Abstract
Duplication of the simian virus 40 (SV40) genome is the best understood eukaryotic DNA replication process to date. Like most prokaryotic genomes, the SV40 genome is a circular duplex DNA organized in a single replicon. This small viral genome, its association with host histones in nucleosomes, and its dependence on the host cell milieu for replication factors and precursors led to its adoption as a simple and powerful model. The steps in replication, the viral initiator, the host proteins, and their mechanisms of action were initially defined using a cell-free SV40 replication reaction. Although our understanding of the vastly more complex host replication fork is advancing, no eukaryotic replisome has yet been reconstituted and the SV40 paradigm remains a point of reference. This article reviews some of the milestones in the development of this paradigm and speculates on its potential utility to address unsolved questions in eukaryotic genome maintenance.
Keywords: Genome structure, DNA replication origin, T antigen, Helicase, Double hexamer, Replication fork, Hand-off, Polymerase switch, DNA damage signaling
Introduction
The study of bacteriophage and viruses over the past 50 years laid the foundations of modern molecular biology. The physicists, chemists, biologists, and physicians pioneered this frontier with the hope that the relative simplicity of these agents might allow them to serve as tools to understand their vastly more complex infected host cells. The discovery of simple DNA viruses that propagated in mammalian cell nuclei and caused tumors in experimental animals led the way to an explosion of eukaryotic molecular biology and its applications to understanding, treating, and preventing human disease. For pioneering studies of the DNA tumor viruses polyomavirus and SV40 in the 1960’s, Renato Dulbecco was awarded the 1975 Nobel Prize in Physiology or Medicine. Equally importantly, the many young scientists who trained in his laboratory were inspired to pursue and expand on this fruitful approach in ever more exciting new directions. The development of the field and collegial interactions among members of the DNA tumor virus community were greatly fostered by annual meetings sponsored by Cold Spring Harbor Laboratory and Imperial Cancer Research Fund, as well as by review volumes edited by John Tooze beginning in 1973 (Tooze, 1973).
This article reviews some of the fundamental lessons on genome structure, DNA replication, and genome maintenance that these deceptively simple viruses have revealed over the past 4 decades. The utility of these viral paradigms in guiding the investigation of mammalian DNA replication is considered. The article concludes with reflections on how the rapidly growing understanding of host genome maintenance is leading to a re-consideration of how these viruses exploit their host cells.
The SV40 minichromosome: genetic and physical maps linked through DNA sequence
The SV40 genome is a covalently closed circular duplex DNA molecule of 3.6 × 106 Da (Crawford and Black, 1964; Dulbecco and Vogt, 1963; Weil and Vinograd, 1963). Biophysical characterization of superhelical SV40 and polyomavirus DNA provided the first insight into the initially puzzling ability of supercoiled DNA to renature rapidly after exposure to alkali (Vinograd et al., 1965; Weil, 1963; Weil and Vinograd, 1963), its limited uptake of intercalating dyes, e.g. ethidium bromide, and other properties typical of supercoiled DNA. The SV40 genome exists in the virus particle and in infected cells as a mini-chromosome packaged with host cell histones into nucleosomes that closely resemble those of the host chromatin (Bonner et al., 1968; Germond et al., 1975; Griffith, 1975; White and Eason, 1971) (Fig. 1). Purification of SV40 DNA from minichromosomes reveals its negatively supercoiled topology. As we now know, this topology endows chromatin with the capacity to readily denature for initiation of DNA replication or transcription.
Figure 1.
Electron micrograph of an SV40 minichromosome isolated from productively infected cells. (Scale bar 100 nm). (Reprinted from (Griffith, 1975) with permission from AAAS.)
Genetic studies of replication in prokaryotes had led to a potentially general model for control of replication: the replicon model of Jacob, Brenner, and Cuzin (Jacob and Brenner, 1963). The model postulated a cis-acting element, the replicator, recognized by a trans-acting factor, the initiator. This interaction would lead to locally denatured duplex DNA in or near the replicator element and initiation of replication. Each replicator with its initiator would thus govern the replication of the flanking regions of DNA, the replicon. If this model were general, one might expect eukaryotic DNA to be organized into replicons in a similar manner. If SV40 DNA represents a eukaryotic replicon, one would predict a genetically definable viral replicator element and an initiator that recognized it.
In the mind of Daniel Nathans at Johns Hopkins University, the appeal of SV40 as an object for genetic analysis converged with the discovery of the first sequence-specific restriction endonucleases by his colleague Hamilton Smith (Fig. 2). Nathans and colleagues generated the first restriction cleavage map of SV40 DNA, by determining the physical order of the Hind II/III and Hpa I/II sites around the SV40 DNA genome (Danna and Nathans, 1971; Danna, Sack, and Nathans, 1973). A unique restriction cleavage site by Eco RI (Morrow and Berg, 1972) provided a point of reference in the viral genome. By 1972, Danna and Nathans had combined a radiolabeled thymidine pulse-chase approach with their restriction map to determine the physical start site for SV40 DNA replication, the origin, and show that replication proceeded bidirectionally to terminate on the opposite side of the DNA molecule (Danna and Nathans, 1972; Nathans and Danna, 1972). [For a fascinating overview of these discoveries, see (Brownlee, 2005; Roberts, 2005)] This physical map greatly facilitated determination of the 5243 bp sequence of SV40 DNA, the first eukaryotic genome to be completely sequenced (Fiers et al., 1978; Reddy et al., 1978). Moreover, the map and the sequence enabled the classical mutational analysis of the SV40 genome (Chou and Martin, 1974; Tegtmeyer, 1972; Tegtmeyer and Ozer, 1971) to be correlated with nucleotide sequence changes that affected viral DNA replication (temperature-sensitive complementation group A (tsA)), cell transformation (tsA), and virion production (tsB, C, BC, D) [for a personal account, see (Nathans, 1978)].
Figure 2.
Daniel Nathans (left) and Hamilton Smith in the laboratory at Johns Hopkins University. (Reprinted from (Roberts, 2005) with permission from Copyright 2005 National Academy of Sciences, U.S.A.)
With the viral DNA sequence in hand and new restriction endonucleases rapidly emerging in several laboratories, the Nathans lab moved quickly to test the function of the SV40 origin of DNA replication by mutational analysis. They devised site-directed mutagenesis protocols for deletions and base substitutions followed by selection for resistance to cleavage by Bgl I, which has a single recognition site in SV40 DNA at the origin (DiMaio and Nathans, 1980; DiMaio and Nathans, 1982; Shortle and Nathans, 1978). These mutations were mapped by DNA sequencing and shown to render SV40 replication defective when the genome was introduced into host cells, satisfying one criterion for a replicator element. To examine the relationship between the putative replicator element and the tsA gene that was also involved in replication, the Nathans lab carried out a mutational screen for second site revertants of the replication-defective mutant origins. These pseudorevertant mutations were then mapped and shown to reside at positions outside of the origin region and to alter the coding sequence of the tsA gene (Margolskee and Nathans, 1984; Shortle, Margolskee, and Nathans, 1979). The A gene encodes the SV40 large tumor (T) antigen (Tag), a multifunctional protein whose structure and roles in viral DNA replication are reviewed below. Thus, the origin element interacted genetically with a viral gene that regulated the rate of viral DNA replication, providing strong evidence for a replicon model in controlling replication of SV40 DNA. Biochemical investigation of Tag promptly confirmed the interaction, paving the way for new experiments to elucidate the mechanism of SV40 DNA replication.
Further dissection of the viral replicator in multiple laboratories revealed a 64 bp core composed of three elements. A central element contains a palindromic array of four GAGGC pentanucleotides that, as we now know, serve as binding sites for Tag. The binding sites are flanked by an easily denatured imperfect palindrome (EP) on one side and by an AT-rich sequence on the other side. The two flanking elements undergo local distortion or melting during initiation of replication (Borowiec et al., 1990). The early and late promoter elements flank the viral core origin and stimulate its activity in infected cells, as does the viral enhancer element. These auxiliary elements may stimulate initiation of replication from the viral core origin at multiple levels, some of which may reflect the close relationship between origins of replication and transcription. The first level may be by modulating the structure of the core origin DNA to facilitate distortion by Tag, e.g. through intrinsically bent AT-rich DNA sequences, or local modulation of supercoiling. These physical properties of origin DNA are also found in eukaryotic chromosomal origins of replication and are thought to be important for binding of the origin recognition complex ORC (Remus, Beall, and Botchan, 2004). Proteins bound to the auxiliary elements may also facilitate Tag assembly on the core origin DNA, Tag remodeling into an active helicase, or recruitment of host replication proteins. For example, chromatin remodeling to generate a nucleosome-free origin region and histone modifications are likely to be important for initiation at the SV40 origin and at chromosomal origins (Saragosti, Moyne, and Yaniv, 1980). Lastly, replication of the viral minichromosome appears to take place in specific subnuclear domains (Ishov and Maul, 1996; Staufenbiel and Deppert, 1983; Tang et al., 2000), but how the minichromosome is targeted to these sites remains poorly defined.
Given the sequence specificity of the SV40 core origin and the dependence of the virus on host cell proteins for DNA replication, it was tempting to imagine that chromosomal origins of replication might also be composed of modules with defined sequences that could be recognized by initiator proteins. Although origins in budding yeast fulfill this expectation to some extent, no common consensus sequence has been found in either the genetically defined replicators or the start sites of replication in higher eukaryotic genomes (Aladjem, 2007; Aladjem and Fanning, 2004; Bell, 2002).
SV40 large T antigen (Tag): the initiator protein in viral DNA replication
Tag was first detected as a 90–100 kDa polypeptide from infected cell extracts that reacted with the serum of rodents bearing tumors induced by SV40 injection (Rundell et al., 1977). Initially it was uncertain whether Tag was in fact encoded by the A gene in the SV40 genome since small deletions in that gene failed to reduce the apparent mass of the immunoreactive protein. This conundrum was resolved through the combined efforts of the Tegtmeyer, Crawford and Berg labs (Fig. 3) (Crawford et al., 1978), providing the first hint that Tag was expressed from one of the three alternatively spliced early SV40 transcripts (see Yaniv article in this issue).
Figure 3.
Peter Tegtmeyer (left) in discussion at a DNA Tumor Virus meeting, University of Wisconsin-Madison (Courtesy of K. Rundell).
Taking advantage of the high level expression of a Tag-related adenovirus-SV40 hybrid protein D2, Robert Tjian succeeded in a classical purification of the first native, biochemically active form of Tag (Tjian, 1978). This key achievement was the first step toward defining the mechanism of SV40 DNA replication. Tjian showed that purified D2 bound to DNA and was capable of specifically protecting SV40 origin DNA sequences against nuclease digestion, confirming the genetic interactions between the SV40 origin and the A gene encoding Tag. Moreover, D2 bound sequentially to several elements of the viral origin DNA in a manner suggesting possible multimerization of the protein on the DNA to protect up to 120 bp. Subsequent work in multiple laboratories led to definition of the pentanucleotide GAGGC as the fundamental recognition motif specifically bound by Tag, either as a tandem repeat separated by 7 bp of intrinsically bent DNA (site I) or, in the core origin, as a 27 bp palindromic arrangement of 4 pentanucleotides separated by 1 bp (site II) (reviewed by (Borowiec et al., 1990; Challberg and Kelly, 1989; Fanning and Knippers, 1992; Stillman, 1989)).
Purified D2 and Tag were soon shown to display a second biochemical activity: the ability to bind and hydrolyze Mg-ATP/dATP, an activity stimulated by single-stranded DNA (Cole et al., 1986; Giacherio and Hager, 1979). Although this behavior suggested that Tag might have DNA helicase activity, it could not be convincingly demonstrated. Using Tag purified from infected cells by immunoaffinity chromatography on monoclonal antibody resins (Deppert, Gurney, and Harrison, 1981; Dixon and Nathans, 1985; Harlow et al., 1981; Simanis and Lane, 1985) and a clever new helicase assay (Hubscher and Stalder, 1985), the Knippers lab showed that highly purified Tag could unwind partial duplex DNA with 3′ to 5′ polarity in an ATP-hydrolysis dependent manner (Fig. 4) (Stahl, Droge, and Knippers, 1986). Moreover, monoclonal antibodies against specific regions of Tag inhibited helicase activity and mutations in Tag that reduced ATPase activity also reduced helicase activity (Stahl, Droge, and Knippers, 1986). The helicase activity of Tag was quickly confirmed in several other laboratories (reviewed in (Fanning and Knippers, 1992)).
Figure 4.

Rolf Knippers in his laboratory at the University of Konstanz, Germany, in the 1980’s. (Courtesy of M. Baack).
Despite this important step in understanding the role of Tag in viral DNA replication, it remained unclear how the helicase activity of Tag could unwind duplex DNA from the origin. The Hurwitz laboratory, collaborating with several others, discovered that Tag assembles into a multimer on the viral origin in an ATP-binding dependent manner to form a bilobed double hexameric structure that distorts the duplex DNA locally (Borowiec et al., 1990; Dodson et al., 1987; Mastrangelo et al., 1989). In the presence of Mg-ATP, a single-stranded DNA binding protein, and topoisomerase I, Tag generated a theta-like structure with single-stranded bubbles of varying sizes with the origin always at the center of the bubble (Dean et al., 1987a; Dean et al., 1987b; Dodson et al., 1987). A large protein complex likely to be Tag hexamer was often observed at the junctions of the single-stranded bubble with duplex DNA, suggesting that two diverging Tag hexamers unwound the duplex at a similar rate. A few years later, under different experimental conditions, active unwinding complexes of double hexamer were visualized with two loops of ssDNA emanating from the double hexamer (Fig. 5). These images suggested a more sophisticated bidirectional unwinding mechanism in which duplex parental DNA is reeled into the double hexamer, coordinately from both sides, and the unwound template is spooled out for replication (Wessel, Schweizer, and Stahl, 1992). This type of unwinding intermediate, not observed in initiation of prokaryotic replication by 5′ to 3′ helicases (Fang, Davey, and O’Donnell, 1999), implied some kind of functional contacts between Tag hexamers. Biochemical, genetic, and structural data (Meinke, Bullock, and Bohm, 2006; Moarefi et al., 1993; Smelkova and Borowiec, 1998; Valle et al., 2006; Valle et al., 2000; Virshup, Russo, and Kelly, 1992; Weisshart et al., 1999) provide evidence for functional interactions of Tag residues 102–259 in one hexamer with the corresponding residues in the other hexamer, strongly supporting this model of bidirectional unwinding as physiologically relevant (reviewed in (Bullock, 1997; Fanning, 1994; Simmons, 2000). This model suggests that both replication forks may assemble, at least initially, on the Tag double hexamer and that progression of the two forks may be coupled (Falaschi, 2000). However, this possibility has not been addressed experimentally.
Figure 5.
Active unwinding from the SV40 origin by a Tag double hexamer. Tag was incubated under unwinding conditions with a duplex plasmid DNA fragment containing the SV40 origin 1.1 or 1.5 kb from the ends. The reaction was terminated by glutaraldehyde, the sample was purified, spread, negatively stained, and visualized by electron microscopy. Two single-stranded loops bound to single-stranded DNA binding protein. emanate from a Tag double hexamer (see inset). (Scale bar 100 nm) The double hexamer and the loops dissociated into a theta-like structure after treatment with EDTA (not shown). (Reprinted from (Wessel, Schweizer, and Stahl, 1992) with permission from Copyright 1992 American Society for Microbiology.)
In parallel with studies of Tag helicase activity, a trio of laboratories embarked on a major quest to establish a cell-free SV40 DNA system that could be used to identify the host proteins necessary to replicate viral DNA. Early work with replicating SV40 nucleoprotein complexes isolated from infected cells had already been shown to complete replication in cell-free extracts (DePamphilis, Beard, and Berg, 1975; Edenberg, Waqar, and Huberman, 1976) and DNA polymerase alpha-primase had been identified as a key activity (Otto and Fanning, 1978; Waqar, Evans, and Huberman, 1978). Joachim Li and Thomas Kelly (Li and Kelly, 1984)were the first to succeed in using purified SV40 origin DNA and extracts from primate cells supplemented with immunopurified Tag to replicate a DNA template from the viral origin, followed quickly by independent studies in the Stillman and Hurwitz laboratories (Li and Kelly, 1984; Stillman and Gluzman, 1985; Wobbe et al., 1985) (Fig. 6, 7). Countless hours of painstaking work in cold rooms in these and other labs resulted in the purification of ten human proteins that, together with Tag, are sufficient to reconstitute the replication of duplex plasmid DNA in an origin-dependent reaction (Waga and Stillman, 1994; Waga and Stillman, 1998).
Figure 6.
Thomas J. Kelly (left) and Bruce Stillman (right) at a reception during the 1994 Cold Spring Harbor Symposium (Courtesy of Cold Spring Harbor Archives).
Figure 7.
A minimal set of replication proteins at a eukaryotic fork. MCM helicase substitutes here for Tag; the PCNA clamp loader RFC and topoisomerases are not shown. (Reprinted from (Garg and Burgers, 2005) with permission from Copyright 2005 Taylor & Francis.)
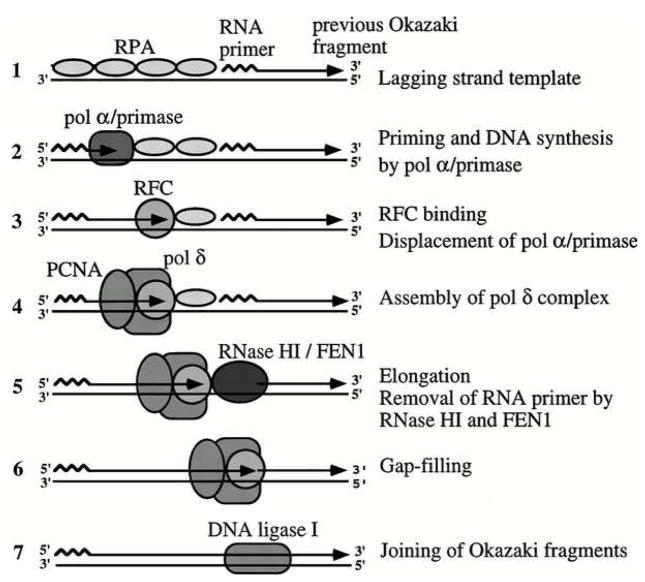
To elucidate the basic mechanisms of replication, sub-reactions for individual steps were reconstituted with purified proteins. Three stages of the replication process have been reconstituted and studied in detail: initiation, elongation, and Okazaki fragment maturation. Four proteins (Tag, RPA, DNA polymerase alpha-primase, and topoisomerase I) are sufficient to reconstitute initiation of replication at the viral origin (Matsumoto, Eki, and Hurwitz, 1990) reviewed by (Borowiec et al., 1990; Bullock, 1997). After primer synthesis, the clamp-loader replication factor C (RFC) orchestrates a switch from DNA polymerase alpha-primase to the more processive DNA polymerase delta (Tsurimoto, Melendy, and Stillman, 1990; Tsurimoto and Stillman, 1991; Weinberg et al., 1990) (Fig. 8). RPA contacts with RFC appear to play a role in displacing DNA polymerase alpha-primase from the template (Yuzhakov et al., 1999). RFC bound to ATP binds the sliding clamp PCNA, cracks open the ring, and RFC contact with primer-template triggers ATP hydrolysis, releasing a closed PCNA complex loaded at the primer-terminus and able to bind and position polymerase delta for primer extension (Indiani and O’Donnell, 2006).
Figure 8.
Linking the polymerase switch to Okazaki fragment processing. The mechanism of elongation of a primed DNA template on the leading strand or for each Okazaki fragment on the lagging strand was studied in reactions reconstituted with purified proteins (steps 1–4). The mechanism of Okazaki fragment maturation was also elucidated in reconstituted reactions (steps 5–7). (Reprinted from (Waga and Stillman, 1998) with permission from Annual Reviews.) (For a current view of Okazaki fragment processing, see (Rossi et al., 2008))
Despite the simplicity of the SV40 replication fork and several key differences relative to host replication forks and their intricate regulatory wiring, the SV40 fork continues to serve as a useful paradigm for host forks (Fig. 7, 8), where new challenges await. Structures of many of these host replication proteins and their domains have now been determined by X-ray crystallography (for examples, see (Bowman, O’Donnell, and Kuriyan, 2004; Fanning, Klimovich, and Nager, 2006; Garg and Burgers, 2005; Indiani and O’Donnell, 2006; Pascal et al., 2004), opening the possibility of developing an atomic level understanding of replication fork operation in eukaryotes. Definition of how these exchanges of proteins and coupling of leading and lagging strand replication are accomplished will require more complete structural information on DNA polymerases, definition of protein interactions required for hand-off reactions, and single molecule studies.
Seeing is believing: structures of SV40 Tag
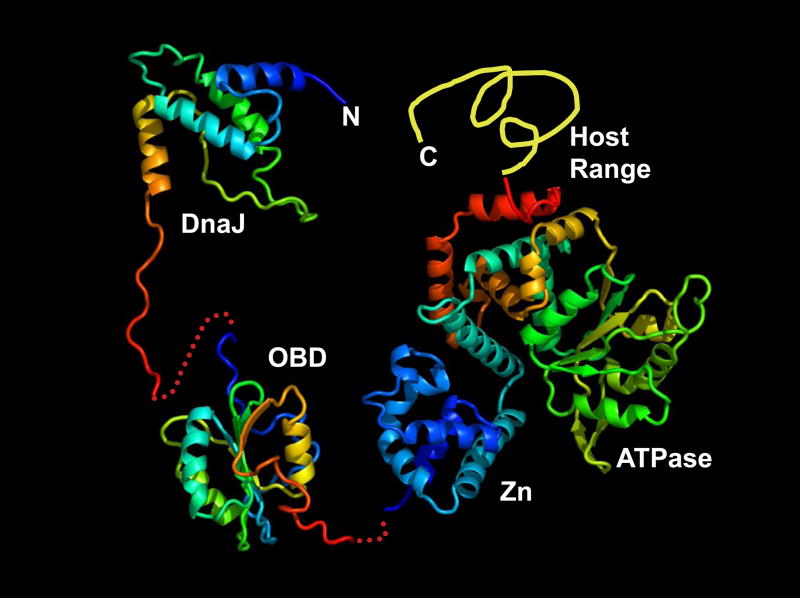
Mapping of functional domains of Tag by molecular genetics and biochemistry provided the first hints that SV40 Tag is a multi-domain protein with a bewildering number of functions (Fanning, 1992). Mapping of structured domains by limited proteolysis correlated to some extent with the functional domain map (Schwyzer et al., 1980), but an atomic structure of Tag remained elusive until the focus shifted to the protein domains. At last, the origin DNA binding domain (OBD) was determined using NMR in the Bullock and Bachovchin laboratories (Fig. 9) (Luo et al., 1996). These structural studies, together with previously mapped mutations that affected origin DNA binding, provided important insight into the mechanism of origin recognition by Tag and assembly of the double hexamer (Bullock, 1997; Joo et al., 1998). The next domain to come into view was the DnaJ co-chaperone domain (Fig. 9) in complex with a fragment of the retinoblastoma tumor suppressor protein ((Kim, Ahn, and Cho, 2001); reviewed by(Hennessy et al., 2005; Sullivan and Pipas, 2002)). Although the DnaJ function is not required for SV40 replication in cell-free reactions (Weisshart et al., 1996), its functions are essential for DNA replication in productively infected cells (Campbell et al., 1997). In a key advance for understanding the mechanism of initiation of viral DNA replication, the first crystal structure of the Tag helicase domain was determined in Xiaojiang Chen’s laboratory (Fig. 10) (Li et al., 2003). The structured domain (residues 251–627) consists of the zinc-binding and ATPase/AAA+ subdomains, comprising about two-thirds of the protein (Fig. 9). The origin DNA binding domain and the DnaJ domain are tethered to one another and to the helicase domain through flexible linkers (Fig. 9). The structure of the host range domain (residues 628–708) remains unknown (see article by Pipas in this issue). These structured domains correlate very well with the functional domains of Tag deduced by biochemistry and molecular genetics (see (Fanning and Knippers, 1992; Weisshart et al., 2004). Remarkably, the Tag helicase structure is closely related to that of the chromosomal replicative helicase, as evidenced by crystal structures of archaeal MCM helicase domains ((Fletcher et al., 2003; Gomez-Llorente et al., 2005); reviewed by (Sclafani and Holzen, 2007)).
Figure 9.
Modular organization of Tag domains and linkers. Atomic structures of DnaJ (Kim, Ahn, and Cho, 2001), OBD (Luo et al., 1996), and helicase (Zn and ATPase/AAA+) domains (Li et al., 2003) are shown approximately to scale with the intervening peptides as dotted lines. The structure of the host-range domain has not been determined. (Courtesy of X.S. Chen.)
Figure 10.
Crystallographer Xiaojiang S. Chen, University of Southern California.
The Tag structures available so far suggest a modular molecule that likely operates dynamically to coordinate origin DNA binding and local distortion by the double hexamer, and re-organization of the double hexamer into a bidirectional unwinding machine (Bochkareva et al., 2006; Gai et al., 2004; Meinke, Bullock, and Bohm, 2006; Meinke et al., 2007; Valle et al., 2006). Re-examination of the initiation reaction and mapping of protein-protein interactions at the atomic level is beginning to provide insight into the dynamic nature of the initiation process. During initiation of replication, RPA association with Tag-OBD couples origin DNA unwinding to loading of RPA on the emerging template (Jiang et al., 2006). Similarly, Tag-OBD-mediated remodeling of RPA-ssDNA complexes facilitates the ssDNA hand-off to the DNA polymerase alpha-primase positioned on the Tag helicase domain (Arunkumar et al., 2005). Nevertheless, until the relative positions of the Tag domains in the double hexamer and the path of the DNA template through the protein during various functional states of replication can be visualized, our understanding of the SV40 replication process is incomplete. One successful path toward this ultimate goal began by analyzing electron micrographs of Tag double hexamers on origin DNA with image processing algorithms (Valle et al., 2000). Recent advances in cryo-electron microscopy of wild type and mutant Tag on origin DNA, new image classification algorithms, and fitting of new atomic structures of Tag may allow this goal to be reached.
The SV40 replication paradigm: a perspective
Much effort has been aimed at elucidating in detail the operation of the SV40 replisome with the hope that general principles can be discerned that will increase our understanding of mammalian chromosomal replication. The bidirectional “replication factory” organization of the SV40 replisome appears to be a simple example of those that duplicate bacterial and eukaryotic chromosomes (Kitamura, Blow, and Tanaka, 2006; Meister, Taddei, and Gasser, 2006). However, SV40 and host DNA replication mechanisms differ in several major features. SV40 encodes its own DNA helicase, whereas chromosomal replication depends on the Cdc45/Mcm2–7/GINS helicase ((Moyer, Lewis, and Botchan, 2006) and references therein). DNA polymerase epsilon is important for chromosomal replication (Seki et al., 2006; Shikata et al., 2006), but SV40 replication does not utilize it in vivo or in vitro (Pospiech et al., 1999; Zlotkin et al., 1996). SV40 DNA replicates during the S/G2 phase of the cell cycle, but DNA polymerase alpha-primase phosphorylated by cyclin-dependent kinase is unable to support replication of viral DNA in a cell-free reaction ((Ott et al., 2002) and references therein). Importantly, SV40 and polyomavirus infection induces DNA damage signaling that promotes viral DNA replication (Dahl, You, and Benjamin, 2005; Shi et al., 2005; Wu et al., 2004; Zhao et al., 2008), but ordinarily inhibits chromosomal replication and cell cycle transitions (Cimprich and Cortez, 2008; Lavin, 2008). Because of these differences, one of the most critical open questions is whether the SV40 replisome operates as a streamlined mimic of host chromosomal replication or rather, a host DNA repair pathway. The unanticipated role of DNA damage signaling in viral DNA replication raises the question of whether other undiscovered factors and mechanisms might participate in SV40 replication in infected cells, including the Hsc70 interaction with the Tag DnaJ domain, sister cohesion and chromosome segregation, subnuclear positioning of viral genomes, interaction with the ubiquitin-proteasome system, and chromatin modification. The SV40 model, with its rich history and wealth of experimental tools, may have an exciting future.
Acknowledgments
EF thanks her mentors, lab members, colleagues, collaborators, and fellow travelers along the SV40 road for fruitful cooperation, companionship, guidance, and inspiration. B. Stillman, C. Clark, K. Rundell, X.S. Chen, and M. Baack kindly provided photographs. Financial support from NIH (GM52948 to EF and P30 CA68485 to the Vanderbilt-Ingram Cancer Center) and Vanderbilt University are gratefully acknowledged.
References
- Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8(8):588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- Aladjem MI, Fanning E. The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep. 2004;5(7):686–91. doi: 10.1038/sj.embor.7400185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunkumar AI, Klimovich V, Jiang X, Ott RD, Mizoue L, Fanning E, Chazin WJ. Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome. Nat Struct Mol Biol. 2005;12(4):332–9. doi: 10.1038/nsmbXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16(6):659–72. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Martynowski D, Seitova A, Bochkarev A. Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA. Embo J. 2006;25(24):5961–9. doi: 10.1038/sj.emboj.7601452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J, Dhamus ME, Fambrough D, Huang RC, Marushige K, Tuan DY. The biology of isolated chromatin. Chromosomes, biologically active in the test tube, provide a powerful tool for the study of gene action. Science. 1968;159(810):47–56. [PubMed] [Google Scholar]
- Borowiec JA, Dean FB, Bullock PA, Hurwitz J. Binding and unwinding--how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60(2):181–4. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429(6993):724–30. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Brownlee C. Danna and Nathans: Restriction enzymes and the boon to modern molecular biology. Proc Natl Acad Sci U S A. 2005;102(17):5909. doi: 10.1073/pnas.0502760102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock PA. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32(6):503–68. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- Campbell KS, Mullane KP, Aksoy IA, Stubdal H, Zalvide J, Pipas JM, Silver PA, Roberts TM, Schaffhausen BS, DeCaprio JA. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11 (9):1098–110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- Challberg MD, Kelly TJ. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Chou JY, Martin RG. Complementation analysis of simian virus 40 mutants. J Virol. 1974;13(5):1101–9. doi: 10.1128/jvi.13.5.1101-1109.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9(8):616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CN, Tornow J, Clark R, Tjian R. Properties of the simian virus 40 (SV40) large T antigens encoded by SV40 mutants with deletions in gene A. J Virol. 1986;57(2):539–46. doi: 10.1128/jvi.57.2.539-546.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford LV, Black PH. The Nucleic Acid of Simian Virus 40. Virology. 1964;24:388–92. doi: 10.1016/0042-6822(64)90176-x. [DOI] [PubMed] [Google Scholar]
- Crawford LV, Cole CN, Smith AE, Paucha E, Tegtmeyer P, Rundell K, Berg P. Organization and expression of early genes of simian virus 40. Proc Natl Acad Sci U S A. 1978;75(1):117–21. doi: 10.1073/pnas.75.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J, You J, Benjamin TL. Induction and utilization of an ATM signaling pathway by polyomavirus. J Virol. 2005;79(20):13007–17. doi: 10.1128/JVI.79.20.13007-13017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K, Nathans D. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. Proc Natl Acad Sci U S A. 1971;68(12):2913–7. doi: 10.1073/pnas.68.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna KJ, Nathans D. Bidirectional replication of Simian Virus 40 DNA. Proc Natl Acad Sci U S A. 1972;69(11):3097–100. doi: 10.1073/pnas.69.11.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna KJ, Sack GH, Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973;78(2):363–76. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Dean FB, Borowiec JA, Ishimi Y, Deb S, Tegtmeyer P, Hurwitz J. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proc Natl Acad Sci U S A. 1987a;84(23):8267–71. doi: 10.1073/pnas.84.23.8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Bullock P, Murakami Y, Wobbe CR, Weissbach L, Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci U S A. 1987b;84(1):16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML, Beard P, Berg P. Synthesis of Superhelical Simian Virus 40 Deoxyribonucleic Acid in Cell Lysates*. J Biol Chem. 1975;250(11):4340–7. [PubMed] [Google Scholar]
- Deppert W, Gurney EG, Harrison RO. Monoclonal antibodies against simian virus 40 tumor antigens: analysis of antigenic binding sites, using adenovirus type 2-simian virus 40 hybrid viruses. J Virol. 1981;37(1):478–82. doi: 10.1128/jvi.37.1.478-482.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D, Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980;140(1):129–42. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- DiMaio D, Nathans D. Regulatory mutants of simian virus 40. Effect of mutations at a T antigen binding site on DNA replication and expression of viral genes. J Mol Biol. 1982;156(3):531–48. doi: 10.1016/0022-2836(82)90265-0. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Nathans D. Purification of simian virus 40 large T antigen by immunoaffinity chromatography. J Virol. 1985;53(3):1001–4. doi: 10.1128/jvi.53.3.1001-1004.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M, Dean FB, Bullock P, Echols H, Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987;238(4829):964–7. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- Dulbecco R, Vogt M. Evidence for a Ring Structure of Polyoma Virus DNA. Proc Natl Acad Sci U S A. 1963;50:236–43. doi: 10.1073/pnas.50.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Waqar MA, Huberman JA. Subnuclear systems for synthesis of simian virus 40 DNA in vitro. Proc Natl Acad Sci U S A. 1976;73(12):4392–6. doi: 10.1073/pnas.73.12.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaschi A. Eukaryotic DNA replication: a model for a fixed double replisome. Trends Genet. 2000;16(2):88–92. doi: 10.1016/s0168-9525(99)01917-4. [DOI] [PubMed] [Google Scholar]
- Fang L, Davey MJ, O’Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4(4):541–53. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- Fanning E. Simian virus 40 large T antigen: the puzzle, the pieces, and the emerging picture. J Virol. 1992;66(3):1289–93. doi: 10.1128/jvi.66.3.1289-1293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E. Control of SV40 DNA replication by protein phosphorylation: a model for cellular DNA replication? Trends Cell Biol. 1994;4(7):250–5. doi: 10.1016/0962-8924(94)90123-6. [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34(15):4126–37. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- Fiers W, Contreras R, Haegemann G, Rogiers R, Van de Voorde A, Van Heuverswyn H, Van Herreweghe J, Volckaert G, Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978;273(5658):113–20. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10(3):160–7. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119(1):47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40(2):115–28. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- Germond JE, Hirt B, Oudet P, Gross-Bellark M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975;72(5):1843–7. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacherio D, Hager LP. A poly(dT)-stimulated ATPase activity associated with simian virus 40 large T antigen. J Biol Chem. 1979;254(17):8113–6. [PubMed] [Google Scholar]
- Gomez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM, San Martin C. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J Biol Chem. 2005;280(49):40909–15. doi: 10.1074/jbc.M509760200. [DOI] [PubMed] [Google Scholar]
- Griffith JD. Chromatin Structure: Deduced from a Minichromosome. Science. 1975;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Harlow E, Crawford LV, Pim DC, Williamson NM. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39(3):861–9. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14(7):1697–709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher U, Stalder HP. Mammalian DNA helicase. Nucleic Acids Res. 1985;13(15):5471–83. doi: 10.1093/nar/13.15.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, O’Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006;7(10):751–61. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134(4):815–26. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Brenner S. [On the regulation of DNA synthesis in bacteria: the hypothesis of the replicon.] C R Hebd Seances Acad Sci. 1963;256:298–300. [PubMed] [Google Scholar]
- Jiang X, Klimovich V, Arunkumar AI, Hysinger EB, Wang Y, Ott RD, Guler GD, Weiner B, Chazin WJ, Fanning E. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. Embo J. 2006;25(23):5516–26. doi: 10.1038/sj.emboj.7601432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo WS, Kim HY, Purviance JD, Sreekumar KR, Bullock PA. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol Cell Biol. 1998;18(5):2677–87. doi: 10.1128/mcb.18.5.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Ahn BY, Cho Y. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. The EMBO Journal. 2001;20(1):295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125(7):1297–308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9(10):759–69. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423(6939):512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci U S A. 1984;81(22):6973–7. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Sanford DG, Bullock PA, Bachovchin WW. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat Struct Biol. 1996;3(12):1034–9. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- Margolskee RF, Nathans D. Simian virus 40 mutant T antigens with relaxed specificity for the nucleotide sequence at the viral DNA origin of replication. J Virol. 1984;49(2):386–93. doi: 10.1128/jvi.49.2.386-393.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo IA, Hough PV, Wall JS, Dodson M, Dean FB, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338(6217):658–62. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Eki T, Hurwitz J. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc Natl Acad Sci U S A. 1990;87(24):9712–6. doi: 10.1073/pnas.87.24.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke G, Bullock PA, Bohm A. Crystal structure of the simian virus 40 large T-antigen origin-binding domain. J Virol. 2006;80(9):4304–12. doi: 10.1128/JVI.80.9.4304-4312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke G, Phelan P, Moine S, Bochkareva E, Bochkarev A, Bullock PA, Bohm A. The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PLoS Biol. 2007;5(2):e23. doi: 10.1371/journal.pbio.0050023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Taddei A, Gasser SM. In and out of the replication factory. Cell. 2006;125(7):1233–5. doi: 10.1016/j.cell.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Moarefi IF, Small D, Gilbert I, Hopfner M, Randall SK, Schneider C, Russo AA, Ramsperger U, Arthur AK, Stahl H, et al. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J Virol. 1993;67(8):4992–5002. doi: 10.1128/jvi.67.8.4992-5002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JF, Berg P. Cleavage of Simian virus 40 DNA at a unique site by a bacterial restriction enzyme. Proc Natl Acad Sci U S A. 1972;69(11):3365–9. doi: 10.1073/pnas.69.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103(27):10236–41. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans D. Restriction Endonucleases, Simian Virus 40, and the New Genetics. Nobel Lecture. 1978 December 8; doi: 10.1126/science.228393. [DOI] [PubMed] [Google Scholar]
- Nathans D, Danna KJ. Specific origin in SV40 DNA replication. Nat New Biol. 1972;236(68):200–2. doi: 10.1038/newbio236200a0. [DOI] [PubMed] [Google Scholar]
- Ott RD, Rehfuess C, Podust VN, Clark JE, Fanning E. Role of the p68 subunit of human DNA polymerase alpha-primase in simian virus 40 DNA replication. Mol Cell Biol. 2002;22(16):5669–78. doi: 10.1128/MCB.22.16.5669-5678.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto B, Fanning E. DNA polymerase alpha is associated with replicating SV40 nucleoprotein complexes. Nucleic Acids Res. 1978;5(5):1715–28. doi: 10.1093/nar/5.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432(7016):473–8. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- Pospiech H, Kursula I, Abdel-Aziz W, Malkas L, Uitto L, Kastelli M, Vihinen-Ranta M, Eskelinen S, Syvaoja JE. A neutralizing antibody against human DNA polymerase epsilon inhibits cellular but not SV40 DNA replication. Nucleic Acids Res. 1999;27 (19):3799–804. doi: 10.1093/nar/27.19.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VB, Thimmappaya B, Dhar R, Subramanian KN, Zain BS, Pan J, Ghosh PK, Celma ML, Weissman SM. The genome of simian virus 40. Science. 1978;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. Embo J. 2004;23(4):897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ. How restriction enzymes became the workhorses of molecular biology. Proc Natl Acad Sci U S A. 2005;102(17):5905–8. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi ML, Pike JE, Wang W, Burgers PMJ, Campbell JL, Bambara RA. Pif1 Helicase Directs Eukaryotic Okazaki Fragments toward the Two-nuclease Cleavage Pathway for Primer Removal. J Biol Chem. 2008;283(41):27483–27493. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell K, Collins JK, Tegtmeyer P, Ozer HL, Lai CJ, Nathans D. Identification of simian virus 40 protein A. J Virol. 1977;21(2):636–46. doi: 10.1128/jvi.21.2.636-646.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saragosti S, Moyne G, Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Schwyzer M, Weil R, Frank G, Zuber H. Amino acid sequence analysis of fragments generated by partial proteolysis from large simian virus 40 tumor antigen. J Biol Chem. 1980;255(12):5627–34. [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–80. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Akita M, Kamimura Y, Muramatsu S, Araki H, Sugino A. GINS is a DNA polymerase epsilon accessory factor during chromosomal DNA replication in budding yeast. J Biol Chem. 2006;281(30):21422–32. doi: 10.1074/jbc.M603482200. [DOI] [PubMed] [Google Scholar]
- Shi Y, Dodson GE, Shaikh S, Rundell K, Tibbetts RS. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J Biol Chem. 2005;280(48):40195–200. doi: 10.1074/jbc.C500400200. [DOI] [PubMed] [Google Scholar]
- Shikata K, Sasa-Masuda T, Okuno Y, Waga S, Sugino A. The DNA polymerase activity of Pol epsilon holoenzyme is required for rapid and efficient chromosomal DNA replication in Xenopus egg extracts. BMC Biochem. 2006;7:21. doi: 10.1186/1471-2091-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D, Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978;75(5):2170–4. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle DR, Margolskee RF, Nathans D. Mutational analysis of the simian virus 40 replicon: pseudorevertants of mutants with a defective replication origin. Proc Natl Acad Sci U S A. 1979;76(12):6128–31. doi: 10.1073/pnas.76.12.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V, Lane DP. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144(1):88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- Simmons DT. SV40 large T antigen functions in DNA replication and transformation. Adv Virus Res. 2000;55:75–134. doi: 10.1016/s0065-3527(00)55002-7. [DOI] [PubMed] [Google Scholar]
- Smelkova NV, Borowiec JA. Synthetic DNA replication bubbles bound and unwound with twofold symmetry by a simian virus 40 T-antigen double hexamer. J Virol. 1998;72 (11):8676–81. doi: 10.1128/jvi.72.11.8676-8681.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl H, Droge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. Embo J. 1986;5(8):1939–44. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staufenbiel M, Deppert W. Different structural systems of the nucleus are targets for SV40 large T antigen. Cell. 1983;33(1):173–81. doi: 10.1016/0092-8674(83)90346-x. [DOI] [PubMed] [Google Scholar]
- Stillman B. Initiation of eukaryotic DNA replication in vitro. Annu Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- Stillman BW, Gluzman Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol Cell Biol. 1985;5(8):2051–60. doi: 10.1128/mcb.5.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Pipas JM. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol Mol Biol Rev. 2002;66(2):179–202. doi: 10.1128/MMBR.66.2.179-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Bell P, Tegtmeyer P, Maul GG. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J Virol. 2000;74(20):9694–700. doi: 10.1128/jvi.74.20.9694-9700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J Virol. 1972;10 (4):591–8. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P, Ozer HL. Temperature-sensitive mutants of simian virus 40: infection of permissive cells. J Virol. 1971;8(4):516–24. doi: 10.1128/jvi.8.4.516-524.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R. The binding site on SV40 DNA for a T antigen-related protein. Cell. 1978;13(1):165–79. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- Tooze J. The Molecular Biology of Tumour Viruses. Cold Spring Harbor Laboratory; N.Y: 1973. [Google Scholar]
- Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346(6284):534–9. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266(3):1961–8. [PubMed] [Google Scholar]
- Valle M, Chen XS, Donate LE, Fanning E, Carazo JM. Structural basis for the cooperative assembly of large T antigen on the origin of replication. Journal of Molecular Biology. 2006;357(4):1295–1305. doi: 10.1016/j.jmb.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Valle M, Gruss C, Halmer L, Carazo JM, Donate LE. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol Cell Biol. 2000;20(1):34–41. doi: 10.1128/mcb.20.1.34-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J, Lebowitz J, Radloff R, Watson R, Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965;53(5):1104–11. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup DM, Russo AA, Kelly TJ. Mechanism of activation of simian virus 40 DNA replication by protein phosphatase 2A. Mol Cell Biol. 1992;12(11):4883–95. doi: 10.1128/mcb.12.11.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369(6477):207–12. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–51. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Waqar MA, Evans MJ, Huberman JA. Effect of 2′,3′-dideoxythymidine-5′-triphosphate on HeLa cell in vitro DNA synthesis: evidence that DNA polymerase alpha is the only polymerase required for cellular DNA replication. Nucleic Acids Res. 1978;5(6):1933–46. doi: 10.1093/nar/5.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil R. The denaturation and the renaturation of the DNA of polyoma virus. Proc Natl Acad Sci U S A. 1963;49:480–7. doi: 10.1073/pnas.49.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil R, Vinograd J. The Cyclic Helix and Cyclic Coil Forms of Polyoma Viral DNA. Proc Natl Acad Sci U S A. 1963;50:730–8. doi: 10.1073/pnas.50.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DH, Collins KL, Simancek P, Russo A, Wold MS, Virshup DM, Kelly TJ. Reconstitution of simian virus 40 DNA replication with purified proteins. Proc Natl Acad Sci U S A. 1990;87(22):8692–6. doi: 10.1073/pnas.87.22.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshart K, Bradley MK, Weiner BM, Schneider C, Moarefi I, Fanning E, Arthur AK. An N-terminal deletion mutant of simian virus 40 (SV40) large T antigen oligomerizes incorrectly on SV40 DNA but retains the ability to bind to DNA polymerase alpha and replicate SV40 DNA in vitro. J Virol. 1996;70(6):3509–16. doi: 10.1128/jvi.70.6.3509-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshart K, Friedl S, Taneja P, Nasheuer HP, Schlott B, Grosse F, Fanning E. Partial proteolysis of simian virus 40 T antigen reveals intramolecular contacts between domains and conformation changes upon hexamer assembly. J Biol Chem. 2004;279(37):38943–38951. doi: 10.1074/jbc.M406159200. [DOI] [PubMed] [Google Scholar]
- Weisshart K, Taneja P, Jenne A, Herbig U, Simmons DT, Fanning E. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J Virol. 1999;73(3):2201–11. doi: 10.1128/jvi.73.3.2201-2211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66(2):804–15. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M, Eason R. Nucleoprotein complexes in simian virus 40-infected cells. J Virol. 1971;8(4):363–71. doi: 10.1128/jvi.8.4.363-371.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe CR, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci U S A. 1985;82 (17):5710–4. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Avni D, Chiba T, Yan F, Zhao Q, Lin Y, Heng H, Livingston D. SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev. 2004;18(11):1305–16. doi: 10.1101/gad.1182804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhakov A, Kelman Z, Hurwitz J, O’Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. Embo J. 1999;18(21):6189–99. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Madden-Fuentes RJ, Lou BX, Pipas JM, Gerhardt J, Rigell CJ, Fanning E. Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in Simian virus 40-infected primate cells. J Virol. 2008;82(11):5316–28. doi: 10.1128/JVI.02677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotkin T, Kaufmann G, Jiang Y, Lee MY, Uitto L, Syvaoja J, Dornreiter I, Fanning E, Nethanel T. DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. Embo J. 1996;15(9):2298–305. [PMC free article] [PubMed] [Google Scholar]