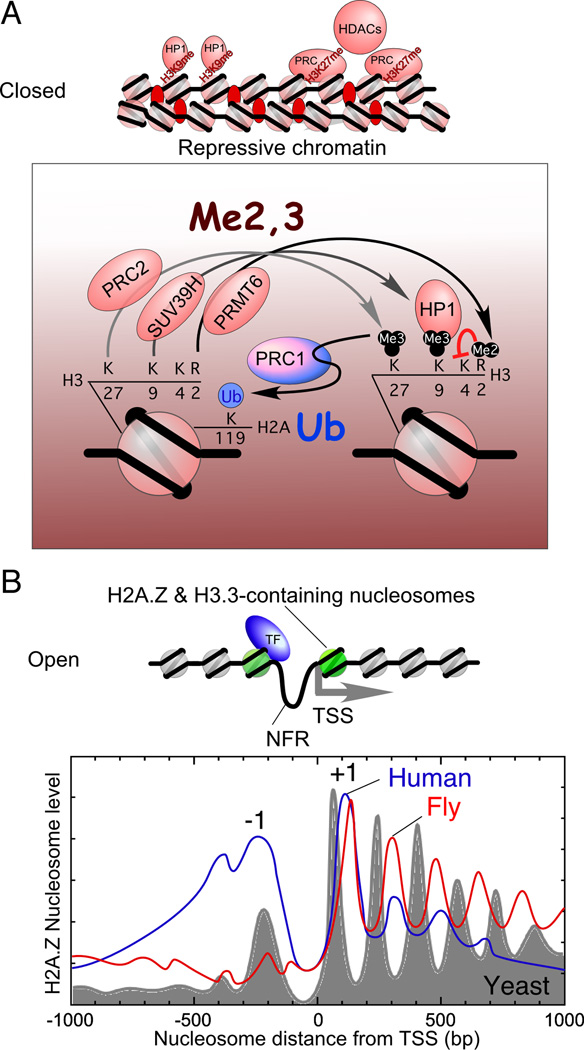
Figure 2. The organization of nucleosomes throughout the genome.
(A) Model for closed chromatin. Repressive chromatin is shown in a closed state that is representative of the 30 nm chromatin fiber. Nucleosome-free regions (NFRs) are absent. The resident canonical histones are methylated at a number of sites including H3K9 and/or H3K27. These sites are bound by HP1 and the PRC complexes, respectively. Compaction is mediated by linker histone H1. These repressive entities recruit histone deacetylases (HDACs) to remove activating acetyl marks on histones.
(B) The promoters of most genes reside in an open chromatin state in which they are competent to undergo activation. Open chromatin is represented as a beads-on-a-string configuration in which a transcription factor (e.g. Reb1 or Abf1, indicated as “TF”) binds to its cognate site, and helps to maintain the NFR as well as to promote the replacement of canonical histones H3 and H2A, with H3.3 and H2A.Z, respectively. (In yeast, H3.3 is the same as H3.) Note that this is a composite representation and may not reflect the disposition of factors at any given gene. Shown are the frequency distributions for H2A.Z-containing nucleosomes relative to the transcriptional start site (TSS) of all genes in budding yeast, fly, and human CD4+ T cells determined by ChIP-seq (Albert et al., 2007; Mavrich et al., 2008b; Schones et al., 2008).
Except in Saccharomyces, this closed repressive chromatin structure is often associated with methylation of histone H3 at lysine 9 (H3K9) by SUV39H (SUppressor of Variegation 3–9 Homolog) and concomitant binding of HP1 (Heterochromatin Protein 1) to that methyl mark (Figure 2A). The repressive state may also or instead be associated with methylation at H3K27 by Polycomb Repressive Complex 2 (PRC2) and the concomitant binding of PRC1, which ubiquitylates H2AK119 (Kouzarides, 2007; Levine et al., 2004; Motamedi et al., 2008; Trojer et al., 2009). Dimethylation of H3R2, by PRMT6 (Protein aRginine MethylTransferase 6) (Iberg et al., 2008), provides an additional repressive mark that serves to prevent formation of the activating trimethyl H3K4 mark (Kirmizis et al., 2007). Whether and how these repressive marks and their cognate factors promote or maintain the closed state or are simply indicators of the closed state remains to be determined. Indeed small noncoding RNAs may be instrumental in establishing repressive chromatin environments (Goodrich and Kugel, 2008; Grewal and Elgin, 2007). The H3K9 and K27 methyl marks might function cooperatively or independently in repression, and may do so in part by recruiting repressive DNA methylases and histone deacetylases that remove activating histone acetylation marks.
Surprisingly, many genes in human embryonic stem cells that contain the methyl K27 repressive mark also contain a methyl mark on H3K4 that is associated with active genes (Bernstein et al., 2006). These so-called bivalent genes may be in a repressed state but potentiated for rapid or well-timed activation that is key for coordinated development of multi-cellular organisms. It remains to be determined whether the active H3K4 methyl mark on bivalent genes is established co-transcriptionally or is placed by some other mechanism, and whether components that recognize the bivalent marks co-exist on the same gene. Interestingly, PRC1 which reads the repressive H3K27 methyl mark copurifies with TAFs (TATA-binding protein Associated Factors) (Saurin et al., 2001), of which at least one (TAF3) reads the active H3K4 methyl mark (Vermeulen et al., 2007). TAFs are components of the general transcription machinery (see below). These and other studies described below suggest that at some genes the transcription machinery may be present at repressed genes. However, in general, the transcription machinery is recruited only when the gene is to be transcribed.
Most genes have a canonical “open” organization of nucleosomes
Chromatin contains a repeating array of nucleosomes that are spaced roughly every 160–200 base pairs throughout the genome. The impact of nucleosomes on gene regulation was generally underappreciated until key experiments, nearly two decades ago, found that the presence of nucleosomes inhibited transcription initiation in vitro (Lorch et al., 1987), indicating that nucleosomes are physical barriers to transcription. In addition, depletion of histones in the budding yeast Saccharomyces led to a global increase in transcription (Han and Grunstein, 1988; Wyrick et al., 1999), thus providing in vivo evidence that nucleosomes can repress transcription (although many other genes were also activated).
The position of nucleosomes at several model genes, such as PHO5, SUC2, GAL1, HMRa, and RNR3, have been mapped in Saccharomyces based upon the principle that nucleosomes protect the underlying DNA from digestion by exogenously added micrococcal nuclease (MNase). These early gene-specific maps provided a glimpse into how the location of nucleosomes across a gene might impact promoter access and transcription (Almer and Horz, 1986; Li and Reese, 2001; Lohr, 1997; Perez-Ortin et al., 1987; Ravindra et al., 1999). The study of such model genes has proven indispensable in guiding our understanding of the interplay of nucleosomes and the transcription machinery. For example, work on the PHO5 gene demonstrated that nucleosomes regulate transcription through occlusion of the TATA and UAS (Upstream Activating Sequence) promoter elements and that nucleosome disruption is critical for activation (Lohr, 1997; Martinez-Campa et al., 2004).
The recent advances in genome-wide mapping technologies have provided a much clearer picture of the genomic nucleosome landscape (reviewed by Jiang and Pugh, 2009). With the increased tiling density of microarray probes, examining the genome-wide structure of chromatin at single nucleosome resolution is becoming routine. Using high-density tiling over a portion of the yeast genome, Rando and colleagues (Yuan et al., 2005) discovered that a nucleosome-free region (NFR) was a common feature of promoters (Figure 2B). The “−1” and “+1” nucleosomes reside in canonical locations upstream and immediately downstream of the NFR, respectively. Thus, nucleosomes are not stochastically dispersed along chromosomal DNA but instead are, by design, positioned at specific distances from the transcription start site (TSS) so as to regulate transcription. Whether individual nucleosomes play specific roles in regulating gene expression is not known, and will likely be an active area of investigation in the future. As “gate-keepers” of the NFR at promoters, the −1 and +1 nucleosomes are well-positioned to have significant regulatory potential.
It is remarkable that upon aligning nucleosome positions throughout the yeast, fly, and human genomes to the TSS a predominant nucleosome organization is apparent (Figure 2B). The existence of a promoter NFR and the uniform nucleosome positioning relative to the TSS, from gene-to-gene, are two features that are evolutionarily conserved. Although these salient features are remarkably similar from yeast to human, seemingly subtle species-specific differences are evident, but have important mechanistic implications for transcription initiation. For example, in yeast the TSS is tucked in the upstream border of the +1 nucleosome, suggesting that this nucleosome potentially regulates access to the TSS (Albert et al., 2007). However, the +1 nucleosome in metazoans is shifted further downstream of the TSS compared to budding yeast, leaving the transcription start site accessible (Mavrich et al., 2008b; Schones et al., 2008). This downstream +1 nucleosome might be better situated to regulate transcription elongation. Indeed, Pol II transcription pauses at the first nucleosome. The −1, NFR, +1 arrangement provides the stage upon which sequence-specific regulators read the genome to direct transcriptional programs.
What creates an NFR is not fully known, although substantial evidence points to the presence of nucleosome-excluding poly-dA:dT tracts in the region (Anderson and Widom, 2001; Iyer and Struhl, 1995; Mavrich et al., 2008a; Yuan et al., 2005), and the presence of sequence-specific factors like Reb1 (RNA polymerase I Enhancer Binding protein 1) (Raisner et al., 2005). Other sequence-specific regulators, such as Rsc3 (Remodel the Structure of Chromatin 3), can influence the nucleosome density in the NFR at some promoters (Badis et al., 2008). In addition, chromatin remodeling complexes are likely to expand and contract the boundaries of the NFR, by using the energy of ATP hydrolysis to reposition nucleosomes (Whitehouse et al., 2007).
Sequence-specific factors direct transcription programs from specific sets of genes
Cis-regulatory elements
Transcription programs are governed by trans-acting sequence-specific factors that control transcription by binding to cis-regulatory elements (CREs) (Table 1). Depending on their impact on transcription, these sites are collectively called upstream activating/repressing sequences (UAS/URS) in yeast or enhancers in metazoans. The number of promoters targeted by any given sequence-specific regulator ranges from a few to several hundred (or several thousand in metazoans). For example, the galactose regulator Gal4 binds to only ten promoters (Ren et al., 2000), whereas Rap1 (Repressor Activator Protein 1) binds to over 300 promoters in the Saccharomyces genome (Buck and Lieb, 2006). The activity and subcellular localization of these factors are controlled by internal and external environmental cues, often using phosphorylation or targeted proteolysis as a molecular switch between active and inactive states.
Table 1.
Common DNA elements
| DNA Element | Acronym meaning | Description | Bound Protein | Species specificity |
|---|---|---|---|---|
| Core promoter elements | ||||
| NFR | Nucleosome Free Region | ~140 bp regions present at the beginning and end of genes that lack nucleosomes, rich in poly-dA:dT tracts | n.a. | Yeast and Metazoans |
| TSS | Transcription Start Site | First transcribed nucleotide of the RNA transcript, typically an A | Pol II | Yeast and Metazoans |
| TATA | -- | Located ~60 bp upstream (25–30 bp in metazoans) of the transcription start site, site of PIC assembly, ~20% of genes contain a TATA box in yeast | TBP | Yeast and Metazoans |
| BRE | TFIIB Recognition Element | Two distinct motifs flank the site where the TATA box typically resides, helps to orient the directionality of the PIC | TFIIB | Metazoans |
| Inr | Initiator | Immediately adjacent to the TSS, can accurately direct initiation alone or with the TATA box | Taf1,Taf2 | Metazoans |
| MTE | Motif Ten Element | Located 20 bp downstream of TSS, functions with Inr to enhance transcription, can substitute for TATA | n.a. | Metazoans |
| DPE | Downstream Promoter Element | Located 30 bp downstream of the TSS, found in many TATA-less Drosophila promoters | Taf6, Taf9 | Metazoans |
| Upstream elements | ||||
| UAS | Upstream Activating Sequence | Recognized by activators to stimulate transcription through recruitment of coactivator complexes | Activators | Yeast |
| Enhancer | -- | Functionally similar to the UAS but often located several thousand bp away from the corresponding promoter | Activators | Metazoans |
| URS | Upstream Repressing Sequence | Recognized by repressors to negatively regulate transcription | Repressors | Yeast |
While UAS/URSs are typically found several hundred base pairs upstream of the translation start site in Saccharomyces (Harbison et al., 2004), other elements such as the TATA-box are present in the core promoter region and are typically 30–60 bp from the transcription start site (Table 1). Phylogenetic analysis of six Saccharomyces species reveals that about 20% of the 5,700 yeast genes contain a TATA-box element (Basehoar et al., 2004). In metazoans, additional core promoter elements exist that interact with various components of the basal transcription machinery, such as the Initiator (Inr), downstream promoter element (DPE), motif ten element (MTE), and TFIIB recognition element (BRE) (Smale and Kadonaga, 2003; Thomas and Chiang, 2006). Some operate in lieu of a TATA box, while most promoters seem to lack any recognizable core promoter element.
Gene regulatory networks
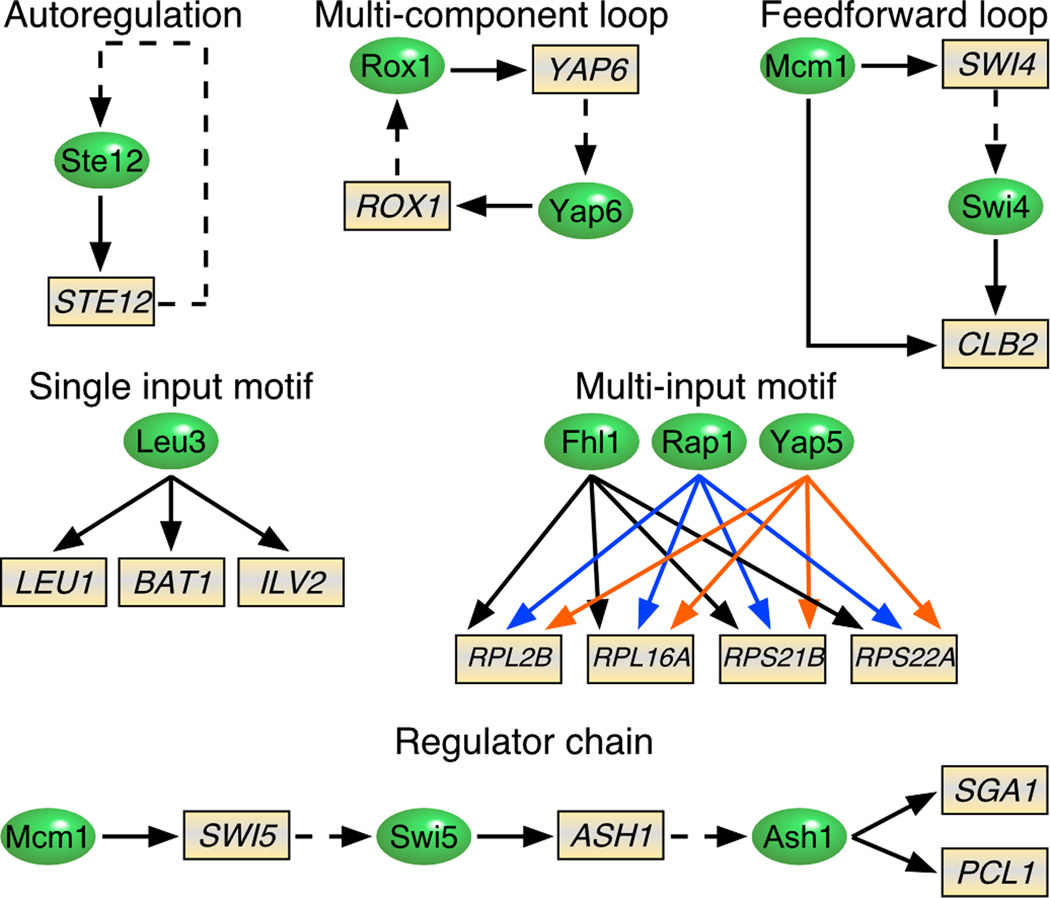
How are sequence-specific regulators organized in the genome to allow global control of gene expression programs? The topology of regulatory circuits that underpin expression programs is best understood in yeast since the genomic binding locations for many of the ~120 sequence-specific regulators are known (Lee et al., 2002). Analysis of the compendium of regulator-gene interactions identified six basic network motifs, each providing unique regulatory advantages (Figure 3). For example, the “single-input” motif ensures the concerted expression of the leucine biosynthetic genes by Leu3, whereas the “multiple-input” motif can integrate several signaling pathways to coordinate the expression of a set of genes under different conditions. The prototypical metazoan interferon beta enhancesome (Panne, 2008) may be regarded as a type of multiple-input motif. However, it is not known whether all multiple input motifs require concurrent binding of sequence-specific regulators for expression of the target gene, as in the case of the interferon beta gene. Regulation of the cell cycle by sequence-specific regulators exemplifies the “regulator chain” motif in which transcriptional events are ordered in a temporal sequence in accordance with temporal nature of the cell cycle.
Figure 3. Regulatory networks controlled by sequence-specific transcriptional regulators.
Regulatory circuits are composed of simple network motifs. Specific examples for six regulatory motifs are shown. Sequence-specific regulators and target genes are indicated by ovals and rectangles, respectively. Solid arrows denote binding of an activator to a gene promoter. Dashed arrows designate genes encoding a sequence-specific regulator. In the multi-input motif, for clarity arrows associated with the each factor are colored differently. The illustration is modified from (Lee et al., 2002).
The simple regulator chain motif has been expanded to construct a global pyramid-shaded hierarchical network (Yu and Gerstein, 2006). This hierarchical network describes a “chain-of-command” organization with a few master regulators at the top, which tend to have the maximal influence on global expression levels. The hierarchical regulatory structure is a decision making scheme that allows for the convergence of multiple internal and external stimuli to precisely modulate the expression of select groups of genes. For example, in yeast oxygen and heme levels activate expression of Mot3, a master regulator for aerobic growth. Mot3 activates Gcn4 (General Control Nonderepressible 4), which in turn activates Put3 (Proline Utilization 3) and Uga3 (Utilization of GAba 3), which in turn activates many genes involved in proline and nitrogen metabolism.
Sequence-specific regulators as orchestrators
While the genome-wide locations for many sequence-specific regulators have been determined in Saccharomyces, how these factors are specifically contributing to transcription is less clear. Whether a sequence-specific regulator acts as an activator or repressor may depend on its genomic context and what co-regulators they recruit. Sequence-specific regulators orchestrate multiple aspects of transcription through direct recruitment of: i) chromatin remodeling complexes, ii) general transcription factors, iii) chromatin modifying complexes, and iv) Pol II via the Mediator complex (Brown et al., 2001; Cosma et al., 1999; Garbett et al., 2007; Goldmark et al., 2000; Green, 2005; Larschan and Winston, 2001; Neely et al., 2002; Nourani et al., 2004; Park et al., 2000; Yudkovsky et al., 1999). Each of these aspects will be discussed below.
ATP-driven machines remodel the DNA on nucleosomes
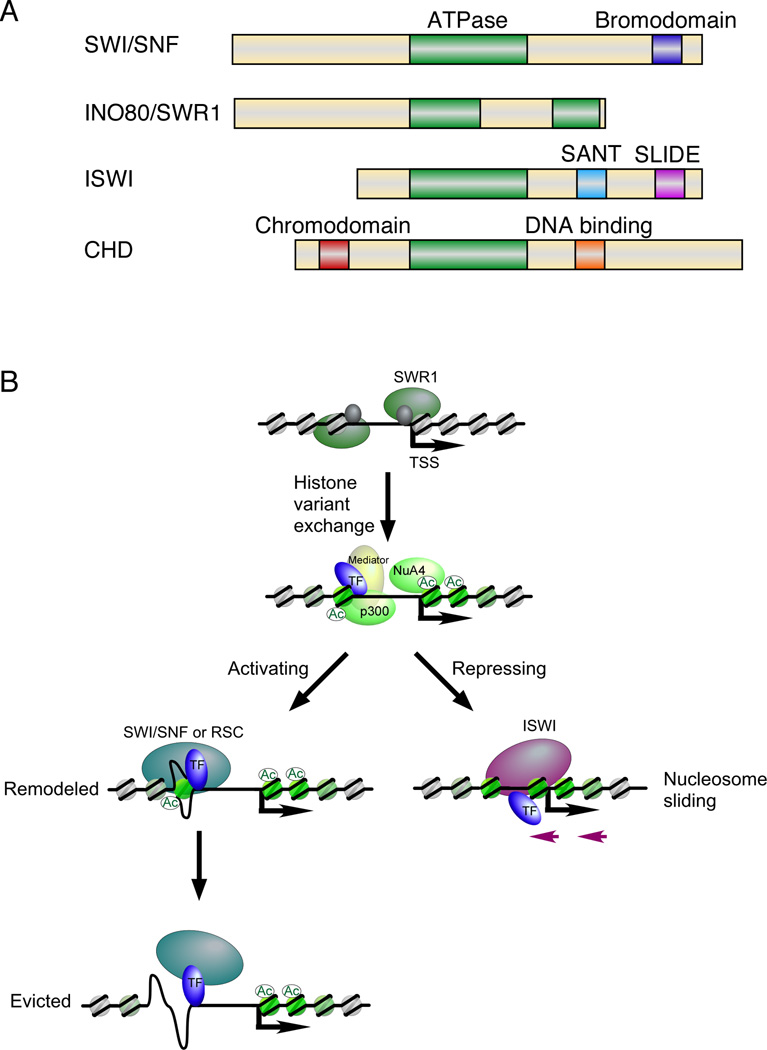
Early work on model genes demonstrated that transcriptional activation involves the movement of nucleosomes (Almer and Horz, 1986; Li and Reese, 2001; Lohr, 1997; Perez-Ortin et al., 1987; Ravindra et al., 1999). However, the existence of chromatin remodeling complexes was unknown until the early 1990s (Hirschhorn et al., 1992), despite the isolation of genes for key subunits (e.g. SWI2/SNF2) via genetic screens (Neigeborn and Carlson, 1984; Stern et al., 1984). Chromatin remodeling complexes fall into four families based upon sequence conservation (Figure 4A) (Bao and Shen, 2007b): SWI/SNF, INO80/SWR1, ISWI, and CHD.
Figure 4. Chromatin remodeler families and conserved domains.
(A) Domain organization of the ATPase subunit of chromatin remodeling complexes. All four families share an evolutionarily conserved Snf2-like ATPase domain belonging to the DEAD/H-box helicases. The catalytic subunit of the SWI/SNF family contains a bromodomain at the C-terminus that binds acetylated lysines. The INO80/SWR1 family is distinct from the other three families by having a split ATPase domain. The ATPase subunit of the ISWI family of remodelers harbors a SANT and SLIDE domain at the C-terminus, which are thought to bind histone tails and linker DNA, respectively. The CHD family contains an N-terminal chromodomain, which binds methylated lysines, and a C-terminal DNA-binding domain. The illustration is modified from (Tsukiyama, 2002).
(B) In the “open” chromatin configuration, chromatin remodeling complexes use the energy of ATP hydrolysis to dissociate DNA from the histone surface so that histone variants (H2A.Z and H3.3, shown in green) may be exchanged in, or that DNA binding sites may become exposed (activating pathway) or the sites may be covered (repressing pathway). Some of this may be facilitated by histone acetylation (p300 and NuA4 examples are indicated). The presence of an NFR may allow partial assembly of the PIC prior to nucleosome remodeling or eviction. Note that this is a composite representation and may not reflect the disposition or order of remodeling at any given gene. The absence of Mediator and other components in the later steps is only for clarity.
Since chromatin remodelers generally lack the intrinsic ability to target specific genes, sequence-specific regulators are likely to directly recruit these complexes to promoter regions (Figure 4B). How chromatin remodeling complexes are targeted to specific nucleosomes and their distinct roles remains an active area of investigation.
Chromatin remodeling complexes utilize the power of ATP hydrolysis to alter the structure, position, or composition, of nucleosomes (Figure 4B) (reviewed by Flaus and Owen-Hughes, 2004; Saha et al., 2006; Tsukiyama, 2002). Based on numerous in vitro biochemical and single molecule optical trap studies, the current view for the mechanism of chromatin remodeling is that the DNA wound around the nucleosome forms a loop that is translocated by the mechanical power generated by these ATPase motors, resulting in the histone octamer either sliding along DNA or being altogether evicted from the DNA (reviewed by Cairns, 2007; Saha et al., 2006). The mechanistic details will vary among the different families. The genomic context of such remodeling activity influences the accessibility of DNA at promoters and in some cases suppresses cryptic initiation sites within the body of the gene (Whitehouse et al., 2007).
The nucleosomes surrounding promoter regions tend to be highly dynamic (Dion et al., 2007), which suggests that chromatin remodeling complexes may be constitutively present and active at many promoters even when the promoter is largely quiescent. Since the positions of the −1 and +1 nucleosomes may largely influence the nucleosome architecture internal to genes (Mavrich et al., 2008a), and that the −1/+1 nucleosomes control the accessibility of key promoter elements, it is likely that these nucleosomes will be prime targets of chromatin remodeling complexes (Figure 4B) (Venters and Pugh, 2009).
Conceivably, remodeling complexes that reposition nucleosomes could cause adjacent nucleosomes to reposition as well, such as through steric clashes. However, whether a remodeling complex individually targets each nucleosome in an array, or whether targeting of a single nucleosome in an array is sufficient to move an entire array is not known. Answers to this question are key to understanding whether a remodeling complex that repositions nucleosomes on an entire gene needs to focus on a single linchpin nucleosome or needs to cover the entire domain.
SWI/SNF family and its relationship to histone acetylation
Classic genetic selections for mating-type switching deficient and sucrose non-fermenting phenotypes identified the SWI2/SNF2 gene, which encodes the catalytic subunit of the SWI/SNF complex (Neigeborn and Carlson, 1984; Stern et al., 1984). Genetic and molecular evidence with Swi2/Snf2 and Snf5 mutants later showed that the SWI/SNF complex alters the structure of chromatin through sliding and/or ejecting nucleosomes, independent of transcription (Hirschhorn et al., 1992; Lorch et al., 2006).
The SWI/SNF family of chromatin remodelers, which also includes the RSC (Remodels Structure of Chromatin) complex, is generally viewed as a positive regulator of transcription (although some genes are negatively regulated) (Angus-Hill et al., 2001; Sudarsanam et al., 2000). Location profiling by ChIP-chip finds RSC at the promoters of several hundred genes (Damelin et al., 2002; Ng et al., 2002), suggesting a role in transcription initiation and nucleosome organization. Consistent with this notion, RSC mutants perturb the translational setting of promoter nucleosomes (Parnell et al., 2008). SWI/SNF, in particular, might have additional functions in transcription elongation (Schwabish and Struhl, 2007).
Promoter nucleosomes tend to be hyper-acetylated, and this may help retain bromodomain-containing chromatin remodeling complexes such as SWI/SNF and RSC (Figure 4B) (Hassan et al., 2001; Hassan et al., 2002; reviewed by Ruthenburg et al., 2007). Bromodomains bind to acetylated lysines. Given that acetylation may also diminish the electrostatic histone lysine-DNA interactions, as well as disrupt higher order compaction, prior acetylation of nucleosomes might facilitate nucleosome remodeling and dismantling during gene activation. Acetylation may also be instrumental in nucleosome re-assembly directed by remodeling complexes. For example, NuA4 (NUcleosome Acetyltransferase of H4)-directed acetylation of H2A.Z is important for its deposition into promoter nucleosomes (Keogh et al., 2006). H3K56 acetylation by Rtt109 (Regulator of Ty1 Transposition 109) (Schneider et al., 2006), and other histone acetylation events by Hat1/HAT-B (Histone AcetylTransferase 1) (Parthun et al., 1996), may be important for histone deposition. Other key acetyltransferase complexes associated with chromatin remodeling include SAGA (Spt-Ada-Gcn5 Acetyltransferase) and p300 (Sterner and Berger, 2000). As we will discuss below, SAGA has multiple roles in the transcription cycle beyond nucleosome acetylation.
In vitro transcription studies show that nucleosome acetylation by NuA4 stimulates the activity of RSC and enhances passage of Pol II through a nucleosome (Carey et al., 2006). Both RSC and SWI/SNF are directed to some promoters through interactions with activators (Cosma et al., 1999; Yudkovsky et al., 1999). RSC also functions at many Pol I and Pol III promoters (Damelin et al., 2002; Ng et al., 2002), which may be the reason why RSC is essential for viability in yeast whereas SWI/SNF is not (Cairns et al., 1996).
INO80/SWR1 family
The INO80/SWR1 (INOsitol requiring 80/Sick With Rat8 ts 1) family of remodelers is unique in that it contains a split ATPase domain (Bao and Shen, 2007a). The INO80 gene was identified in a screen for genes required for activating the inositol synthetase gene (INO1), which synthesizes a compound required for several secondary messenger signaling pathways (Ebbert et al., 1999). The INO80 complex plays a broader role in genome regulation than many other remodeling complexes in that it participates in transcription activation, DNA repair, and resolving stalled replication forks (Shen et al., 2000; Shimada et al., 2008). Understanding how INO80 is targeted to sites of transcription, repair and replication will be of interest because unlike other remodeling complexes in yeast, INO80 lacks known histone recognition modules.
The SWR-C/SWR1 complex is a chromatin remodeler that alters the composition (as opposed to the position) of nucleosomes. The SWR1 complex uses ATP hydrolysis to replace H2A with H2A.Z in promoter nucleosomes (Guillemette et al., 2005; Li et al., 2005; Raisner et al., 2005; Zhang et al., 2005). H2A.Z is thought to promote transcription by destabilizing nucleosomes (Krogan et al., 2004; Zhang et al., 2005). Indeed, H2A.Z is associated with an open chromatin state rather than the closed state (Figure 2B), although the presence of H2A.Z does not suffice to create a transcriptionally active state. In mammalian cells, H2A.Z is required for lineage commitment by embryonic stem cells (Creyghton et al., 2008), suggesting that altering the composition of chromatin with histone variants plays an important role in development, perhaps by helping commit specific genes to an activated state.
ISWI family
In contrast to the SWI/SNF and INO80 families, the Imitation SWItch (ISWI) family of remodelers tends to negatively regulate transcription. For example, genome-wide expression profiling and DNase I sensitivity studies in Saccharomyces found that the ISW2 complex in concert with the histone deacetylase Rpd3 (Reduced Potassium Dependency 3) represses meiotic genes by creating a repressive nucleosome arrangement (Fazzio et al., 2001). ISW2 is recruited to repressive loci by Ume6, a key sequence-specific regulator of early meiotic genes (Goldmark et al., 2000). ISW2 might also cooperate with other repressive complexes, such as TUP1-SSN6 (deoxyThymidine monophosphate UPtake 1 and Suppressor of Sucrose Nonfermentor 6), to maintain repressive states (Zhang and Reese, 2004). Since ISW2 may be important in maintaining a targeted closed deacetylated state (in cooperation with Rpd3), it makes sense that these complexes lack bromodomains, as they would antagonize the generally open acetylated state that exists throughout the euchromatic genome. As a result, ISW2 may depend more on sequence-specific regulators for recruitment than on recognition of histone modifications.
CHD family
The intracellular role of the CHD (Chromatin organization modifier, Helicase, and DNA-binding domains) family of chromatin remodelers is the least understood of the four families of remodelers. Expression profiling in a Chd1 yeast mutant has shown that few genes are affected (Tran et al., 2000), suggesting that Chd1 may operate in parallel pathways with other chromatin remodelers or is targeted to few genes. Biochemical purification of the SAGA complex from S. cerevisiae led to the identification of Chd1 as one of its components (Pray-Grant et al., 2005). To what extent Chd1 co-exists with SAGA at specific genes and functions with SAGA is currently not understood. A defining feature of the CHD family is that it contains a chromodomain, which binds to methylated lysines. Indeed, Chd1 interacts with methylated H3K4 in vitro (Biswas et al., 2007; Flanagan et al., 2005; Pray-Grant et al., 2005). However, there is disagreement between the difference in binding specificity for the yeast and human counterparts. The in vivo significance of Chd1 binding to H3K4 awaits further investigation, and its potential connection with the acetylation and de-ubiquitylation activities associated with SAGA remain largely unexplored. Since histone acetylation, chromatin remodeling, recognition of H3K4 trimethyl marks, and de-ubiquitylation are all associated with transcription, there may be some logic for these multiple activities being associated with a single SAGA complex.
General transcription factors assemble into the transcription pre-initiation complex
Mediator plays a key early role
While chromatin remodeling complexes may keep the chromatin in the promoter region dynamic, the many facets of the remodeling process may not be driven to completion until the Mediator complex and the GTFs assemble into a pre-initiation complex. Mediator is a large complex of proteins involved in the many aspects of transcription (reviewed by Biddick and Young, 2005; Kornberg, 2005). First discovered as a biochemical entity that mediated transcriptional activation in vitro (Kim et al., 1994), Mediator may be recruited early during PIC assembly through direct interactions with activators (illustrated in Figure 4B) (Natarajan et al., 1999). One example of how Mediator might regulate PIC assembly is through recruitment of the p300 histone acetyltransferase to promoters. This recruitment blocks PIC assembly until p300 has acetylated its targets (histones and itself), which then induces p300 to dissociate from the promoter (Black et al., 2006). In essence, Mediator and p300 create a checkpoint to ensure that PIC assembly does not proceed until certain acetylation events are completed.
TBP is a highly regulated nucleator of PIC assembly in the NFR
Some thirty years ago, biochemical fractionation of crude cell extracts by the Roeder lab led to the identification of general transcription factors that accurately initiated transcription in vitro at a minimal core promoter (Thomas and Chiang, 2006). These GTFs were first isolated from mammalian cells (Matsui et al., 1980) and later the corresponding factors were identified in yeast (Sayre et al., 1992). The GTFs include TFIIA, -B, -D, -E, -F, and -H. In contrast to sequence-specific regulators that are targeted to a discrete set of genes, GTFs, as their name suggests, are broadly utilized by the cell at many genes and typically have minimal built-in gene specificity. These components of the basal transcription machinery function at most genes to assist in the loading and release of RNA polymerase II at the TSS.
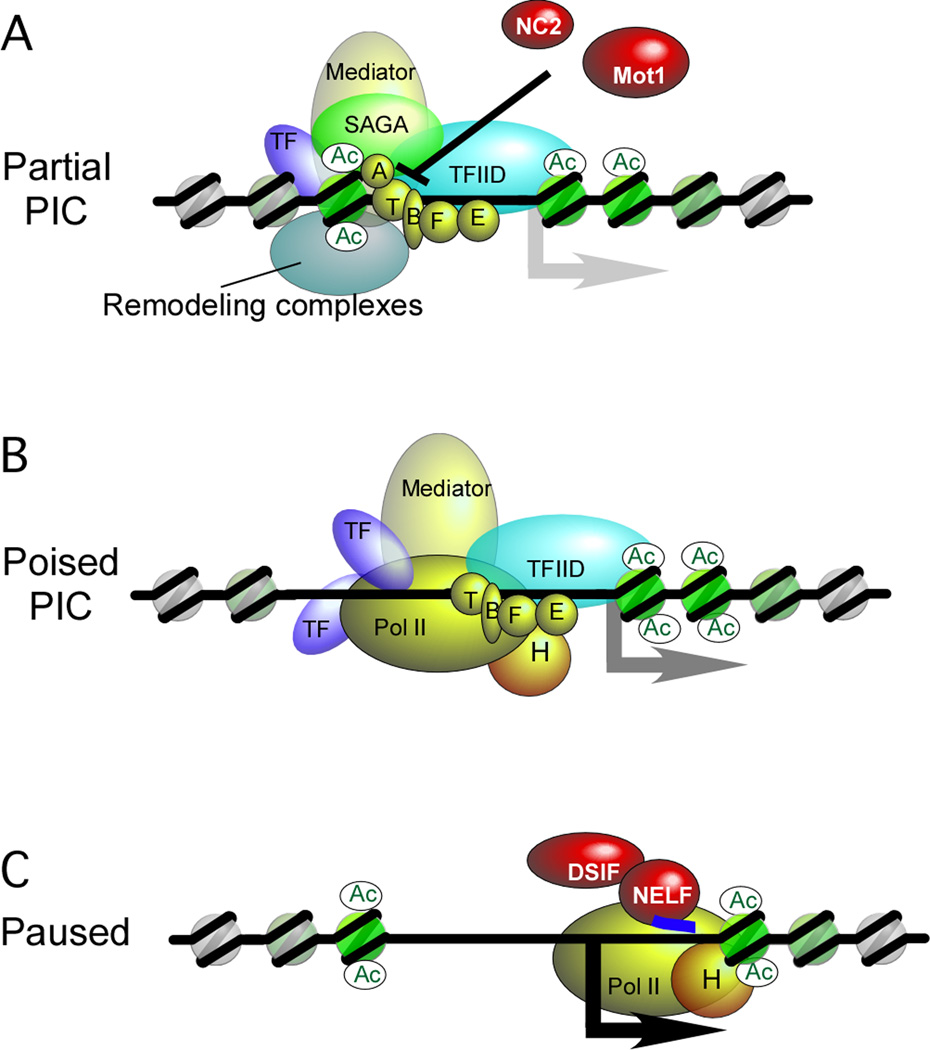
TFIID (Transcription Factor II D) is a large multisubunit complex that contains TBP (TATA Binding Protein) and TAFs (TBP-Associated Factors). Paradoxically, TFIID is largely recruited to TATA-less “housekeeping” promoters, which represent the vast majority of all genes (Basehoar et al., 2004). TBP also exists free of the TFIID complex, and is delivered to TATA-containing promoters via the SAGA complex (Sermwittayawong and Tan, 2006). TBP is removed from such promoters by the combined direct action of NC2 (Negative Cofactor 2) and the TBP-dependent ATPase Mot1 (Modifier of Transcription 1) (reviewed by Lee and Young, 1998; Pugh, 2000). TFIIA and TFIIB interact with TBP and promote PIC formation by stabilizing TBP/DNA interactions and counteracting the effects of NC2 and Mot1. At least at some genes, GTFs assemble into partial PICs that are relatively depleted of TFIIH and Pol II (Figure 5A). Partial PICs might represent regulated intermediates (for example, in the coupling of responses to heat shock and oxidative stress, in which the former causes the latter) (Zanton and Pugh, 2006). Interestingly, partial PICs can assemble in the NFR without apparent loss of the surrounding nucleosomes. Only when full PICs (containing Pol II) are assembled is the −1 nucleosome removed (Venters and Pugh, 2009).
Figure 5. Assembly of the pre-initiation complex.
Two forms of pre-initiation complexes and an early elongation complex are shown: A) Partial PIC (Zanton and Pugh, 2006), B) Poised PIC (Martens et al., 2001; Radonjic et al., 2005; Sekinger and Gross, 2001), and C) Paused Pol II complex (Lee et al., 2008; Muse et al., 2007; Zeitlinger et al., 2007).
(A) A partial PIC contains GTFs assembled in the context of resident nucleosomes, but is relatively depleted of TFIIH and Pol II.
(B) A poised PIC contains Pol II and TFIIH in addition to the GTFs and exists in the context of an evicted -1 nucleosome. The poised PIC has not yet cleared the promoter. In vivo, such complexes may be undergoing abortive initiation events where very short transcripts are released and degraded.
(C) A paused Pol II complex typically occurs 30–50 nucleotides after the TSS. Negative elongation factor (NELF) and other factors (not shown) bound to Pol II help create the paused state. The +1 nucleosomes might also contribute to pausing by creating a barrier. Many initiation and regulatory factors may be retained at the promoter after Pol II has cleared the area, which might promote subsequent rounds of transcription (not shown).
Although TBP can replace TFIID in biochemically reconstituted transcription assays, it is the TAFs that are generally required for sequence-specific activators to promote maximal transcription (Pugh and Tjian, 1990; Reese et al., 1994), suggesting that TAFs may interact with some activators. As one example, Rap1 is an activator that directly interacts with TFIID in vitro, and this interaction is important in driving transcription of the highly-transcribed ribosomal protein genes (Garbett et al., 2007; Mencia et al., 2002).
The physiological importance of TBP is underscored by the resources that the cell devotes to regulating its activity. Mot1 and NC2 together regulate the genomic distribution of TBP, but through different mechanisms. Mot1 might couple ATP hydrolysis to localized DNA translocation, which is then used to disrupt TBP/DNA interactions (Auble et al., 1994). In this way Mot1 would act negatively in transcription. However, Mot1 also acts positively at some genes, possibly by removing improperly assembled TBP that might bind in a reverse orientation or at inappropriate sites due to TBP’s intrinsic weak specificity for TATA (Muldrow et al., 1999; Sprouse et al., 2008b). In contrast, NC2 clamps TBP to DNA, and blocks further PIC assembly by sterically interfering with TFIIB binding (Inostroza et al., 1992). Taken together, while TBP binding to DNA is intrinsically stable, its interaction in vivo may be highly dynamic (Sprouse et al., 2008a). Stability appears to be driven by TFIIA and TFIIB (perhaps in cooperation with other GTFs and regulatory factors), and instability might be driven first by NC2 counteracting TFIIB (and TFIIA), which then allows Mot1 to induce dissociation. It will be interesting to learn whether loss of NC2 or Mot1, in the presence of sufficient amounts of TBP, causes TBP to accumulate at NFRs in the genome. This issue gets at whether NFRs are actually protein-free and thus capable of binding proteins, or whether they are already occupied by other unknown factors and thus inaccessible.
Atomic resolution structures of complexes containing either TFIIA or TFIIB bound to TBP-TATA DNA reveals that TFIIA binds upstream of TBP (away from the TSS) (Nikolov et al., 1995; Tan et al., 1996), whereas TFIIB resides mostly downstream of TBP but also straddles TBP thereby clamping the DNA on both sides of TBP. TFIIB and TFIIF are required for association of Pol II with the PIC (Figure 5B), and this is supported by the Pol II-TFIIB crystal and Pol II-TFIIF cryo EM structures (Bushnell et al., 2004; Chung et al., 2003).
TFIIE and TFIIH regulate transcription initiation
The TFIIE and TFIIH complexes work together to modulate the activity of Pol II and facilitate promoter clearance. Step-wise assembly of the PIC in vitro indicates that TFIIE associates after Pol II but before TFIIH (Buratowski et al., 1989), although other studies indicate that TFIIE can associate with a partial PIC in the absence of Pol II (Yokomori et al., 1998). Recent two-hybrid and ChIP experiments in a Mediator mutant yeast strain suggest that Mediator facilitates the incorporation of TFIIH into the PIC through direct interactions (Esnault et al., 2008). TFIIE also recruits TFIIH and regulates TFIIH’s helicase and kinase activities. The ATP-dependent helicase activity is important for DNA strand separation (promoter melting) so that an open promoter complex with Pol II may form (Wang et al., 1992). The Kin28 (KINase 28) subunit of TFIIH mediates the transition from transcription initiation to elongation through phosphorylation of serines in the 5th position of the highly repeated YSPTSPS motif in the CTD (Carboxy-Terminal Domain) of the largest Pol II subunit (Valay et al., 1995). Together the GTFs function as a cohort of factors to recognize core promoters, assemble the starting platform for transcription, recruit Pol II, and facilitate the transition from Pol II initiation to elongation.
In Saccharomyces, the transition from transcription initiation to elongation has been suggested to be rapid, with little post-recruitment regulation except in isolated examples (Martens et al., 2001; Radonjic et al., 2005; Sekinger and Gross, 2001; Wade and Struhl, 2008). However, recent findings have challenged this notion. Pol II has been found to be relatively enriched in promoter regions compared to transcribed regions across the Saccharomyces genome (Venters and Pugh, 2009), suggesting that the conversion of the PIC into a transcription elongation complex may be at least partially rate-limiting at many yeast genes and thus targeted for regulation. This is consistent with post-recruitment regulation mechanisms identified in yeast (Zhang et al., 2008). In contrast, Pol II is found more-or-less uniformly throughout highly transcribed genes, suggesting that initiation is not rate-limiting at those genes.
Pausing of Pol II immediately after initiation is widespread
The transcription cycle involves recruitment of Pol II to promoters, then transcription initiation, elongation, and termination (Wade and Struhl, 2008). Any of these steps can potentially be rate-limiting. Historically, unexpressed genes have been thought to be rate-limited by steps leading up to Pol II recruitment (Ptashne and Gann 1997). However, this notion has recently been challenged by the finding of Pol II at most genes, from yeast to humans (Guenther et al., 2007; Steinmetz et al., 2006), which taken at face value suggests that the expression of many or most genes is rate-limited after Pol II recruitment (Core and Lis, 2008; Margaritis and Holstege, 2008; Price, 2008; Struhl, 2007; Wade and Struhl, 2008). This latter view cannot be entirely correct because lowly expressed genes generally have less bound Pol II than highly expressed genes. Thus, detection of Pol II at genes is not equivalent to full occupancy, and so it is likely that steps leading up to PIC assembly and steps subsequent to Pol II recruitment will both regulate the transcription cycle.
Recent studies find that Pol II is paused at the 5’ end of ~10% of all Drosophila genes (Lee et al., 2008; Muse et al., 2007; Zeitlinger et al., 2007), suggesting that Pol II pausing during an early elongation step is rate-limiting in the expression of many genes (Figure 5C). This pausing involves the NELF (Negative ELongation Factor) and DSIF (DRB Sensitivity Inducing Factor) complexes. Phosphorylation of NELF and DSIF by P-TEFb (Positive Transcription Elongation Factor b) promotes the release of the paused polymerase (Peterlin and Price, 2006). P-TEFb also phosphorylates the 2nd serine in the Pol II CTD repeat (in contrast to TFIIH phosphorylating the 5th serine) rendering Pol II elongation competent. In mammals, most genes are also enriched with Pol II at their 5′ ends (Guenther et al., 2007), but it remains unclear whether this reflects post-recruitment regulation of a poised PIC that is not yet transcriptionally engaged (Figure 5B) and/or a paused Pol II that is transcriptionally engaged (Figure 5C).
Pol II CTD serine 5 phosphorylation during initiation sets off a cascade of events that regulate the nascent RNA and the underlying chromatin
The Pol II CTD coordinates events during the transcription cycle by recruiting proteins involved in histone modifications, elongation, termination, and mRNA processing (reviewed by Egloff and Murphy, 2008; Meinhart et al., 2005; Phatnani and Greenleaf, 2006). Numerous proteins recognize and bind to a specific phosphorylation pattern of the CTD (Figure 6), which dynamically changes during the transcription cycle. The number of YSPTSPS heptapeptide repeats of the CTD vary from 26 in Saccharomyces to 52 in human. Since there are five potential phosphorylation sites in each CTD repeat, a CTD code has been suggested (Buratowski, 2003). Only serine phosphorylation at the 5th and 2nd position have been well characterized. Recently, serine 7 was also discovered to be phosphorylated in human cells, and has been implicated in transcription of small nucleolar RNAs (Egloff et al., 2007). Whether serine 7 has a role in other Pol II genes remains unclear.
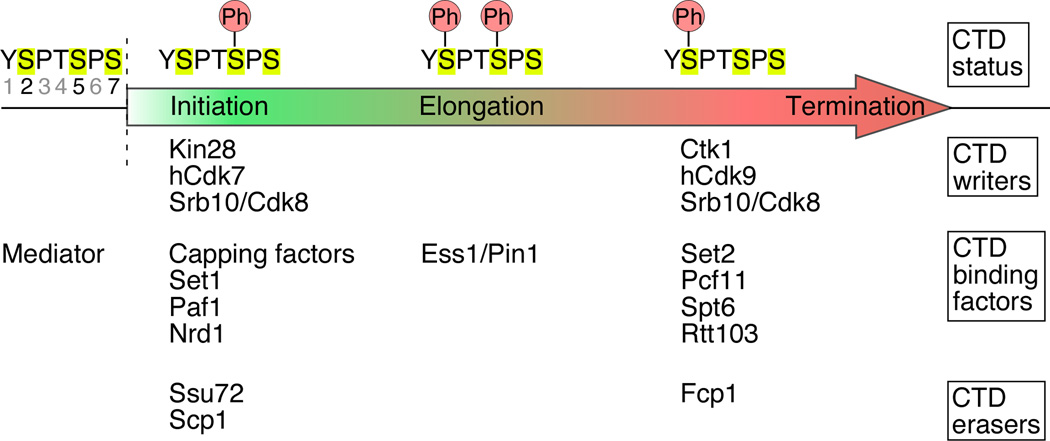
Figure 6. Writers, readers, and erasers of the Pol II CTD code.
Writers (kinases), readers, and erasers (phosphatases) of the CTD code are listed under the different phosphorylation (Ph) statuses of the CTD (single amino acid code YSPTSPS). This phosphorylation status changes from the 5’ end of genes to the 3’ end. Y1, T4, and S7 may also be phosphorylated in vivo (Baskaran et al., 1993; Egloff et al., 2007; Zhang and Corden, 1991), but their function remains to be deciphered.
During PIC assembly, a dephosphorylated Pol II is recruited to promoters through interactions with TFIIB and Mediator (Myers et al., 1998). One of first steps in transcription initiation is the phosphorylation of serine 5 of the CTD by Kin28 (Komarnitsky et al., 2000). This Pol II mark recruits a number of factors including the mRNA capping enzyme, an RNA surveillance complex called Nrd1-Nab3 (Nuclear pre-mRNA Down-regulation 1 and Nuclear polyAdenylated RNA-Binding 3), and the PAF (Polymerase II-Associated Factor) complex (Figure 6 and Figure 7A) (Hampsey and Reinberg, 2003; Komarnitsky et al., 2000; Ng et al., 2003b; Vasiljeva et al., 2008). The 7-methyl guanylyl cap protects the nascent RNA from degradation, and marks the mRNA for transport to the cytoplasm and ultimately translation. The Nrd1-Nab3 complex may track with Pol II and the RNA to ascertain whether the emerging RNA contains short Nrd1 binding motifs (e.g. GUA and UCUU, (Carroll et al., 2004)), which may be evolutionarily depleted in mRNAs, but present in inappropriately-transcribed and thus nonfunctional regions of the genome. The Nrd1-Nab3 complex promotes termination of these latter transcripts, and directs them through the nuclear exosome pathway for degradation.
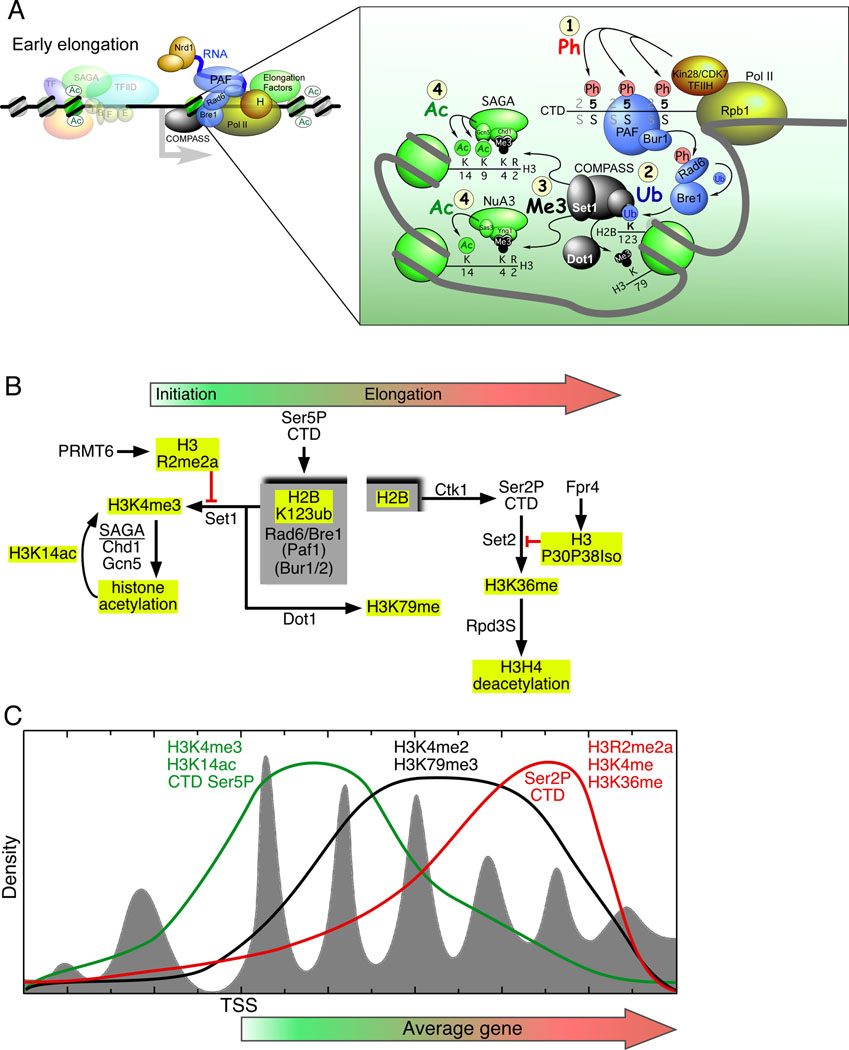
Figure 7. Histone crosstalk and the distribution of post-translational modifications across genes.
(A) Model for early elongation. Early elongation includes promoter proximal pausing, elongation factors bound to the CTD, and a cascade of histone modifications (shown in the inset and numbered 1–4). Abbreviations include: Ph, phosphorylation; Ub, ubiquitylation; Me3, trimethylation; Ac, acetylation. See text for an explanation of the model.
(B) Post-translational modification network. At the hub of this histone modification network is a cycle of ubiquitylation and deubiquitylation on histone H2B, demarcated in the gray boxes. Solid arrows and red lines connect the interdependencies of post-translational modifications. The red line indicates that a particular modification blocks the subsequent modification. Histone tail modifications are highlighted with yellow boxes. Marks generally associated with initiation are shown toward the left, whereas modifications linked to elongation are shown toward the right (with the exception of H3R2me2a, which occurs during elongation to block H3K4me3), as designated by the filled arrow at the top of the panel.
(C) Model for the distributions of histone modifications and phosphorylation of the Pol II CTD in relation to gene length. The nucleosome distribution relative to the TSS is shown in gray fill. The green, black, and red traces model the genome-wide distribution of the indicated histone modifications and CTD phosphorylation state.
The PAF complex connects serine 5 phosphorylation of the CTD to a network of histone modifications. The multitude of interactions with serine 5 phosphorylated CTD may explain the highly repetitive nature of the heptad motif. While each complex could, in principle, interact with a subset of phosphorylated repeats there is very little known about the arrangement of factors on the CTD. The PAF complex, in conjunction with the Bur1/2 (Bypass UAS Requirement 1 and 2) kinase, engages the serine 5 phosphorylated CTD and promotes recruitment of the Rad6 (RADiation sensitive 6) and Bre1 (BREfeldin A sensitivity 1) E2 and E3 ubiquitin ligases (Figure 7A), to set off a series of histone modification crosstalks, known as the histone code.
Pol II phosphorylation is connected to writers, readers, and erasers of the histone code
The chromatin landscape is peppered with numerous posttranslational modifications (or marks) to histone tails (reviewed by Berger, 2007; Kouzarides, 2007; Lee and Shilatifard, 2007; Millar and Grunstein, 2006; Shilatifard, 2008; Weake and Workman, 2008). Histone tails consist of 20–40 amino-terminal amino acids of each histone, which extend from the globular nucleosome core into the surrounding solvent. The best studied of the histone marks include acetylation, methylation, and ubiquitylation. These marks have been proposed to function by i) directly changing the structure of chromatin by altering electrostatic or internucleosomal contacts, and/or ii) providing docking sites for proteins (“readers”). For each mark written, there is a corresponding complex that is capable of erasing it, consistent with the dynamic nature of chromatin marks (Bannister and Kouzarides, 2005; Klose and Zhang, 2007; Shi and Whetstine, 2007). Some histone modifications, such as methylated H3K9/27, are stably maintained through multiple cell divisions and reflect epigenetic processes where the modifications become self-propagating (reviewed by Bibikova et al., 2008; Lunyak and Rosenfeld, 2008). Whether such modifications are the cause or consequence of epigenetic inheritance remains to be demonstrated.
From Pol II CTD serine-5 phosphorylation to H3K4 methylation
It is becoming increasingly clear that individual histone marks influence the writing and erasing of other marks, creating a network of histone crosstalk during transcription. Recent reports support a model wherein transcription initiation and elongation accompanies a dynamic cycle of coupled histone modifications, perhaps exemplified by the events associated with the ubiquitylation of H2BK123 (Figure 7A and B). Once Ser5 phosphorylation of the Pol II CTD has recruited the PAF and Bur1/2 complexes, then at least Bur1 phosphorylates Rad6 (Wood et al., 2005), and this stimulates the Rad6 E2 ligase activity to mono-ubiquitylate Bre1. Bre1 then passes the ubiquitin onto H2BK123 as an E3 ligase (Kao et al., 2004; Laribee et al., 2005; Ng et al., 2003a; Robzyk et al., 2000; Wood et al., 2003, 2005). H2BK123ub then directs trimethylation of H3K4 by COMPASS/Set1 (Complex Of Proteins ASsociated with Set1/Su(var)3–9, Enhancer of zeste, Trithorax 1) and K79 by Dot1 (Disruptor Of Telomeric silencing 1) (Briggs et al., 2002; Dover et al., 2002; Nakanishi et al., 2008; Sun and Allis, 2002; Wood et al., 2003).
Trimethylation of H3K4 also requires acetylation at H3K14 (Nakanishi et al., 2008), which presumably occurs during PIC assembly or co-transcriptionally using acetyltransferases (such as Elongator) that translocate with Pol II. Since acetylation might enhance nucleosome eviction during transcription, the H3K4me3 mark might sense local acetylation levels through K14. Although it is not entirely clear what the H3K4me3 mark is doing, conceivably it might bind to “readers” such as the TAF3 subunit of TFIID, the Chd1 component of SAGA, or the Yng1 (Yeast homolog of mammalian Ing1 1) subunit of NuA3 to help maintain PIC assembly and the active acetylated state.
Transcription-associated H3K4me3 occurs at the beginning of genes (Liu et al., 2005; Pokholok et al., 2005), but ultimately transitions into dimethylation (H3K4me2) less than 1 kb downstream of the TSS (Figure 7C). The transition might occur through alterations in the specificity of the methyltransferase active site of Set1, which could be linked to the phosphorylation status of the CTD (i.e. a switch from serine 5 to serine 2 phosphorylation triggers a switch from H3K4me3 to me2), but this remains to be determined. H3K79me3, which is also linked to CTD serine-5 phosphorylation, is maintained throughout the body of the gene, and is linked to transcription and telomere silencing (Lacoste et al., 2002; Pokholok et al., 2005).
From CTD serine-2 phosphorylation to H3K36 methylation
As elongation proceeds from the 5′ end of the gene towards the 3′ end, a phosphorylation switch occurs on the CTD from serine-5 to serine-2 (Figure 6 and Figure 7C) (Komarnitsky et al., 2000), although it is not clear to what extent one event triggers the other. Ctk1 (Carboxy-Terminal domain Kinase 1) in yeast and P-TEFb in higher eukaryotes phosphorylate serine-2 (Lee and Greenleaf, 1997; Marshall et al., 1996; Sterner et al., 1995; Wyce et al., 2007), and thus are regulators of elongation.
The CTD serine-5 phosphorylation mark is erased by the Ssu72 (Suppressor of SUa7 2) phosphatase (and possibly other phosphatases such as Scp1 (S. cerevisiae CalPonin 1, which is a calcium binding protein)) (Hausmann et al., 2005; Krishnamurthy et al., 2004; Yeo et al., 2003). Ssu72 may be responsible for many aspects of the transcription cycle: 1) It associates with TFIIB in the PIC; 2) It is required for the transition from serine-5 to serine-2 phosphorylation; and 3) It is part of the cleavage and polyadenylation complex that operates during transcription termination (Dichtl et al., 2002a).
Remarkably, the CTD appears to regulate its own structural status through prolines located in the YSPTSPS heptapeptide repeat. Prolines are the only natural amino acid that can exist in structurally distinct cis and trans configurations, which result in altered paths of the CTD polypeptide chain. Studies using surface plasmon resonance demonstrate that the Ess1 (ESSential 1) proline isomerase preferentially associates with the doubly phosphorylated CTD at serines 2 and 5 (Phatnani et al., 2004). The implications for regulating the cis/trans configuration of the two prolines in the CTD are unclear at present, but conceivably isomerization of the prolines could regulate dephosphorylation of the CTD or the binding of proteins that recognize the phosphorylation patterns on the CTD. Indeed, genetic studies suggest that the role of Ess1 may be to oppose the functions of the CTD kinases (Wilcox et al., 2004). Given the two to four dozen YSPTSPS heptad repeats arrayed on a single complex of Pol II, with two prolines and three serine phosphorylation sites in each repeat, one can envision an extensive CTD code having positional, structural, and chemical information.
As Pol II transcribes into the body of a gene, PAF-stimulated H2BK123ub marks continue to be generated, possibly followed by H3K4me2 marking. However, H2BK123ub blocks nucleosomal association of Ctk1 and subsequent serine 2 phosphorylation of the CTD (Wyce et al., 2007), which represents the elongation state of Pol II. Current models suggest that the SAGA complex may travel with Pol II and promote deubiquitylation of H2BK123ub through its Ubp8 (UBiquitin-specific processing Protease 8) subunit (Henry et al., 2003), although not all genes may partake in this process (Shukla et al., 2006). Removal of the H2BK123ub mark by Ubp8 may allow turnover of the H3K4 methyl mark via demethylases (and perhaps resetting the chromatin state for a new round of transcription), but this remains to be tested. Alternatively, removal of the H3K4 mark might occur through dynamic histone subunit exchange with unmethylated copies. Importantly, loss of the H2BK123ub mark allows Ctk1 to phosphorylate serine-2 of the CTD.
Not only does H2BK123 ubiquitylation/deubiquitylation serve as a regulatory switch in a network of histone modifications (Weake and Workman, 2008), but H2BK123ub also regulates nucleosome dynamics during elongation. For instance, loss of H2B ubiquitylation and deletion of SPT16 (SuPpressor of Ty 16) results in lower levels of histone occupancy at the GAL1 locus, suggesting that H2BK123ub and FACT (FAcilitates Chromatin Transcription) (via the Spt16 subunit) regulate nucleosome reassembly in the wake of elongating Pol II (Fleming et al., 2008).
Serine-2 phosphorylation triggers a second wave of communication between the CTD and chromatin, ultimately impinging upon H3K36 methylation. Whereas the PAF complex may have been the connector from the serine-5 phosphorylation mark, the H3K36 methylase Set2 appears to be the connector in this second phase. Set2 binds to the CTD phosphorylated at serine-2 (Figure 6 and 8A) (Kizer et al., 2005; Li et al., 2003; Li et al., 2002; Xiao et al., 2003), and catalyzes di and trimethylation of H3K36. Whether di versus trimethylation of K36 serve distinct purposes is not known. Nonetheless, H3K36me2 appears to be a mark of actively transcribed genes (Rao et al., 2005), although its function may be to repress transcription that might arise promiscuously or cryptically in the ORF.
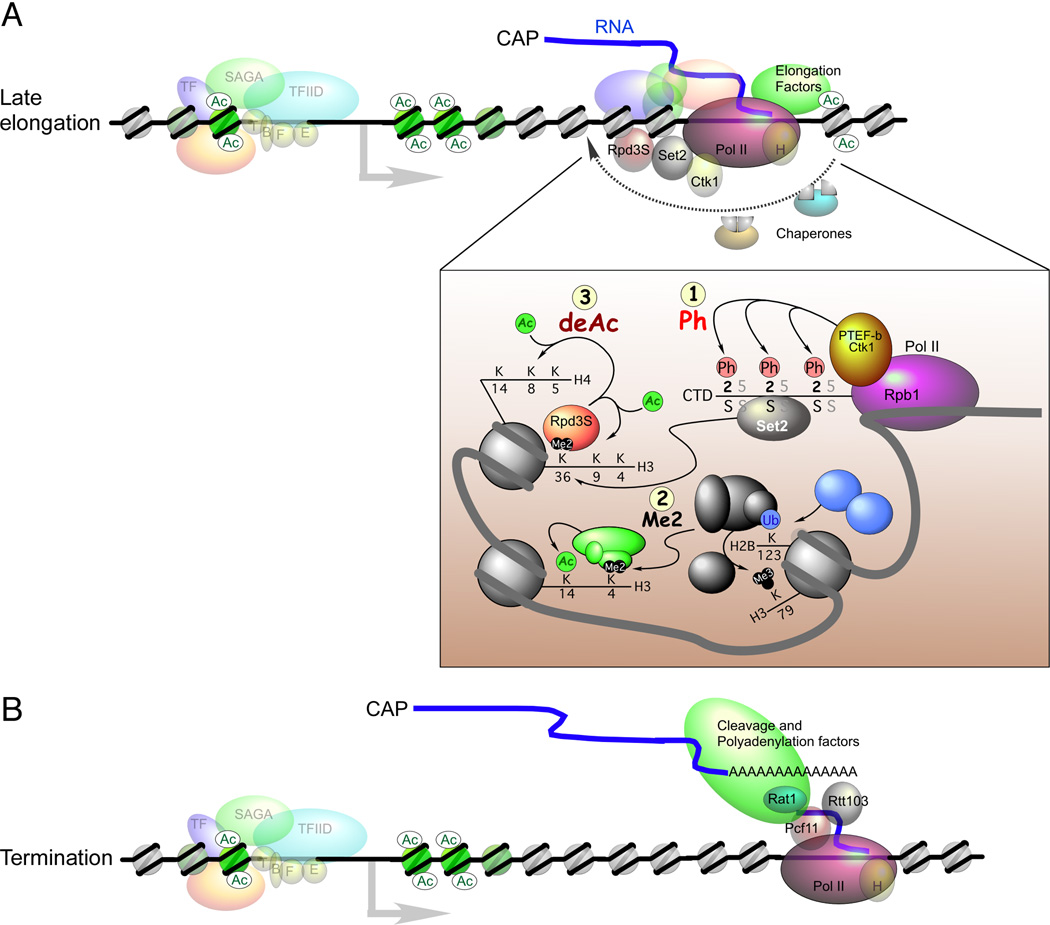
Figure 8. Models for late elongation and transcription termination.
(A) During late stages of the transcription cycle the phosphorylation pattern on the Pol II CTD changes from serine-5 to serine-2, with the latter being recognized by a different set of proteins. The histone modifications that are most prominent during late elongation are highlighted in the inset and numbered 1–3. Also shown are the continued modifications of H3 that occur throughout the gene.
(B) Termination of Pol II transcription is accompanied by cleavage and polyadenylation factors that bind to the serine-2 phosphorylated form of the Pol II CTD. See text for details.
Isomerization of prolines 30 and 38 in the H3 tail by Fpr4 (FKBP Proline Rotamase 4) negatively regulates Set2 methylation at neighboring K36 (Nelson et al., 2006), suggesting that the secondary structure of the H3 tail provides an additional dimension of regulation to transcription, as was seen in proline isomerization of the Pol II CTD. The RPD3S (RPD3-Small) histone deacetylase complex then binds to H3K36me2 (Figure 8A) through combinatorial domain recognition (Li et al., 2007b; Li et al., 2009; Rao et al., 2005). RPD3S then deacetylates nucleosomes which returns the nucleosomes back to their transcriptionally impervious state, and thus suppressing cryptic transcription that might initiate at inappropriate sites (Carrozza et al., 2005; Li et al., 2007c).
Histone modification crosstalk may create positive feedback loops. For example, H3K4me3 and H3K14ac may reinforce each other. In this loop, SAGA binds to H3K4me3 to acetylate H3K14, and H3K14ac promotes H3K4me3 (Figure 7A and B). The SAGA complex is functionally similar to TFIID in that both have TAFs and both deliver TBP to promoters. However, SAGA is clearly involved in other capacities in transcription, in that it is associated with an acetyltransferase (Gcn5, although higher eukaryotic TAF1 is an acetyltransferase), a deubiquitinase (Ubp8), and an ATP-dependent chromatin remodeler (Chd1).
Despite such intricate crosstalk of histone modifications, neither Rad6/Bre1, Set1, Set2, Dot1, nor individual histone tails are essential for viability in budding yeast (Giaever et al., 2002; Kayne et al., 1988; Megee et al., 1990; Morgan et al., 1991; Nakanishi et al., 2008). This begs the question as to why these marks exist. One plausible explanation might be that, while the marks are not essential for regulated transcription, they impart robustness and diminish stochasticity in expression (e.g., perhaps by rapidly restoring the canonical nucleosome architecture once Pol II has passed) such that responses to activating signals can be coordinated in a population of cells. Rapid and well-timed execution of genetic programs is particularly evident in embryonic development where precisely coordinated cell division and expression is critical. The maintenance of histone modification states is particularly critical to stem cell differentiation (reviewed by Bibikova et al., 2008; Lunyak and Rosenfeld, 2008).
Many questions remain regarding the purpose and interdependencies of readers, writers, and erasers of the chromatin landscape. For example, what role do the mono-, di- and tri-methylated states serve? In other words, do mono- and di- simply represent unfinished modifications or are they functionally instructive? The deubuitination activity of Ubp8 is housed in the SAGA complex, which also contains a histone acetyltransferase in Gcn5 and an ATP-dependent remodeling activity in Chd1. How then does SAGA parse these disparate activities in the same complex? Since several chromatin remodelers contain histone-binding modules that recognize acetylated and methylated lysines, what specific residues or combination of residues are recognized by these chromodomains and bromodomains?
Regulation of transcription termination
As Pol II transcription proceeds toward the 3′-end of genes serine-2 phosphorylation of the CTD predominates over serine-5 phosphorylation, thereby enhancing the recruitment (or retention) of the 3′-end RNA processing machinery (Ahn et al., 2004), such as Pcf11 (Protein 1 of Cleavage and polyadenylation Factor I 11), Spt6, Rtt103, and Cft1 (Cleavage Factor Two 1) (Figure 8B) (Dichtl et al., 2002b; Kim et al., 2004; Licatalosi et al., 2002; Yoh et al., 2007). Binding of termination factors may also be dependent on the proline conformations of the CTD, since two temperature sensitive Ess1 mutants are defective in pre-mRNA 3′-end formation (Hani et al., 1999).
Pcf11 is a component of the cleavage and polyadenylation factor and is involved in transcription termination. Biochemical studies in Drosophila demonstrate that Pcf11 terminates transcription by dismantling elongation complexes through mediating interactions between the CTD and nascent RNA (Zhang and Gilmour, 2006). Rtt103 specifically associates with phosphorylated serine-2 of the CTD, and is involved in transcription termination through interactions with the Rat1 (Ribonucleic Acid Trafficking 1) exonuclease, which degrades RNA downstream of the mRNA cleavage site (Kim et al., 2004).
It is now evident that all stages of the transcription cycle from initiation, to pausing, to elongation, and finally termination, are regulated by Pol II CTD serine 5 and 2 phosphorylation and interdigitated proline isomerization. These Pol II marks control the coming and going of histone modifying complexes and pausing/termination factors. As a result, Pol II rapidly and efficiently navigates nucleosomal barriers, producing full length accurately initiated and accurately terminated transcripts.
Noncoding transcription can regulate genes
With the increasing sensitivity of RNA-sequencing and array-based technologies to study the levels of RNA transcripts, recent studies have reported detectable levels of transcription from the vast majority of the genome from yeast to human (David et al., 2006; Dutrow et al., 2008; Kapranov et al., 2007; Mortazavi et al., 2008; Nagalakshmi et al., 2008; Sultan et al., 2008; Wilhelm et al., 2008), albeit most genes are transcribed at a relatively low level. The remarkable sensitivity of RNA detection methods also revealed the presence of antisense transcripts from coding regions (being transcribed in the opposite direction of the a “sense” transcript) and cryptic unstable transcripts (CUTs) from intergenic regions in budding and fission yeast (Dutrow et al., 2008; Neil et al., 2009; Xu et al., 2009). Stable antisense transcripts are generally detected in wild type strains, whereas CUTs are rapidly degraded by the exosome and thus only detectable in an exosome mutant (reviewed by Houseley et al., 2006). It has been suggested that as much as 90% of Pol II initiation is biological noise (Struhl, 2007), which begs the question: Are these antisense and cryptic transcripts physiologically relevant?
Several recent studies in S. cerevisiae provide clues regarding the functional roles of antisense transcripts and CUTs. For example, the SRG1 CUT generated in the sense direction upstream of the serine biosynthetic gene SER3 regulates its transcription through a transcription-interference mechanism that abrogates utilization of the SER3 promoter elements (Martens et al., 2004). The nucleotide biosynthesis genes, URA2 and IMD2, also appear to use CUTs arising upstream of the promoter to repress transcription (Davis and Ares, 2006; Kopcewicz et al., 2007; Thiebaut et al., 2008). The mechanistic basis for these sense-directed repressive CUTs is now being worked out. Their regulation of amino acid and nucleotide biosynthesis genes provides an opportunity for these end-product metabolites to provide feedback regulation on their respective CUTs. For example, serine binds Cha4 (Catabolism of Hydroxy Amino acids 4) which then binds to and activates the SRG1 promoter (Martens et al., 2005). At IMD2, high cellular guanine nucleotide levels favor Pol II initiation at upstream CUTs, which have G at their +1 transcription start sites instead of the normal A (Kuehner and Brow, 2008). Start site selection tends to be highly nucleotide concentration dependent.
GAL10 provides an example of repression via antisense transcription. The Reb1 activator appears to create an NFR near the 3′-end of the GAL10 coding region to facilitate production of antisense transcription under GAL10-repressive conditions (glucose present, and galactose absent), which alters the chromatin across the GAL10 locus (Houseley et al., 2008). Specifically, GAL10 antisense transcription leads to recruitment of the Set2 methyltransferase and the RPD3S histone deacetylase complex presumably via H3K36me2, which then maintains the chromatin in a transcriptionally impervious deacetylated state.
A somewhat different mechanism for a repressive antisense RNA occurs at PHO84, which codes for a phosphate uptake transporter. Under PHO84-repressive conditions (phosphate present), deletion or down-regulation of the nuclear exosome leads to stabilization of a PHO84 antisense transcript, which by some unknown mechanism promotes recruitment of the HDA1 (Histone DeAcetylase 1) complex to create a deacetylated state (Camblong et al., 2007). These findings suggest that the production of noncoding RNAs can repress transcription of nearby genes through altering the chromatin environment with histone deacetylases.
Antisense transcripts can also positively regulate transcription. For example, abolishing production of a transcript antisense to the PHO5 promoter actually delays chromatin remodeling of promoter nucleosomes and recruitment of Pol II to the promoter (Uhler et al., 2007). The authors propose that the act of PHO5 antisense transcription enhances the chromatin plasticity, which positively regulates PHO5 transcription. This notion would seem to be at odds with the mechanisms of antisense regulation of PHO84 and GAL10.
Although the genome-wide distribution of antisense transcripts has been described (Perocchi et al., 2007), a comprehensive functional study of CUTs is lacking. Understanding the role of more antisense transcripts and CUTs will provide additional insight into how genes are regulated using disparate and seemingly conflicting strategies to fine tune gene activity.
Is the transcription cycle basically the same at all mRNA genes?
One can understand and appreciate that each of the thousands of genes in a genome make unique contributions to cellular physiology, and thus should be uniquely regulated. However, this does not seem to be entirely the case. For certain, virtually every mRNA gene is unique to some degree in its promoter, coding, and termination regions. Although sequence-specific transcription factors provide combinatorial diversity in gene regulation, the mechanistic aspects of the regulated transcription cycle (described in this review) seem unlikely to be combinatorially controlled, although no less intricate. Perhaps the mechanistic constancy of the transcription cycle may be metaphorically related to the automobile engine. That is, engines from different manufactures pretty much run the same way. Thus, two different transcription complexes may contain protein variants that elicit each mechanistic step. But fundamentally, the transcription machinery at all mRNA genes in eukaryotes runs basically the same way. Thus, most genes have the same basic chromatin organization, and once activators are activated, they direct chromatin remodeling events, PIC assembly, and early events in transcription initiation and elongation in basically the same way.
Yet it is the intricacies of eukaryotic gene regulation that are so remarkable. Why are there so many mechanistic steps? Why can’t an activated activator bind to its target promoter and simply recruit an RNA polymerase juggernaut? Why reposition nucleosomes to cover up certain transcription factor binding sites? Why are more than 50 proteins needed for PIC assembly, when in principle Pol II should have unfettered access to nucleosome-free promoter regions? Why are there so many histone modifications, and why does the Pol II CTD need to coordinate networks of modifications that occur during transcription? Some of these questions have been debated in the literature, but most of these issues will likely remain enigmatic until the mechanistic details of eukaryotic transcriptional regulation are more fully worked out.
Archaebacteria successfully employ a simplified stripped-down version of the eukaryotic transcription machinery, and so the eukaryotic embellishment is not a fundamental constant of gene regulation. It may be widely accepted that the combination of genome duplication and evolution into diverse and/or multicellular environments that are characteristic of eukaryotes has driven the enormous number and combinatorial diversity of sequence-specific transcription factors. However, the basis of the mechanistic complexity of a regulated transcription cycle is not obvious (to us). For transcription, evolution does not have the luxury of de novo design and is forced to build on pre-existing states, which over the evolutionary history of eukaryotes is wrought with enormous changes in selective pressure caused by punctuated environmental changes. Thus, in this case, complexity might beget more complexity, leaving us with mechanisms (as illustrated here) that appear to be a Rube Goldberg design (performing a simple task using an overly complex apparatus).
Acknowledgement
This work was supported by NIH/NIEHS grant ES013768.
References
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Almer A, Horz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. Embo J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- Auble DT, Hansen KE, Mueller CG, Lane WS, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X. INO80 subfamily of chromatin remodeling complexes. Mutat Res. 2007a;618:18–29. doi: 10.1016/j.mrfmmm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Shen X. SnapShot: chromatin remodeling complexes. Cell. 2007b;129:632. doi: 10.1016/j.cell.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Baskaran R, Dahmus ME, Wang JY. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci U S A. 1993;90:11167–11171. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. Gene regulation. Local or global? Nature. 2000;408:412–413. 415. doi: 10.1038/35044160. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2:123–134. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Biddick R, Young ET. Yeast mediator and its role in transcriptional regulation. C R Biol. 2005;328:773–782. doi: 10.1016/j.crvi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Biswas D, Dutta-Biswas R, Stillman DJ. Chd1 and yFACT act in opposition in regulating transcription. Mol Cell Biol. 2007;27:6279–6287. doi: 10.1128/MCB.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet. 2006;38:1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chung WH, Craighead JL, Chang WH, Ezeokonkwo C, Bareket-Samish A, Kornberg RD, Asturias FJ. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol Cell. 2003;12:1003–1013. doi: 10.1016/s1097-2765(03)00387-3. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ Is Enriched at Polycomb Complex Target Genes in ES Cells and Is Necessary for Lineage Commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M, Simon I, Moy TI, Wilson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR, Silver PA. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol Cell. 2002;9:563–573. doi: 10.1016/s1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol Cell. 2002a;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Sadowski M, Hubner W, Weiser S, Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. Embo J. 2002b;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbert R, Birkmann A, Schuller HJ. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol Microbiol. 1999;32:741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr Opin Genet Dev. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol Cell Biol. 2007;27:297–311. doi: 10.1128/MCB.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF. From Bacteria to Humans, Chromatin to Elongation, and Activation to Repression: The Expanding Roles of Noncoding RNAs in Regulating Transcription. Crit Rev Biochem Mol Biol. 2008:1. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR. Eukaryotic transcription activation: right on target. Mol Cell. 2005;18:399–402. doi: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant Histone H2A.Z Is Globally Localized to the Promoters of Inactive Yeast Genes and Regulates Nucleosome Positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Hani J, Schelbert B, Bernhardt A, Domdey H, Fischer G, Wiebauer K, Rahfeld JU. Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3'-end formation of a pre-mRNA in Saccharomyces cerevisiae. J Biol Chem. 1999;274:108–116. doi: 10.1074/jbc.274.1.108. [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Hausmann S, Koiwa H, Krishnamurthy S, Hampsey M, Shuman S. Different strategies for carboxyl-terminal domain (CTD) recognition by serine 5-specific CTD phosphatases. J Biol Chem. 2005;280:37681–37688. doi: 10.1074/jbc.M505292200. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish DR, Burgoyne LA. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973;52:504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA Modulates Histone Modification and mRNA Induction in the Yeast GAL Gene Cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- Inostroza JA, Mermelstein FH, Ha I, Lane WS, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. Embo J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 2006;16:1505–1516. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kayne PS, Kim UJ, Han M, Mullen JR, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim TH, Ren B. Genome-wide analysis of protein-DNA interactions. Annu Rev Genomics Hum Genet. 2006;7:81–102. doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopcewicz KA, O'Rourke TW, Reines D. Metabolic regulation of IMD2 transcription and an unusual DNA element that generates short transcripts. Mol Cell Biol. 2007;27:2821–2829. doi: 10.1128/MCB.02159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci U S A. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- Levine SS, King IF, Kingston RE. Division of labor in polycomb group repression. Trends Biochem Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007a;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007b;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007c;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. Histone H3 lysine 36 di-methylation (H3K36ME2) is sufficient to recruit the Rpd3S histone deacetylase complex and to repress spurious transcription. J Biol Chem. 2009 doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Reese JC. Ssn6-Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J Biol Chem. 2001;276:33788–33797. doi: 10.1074/jbc.M104220200. [DOI] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3' end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. Nucleosome transactions on the promoters of the yeast GAL and PHO genes. J Biol Chem. 1997;272:26795–26798. doi: 10.1074/jbc.272.43.26795. [DOI] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Rosenfeld MG. Epigenetic regulation of stem cell fate. Hum Mol Genet. 2008;17:R28–R36. doi: 10.1093/hmg/ddn149. [DOI] [PubMed] [Google Scholar]
- Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Martens C, Krett B, Laybourn PJ. RNA polymerase II and TBP occupy the repressed CYC1 promoter. Mol Microbiol. 2001;40:1009–1019. doi: 10.1046/j.1365-2958.2001.02445.x. [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Campa C, Politis P, Moreau JL, Kent N, Goodall J, Mellor J, Goding CR. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol Cell. 2004;15:69–81. doi: 10.1016/j.molcel.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Matsui T, Segall J, Weil PA, Roeder RG. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008a;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008b;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- Mencia M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Mittman BA, Smith MM. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldrow TA, Campbell AM, Weil PA, Auble DT. MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol Cell Biol. 1999;19:2835–2845. doi: 10.1128/mcb.19.4.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Zhou H, Winston F, Hinnebusch AG. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Brown CE, Howe L, Workman JL. Transcription activator interactions with multiple SWI/SNF subunits. Mol Cell Biol. 2002;22:1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 2003a;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003b;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPAR{gamma}:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- Nourani A, Utley RT, Allard S, Cote J. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. Embo J. 2004;23:2597–2607. doi: 10.1038/sj.emboj.7600230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Panne D. The enhanceosome. Curr Opin Struct Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ. In vivo requirement of activator-specific binding targets of mediator. Mol Cell Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. Embo J. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Perez-Ortin JE, Estruch F, Matallana E, Franco L. Fine analysis of the chromatin structure of the yeast SUC2 gene and of its changes upon derepression. Comparison between the chromosomal and plasmid-inserted genes. Nucleic Acids Res. 1987;15:6937–6956. doi: 10.1093/nar/15.17.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Xu Z, Clauder-Munster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Price DH. Poised polymerases: on your mark…get set…go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Pugh BF. Control of gene expression through regulation of the TATA-binding protein. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- Pugh BF, Gilmour DS. Genome-wide analysis of protein-DNA interactions in living cells. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-4-reviews1013. REVIEWS1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra A, Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol Cell Biol. 1999;19:7944–7950. doi: 10.1128/mcb.19.12.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese JC, Apone L, Walker SS, Griffin LA, Green MR. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Sayre MH, Tschochner H, Kornberg RD. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J Biol Chem. 1992;267:23376–23382. [PubMed] [Google Scholar]
- Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Gross DS. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell. 2001;105:403–414. doi: 10.1016/s0092-8674(01)00329-4. [DOI] [PubMed] [Google Scholar]
- Sermwittayawong D, Tan S. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. Embo J. 2006;25:3791–3800. doi: 10.1038/sj.emboj.7601265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse RO, Karpova TS, Mueller F, Dasgupta A, McNally JG, Auble DT. Regulation of TATA-binding protein dynamics in living yeast cells. Proc Natl Acad Sci U S A. 2008a;105:13304–13308. doi: 10.1073/pnas.0801901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse RO, Wells MN, Auble DT. TATA-binding protein variants that bypass the requirement for Mot1 In Vivo. J Biol Chem. 2008b doi: 10.1074/jbc.M808951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Struhl K, Kadosh D, Keaveney M, Kuras L, Moqtaderi Z. Activation and repression mechanisms in yeast. Cold Spring Harb Symp Quant Biol. 1998;63:413–421. doi: 10.1101/sqb.1998.63.413. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tan S, Hunziker Y, Sargent DF, Richmond TJ. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–151. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Colin J, Neil H, Jacquier A, Seraphin B, Lacroute F, Libri D. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell. 2008;31:671–682. doi: 10.1016/j.molcel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Toth J, Biggin MD. The specificity of protein-DNA crosslinking by formaldehyde: in vitro and in drosophila embryos. Nucleic Acids Res. 2000;28:e4. doi: 10.1093/nar/28.2.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. Embo J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D. Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9A/KMT1C and the jumonji domain containing JMJD2/KDM4 proteins. J Biol Chem. 2009 doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat Rev Mol Cell Biol. 2002;3:422–429. doi: 10.1038/nrm828. [DOI] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci U S A. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh F. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009 doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Carey M, Gralla JD. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Wilcox CB, Rossettini A, Hanes SD. Genetic interactions with C-terminal domain (CTD) kinases and the CTD of RNA Pol II suggest a role for ESS1 in transcription initiation and elongation in Saccharomyces cerevisiae. Genetics. 2004;167:93–105. doi: 10.1534/genetics.167.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M, Lin PS, Dahmus ME, Gill GN. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J Biol Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K, Verrijzer CP, Tjian R. An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription. Proc Natl Acad Sci U S A. 1998;95:6722–6727. doi: 10.1073/pnas.95.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc Natl Acad Sci U S A. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Corden JL. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2290–2296. [PubMed] [Google Scholar]
- Zhang L, Fletcher AG, Cheung V, Winston F, Stargell LA. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol. 2008;28:1393–1403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Reese JC. Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. Embo J. 2004;23:2246–2257. doi: 10.1038/sj.emboj.7600227. [DOI] [PMC free article] [PubMed] [Google Scholar]