Abstract
Objective
Studies of cervical cancer and its immediate precursor, cervical intraepithelial neoplasia 3 (CIN3), have identified genes that often show aberrant DNA methylation and therefore, represent candidate early detection markers. We used quantitative PCR assays to evaluate methylation in five candidate genes (TNFRSF10C, DAPK1, SOCS3, HS3ST2 and CDH1) previously demonstrated as methylated in cervical cancer.
Methods
In this analysis, we performed methylation assays for the five candidate genes in 45 invasive cervical cancers, 12 histologically normal cervical specimens, and 23 liquid-based cervical cytology specimens confirmed by expert review as unequivocal demonstrating cytologic high-grade squamous intraepithelial lesions, thus representing the counterparts of histologic CIN3.
Results
We found hypermethylation of HS3ST2 in 93% of cancer tissues and 70% of cytology specimens interpreted as CIN3; hypermethylation of CDH1 was found in 89% of cancers and 26% of CIN3 cytology specimens. Methylation of either HS3ST2 or CDH1 was observed in 100% of cervical cancer tissues and 83% of CIN3 cytology specimens. None of the five genes showed detectable methylation in normal cervical tissues.
Conclusion
Our data support further evaluation of HS3ST2 and CDH1 methylation as potential markers of cervical cancer and its precursor lesions.
Keywords: cervical cancer, promoter methylation, tumor suppressor genes
INTRODUCTION
In developed countries, cytologic screening has contributed to a reduction in cervical cancer incidence and mortality [1–3]. Although individual cytologic screens are insensitive, repeat testing beginning at an early age permits detection of precursors such as cervical intraepithelial neoplasia 3 (CIN3) prior to invasion. Recognition that this process is inefficient and costly, however, has prompted efforts to develop highly sensitive screening approaches that can be performed at more widely spaced intervals [4].
Hypermethylation of promoter-associated CpG islands is now recognized as a frequent early event in the pathogenesis of many cancers, including cervical cancer. Developing a panel of methylation markers may therefore have value in early detection of cancer precursors, provide added reassurances of safety for women who are candidates for less frequent screens, and predict outcomes of women infected with carcinogenic human papillomavirus infections [5]. In this analysis, we evaluated methylation in cervical cancer and CIN3 using a candidate panel of five genes: TNFRSF10C, an antiapoptotic receptor of the TRAIL (TNF-related apoptosis-inducing ligand) pathway lacking death domain decoys [6]; death-associated protein kinase (DAPK1) [7]; SOCS3, a suppressor of cytokine signaling [8]; heparan sulfate D-glucosamyl 3-O-sulfotransfarase-2 (HS3ST2), which encodes an enzyme involved in the final modification step of heparan sulfate proteoglycans (HSPGs) [9]; and CDH1, an adhesion molecule [10] Previous studies have shown that these genes are frequently methylated in cervical cancers and tumors of other organ sites whereas aberrant methylation was not detected in a limited number of normal cervical tissues tested [6–10]. The aim of the present study was to evaluate methylation of these five a priori candidate genes in a convenience sample of invasive cervical cancer tissues and residual liquid-based cytology specimens confirmed by expert review as CIN3.
MATERIALS AND METHODS
Clinical samples
A total of 45 histologically confirmed cervix cancers (35 squamous cell carcinomas, 10 adenocarcinomas) were snap frozen in liquid nitrogen and stored at −80°C at the Washington University School of Medicine (St. Louis, MO). Sample collection was approved by Washington University in St. Louis Human Studies Committee and informed consent was obtained from all patients. Cervical cancer specimens comprised both biopsies and radical hysterectomies. Over half of the cancers were stage 1; the remaining cancers were equally distributed as stage 2 and stage 3. Residual liquid based cytology specimens collected in PreservCyt (Cytyc, Boxborough, MA) from 23 clinical specimens were collected at the Northwestern University Feinberg School of Medicine (Chicago, IL) and at the University of Rochester Medical Center (Rochester, NY). Expert cytopathologists rigorously confirmed the cytologic interpretations of high-grade squamous intraepithelial lesions equivalent to histologic CIN3. PreservCyt specimens were anonymized, stored at −80°C and subsequently tested under an IRB exemption granted by participating institutions. All tissue and cytology samples were shipped on dry ice to the University of Texas Southwestern Medical Institute, where an additional twelve normal cervix tissues collected anonymously under an IRB exemption and stored at −70°C until DNA extraction were also evaluated.
DNA extraction, bisulfite treatment
Genomic DNA was extracted from cells spun down from 1.5-5 ml of residual PreservCyt and frozen primary tumors as previously described (Serological Corporation, Norcross, GA) [11]. Chemical modification of unmethylated cytosines to uracil within CpG islands using sodium-bisulfite treatment was performed as described previously [12]. The modified DNA was used as a template for subsequent PCR analysis.
Semiquantitative real time PCR
Real time PCR analysis was performed using the Chromo4 MJ Research Real time PCR system (Waltham, MA). Sodium bisulfite-treated genomic DNA was amplified by fluorescence-based real-time methylation-specific primers using TaqMan technology and specifically designed oligonucleotide primers designed to amplify bisulfite-converted DNA. Specifically, primers amplified regions of the genes previously shown to be associated with loss of mRNA or protein expression [13]. PCR assays were performed in a reaction volume of 25 μl. The final reaction mixture contained the forward and reverse primers at 600 nmol/L each; the probe at 200 nmol/L; 200 μmol/L for each of the four nucleotide phosphates; 5.5 mmol/L MgCl2; 1XTaqMan Buffer A; 1U of Hot Star Taq DNA polymerase (Qiagen, Valencia, CA) and 1μl bisulfite-converted genomic DNA (corresponds to 50 ng initial DNA used for conversation). PCR was performed under the following conditions: 95°C for 15 minutes, followed by 50 cycles at 95°C for 15 seconds and 60°c for 1 minute.
The non-methylated form of MYOD1 (uMYOD1) was used as an internal reference standard. Amplification of MYOD1 is independent of its methylation status, whereas the amplification of test genes is proportional to the degree of cytosine methylation within the amplicon. Serial dilution of the Sss1 treated human genomic DNA from normal human lymphocytes was therefore used to create a standard curve. The fluorescence emission intensities (threshold cycle or Ct values) for the five genes and uMYOD1 were calculated using the intercept and the slope of the standard graph. The methylation ratio was defined as the ratio of the fluorescence emission intensity values for the PCR products of the five genes to those of PCR products of uMYOD1 multiplied by 100. The ratio is therefore a measure for the relative level of methylation in an individual sample.
Both quantitative ratio (QR) for methylation (e.g., methylation level) and methylation frequency (percent) in all specimens were evaluated. For QR, the median, average and range were calculated. A QR of 0 was considered methylation below the level of detection. Calculations were made for all cervical cancer tissues, CIN3 cytology specimens, and normal cervix tissues. Cervical cancers were further analyzed by histologic subtype (squamous cell carcinoma and adenocarcinoma).
Statistical Analysis
The frequencies of methylation between any two groups were compared using two-sided Fisher’s exact test. Probability values of P<0.05 were regarded as statistically significant.
RESULTS
94% of squamous cell carcinomas and 90% of adenocarcinomas showed abnormal methylation results for HS3ST2 and CDH1 (Table 1). Assays for the other three genes yielded lower methylation frequencies than HS3ST2 and CDH1 in both tumor types with comparable semi-quantitative levels. Methylation of either HS3ST2 or CDH1 was identified in all cervical cancers.
Table 1.
Quantitative ratio (median, average, and range) for methylation of genes in cervical cancers and precancerous lesions.
| Methylated Gene | Squamous Cell Carcinomaa | Adenocarcinomab | CIN-3c | |||
|---|---|---|---|---|---|---|
| Methylation Frequency | Methylation Frequency | Methylation Frequency | ||||
| HS3ST2 | ||||||
| Median | 100 | 647.43 | 3.15 | |||
| Average | 259.41 | 33/35 (94%) | 791.66 | 9/10 (90%) | 67.82 | 16/23 (70%) |
| Range | 0–1207.82 | 0–2029.16 | 0–891.25 | |||
|
| ||||||
| TNFRSF10C | ||||||
| Median | 0.45 | 0 | 2/10 (20%) | 0 | 4/23 (17%) | |
| Average | 42.56 | 20/35 (57%) | 0.04 | (* a,b=0.043) | 0.36 | (* a,c=0.003) |
| Range | 0–366.44 | 0–0.34 | 0–5.08 | |||
|
| ||||||
| CDH1 | ||||||
| Median | 3.65 | 2.89 | 0 | 6/23 (26%) | ||
| Average | 8.14 | 31/35 (89%) | 4.00 | 9/10 (90%) | 0.65 | (* a,c<0.001) |
| Range | 0–52.97 | 0–11.77 | 0–5.74 | (* b,c<0.001) | ||
|
| ||||||
| DAPK1 | ||||||
| Median | 0.78 | 0 | 0 | |||
| Average | 28.32 | 21/35 (21%) | 11.34 | 3/10 (30%) | 1.06 | 4/23 (17%) |
| Range | 0–171.79 | 0–106.22 | 0–14.53 | |||
|
| ||||||
| SOCS3 | ||||||
| Median | 0 | 0.16 | 0 | |||
| Average | 17.17 | 7/35 (20%) | 77.68 | 5/10 (50%) | 1.47 | 2/23 (9%) |
| Range | 0–537.03 | 0–562.14 | 0–29.5 | |||
The levels of methylation were quantitated using quantitative PCR technique as described in Materials and Methods Quantitative ratios (QR) are ratios of the fluoresecence intensities of the gene and MYOD1. QR 0 means below the level of detection
Statistical significance by Fisher’s test p<0.05
The frequency of methylation and the semi-quantitative levels of methylation when detected were lower for every gene in CIN3 cytology specimens than in cancer. In CIN3 cytology specimens, the frequency of methylation ranged from 9% to 70% for the five genes, with the highest frequencies found for HS3ST2 (70%) and CDH1 (26%). Methylation of at least one of the two genes (HS3ST2, CDH1) was observed in 83% of cytology specimens interpreted as CIN3. Detection of methylation in any of the five genes did not increase sensitivity.
In general, genes that were most frequently methylated also had higher QRs, and thus were most heavily methylated. For both squamous cell and adenocarcinoma, HS3ST2 demonstrated the highest methylation QR. Interestingly, the median HS3ST2 QR observed in invasive adenocarcinoma (647.43) was much higher than that observed for squamous cell carcinoma (100). CDH1 displayed modest QR in both squamous cell (3.65) and adenocarcinoma (2.89). The remaining three genes had low QRs for both squamous cell and adenocarcinomas.
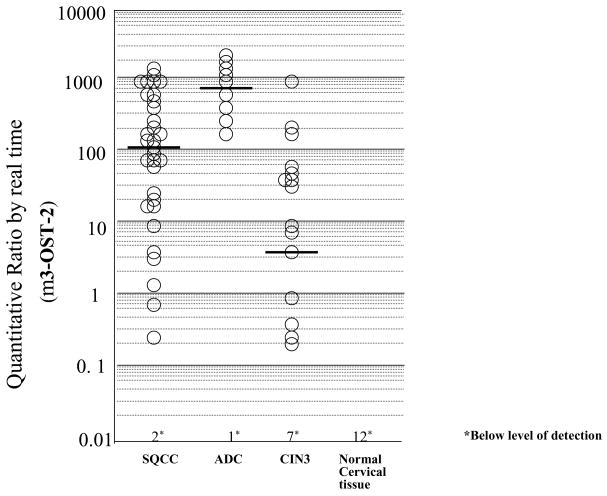
We observed significantly lower median and mean QRs for liquid-based cytology specimens interpreted as CIN3 than cancer tissues for all genes. For CIN3 cytology specimens, HS3ST2 had the highest median QR (3.15); median QRs in CIN3 for the remaining four genes were below the level of detection. Comparison of QR for squamous cell, adenocarcinoma, and CIN3 is illustrated in Figure 1 for HS3ST2. None of the five genes showed detectable methylation in 12 normal cervical tissues.
Figure 1.
Methylation levels of HS3ST2 in squamous cell carcinoma (SQCC), adenocarcinoma (ADC), cervical intraepithelial neoplasia 3 (CIN3), and normal cervical tissue. Real-time analysis was performed as described in “Materials and Methods.” Quantitative ratio is defined as the ratio of the fluorescence emission intensity values for the PCR products of the gene to those of PCR products of uMYOD1 multiplied by 100. The ratio is a measure for the relative level of methylation in an individual sample. Horizontal bars indicate median values.
DISCUSSION
In this analysis, the combined results for PCR assays designed to detect methylation of HS3ST2 or CDH1 were positive in 100% of cervical cancer tissues and 83% of cytology specimens interpreted as CIN3. Methylation was not detected in any of 12 histologically normal tissue specimens for either marker. The two genes are of further interest as heparin sulfate proteoglycans have been demonstrated to mediate binding of the human papillomavirus (HPV) to cell surfaces [14] and e-cadherin has been reported to play an important role in mounting an immune response to HPV infection by mediating the adhesion of dendritic and Langerhans cells [15].
Our results provide support for evaluation of HS3ST2 and CDH1 and other candidate markers in a large epidemiologic study where there are cytology specimens associated with a range of cytologic interpretations and well-defined outcomes determined in prospective follow-up. The equally sensitive detection of methylation of HS3ST2 and CDH1 in squamous cell and adenocarcinoma is potentially important for further follow-up, particularly among adenocarcinoma in situ (AIS) because cytology is insensitive in identifying AIS and adenocarcinomas are suspected to account for a disproportionate percentage of rapidly developing cancers [16].
Although several studies have evaluated methylation markers in cervical cancers [17–20] data for cervical precursor lesions are more limited. Steenbergen et al [21] reported methylation of TSLC1 gene at frequencies of 35% and 58% in precancerous lesions and cervical cancers, respectively. Li et al [22] reported methylation of TSLC1 gene at a frequency of 65% in cervical cancers. Feng et al [23] reported methylation of at least one of the three genes (DAPK1, RARβ and TWIST1) at frequencies of 57% and 74% in CIN3 lesions and cervical cancers, respectively.
Similar to these previous studies, we found that detection of specific methylation markers was generally lower in cytologic specimens showing CIN3 than in cancer tissues. Both lesion severity and specimen type could have contributed to this difference in our study.
Higher levels of methylation in cancer as opposed to CIN3 may reflect progressive accumulation of methylation events as putative precursors progress to carcinoma. This might be expected if methylation confers cells with a relative growth advantage. Although CIN3 represents the most proximate histologic precursor of invasive carcinoma, the peak age of detection for CIN3 lesions is 10–20 years younger than invasive cancer. Therefore, many CIN3 lesions seem to persist and expand for long periods prior to invasion, which itself is not recognized as an inevitable occurrence. These data suggest that assessment of methylation among women infected with carcinogenic HPV types stratified by severity of concurrent lesions, age, HPV type and other factors may be of interest in understanding progression.
We acknowledge that the disparity observed between CIN3 cytology and cancer tissues may reflect inherent differences between the specimen types. Cytology sampling always includes many ”normal” cells and also includes many superficial epithelial cells, whereas cancer tissues may consist of purer populations of tumor cells. Consequently, useful assays must have high analytical sensitivity to detect rare but important changes in samples, yet at the same time not detect random insignificant changes that lack biological consequences. The importance of these concerns is demonstrated by the wide range of semi-quantitative methylation values that were determined in the cytology specimens, even though all had been interpreted by experts as showing definite CIN3.
Finally, we acknowledge inconsistencies in the published literature regarding methylation of the same genes. For example, Kang et al [24] and Yang et al [25] reported DAPK1 methylation of 73% and 60%, respectively, compared to our report of 21%. Yang et al also found DAPK1 methylation more common in SCC than in AC. Such discrepancies underscore many of the technical issues related to assay development. Specifically, in the Kang et al and Yang et al articles, methylation was conducted with a standard/conventional protocol, not real time PCR. Our present manuscript uses real time PCR with TaqMan probe Technology where two additional primers are used (forward and reverse) and a sequence specific probe in the middle makes the assay more specific. Standard or conventional PCR may thus result in higher methylation frequencies due to the lack of specificity. Reasons for discrepancy by cell type are more difficult to explain although the combination of using different assays, having varying and usually small sizes, and geographically different patient populations [26–28] are possibilities.
In summary, iterative cytologic screening has demonstrated effectiveness in reducing cervical cancer incidence and mortality, but burdens resulting from repeated testing have prompted efforts to develop highly sensitive tests that can provide safety with fewer lifetime screens. Identifying abnormal methylation events is a promising avenue of research for developing such tests, but further efforts to identify and validate a clinical useful marker panel are needed.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, and partially funded by CA95713 and CA94141.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pisani P, Bray F, Parkin DM. Estimates of the world-wide prevalence of cancer for 25 sites in the adult population. Int J Cancer. 2002;97:72–81. doi: 10.1002/ijc.1571. [DOI] [PubMed] [Google Scholar]
- 2.Wang SS, Sherman ME, Hildesheim A, Lacey JV, Jr, Devesa S. Cervical adenocarcinoma and squamous cell carcinoma incidence trends among white women and black women in the United States for 1976–2000. Cancer. 2004;100:1035–1044. doi: 10.1002/cncr.20064. [DOI] [PubMed] [Google Scholar]
- 3.Kulasingam SL, Hughes JP, Kiviat NB, Mao C, Weiss NS, Kuypers JM, Koutsky LA. Evaluation of human papillomavirus testing in primary screening for cervical abnormalities: comparison of sensitivity, specificity, and frequency of referral. Jama. 2002;288:1749–1757. doi: 10.1001/jama.288.14.1749. [DOI] [PubMed] [Google Scholar]
- 4.Bulkmans NW, Berkhof J, Bulk S, Bleeker MC, van Kemenade FJ, Rozendaal L, Snijders PJ, Meijer CJ. HPV type-specific clearance rates in cervical screening. Br J Cancer. 2007;96(9):1419–24. doi: 10.1038/sj.bjc.6603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 6.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 7.Kogel D, Prehn JH, Scheidtmann KH. The DAP kinase family of pro-apoptotic proteins: novel players in the apoptotic game. Bioessays. 2001;23:352–358. doi: 10.1002/bies.1050. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson SE, De Souza D, Fabri LJ, Corbin J, Willson TA, Zhang JG, Silva A, Asimakis M, Farley A, Nash AD, Metcalf D, Hilton DJ, Nicola NA, Baca M. Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proc Natl Acad Sci U S A. 2000;97:6493–6498. doi: 10.1073/pnas.100135197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto K, Asada K, Fukutomi T, Okochi E, Yagi Y, Hasegawa T, Asahara T, Sugimura T, Ushijima T. Methylation-associated silencing of heparan sulfate D-glucosaminyl 3-O-sulfotransferase-2 (3-OST-2) in human breast, colon, lung and pancreatic cancers. Oncogene. 2003;22:274–280. doi: 10.1038/sj.onc.1206146. [DOI] [PubMed] [Google Scholar]
- 10.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 11.Shivapurkar N, Takahashi T, Reddy J, Zheng Y, Stastny V, Collins R, Toyooka S, Suzuki M, Parikh G, Asplund S, Kroft SH, Timmons C, McKenna RW, Feng Z, Gazdar AF. Presence of simian virus 40 DNA sequences in human lymphoid and hematopoietic malignancies and their relationship to aberrant promoter methylation of multiple genes. Cancer Res. 2004;64:3757–3760. doi: 10.1158/0008-5472.CAN-03-3307. [DOI] [PubMed] [Google Scholar]
- 12.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shivapurkar N, Stastny V, Suzuki M, Wistuba II, Li L, Zheng Y, Feng Z, Hol B, Prinsen C, Thunnissen FB, Gazdar AF. Application of a methylation gene panel by quantitative PCR for lung cancers. Cancer Lett. 2006 doi: 10.1016/j.canlet.2006.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J Virol. 2001;75(3):1565–70. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humbert P, Caberg JH, Gilles C, Bousarghin L, Franzen-Detrooz E, Boniver J, Delvenne P. E-cadherin-dependent adhesion of dendritic and Langerhans cells to keratinocytes is defection in cervical human papillomavirus-associated (pre)neoplastic lesions. J Pathol. 2005;206(3):346–55. doi: 10.1002/path.1771. [DOI] [PubMed] [Google Scholar]
- 16.Hildesheim A, Hadjimichael O, Schwartz PE, Wheeler CM, Barnes W, Lowell DM, Willett J, Schiffman M. Risk factors for rapid-onset cervical cancer. Am J Obstet Gynecol. 1999;180:571–577. doi: 10.1016/s0002-9378(99)70256-5. [DOI] [PubMed] [Google Scholar]
- 17.Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res. 2001;7:1982–1986. [PubMed] [Google Scholar]
- 18.Sova P, Feng Q, Geiss G, Wood T, Strauss R, Rudolf V, Lieber A, Kiviat N. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:114–123. doi: 10.1158/1055-9965.EPI-05-0323. [DOI] [PubMed] [Google Scholar]
- 19.Shivapurkar N, Toyooka S, Toyooka KO, Reddy J, Miyajima K, Suzuki M, Shigematsu H, Takahashi T, Parikh G, Pass HI, Chaudhary PM, Gazdar AF. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int J Cancer. 2004;109:786–792. doi: 10.1002/ijc.20041. [DOI] [PubMed] [Google Scholar]
- 20.Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF. Aberrant methylation during cervical carcinogenesis. Clin Cancer Res. 2001;7:584–589. [PubMed] [Google Scholar]
- 21.Steenbergen RD, Kramer D, Braakhuis BJ, Stern PL, Verheijen RH, Meijer CJ, Snijders PJ. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004;96:294–305. doi: 10.1093/jnci/djh031. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Zhang Z, Bidder M, Funk MC, Nguyen L, Goodfellow PJ, Rader JS. IGSF4 promoter methylation and expression silencing in human cervical cancer. Gynecol Oncol. 2005;96:150–158. doi: 10.1016/j.ygyno.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 23.Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi L, Lu H, McIntosh MW, Young AM, Kiviat NB. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97:273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]
- 24.Kang S, Kim JW, Kang GH, Park NH, Song YS, Kang SB, Lee HP. Polymorphism in folate- and methionine-metabolizing enzyme and aberrant CpGisland hypermethylation in uterine cervical cancer. Gyn Oncol. 2005;97:173–80. doi: 10.1016/j.ygyno.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 25.Yang HJ, Liu VW, Wang Y, Chan KY, Tsang PC, Khoo US, Cheung AN, Ngan HY. Detection of hypermethylated genes in tumor and plasma of cervical cancer patients. Gyn Oncol. 2004;93:435–40. doi: 10.1016/j.ygyno.2004.01.039. [DOI] [PubMed] [Google Scholar]
- 26.Chan AO, Soliman AS, Zhang Q, Rashid A, Bedeir A, Houlihan PS, Mokhtar N, Al-Masri N, Ozbek U, Yaghan R, Kandilci A, Omar S, Kapran Y, Dizdaroglu F, Bondy ML, Amos CI, Issa JP, Levin B, Hamilton SR. Differing DNA methylation patterns and gene mutation frequencies in colorectal carcinomas from Middle Eastern countries. Clinical Cancer Res. 2005;11:8281–8287. doi: 10.1158/1078-0432.CCR-05-1000. [DOI] [PubMed] [Google Scholar]
- 27.Yang et al. American Journal of Pathology. 2003;163:1101–1107. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang B, Guo M, Herman JG, Clark DP. Aberrant Promoter Methylation Profiles of Tumor Suppressor Genes in Hepatocellular Carcinoma. Am J Pathol. 2003;163:1101–1107. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the Epidermal Growth Factor Receptor Gene in Lung Cancer: Biological and Clinical Implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]