Abstract
The LONI Pipeline is a graphical environment for construction, validation and execution of advanced neuroimaging data analysis protocols (Rex et al., 2003). It enables automated data format conversion, allows Grid utilization, facilitates data provenance, and provides a significant library of computational tools. There are two main advantages of the LONI Pipeline over other graphical analysis workflow architectures. It is built as a distributed Grid computing environment and permits efficient tool integration, protocol validation and broad resource distribution. To integrate existing data and computational tools within the LONI Pipeline environment, no modification of the resources themselves is required. The LONI Pipeline provides several types of process submissions based on the underlying server hardware infrastructure. Only workflow instructions and references to data, executable scripts and binary instructions are stored within the LONI Pipeline environment. This makes it portable, computationally efficient, distributed and independent of the individual binary processes involved in pipeline data-analysis workflows. We have expanded the LONI Pipeline (V.4.2) to include server-to-server (peer-to-peer) communication and a 3-tier failover infrastructure (Grid hardware, Sun Grid Engine/Distributed Resource Management Application API middleware, and the Pipeline server). Additionally, the LONI Pipeline provides three layers of background-server executions for all users/sites/systems. These new LONI Pipeline features facilitate resource-interoperability, decentralized computing, construction and validation of efficient and robust neuroimaging data-analysis workflows. Using brain imaging data from the Alzheimer's Disease Neuroimaging Initiative (Mueller et al., 2005), we demonstrate integration of disparate resources, graphical construction of complex neuroimaging analysis protocols and distributed parallel computing. The LONI Pipeline, its features, specifications, documentation and usage are available online (http://Pipeline.loni.ucla.edu).
Keywords: LONI Pipeline, software tools, resources, workflows, tool interoperability, data provenance, tool integration, neuroimaging
Introduction
Modern tools for image processing employ large amounts of heterogeneous data, diverse computational resources and distributed web-services (Toga and Thompson, 2007). Efficient analysis protocols combine diverse data, software tools and network infrastructure to obtain, analyze and disseminate results. Construction of such analysis protocols are significantly enhanced by a graphical workflow interface that provides high-level manipulation of the analysis sequence while hiding many of its technical details. In this manuscript, we discuss the challenges of development, maintenance and dissemination of integrated resources including data, software tools and web-services, as platform-independent, agile and scalable frameworks. We demonstrate the development and utilization of the LONI Pipeline environment for combining of user computational and biological expertise with disparate resources and Grid infrastructures. Version 4 of the LONI Pipeline, extends the previous implementation of this environment (Rex et al., 2003).
To provide an extensible framework for interoperability of diverse computational resources the LONI Pipeline employs a decentralized infrastructure, where tools, services and data are linked through an external resource-mediating-layer. This approach requires no modifications of existing tools to enable their interoperability with other computational counterparts. A new XML schema forms the backbone for the inter-resource-mediating-layer. Each XML resource (module) description includes important information about the resource location, the proper invocation protocol (i.e., I/O types, parameter specifications, etc.), run-time controls and data-types. Also included are auxiliary meta-data about the resource state, specifications, history, authorship and bibliography. This infrastructure1 facilitates the integration of disparate resources and provides a natural and comprehensive data provenance (MacKenzie-Graham et al., 2008a). The LONI Pipeline also enables the broad dissemination of resource meta-data descriptions via web-services and the constructive utilization of multidisciplinary expertise by experts, novice users and trainees.
There are a number of efforts to develop environments for tool integration, interoperability and meta-analysis (Rex et al., 2004). There is a clear community need to establish efficient tool interoperability, which enables new types of analyses and facilitates new applications (Dinov et al., 2008). Taverna (Oinn et al., 2005) is an open-source, platform-independent graphical workflow environment, which enables linking tools after explicit rebuilding. It is mainly employed for bioinformatics applications via myGRID infrastructure. Kepler (Ludäscher et al., 2006) is another scientific workflow environment used for various applications, which also requires rebuilding each executable to link it with the core libraries. The Triana (Churches et al., 2006) workflow environment enables external data storage which significantly improves the efficiency and robustness of its user interface and optimizes the system requirements (e.g., low memory demands). VisTrails (Callahan et al., 2006) addresses the problem of visualization from a data management perspective where imaging data and meta-data are represented as a conjoint visualization product. Swift is another graphical workflow environment, which uses a scripting language, SwiftScript, to enable concise high-level specification of workflows based on various applications using large quantities of data. The Swift engine provides efficient execution of these workflows on sequential or parallel computers or distributed grids (Stef-Praun et al., 2007). There are a number of other graphical workflow environments that are proposed, tested and validated for specific applications, types of users, scientific areas or hardware infrastructures (Bowers et al., 2008).
Compared to other graphical workflow environments, the LONI Pipeline offers several advantages. It facilitates the back-end integration with distributed Grid-enabled and client-server infrastructures, and provides an efficient and robust framework for deployment of new resources to the community (new tools need not be recompiled, migrated or altered in anyway to be made functionally available to the community). The choice of a particular analysis workflow infrastructure always depends on the application domain, the type of user, types of access to resources (e.g., computational framework, human or machine resource interface, database, etc.), as well as, the desired features and functionalities (Bitter et al., 2007). There are inevitable similarities between the LONI Pipeline and other such environments. These include the graphical interface provided by most workflow environments, which facilitates the design of analysis protocols and improves the usability of these graphical protocols. Visual interfaces present complex analysis protocols in an intuitive manner and improve the management of technical details. Most of the graphical workflow environments provide the ability to save, load and distribute protocols through servers, SOAP/WSDL/XML or other means.
The LONI Pipeline addresses the specific needs of the neuroimaging and computational neuroscience community, but its general goals of providing portability, transparency, intuitiveness and abstraction from Grid mechanics make it appealing in other fields. The Pipeline is a dynamic resource manager, treating all resources as well-described external applications that may be invoked with standard remote execution protocols. The LONI Pipeline XML description protocol allows any command-line driven process, web-service or data-server to be accessed within the environment by reference, with dependencies validated (checked) dynamically on-demand. There is no need to reprogram, revise or recompile external resources to make them usable within the LONI Pipeline. One side effect of this design choice is that to all external Pipeline server installations require complete installations of all software tools, services and data as currently available on the LONI Grid. This however, is only required, if a remote Pipleine server must mirror all tools and resources as available on the LONI Grid. In most situations, each site has specific suits of tools that they utilize to meet their computational needs. This design reduces the integration/utilization costs of including new resources within the LONI Pipeline environment. This approach provides the benefit of quick and easy management of large and disparately located resources and data. In addition, this choice significantly minimizes the user/client machine hardware and software requirements (e.g., memory, storage, CPU). Finally, a key difference between the LONI Pipeline and some other environments is its management of distributed resources via its client-to-server infrastructure and its ability to export automated makefiles/scripts. These allow the LONI Pipeline to provide processing power independently of the available computational environment (e.g., SOLARIS, LINUX, Grid, mainframe, desktop, etc.). The LONI Pipeline servers communicate and interact with clients and facilitate secure transfer of processes, instructions, data and results via the Internet.
Version 4 of the LONI Pipeline introduces several important improvements and extensions of the previous version of the LONI Pipeline (Rex et al., 2003). These include a 3-tier failover mechanism for Grid hardware, Sun Grid Engine (SGE)/Distributed Resource Management Application API (DRMAA) middleware, and the Pipeline server, as well as client-server communication, makefile/script export and data provenance model. LONI Pipeline v.4 also includes a new more functional and robust graphical user interface and a significantly increased library of tools. V.4 also simplifies the inclusion of external data display modules and facilitates remote database connectivity (e.g., LONI Imaging Data Archive2, BIRN Storage Resource Broker3, XNAT4, etc.)
Materials and Methods
The main goal of developing the LONI Pipeline was to provide a robust and extensible infrastructure for computational neuroscience enabling efficient data utilization, construction of reliable analysis workflows, and provide the means for wide dissemination and validation of research protocols and scientific findings. The LONI Pipeline developments are subdivided into several complementary goals:
Efficient Distributed Computing: Facilitate the integration of disparate, heterogeneous and multi-platform implementations of software tools, database protocols and remote web-services. The LONI Pipeline client-server communication protocol allows blending of resources that are built on remote server architectures to be accessed by the pipeline clients. This greatly lowers the usability requirements for the general user. In addition, we need a flexible export of available pipeline workflows into makefiles and bash scripts that can be submitted virtually to any computational architecture.
Design a robust 3-tier failover mechanism for the LONI Grid: This included the three layers of the Grid submission protocol – Sun Grid Engine (SGE), Distributed Resource Management Application API5, and the Pipeline job handling server. These three layers of background-server executions enable various types of users and systems to utilize the Pipeline environment in any of these three execution modes: single machines, or main-frames with only one queue job submission protocol; Globus Grid infrastructure, and SGE Grid Infrastructure.
Provenance: LONI Pipeline includes a provenance manager, which enables tracking data, workflow and execution history of all processes. This functionality improves the communication, reproducibility and validation of newly proposed experimental designs, scientific analysis protocols and research findings. This includes the ability to record, track, extract, replicate and evaluate the data and analysis provenance to enable rigorous validation and comparison of classical and novel design paradigms.
Tool Discovery: Enable expert researchers to quickly design, test and validate novel experimental designs and data analysis protocols. This is achieved via a dynamic, responsive and intelligent graphical user interface for tool exploration and construction of draft pipeline workflows.
Friendly Graphical User Interface: Create a robust environment for tool interoperability, Grid integration and low-cost interactive user interface. For maximum portability, scalability and efficiency, this environment is built in Java and utilizes XML for storing and communication of meta-data, and descriptors for tools and services.
The LONI Pipeline execution environment controls the local and remote server connections, module communication, process management, data transfers and Grid mediation. The XML descriptions of individual modules, or networks of modules, may be constructed, edited and revised directly within the LONI Pipeline graphical user interface, as well as saved or loaded from disk or the LONI Pipeline server. These workflows completely describe new methodological developments and allow validation, reproducibility, provenance and tracking of data and results. The core six types of Pipeline specifications are summarized below.
Types of tools and services that can be integrated within the LONI Pipeline
The development and utilization of the LONI Pipeline environment is focused on neuroimaging data and analysis protocols. However, by design, the LONI Pipeline software architecture is domain agnostic and has been adopted in other research and clinical fields, e.g., bioinformatics (Dinov et al., 2008). There are two major types of resources that may be integrated within the LONI Pipeline. The first one is data, in terms of databases, data services and file systems. The second type of pipelineable resources includes stand-alone tools, comprising local or remote binary executables and services with well-defined command line syntax. This flexibility permits efficient resource integration, tool interoperability and wide dissemination.
General LONI Pipeline specifications including grid integration
The LONI Pipeline routinely executes thousands of simultaneous jobs on our symmetric multiprocessing systems (SMP) and on DRMAA6 clusters. On SMP systems, the LONI Pipeline can detect the number of available processing units and scale the number of simultaneous jobs accordingly to maximize system utilization and prevent system crashes. For computer clusters, a grid engine implementing DRMAA, with Java bindings, may be used to submit jobs for processing, and a shared file system is used to store inputs and outputs from individual jobs. Later, we will extend the scope of the LONI Pipeline server to interact and submit jobs to other Grid infrastructures, e.g., Condor, Globus, etc.
The LONI Pipeline environment has been integrated with UNIX authentication using Pluggable Authentication Module (PAM), to enable a username and password challenge-response authentication method using existing credentials. A dependency on the underlying security and encryption system of the LONI Pipeline server's host machine offers maximum versatility in light of the diverse policies governing system authentication and access control.
Using Java binding to DRMAA interface, we have integrated the LONI Pipeline environment with the SUN Grid Engine (SGE), a free, well-engineered distributed resource manager (DRM) that simplifies the processing and management of submitted jobs on the grid. It is important to note, however, that other DRMs such as Condor, LSF and PBS/Torque could be made compatible with the LONI Pipeline environment using the same interface. DRMAA's Java foundation allows jobs to be submitted from the LONI Pipeline to the compute grid without the use of external scripts and provides significant job control functionality internally. We accomplished several key goals with the LONI Pipeline-DRMAA-SGE integration:
the parallel nature of the LONI Pipeline environment is enhanced by allowing for both horizontal (across compute nodes) and vertical (across CPUs on the same node) processing parallelization;
the LONI Pipeline's client-server functionality can directly control a large array of computational resources with DRMAA over the network, significantly increasing its versatility and efficacy;
facilitate the use of a heterogeneous set of neuroimaging software tools in pipelines involving large number of datasets and multiple processing tools;
the overall usability of grid resources is improved by the intuitive graphical interface offered by the LONI Pipeline environment, and
the ability to display interim results from user-specified modules, which can be used for visual inspection of the outputs of various tools (interactive outcome checking).
LONI Pipeline data provenance
In neuroimaging studies, data provenance, or the history of how the data were acquired and subsequently processed, is often discussed but seldom implemented (MacKenzie-Graham et al., 2008b). Recently, several groups have proposed provenance challenges in order to evaluate the status of various provenance models (Miles et al., 2006). For instance, collecting provenance information from a simple neuroimaging workflow (Zhao et al., 2007) and documenting each system's ability to respond to a set of predefined queries. Some of the existing provenance systems are designed as mechanisms for capturing provenance in neuroimaging (MacKenzie-Graham et al., 2008a; Zhao et al., 2007). It is difficult to provide systematic, accurate and comprehensive capture of provenance information with minimal user intervention. The processes of data provenance and curation are significantly automated via the LONI Pipeline. Each dataset has a provenance file (*.prov) that is automatically updated by the LONI Pipeline, based on the protocols used in the data analysis. This data processing history reflects sequentially the steps that a dataset goes through and provides a detailed record of the types of tools, versions, platforms, parameters, control and compilation flags. The data provenance can be imported and exported by the LONI Pipeline, which enables utilization internally by other Pipeline workflows or by external resources (e.g., databases, workflow environments).
Provenance can be used for determining data quality, for result interpretation, and for protocol interoperability (Simmhan et al., 2005; Zhao et al., 2007). It is imperative that the provenance of neuroimaging data be easily captured and readily accessible (MacKenzie-Graham et al., 2008b). For instance, increasingly complex analysis workflows are being developed to extract information from large cross-sectional or longitudinal studies in multiple sclerosis (Liu et al., 2005), Alzheimer's disease (Fleisher et al., 2005), autism (Langen et al., 2007), depression (Drevets, 2001), schizophrenia (Narr et al., 2007), and studies of normal populations (Gogtay et al., 2006). The implementation of the complex workflows associated with these studies requires provenance-based quality control to ensure the accuracy, reproducibility, and reusability of the data and analysis protocols.
We designed the provenance framework to take advantage of context information that can be retrieved and stored while data is being processed within the LONI Pipeline environment (MacKenzie-Graham et al., 2008b). Additionally, the LONI Provenance Editor is a self-contained, platform-independent application that automatically extracts provenance information from image headers (such as a DICOM images) and generates an XML data provenance file with that information. The Provenance Editor7 allows the user to edit the meta-data prior to saving the provenance file, correcting inaccuracies or adding additional information. This provenance information is stored in.prov files, XML formatted files that contain the meta-data and processing provenance and follows the XSD definition8. Then the data provenance is expanded by the LONI Pipeline to include the analysis protocol, the specific binaries used for analysis, and the environment that they were run in. The LONI Pipeline dramatically improves compliance by minimizing the burden on the provenance curator. This frees the user to focus on performing neuroimaging research rather than on managing provenance information.
LONI Pipeline intelligence
Construction of elaborate, functional and valid workflows within the LONI Pipeline environment requires deep understanding of the research goals, tool specifications and neuroscientific expertise. To enhance the usability of this environment, we developed an intelligent LONI Pipeline component. It has two complementary features – constructive and validating. The pipeline constructive intelligent feature uses the spectra of available module descriptors and pipeline workflows to automatically generate valid versions of new graphical protocols according to a set of user-specified keywords. This intelligence feature uses a grammar on the set of XML module and pipeline descriptions to determine the most appropriate analysis protocol, and its corresponding module inputs and outputs, according to the keywords provided by the user. Then, it exports a.pipe file, which contains a draft of the desired analysis protocol, Figure 1.
Figure 1.
LONI Pipeline intelligence component uses key-words to automatically generate a hierarchical interface and the complete analysis workflow, which represents the proposed study protocol, using semantic natural language processing of language grammar. The image insert shows the graphical user interface invoking the pipeline intelligence grammar view.
The pipeline validating intelligence feature offers interactive support for running or modifying existent pipeline workflows. This feature contextually monitors the consistency of the data types, parameter matches, validity of the analysis protocol, and ensures optimal job-submission (e.g., order of module execution). The LONI Pipeline intelligence component reduces the need to review in details of, and double check modifications of new or existing workflows. Still, users control the processes of saving workflows and module descriptions, data input and output, and the scientific design of their experiments. This functionality significantly improves usability and facilitates scientific exploration.
LONI Pipeline graphical and scripting interfaces
Pipeline workflows (.pipe files) may be constructed in many different ways (e.g., using text editors) and these protocols may be executed in a batch mode without involving the LONI Pipeline graphical user interface (GUI). However, the LONI Pipeline GUI significantly aids most users in designing and running analysis workflows. A library of available tools for usage is presented on the left hand side of the LONI Pipeline client window. Users may search for, drag and drop these tools onto the main canvas to create or revise a workflow. Connections between the nodes are used to represent the piping of output from one program to another. This is accomplished without requiring the user to specify file paths, server locations or command line syntax. Pipeline workflows may be constructed and executed with data dynamically flowing (by reference) within the workflow. This enables trivial inclusion of pipeline protocols in external scripts and integration into other applications. Currently, the LONI Pipeline allows exporting of any workflow from XML (*.pipe) format to a makefile or a bash script for direct or queuing execution.
Functionality and usability
In the past 3 years, we have gone through several cycles of design, implementation, analysis and re-design stages of the new LONI Pipeline. During this process a number of usability issues were addressed. These included the editing and usage modes of the graphical user interface, state specific menus, pop-up and information dialogs, the handling of local and global variables within the pipeline, the integration of data sources and executable module nodes, data type checking and workflow validation, client connect and disconnects, job management and client-server communications. All of these were critical in improving the usability of the LONI Pipeline and are necessary before the execution of any data analysis workflow.
The core LONI Pipeline functionality is based on our prior experience (Rex et al., 2003), user feedback and information technology advancements over the past several years. The current LONI Pipeline functionality includes – tool discovery engine, plug-in interface for meta algorithm design, grid interface, secure user authentication, data transfers and client-server communications, graphical and batch-mode execution, encapsulation of tools, resources and workflows, and data provenance.
Results
LONI Pipeline can be used to construct a wide variety of processing and analysis workflows. Here we demonstrate the utilization of the LONI Pipeline to conduct and validate new (semi)automated, robust and user-friendly protocols for (1) regional parcellation and volume extraction, (2) population-based atlas construction, and (3) the analysis of multiple population cohorts. In the following sections, we discuss the graphical Pipeline workflows for each of these applications:
Brain parcellation
Regional parcellation of distinct brain regions is often needed to perform region-of-interest-based analyses between healthy as compared to diseased subjects. Manual region drawing can be labor intensive, prone to errors, and have poor reproducibility. Figure 2 illustrates a pipeline workflow constructed to automatically extract 3D masks of 56 regions of interest using Brain Parser (Tu et al., 2008)9. These regions can be then be used to examine regionally specific shape characteristics among other variables of interest to the neuroimaging community. The ability to automatically obtain robust 3D masks of various brain regions is a critical step in many brain mapping studies.
Figure 2.
Using a robust executable entitled Brain Parser (Tu et al., 2008), LONI Pipeline can be used to extract 56 predefined ROI masks from any input brain image volume (inserts).
Brain atlas construction
Brain atlasing is a major research effort in the field and the development of efficient workflows to take large numbers of T1-weighted anatomical images, spatially warp them into a common space, and then to pool them to result in a representative atlas is often a complex process. Development of efficient workflows and utilizing a large-scale computational Grid, based at LONI, permits streamlined and rapid atlas creation in normal subjects as well as in disease, Figure 3. Using Automated Image Registration (Woods et al., 1998), we constructed a workflow to systematically create a whole brain atlas for use in describing the average brain anatomical structure in patients drawn from the ADNI series of Alzheimer's subject MRI data contained in the LONI Image Data Archive (IDA) (Mueller et al., 2005). Such atlases characterize “mean” population features such as shape, regional area, sulcal anatomy, etc.
Figure 3.
LONI Pipeline workflow for constructing a population-based whole brain anatomical atlas in Alzheimer's Disease patients (insets).
Structural analysis of Alzheimer's Disease (AD) neuroimaging study
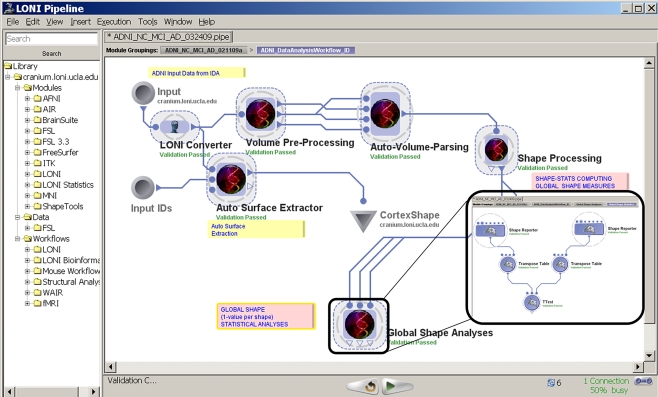
We used brain imaging data from the Alzheimer's Disease Neuroimaging Initiative, ADNI (Mueller et al., 2005), to demonstrate the processes of construction, validation and execution of integrated workflow analysis protocols. This AD pipeline workflow represents a complex neuroimaging analysis protocols based on disparate tools, data and distributed parallel-computing infrastructure. Figure 4 demonstrates this Alzheimer's disease Pipeline workflow. The left-panel in the Pipeline environment contains some predefined module definitions and complete workflows. The user may drag-and-drop these in the main workflow canvas to design new analysis protocols. The central workflow canvas shows that main six steps of the AD data analysis. These include data conversion, volumetric data pre-processing, automated extraction of regions of interest, shape processing, global shape analysis and automated cortical surface extraction. Each of these steps is itself a nested collection of groups of modules, a nested pipeline workflow, which contains a series of processing steps. The insert-figure illustrates the 3-level deep nested processing part of the Global Shape Analysis node (see the top-level tabs of the insert).
Figure 4.
An Alzheimer's Disease (AD) Pipeline workflow. The left-panel contains some predefined Pipeline module definitions including some complete workflows. The central-panel shows the main six steps of the data analysis – data conversion, pre-processing, automated extraction of regions of interest, shape processing, global shape analysis and automated cortical surface extraction. Each of these steps is itself a nested collection of modules, a pipeline, which contains a series of processing steps. The insert-figure illustrates the 3-level deep nested processing part of the Global Shape Analysis node (see the top-level tabs of the insert).
This pipeline workflow demonstrates the entire data processing and analysis protocol, from retrieval of the data from the LONI Imaging Data Archive10, through the data manipulation, shape processing, generation of derived data (e.g., global shape measures like curvature, fractal dimension, surface area, etc.), to the final statistical analysis. In this case, the study design included three age-matched populations – asymptomatic subjects (NC), minor cognitive impairment (MCI), and Alzheimer's disease (AD) patients. There were five males and five females for each group and each subject was scanned several times longitudinally. A total of 104 brain volumes were automatically processed in about 26 h. The time of workflow completion depends on the study and workflow designs, number of subjects, and general hardware infrastructure specifications (e.g., system characteristics and user demand). The results of this completely automated pipeline workflow included cortical surface representations (shapes) for each subject, parcellations of the raw MRI brain scans into 56 regions of interest (labels), surface models for each of the 56 regions for each subject and time-point, and global statistical mapping identifying the NC, MCI and AD group differences for each of the 56 regions.
Figure 5 depicts the shape-curvature measure for five regions of interest (ROI's) at two time-points – baseline (blue) and 12-month follow-up (green) for the cohort of normal subjects (NC). Figure 6 compares the shape measures for one region (right Superior Frontal Gyrus) across all three cohorts, at baseline (time = 0). Notice the consistent decrease of shape and volume measures, for both time points, going from NC (asymptomatic) to MCI and AD (most effected individuals).
Figure 5.
NC shape-curvature measure for 5 ROI's at two times (T1, baseline, blue, and T2, 12-month follow-up, green). L_Caudate and R_Caudate, left and right caudate, L_Hippo and R_Hippo, left and right hippocampus, R_SupFrontalGyrus, right superior frontal gyrus.
Figure 6.
Comparison of the four volume and shape measures for both times (baseline and 12-month follow up) across the three cohorts for one region of interest – the Right Superior Frontal Gyrus. T1 and T1 labels represent baseline and follow-up time scans. The statistics signature vector includes MeanCurv, average global shape mean curvature; MeanSurf, total shape surface area; MeanFract, mean global shape fractal dimension; and MeanVol, volume of the inside region of the shape. The three different cohorts, NC, MCI and AD, are colored in blue, green and red, respectively.
Discussion
Interactive workflow environments for automated data analysis are critical in many research studies involving complex computations and large datasets (Kawas et al., 2006; Myers et al., 2006; Oinn et al., 2005; Taylor et al., 2006). There are three distinct necessities that underlie the importance of such graphical frameworks for management of novel analysis strategies – high data volume and complexity, sophisticated study protocols and demands for distributed computational resources. These three fundamental needs are evident in most modern neuroimaging, bioinformatics and multidisciplinary studies.
The LONI Pipeline environment aims to provide distributed access to varieties of computational resources via its graphical interface. The ability of investigators to share, integrate, collaborate and expand resources will increase the statistical power in studies involving heterogeneous datasets and complex analysis protocols. New challenges that emerge from our increased abilities to utilize computational resources and hardware infrastructure include the need to assure reliability and reproducibility of identically analyzed data, and the desire to continually lower the costs of employing and sharing data, tools and services. The LONI Pipeline environment attempts to provide the means to address these difficulties by providing secure integrated access to resource visualization, databases and intelligent agents.
The LONI Pipeline already has been used in a number of neuroimaging applications including health (Sowell et al., 2007), disease (Thompson et al., 2003), animal models (MacKenzie-Graham et al., 2006), volumetric (Luders et al., 2006), functional (Rasser et al., 2005), shape (Narr et al., 2007) and tensor-based (Chiang et al., 2007) studies. The LONI Pipeline infrastructure improved consistency, reduced development and execution times, and enabled new functionality and usability of the analysis protocols designed by expert investigators in all of these studies. Perhaps the most powerful feature provided by the LONI Pipeline environment is the ability to quickly communicate new protocols, data, tools and service resources, findings and challenges to the wider community.
The main LONI Pipeline page11 provides links to the forum, support, video tutorials and usage. There are examples demonstrating how to describe individual modules and construct integrated workflows. Version information, download instructions and server/forum account information is also available on this page. There are example pipeline workflows and the XSD schema definition12 for the.pipe format used for module and workflow XML description. Users may either install Pipeline servers on their own hardware systems, or they may use some of the available Pipeline servers. The primary LONI Pipeline server is cranium.loni.ucla.edu. It utilizes a CentOS-based compute cluster comprised of approximately 800 Core 2, 2.4GHz, 8GB RAM, AMD Opteron processors. Each dual-processor compute node has eight gigabytes of memory to accommodate memory-intensive neuroimaging applications. We selected SUN Grid Engine v6, bound by DRMAA, as the LONI Pipeline distributed resource manager. A highly-optimized non-blocking Cisco Gigabit network provides the connectivity infrastructure with sixteen terabytes of fault-tolerant, clustered storage from Isilon Systems acting as a cache file system for the LONI Pipeline environment. Users may obtain accounts on this Grid13.
In general, some practical difficulties in validating new LONI Pipeline workflows may be caused by unavailability of the initial raw data, differences of hardware infrastructures or variations in compiler settings and platform configurations. Such situations require analysis workflow validation by teams of experts capable of validating the input, output and state of each module within the pipeline workflow. Further LONI Pipeline validation would require comparison between synergistic workflows that are implemented using different executable modules or module specifications. For example, one may be interested in comparing similar analysis workflows by choosing different sets of imaging filters, reconfiguring computation parameters or manipulating the resulting outcomes, e.g., file format, (Bitter et al., 2007). Such studies contrasting the benefits and limitations of each resource or processing workflow aid both application developers and general users in the decision of how to design and utilize module and pipeline definitions to improve resource usability.
A significant challenge in computational neuroimaging studies is the problem of reproducing findings and validating analyses described by different investigators. Frequently, methodological details described in research publications may be insufficient to accurately reconstruct the analysis protocol used to study the data. Such methodological ambiguity or incompleteness may lead to misunderstanding, misinterpretation or reduction of usability of newly proposed techniques. The LONI Pipeline mediates these difficulties by providing clear, functional and complete record of the methodological and technological protocols for the analysis.
Even though the LONI Pipeline was designed and tested to solve neuroimaging problems, its generic architecture will permit applications in other fields, where computationally intense tasks are performed and there is a need of resource interoperability. Its light-weight and platform-independent design and its low memory requirements make the LONI Pipeline potentially useful in many research fields relying on the integration of large and heterogeneous processing protocols. For example, the LONI Pipeline was recently used in conjunction with a number of bioinformatics data processing and analysis protocols (Dinov et al., 2008). We are also working on several new features of the LONI Pipeline including web-service-based client interface, direct integration with external resource archives (e.g., http://www.ncbcs.org/biositemaps, http://NeuroGateway.org, etc.) and interface enhancements using intelligent plug-in components.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by the National Institutes of Health through the NIH Roadmap for Medical Research, Center for Computational Biology Grant U54 RR021813, NIH/NCRR 5 P41 RR013642 and NIH/NIMH 5 R01 MH71940. The authors are also indebted to members of the Laboratory of Neuro Imaging, and various collaborators and users for their patience with testing the LONI Pipeline. Arash Payan, Jia-Wei Tam, Celia Cheung, Cornelius Hojatkashani and Jagadeeswaran Rajendiran contributed to the implementation of Pipeline V.4.
Footnotes
References
- Bitter I., Van Uitert R., Wolf I., Ibanez L. A., Kuhnigk J. M. A., Kuhnigk J. M. (2007). Comparison of four freely available frameworks for image processing and visualization that use ITK. Trans. Vis. Comput. Graph. 13, 483–493 10.1109/TVCG.2007.1001 [DOI] [PubMed] [Google Scholar]
- Bowers S., McPhillips T., Ludäscher B. (2008). Provenance in collection-oriented scientific workflows. Concur. Comput. Prac. Exp. 20, 519–529 10.1002/cpe.1226 [DOI] [Google Scholar]
- Callahan S., Freire J., Santos E., Scheidegger C. E., Silva C. T., Vo H. T. (2006). VisTrails: Visualization Meets Data Management, in Proceedings of the 2006 ACM SIGMOD International Conference on Management of Data. Chicago, IL, ACM [Google Scholar]
- Chiang M.-C., Dutton R. A., Hayashi K. M., Lopez O. L., Aizenstein H. J., Toga A. W., Becker J. T., Thompson P. M. (2007). 3D pattern of brain atrophy in HIV/AIDS visualized using tensor-based morphometry. NeuroImage 34, 44–60 10.1016/j.neuroimage.2006.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churches D., Gombas G., Harrison A., Maassen J., Robinson C., Shields M., Taylor I., Wang I. (2006). Programming scientific and distributed workflow with Triana services. Concur. Comput. Prac. Exp. 18, 1021–1037 10.1002/cpe.992 [DOI] [Google Scholar]
- Dinov I. D., Rubin D., Lorensen W., Dugan J., Ma J., Murphy S., Kirschner B., Bug W., Sherman M., Floratos A., Kennedy D., Jagadish H. V., Schmidt J., Athey B., Califano A., Musen M., Altman R., Kikinis R., Kohane I., Delp S., Parker D. S., Toga A. W. (2008). iTools: a framework for classification, categorization and integration of computational biology resources. PLoS ONE 3, e2265. 10.1371/journal.pone.0002265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets W. C. (2001). Neuroimaging and neuropathological studies of depression: implications for the cognitive-emotional features of mood disorders. Curr. Opin. Neurobiol. 11, 240–249 10.1016/S0959-4388(00)00203-8 [DOI] [PubMed] [Google Scholar]
- Fleisher A., Grundman M., Jack C. R., Jr., Petersen R. C., Taylor C., Kim H. T., Schiller D. H. B., Bagwell V., Sencakova D., Weiner M. F., DeCarli C., DeKosky S. T., van Dyck C. H., Thal L. J. (2005). Sex, apolipoprotein E {varepsilon}4 status, and hippocampal volume in mild cognitive impairment. Arch. Neurol. 62, 953–957 10.1001/archneur.62.6.953 [DOI] [PubMed] [Google Scholar]
- Gogtay N., Nugent T. F., Herman D. H., Ordonez A., Greenstein D., Hayashi K. M., Clasen L., Toga A. W., Giedd J. M., Rapoport J. L., Thompson P. M. (2006). Dynamic mapping of normal human hippocampal development. Hippocampus 16, 664–672 10.1002/hipo.20193 [DOI] [PubMed] [Google Scholar]
- Kawas E., Senger M., Wilkinson M. (2006). BioMoby extensions to the Taverna workflow management and enactment software. BMC Bioinformatics 7, 523. 10.1186/1471-2105-7-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen M., Durston S., Staal W., Palmen S., van Engeland H. (2007). Caudate nucleus is enlarged in high-functioning medication-naive subjects with autism. Biol. Psychiatry 62, 262–266 10.1016/j.biopsych.2006.09.040 [DOI] [PubMed] [Google Scholar]
- Liu L., Meier D., Polgar-Turcsanyi M., Karkocha P., Bakshi R., Guttmann C. R. G. (2005). Multiplesclerosis medical image analysis and information management. Neuroimaging 15(4 Suppl.), 103S–117S 10.1177/1051228405282864 [DOI] [PubMed] [Google Scholar]
- Ludäscher B., Altintas I., Berkley C., Higgins D., Jaeger E., Jones M., Lee E. A., Tao J., Zhao Y. (2006). Scientific workflow management and the Kepler system. Concur. Comput. Prac. Exp. 18, 1039–1065 10.1002/cpe.994 [DOI] [Google Scholar]
- Luders E., Narr K. L., Thompson P. M., Rex D. E., Woods R. P., DeLuca H., Jancke L., Toga A. W. (2006). Gender effects on cortical thickness and the influence of scaling. Hum. Brain Mapp. 27, 314–324 10.1002/hbm.20187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie-Grahama A., Tinsleyb M. R., Shaha K. P., Aguilara C., Stricklanda L. V., Bolinea J., Martinc M., Moralesd L., Shattucka D. W., Jacobse R. E., Voskuhld R. R., Toga A. W. (2006). Cerebellar cortical atrophy in experimental autoimmune encephalomyelitis. Neuroimage 32, 1016–1023 10.1016/j.neuroimage.2006.05.006 [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A., Payan A., Dinov I. D., Van Horn J. D., Toga A. W. (2008a). Neuroimaging data provenance using the LONI pipeline workflow environment. LNCS 5272, 208–220 [Google Scholar]
- MacKenzie-Graham A., Van Horn J. D., Woods R. P., Crawford K. L., Toga A. W. (2008b). Provenance in neuroimaging. NeuroImage 42, 178–195 10.1016/j.neuroimage.2008.04.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles S., Groth P., Branco M., Moreau L. (2006). The requirements of using provenance in e-science experiments. J. Grid Comput. 5, 1–25 10.1007/s10723-006-9055-3 [DOI] [Google Scholar]
- Mueller S., Weiner M., Thal L., Petersen R., Jack C., Jagust W., Trojanowski J., Toga A., Beckett L. (2005). Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI). Alzheimers Dement. 1, 55–66 10.1016/j.jalz.2005.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J., Allison T. C., Bittner S., Didier B., Frenklach M., Green W. H., Ho Y. L., Hewson J., Koegler W., Lansing C., Leahy D., Lee M., McCoy R., Minkoff M., Nijsure S., Von Laszewski G., Montoya D., Oluwole L., Pancerella C., Pinzon R., Pitz W., Rahn L. A., Ruscic B., Schuchardt K., Stephan E., Wagner A., Windus T., Yang C. (2006). A collaborative informatics infrastructure for multi-scale science. Cluster Comput. 8, 243–253 10.1007/s10586-005-4092-4 [DOI] [Google Scholar]
- Narr K. L., Bilder R. M., Luders E., Thompson P. M., Woods R. P., Robinsond D., Szeszkod P.R., Dimtcheva T., Gurbani M., Toga A. W. (2007). Asymmetries of cortical shape: effects of handedness, sex and schizophrenia. NeuroImage 34, 939–948 10.1016/j.neuroimage.2006.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinn T., Greenwood M., Addis M., Alpdemir M. N., Ferris J., Glover K., Goble C., Goderis A., Hull D., Marvin D., Li P., Lord P., Pocock M. R., Senger M., Stevens R., Wipat A., Wroe C. (2005). Taverna: lessons in creating a workflow environment for the life sciences. Concur. Comput. Prac. Exp. 18, 1067–1100 10.1002/cpe.993 [DOI] [Google Scholar]
- Rasser P., Johnston P., Lagopoulos J., Ward P. B., Schall U., Thienel R., Bender S., Toga A. W., Thompson P. M. (2005). Functional MRI BOLD response to Tower of London performance of first-episode schizophrenia patients using cortical pattern matching. NeuroImage 26, 941–951 10.1016/j.neuroimage.2004.11.054 [DOI] [PubMed] [Google Scholar]
- Rex D. E., Ma J. Q., Toga A. W. (2003). The LONI pipeline processing environment. Neuroimage 19, 1033–1048 10.1016/S1053-8119(03)00185-X [DOI] [PubMed] [Google Scholar]
- Rex D. E., Shattuck D. W., Woods R. P., Narr K. L., Luders E., Rehm K., Stolzner S. E., Rottenberg D. A., Toga A. W. (2004). A meta-algorithm for brain extraction in MRI. NeuroImage 23, 625–637 10.1016/j.neuroimage.2004.06.019 [DOI] [PubMed] [Google Scholar]
- Simmhan Y. L., Plale B., Gannon D. (2005). A survey of data provenance in escience. ACM SIGMOD Rec. 34, 31–36 10.1145/1084805.1084812 [DOI] [Google Scholar]
- Sowell E., Peterson B. S., Kan E., Woods R. P., Yoshii J., Bansal R., Xu D., Zhu H., Thompson P. M., Toga A. W. (2007). Sex differences in cortical thickness mapped in 176 healthy individuals between 7 and 87 years of age. Cereb. Cortex 17, 1550–1560 10.1093/cercor/bhl066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stef-Praun T., Clifford B., Foster I., Hasson U., Hategan M. S., Small L., Wilde M., Zhao Y. (2007). Accelerating medical research using the swift workflow system. In Studies in Health Technology and Informatics: From Genes to Personalized HealthCare: Grid Solutions for the Life Sciences – Proceedings of HealthGrid 2007, Nicolas Jacq H. M., Blanquer I., Legré Y., Breton V., Hausser D., Hernández V., Solomonides T., Hofmann-Apitius M., eds, pp. 207–216 [PMC free article] [PubMed] [Google Scholar]
- Taylor I., Shields M., Wang I., Harrison A. (2006). Visual grid workflow in Triana. J. Grid Comput. 3, 153–169 10.1007/s10723-005-9007-3 [DOI] [Google Scholar]
- Thompson P., Hayashi K. M., de Zubicaray G., Janke A. L., Rose S. E., Semple J., Herman D., Hong M. S., Dittmer S. S., Doddrell D. M., Toga A. W. (2003). Dynamics of gray matter loss in Alzheimer's disease. J. Neurosci. 23, 994–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga A. W., Thompson P. M. (2007). What is where and why it is important. NeuroImage 37, 1045–1049 10.1016/j.neuroimage.2007.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z., Narr K. L., Dollar P., Dinov I., Thompson P. M., Toga A. W. (2008). Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans. Med. Imaging 27, 495–508 10.1109/TMI.2007.908121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R., Grafton S. T., Holmes C. J., Cherry S. R., Mazziotta J. C. (1998). Automated Image Registration: I. General Methods and Intrasubject, Intramodality Validation. J. Comput. Assist. Tomogr. 22, 139–152 10.1097/00004728-199801000-00027 [DOI] [PubMed] [Google Scholar]
- Zhao J., Goble C., Stevens R., Turi D. (2007). Mining Taverna's semantic web of provenance. Concur. Comput. Prac. Exp. 20, 463–472 10.1002/cpe.1231 [DOI] [Google Scholar]