Abstract
Survivorship experiments play a central role in aging research and are performed to evaluate whether interventions alter the rate of aging and increase lifespan. The accelerated failure time (AFT) model is seldom used to analyze survivorship data, but offers a potentially useful statistical approach that is based upon the survival curve rather than the hazard function. In this study, AFT models were used to analyze data from 16 survivorship experiments that evaluated the effects of one or more genetic manipulations on mouse lifespan. Most genetic manipulations were found to have a multiplicative effect on survivorship that is independent of age and well-characterized by the AFT model “deceleration factor”. AFT model deceleration factors also provided a more intuitive measure of treatment effect than the hazard ratio, and were robust to departures from modeling assumptions. Age-dependent treatment effects, when present, were investigated using quantile regression modeling. These results provide an informative and quantitative summary of survivorship data associated with currently known long-lived mouse models. In addition, from the standpoint of aging research, these statistical approaches have appealing properties and provide valuable tools for the analysis of survivorship data.
Keywords: AFT model, cox, insulin-like growth factor, proportional hazard, survival analysis
1. Introduction
Survivorship patterns are a key indicator of the rate at which aging occurs within a population. Interventions and genetic manipulations found to significantly extend lifespan therefore gain prominence as new models for aging research and receive considerable attention. In mice, this approach has repeatedly been used to identify environmental interventions that increase lifespan (e.g., Baur et al., 2006; Miller et al., 2007), as well as genes for which knockout or over-expression leads to improved longevity (e.g., Brown-Borg et al., 1996; Flurkey et al., 2001; Schriner et al., 2005). Indeed, there has been striking proliferation in the number of genetic manipulations that increase mouse lifespan, with seven new models having emerged in 2007 alone (Conover and Bale, 2007; Dell’agnello et al., 2007; Li and Ren, 2007; Ran et al., 2007; Taguchi et al., 2007; Wu et al., 2007; Yan et al., 2007). It may, in fact, soon be of little interest to identify interventions that simply increase mouse lifespan “significantly”. Rather, more attention may be given to effect size, with greater focus on interventions for which lifespan increases are both “significant” and “large”. It is challenging, however, to generate robust and precise effect size estimates, since most laboratory experiments involve small sample sizes. In addition, a diverse range of metrics are now used by investigators to summarize treatment effects on mouse lifespan, and these are often reported without a confidence interval. This has made it difficult to carry out quantitative comparisons among treatments that increase mouse longevity, and to prioritize models in terms of their significance to aging research.
The percent treatment difference in mean or median lifespan is the most widely used metric for summarizing treatment effects on lifespan (Liang et al., 2003), and has been most commonly cited in abstracts of research reports describing treatments that increase longevity (e.g., Miskin and Masos, 1997; Migllaccio et al., 1999; Blüher et al., 2003; Holzenberger et al., 2003; Schriner et al., 2005; Conti et al., 2006; Conover and Bale, 2007; Taguchi et al., 2007; Yan et al., 2007). The abstract of Yan et al. (2007), for example, reports that mice lacking 5 adenylyl cyclase (AC5) have “increased median lifespan of ~ 30%”. An advantage of this approach is that it is based upon survival times and therefore provides an intuitive measure of treatment effect. At the same time, however, there are limitations, since estimated parameters are not embedded within a statistical model. It is not possible, for example, to calculate a 95% confidence interval for the effect of a given treatment, and records from censored individuals that have been removed from cohorts during an experimental study are ignored. More importantly, the measure does not incorporate covariate variables, which can impact the magnitude and significance of estimated treatment effects. Recently, for example, Taguchi et al. (2007) found that lifespan was significantly influenced by date of birth and the parental IDs associated with mice, and using a statistical modeling approach, accounted for these covariates when evaluating the effect of Irs2 mutations on survivorship (see also Conti et al., 2006). There are, in fact, many factors not fully controlled in most mouse longevity studies (e.g., litter size, parental age, mating status, number of animals per cage, duration of weaning, consumption of calories), and provided that this information is recorded, statistical modeling can be used to calculate treatment effects adjusted for these factors.
The semi-parametric Cox proportional hazards (PH) model is the most common approach for statistical modeling of survivorship data, and has been widely applied in epidemiological studies (Cox, 1972). Interestingly, however, the PH model has only seldom been used to evaluate treatment effects on mouse survivorship (Conti et al., 2006; Taguchi et al., 2007). One explanation is that the PH model does not generate an intuitive summary statistic that is interpreted in terms of survivorship (Keene, 2002). The PH model is based upon the hazard function and summarizes treatment effects in terms of the ratio of age-specific mortality rates in two treatments (i.e., the hazard ratio). Hazard ratios have been an important tool in medical research, but it is clear that biology of aging researchers prefer to visualize experimental results in terms of survival curves, rather than hazard functions. This has likely prevented the PH model from being widely used in experimental aging research, and has compelled many investigators to summarize treatment effects in terms of percent change in median or mean lifespan.
Parametric accelerated failure time (AFT) models provide an alternative to the PH model for statistical modeling of survival data (Wei, 1992). Unlike the PH model, the AFT approach models survival times directly and generates a summary measure that is interpreted in terms of the survival curve (Hutton and Monaghan, 2002; Orbe et al., 2002; Patel et al., 2006; Pourhoseinghol et al., 2007). Suppose that S1 (t) is the survivorship of mice receiving an experimental treatment at time t, while S0 (t) is the survivorship of mice belonging to a control treatment at time t. Within the AFT model framework, the treatment effect is to uniformly shift the survival curve forward or backward, with the extent of shift being determined by the parameter c in the following relationship (Collett, 2003).
| (1) |
If an experimental treatment increases survival, such that the survival curve S1 (t) of the treated mice is shifted forward, the estimated value of c will exceed one. For example, if the treatment increases median lifespan by 30%, the estimated value of c will be approximately 1.30. The AFT model therefore generates an intuitive summary measure, but provides the advantages of statistical modeling. In particular, a confidence interval on c can be calculated, and an adjusted value of c can be estimated that corrects for differences in covariate variables between treatments. The AFT approach requires that experimental data satisfy certain assumptions. In particular, the treatment effect on survivorship should be roughly consistent throughout the lifespan, and survival times are assumed to follow a parametric distribution that must be specified (e.g., exponential, Weibull, log-normal, log-logistic).
The purpose of this study was to evaluate the suitability of AFT models for analysis of survivorship data generated in mouse longevity experiments, and to compare AFT model results with those from other statistical methods. A timely and appropriate context for these evaluations is provided by the proliferating number of survivorship experiments that have demonstrated increased mouse longevity due to genetic manipulation (e.g., Dell’agnello et al., 2007; Taguchi et al., 2007). Previously published datasets are analyzed in uniform fashion using both AFT and PH models, and assumptions associated with each modeling approach are evaluated on a case-by-case basis. Using both approaches, effects of genetic manipulations on longevity are evaluated, and confidence intervals are presented to reflect the uncertainty associated with treatment effects. Quantile regression is also used to evaluate age-dependent treatment effects, as well as effects at late stages of the lifespan (Koenker and Geling, 2001; Wang et al., 2004). These analyses provide a quantitative summary of survivorship data associated with long-lived mouse models, and demonstrate analytical methods that can improve the evaluation of survivorship data upon which aging research models are based.
2. Methods
2.1 Survivorship Datasets
Experimental data were obtained from contact authors of research reports describing genetic manipulations that significantly increase mouse lifespan. In this context, “genetic manipulation” broadly refers to gene knockout mutations, as well as transgenic models in which a specific gene has been over-expressed. A comprehensive effort was made to obtain survivorship data from all such genetic manipulations that have been described, and ultimately, raw survivorship data was obtained from 16 published research studies (Brown-Borg et al., 1996; Miskin and Masos, 1997; Migliaccio et al., 1999; Flurkey et al., 2001; Mitsui et al., 2002; Blüher et al., 2003; Holzenberger et al., 2003; Kurosu et al., 2005; Liu et al., 2005; Schriner et al., 2005; Bonkowski et al., 2006; Conti et al., 2006; Conover and Bale, 2007; Dell’agnello et al., 2007; Ran et al., 2007; Taguchi et al., 2007). In one case, original survivorship data had been lost (Miskin and Masos, 1997), so approximated survival times were obtained from a magnified version of the published survival curve. Accuracy was verified by close agreement between descriptive statistics obtained from approximated survival times and those provided in the original research report.
Each analysis involved a two-treatment survivorship comparison between a (long-lived) experimental cohort and a control cohort. These comparisons are listed in Table 1, along with sample sizes used for experimental and control treatments, and a description of the genetic manipulation applied to experimental groups. For some comparisons, data was available that provided the option of evaluating treatment effects separately by gender, or separately for different genetic backgrounds. In these cases, the simplest model was assumed initially, with similar treatment effects in each gender or on multiple backgrounds. However, if there was statistically significant evidence to suggest that the simplest model was incorrect (e.g., significant gender × treatment interaction effect), treatment effects were estimated separately by gender or genetic background. This approach used the maximal amount of data for estimating treatment effects when such effects are similar by gender or background, but did not ignore interactions between treatment effects and other factors when evidence for such interactions was present.
Table 1.
Genetic manipulations that increase mouse lifespan. Each identifier represents a survivorship comparison between an experimental and control cohort, where experimental mice have undergone a genetic manipulation that promotes longevity. In each row, the identifier associated with each experiment is listed, along with sample sizes of experimental and control cohorts (excluding censored mice), and a description of the genetic manipulation associated with the experimental group. When there was significant evidence that the effects of a mutation on lifespan were gender-dependent, analyses were carried out separately by gender, and separate identifiers corresponding to only male mice (M) or only female mice (F) are listed. Similarly, when there was evidence that the effect of a mutation depended upon genetic background, separate identifiers for each background are listed (S1, S2, etc).
| Identifier | n (Exp.) | n (Control) | Description |
|---|---|---|---|
| Prop1(df/df)a | 32 | 27 | Homozygous mutation of paired like homeodomain factor 1 (Prop1). Ames background. |
| Pit1(dw/dw)b | 24 | 33 | Homozygous mutation of POU domain, class 1, transcription factor 1 (Pit1). DW/J Pit1dw x C3H/HeJ Pit1dw-J background. |
| PappA(−/−)c | 20 | 21 | Homozygous deletion of pregnancy-associated plasma protein A (PappA). Mixed background (C57BL6 and 129SV/E). |
| Clk1(+/−)(S2)d | 9 | 5 | Mice heterozygous for deletion of Clk1. 129Sv/J x Balb/c background. |
| Irs2(+/−)(M)e | 14 | 13 | Mice heterozygous for deletion of insulin receptor substrate 2 (Irs2). C57BL/6J Background. |
| P66(−/−)f | 14 | 14 | Homozygous deletion of src homology 2 domain-containing transforming protein C1 (Shc1). 129/Sv background. |
| Igf1r(+/−)(F)g | 20 | 17 | Mice heterozygous for deletion of insulin-like growth factor I receptor (Igf1r). 129/Sv background. |
| Ghr(−/−)h | 38 | 41 | Homozygous deletion of growth hormone receptor (Ghr). Heterogeneous background. |
| αMUPAi | 33 | 33 | Overexpression of urokinase-type plasminogen activator (uPA) in central nervous system, promoting 20% decrease in food consumption. NIH FVB/N background. |
| bIrs2(+/−)e | 64 | 93 | Brain-specfic, Heterozygous deletion of insulin receptor substrate 2 (Irs2). C57BL/6J Background. |
| Ghrhr(lit/lit)b | 33 | 31 | Homozygous mutation of Ghrhr growth hormone releasing hormone receptor (Ghrhr). C57BL/6 background. |
| MCATj | 62 | 102 | Transgenic mice that overexpress human catalase localized to the mitochondria. B6C3F1 background. |
| Clk1(+/−)(S1)d | 18 | 17 | Mice heterozygous for deletion of mClk1. 129Sv/j and C57BL/6J backgrounds. |
| fIr(−/−)k | 60 | 190 | Fat-specific deletion of insulin receptor (Ir). Mixed background (129Sv, C57Bl/6, FVB). |
| Irs2(+/−)(F)e | 17 | 17 | Mice heterozygous for deletion of insulin receptor substrate 2 (Irs2). C57BL/6J Background. |
| Klothol | 101 | 54 | Transgenic mice that overexpress klotho hormone. C3H background. |
| bIrs2(−/−)e | 46 | 93 | Brain-specific, Homozygous deletion of insulin receptor substrate 2 (Irs2). C57BL/6J Background. |
| TRX-Tgm | 39 | 14 | Transgenic mice overexpressing human thioredoxin (TRX). C57BL/6 background. |
| p66(+/−)f | 8 | 14 | Mice heterozygous for deletion of src homology 2 domain-containing transforming protein C1 (Shc1). 129/Sv background. |
| Hcrt-UCP2n | 85 | 67 | Transgenic mice that overexpress uncoupling protein 2 in hypocretin neurons. C57/BL6 background. |
| Surf1(−/−)o | 41 | 48 | Homozygous deletion of surfeit gene 1 (Surf1). Mixed BDF1 background. |
| Gpx4(+/−)p | 50 | 50 | Mice heterozygous for deletion of glutathione peroxidase 4 (Gpx4). C57BL/6 background. |
2.2 Accelerated Failure Time (AFT) Model
The AFT model was applied to each Table 1 comparison to generate an estimate of the parameter c from Equation (1). The parameter c is here referred to as the “deceleration factor”, since it provides an indication of the degree to which mortality patterns, and perhaps aging, are slowed in the experimental treatment versus on the control treatment (Collett, 2003). For a given comparison, the value of 100(c − 1) serves as an estimate for the percent median lifespan increase in the experimental versus the control treatment (Patel et al., 2006). In fact, the value of 100(c − 1) describes the percent increase in lifespan with respect to any survival time quantile, and not just the median. This broad interpretation of 100(c − 1) is possible, since the AFT model assumes that treatments have a multiplicative effect on survivorship that is consistent throughout the lifespan. Since this is never the case exactly, the value of 100(c − 1) tends to describe the treatment effect “averaged” throughout the lifespan at early, middle and late ages. For example, calculating the percent lifespan increase at the 0.20, 0.40, 0.60 and 0.80 lifespan quantiles, and then averaging these four percentage values, would provide an approximation to the value of 100(c − 1).
The AFT model treats the logarithm of survival time as the response variable and includes an error term that is assumed to follow a particular distribution. Equation (2) shows the log-linear representation of the AFT model for the ith individual, where logTi is the log-transformed survival time, X1…X p are explanatory variables with coefficients β1…βp, εi represents residual or unexplained variation in the log-transformed survival times, while μ and σ are intercept and scale parameters, respectively (Collett, 2003).
| (2) |
In the absence of covariates, a single variable X1 was defined as a 0–1 indicator variable distinguishing between control and experimental treatments, and the deceleration factor was calculated from the coefficient estimate associated with X1 (i.e., ĉ = exp(β̂1)). When covariate data was available, additional terms β2 x2…βp xp were included in the model, where the added variables represented factors such as gender, date of birth and parental ids.
An initial step in fitting an AFT model is determining which distribution should be specified for the survival times Ti (Equation 2). Under the AFT model parameterization, the distribution chosen for Ti dictates the distribution of the error term εi. For instance, if survival times are modeled as a Weibull distribution, the error term is assumed to follow an extreme-value distribution. Likewise, if survival times are modeled using the log-logistic or log-normal distribution, the εi are assumed to be logistic or normal, respectively. For each comparison, preliminary models were fit in which the Ti were modeled using the exponential, Weibull, Gompertz, log-logistic and log-normal distributions, and the appropriate distribution was selected as the one which minimized the Akaike’s Information Criterion (AIC) (Akaike, 1974). In almost every case, the Weibull distribution was the most appropriate based upon the AIC criterion. An exception was the TRX-Tg comparison, for which the log-normal distribution emerged as the most appropriate choice for modeling the Ti. The exact choice of distribution proved to be of limited consequence, and results were similar regardless of the chosen distribution. For example, when deceleration factor estimates were compared between the best and next-best distributions (AIC criterion), estimates differed by only 3.9% on average (range: 0.3% – 11.4%). Moreover, between the best and next-best distributions, statistical significance of deceleration factor estimates differed for only 2 of 22 comparisons (TRX-Tg and p66(+/−)).
Covariate data was available for several comparisons (e.g., gender, date of birth, etc), and in such cases, it was necessary to determine which variables should be included in fitted AFT models. Variable selection was performed using a forward and backward stepwise procedure that searched all possible models to determine which model minimized the AIC (R package: MASS, R function: stepAIC). This approach adds covariates to the model when this improves goodness of fit, but does not generate an overfit model with unnecessary covariates, since the AIC includes a penalty term for each explanatory variable added to the model. Following variable selection, final steps were to fit the selected model, estimate the deceleration factor c, and perform diagnostic analyses to evaluate the adequacy of model fit. Model fit was evaluated based upon a graphical comparison between empirical Kaplan-Meier survival curves and fitted or “predicted” survival curves generated from the final AFT model. Residual analysis was used to evaluate whether certain observations were poorly characterized by the model, and case deletion influence measures were analyzed to determine whether some observations exerted strong influence on parameter estimates.
A central AFT model assumption is that treatments have a multiplicative effect on survival time that is consistent over time. To evaluate the validity of this assumption, quantile-quantile (QQ) plots were constructed for each Table 1 comparison, in which survival time quantiles of experimental treatments were plotted against survival time quantiles of control treatments. When treatment effects are consistent across the lifespan, points in this plot should approximate a straight line (e.g., see Patel et al., 2006). The QQ plot is commonly used and provides the best overall tool for evaluating whether the AFT model is appropriate for a two-sample treatment comparison. However, since the approach is non-quantitative, the AFT model assumption was also evaluated by determining whether estimated Weibull shape parameters differed significantly between experimental and control treatments. This was informative since, provided that survival times follow a Weibull distribution in each treatment, the AFT model is valid if the Weibull shape parameter does not differ between control and experimental treatments. This can be shown based upon the definition of the pth survival time percentile for the Weibull distribution (see Equation 5.9 from Collett, 2003). If control treatment survival times are generated from a Weibull distribution with scale and shape parameters λ0 and γ0, and experimental treatment survival times are generated from a Weibull distribution with scale and shape parameters λ1 and γ1, then the AFT formulation implies that the pth survival time percentile of experimental and control treatments differ by a factor of c.
| (3) |
From Equation (3), it follows that c = (λ1/λ0)1/γ when γ = γ0 = γ1 Consequently, if both experimental and control treatments share a common Weibull shape parameter, c is a constant with value independent of p, as assumed by the AFT model. This suggests that the AFT model assumption is valid when Weibull shape parameters do not differ significantly between experimental and control treatments. This approach serves only as a quantitative complement to diagnostic analysis based upon QQ plots. If sample sizes are small, for example, there may be insufficient statistical power to determine whether Weibull shape parameters differ significantly between control and experimental treatments.
2.3 Cox Proportional Hazard (PH) Model
The Cox PH model was also applied to each Table 1 comparison in order to examine the relationship between AFT model deceleration factors and PH model hazard ratios. The PH approach models hi (t), the hazard function for the ith individual at time t, as the product between a baseline hazard function h0 (t) and a function that depends on coefficients β1,…,βp and explanatory variables X1,…, Xp for the ith individual.
| (4) |
In the absence of covariates, only one variable X1 was included in the model to distinguish between control and experimental treatments (a 0–1 indicator variable), and the hazard ratio (HR) was obtained from the corresponding coefficient estimate (i.e., HR = exp(β̂1) ). Additional covariates were added to PH models for certain comparisons based upon results from AFT model analyses, so that for each Table 1 comparison, PH model covariates were the same as those included in AFT models.
The PH model assumes that the hazard ratio is constant across the lifespan and several steps were taken to evaluate this assumption. Diagnostic plots were constructed in which the log-transformed estimated hazard function for each treatment was plotted against the logarithm of time (i.e., a log-cumulative hazard plot) (Collett, 2003). In this plot, non-proportional hazards are indicated when hazard functions for each treatment are non-parallel, or especially when hazard functions for each treatment intersect. In addition, for each Table 1 comparison, three statistical tests were used to evaluate the proportional hazards assumption (weighted residuals test, score test, smooth test). For each test, departures from proportionality are ultimately detected using residual analysis, but the tests differ with regard to the types of residuals analyzed and how non-proportionality is defined. The weighted residuals test is based upon standardized Schoenfeld residuals, and uses a chi-square distributed test statistic to evaluate whether a relationship exists between residuals and survival times for particular covariate variables. If the proportional hazards assumption is appropriate for a given covariate, there should be no significant relationship between standardized Schoenfeld residuals and survival times (Grambsch and Therneau, 1994) (R package: survival, Function: cox.zph). The score and smooth test procedures generate p-values based on simulation. In the score test, Gaussian distributions are used to approximate the score process expected under the null hypothesis of proportional hazards, where the score process refers to a partial sum process of martingale residuals. A comparison is then made between the observed and expected score process to determine whether significant departure from proportionality exists for a given covariate (Lin et al., 1993) (R package: proptest, Function: scoreproptest). The smooth test is an interesting new approach that combines ideas from Cox (1972) and Lin et al. (1993) to evaluate non-proportionality of individual covariates within a model that may include several covariates (Krauss, 2008) (R package: proptest, Function: smoothproptest). In this procedure, covariates are parametrically modeled as a combination of smooth polynomial basis functions, including artificial time-dependent covariates, which are functionally related to a particular covariate of interest p. If hazards are proportional with respect to covariate p, such time-dependent artificial covariates should not be a significant model effect (Cox, 1972). The smooth test procedure is thus a significance test of artificial time-dependent covariates, where significance is evaluated based upon the score process test of Lin et al. (1993).
2.4 Maximum Lifespan and Quantile Regression
The “maximum lifespan” has been viewed as the most important indicator of whether an experimental treatment influences the aging process (Wang et al., 2004; Flurkey et al., 2007). Following the approach of Wang et al. (2004), “maximum lifespan” is here meant to reference the largest survival times recorded within an experiment, such as the 90th percentile of all survival times in both experimental and control treatments. For evaluating maximum lifespan, Wang et al. (2004) proposed a quantile regression procedure that addresses whether the τth survival time quantile differs significantly between control and experimental treatments. In this method, quantile regression is used to estimate the τth overall survival time quantile (in both experimental and control treatments combined), and each individual is classified according to whether its survival time is greater or lesser than the estimated the τth survival time. This generates a 2-by-2 contingency table in which individuals are classified by treatment (experimental versus control) and survival time (above or below estimated τth survival time), and contingency table analysis is used to evaluate whether there exists a significant relationship between these two dichotomous variables (Redden, 2004; Wang et al., 2004). This approach was applied to each of the comparisons listed in Table 1 with respect to the τ = 0.90 survival time quantile, and statistical significance was based upon the exact unconditional test (score statistic) described by Wang et al. (2004). Simulation analyses have shown that this approach provides good statistical power and an acceptable Type I error rate (α) (α = 0.016 – 0.0586 for nominal α of 0.05) (Wang et al. 2004).
Treatment differences in maximum lifespan were also evaluated based upon coefficient estimates from a quantile regression model (Koenker and Geling, 2001). Coefficient estimates were obtained using the Barrodale and Roberts’s algorithm (Barrodale and Roberts, 1974), with confidence intervals generated by rank inversion (Koenker, 1994) (see R package: quantreg, Function: rq). The basic form of this model is similar to the Equation (2) AFT model, except the primary response variable is Qlog(T) (τ|x), which represents conditional quantile functions of the log-transformed survival times (Koenker, 2005).
| (5) |
The model provides considerable flexibility for evaluating treatment effects at late ages, or at any point in the lifespan, since the coefficients β1,…, βp are free to vary across quantiles (τ∈[0,1]). It is therefore possible to evaluate treatment effects across a wide range of survival time quantiles, while adjusting for the possible influence of covariates. A limitation is that rank inversion confidence intervals for coefficient estimates are asymptotically correct. The method is therefore most appropriate for comparisons with larger sample sizes (e.g., bIrs2(+/−), Hcrt-UCP2). This approach was applied to Table 1 comparisons with at least n = 20 observations in both experimental and control treatments. Simulation analyses showed that type I error rates were slightly inflated for sample sizes in the range of 20 < n < 50 (α = 0.05 – 0.09 for nominal α of 0.05), but were more acceptable for larger experiments with n > 50 (α = 0.05 – 0.065 for nominal α of 0.05).
3. Results
3.1 Absolute lifespan
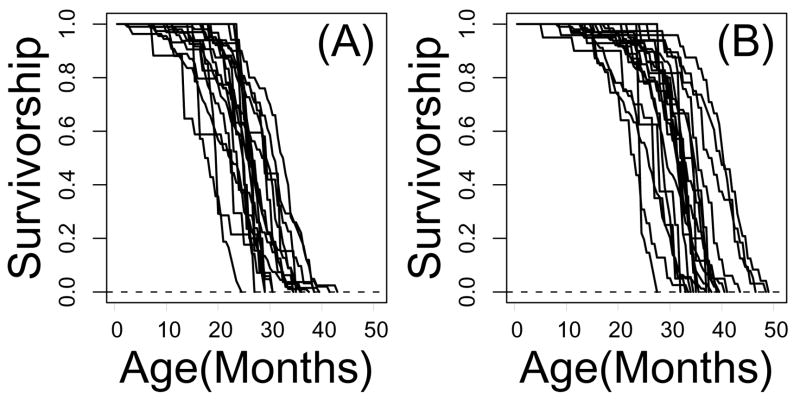
Experiments analyzed in this study were performed in multiple laboratories and utilized several different genetic backgrounds (Table 1). Given these environmental and genetic differences, survivorship patterns varied considerably among control cohorts, and were even more diverse among long-lived experimental mice (Figure 1). Among control cohorts, the TRX-Tg control mice had the lowest median longevity (17.5 months), while the Gpx4(+/−) control mice had the highest median longevity (32.1 months). Strikingly, in terms of absolute lifespan, the control population associated with Gpx4(+/−) mice was longer-lived than several of the “long-lived” experimental populations listed in Table 1. Since control mice from both TRX-Tg and Gpx4(+/−) comparisons were of the same genetic background (C57BL/6), this variability is surprising, and could be due to a wide range of environmental factors (e.g., number of littermates, diet, precautions against airborne infection), or breeding protocols generating mice used in survivorship experiments. Variation in control cohort survivorship should be considered in the context of analyses presented below, which considers the effect of genetic manipulations on survivorship, irrespective of baseline longevity within the control cohort. Based on absolute longevity, for example, Pit1(dw/dw) mice were the longest-lived of all the experimental cohorts (39.8 months), while TRX-Tg transgenic mice were the shortest lived (23.0 months).
Figure 1.
Survivorship curves from control and experimental treatments. Part (A) shows survivorship curves associated with control cohorts from the 18 comparisons listed in Table 1. Part (B) shows survivorship curves associated with (long-lived) experimental cohorts from the 22 comparisons listed in Table 1.
3.2 Accelerated Failure Time (AFT) model
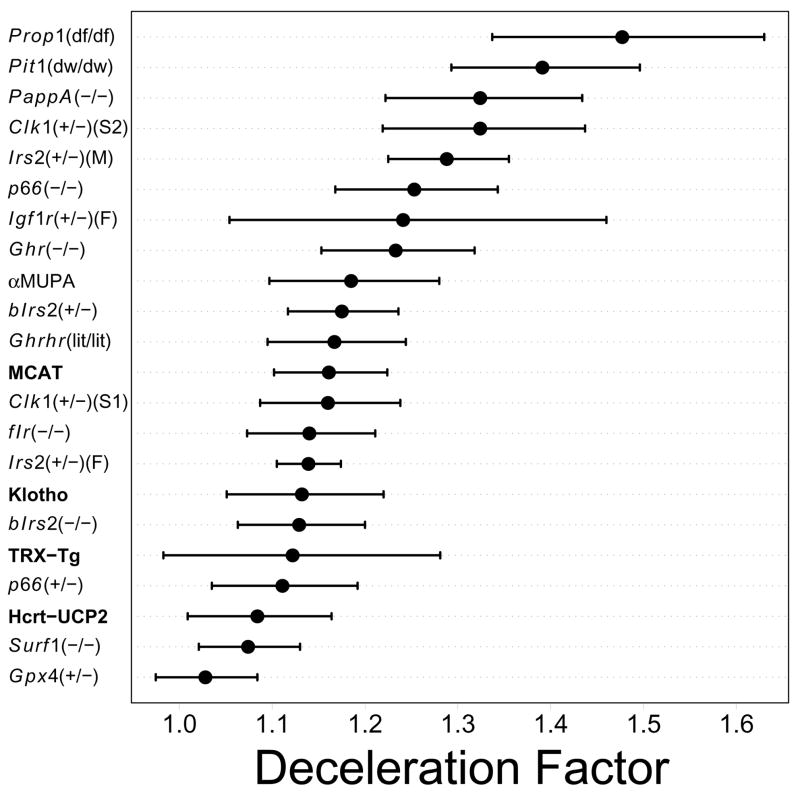
The AFT model was applied to each Table 1 comparison in order to quantify the longevity increase associated with experimental treatments. Overall, the Prop1(df/df) and Pit1(dw/dw) mutations had the strongest effects on survivorship (Table 2 and Figure 2). The Prop1(df/df) mutation had a stronger effect than the Pit1(dw/dw) mutation, but for both mutations, 95% confidence intervals associated with treatment effects overlapped (Prop1(df/df): ĉ = 1.48, 95% CI: 1.31, 1.61) (Pit1(dw/dw): ĉ = 1.39, 95% CI: 1.29, 1.50). A “second tier” of genetic manipulations with strong lifespan effects included PappA(−/−) mice, Clk1(+/−) mice (129Sv/j and C57BL/6J background) and male Irs2(+/−) mice (1.20 < ĉ < 1.40). For all other genetic manipulations, there was considerable overlap in terms of the estimated effect on survivorship, and the effect on survivorship was generally small (1.03 < ĉ < 1.20) (Figure 2). There were some cases in which the effects of genetic manipulations on survivorship were either gender or strain-dependent. For example, the effect of the Irs2(+/−) mutation was significantly greater in males than in females, and the Clk1(+/−) mutation had dissimilar effects on each of two genetic backgrounds (Figure 2).
Table 2.
Deceleration factor and hazard ratio estimates. Genetic manipulations are listed in each row and have been ranked according to their estimated effect on overall survivorship. Rankings are based upon the estimated AFT model deceleration factor ĉ. This value corresponds to the parameter c in S1 (ct) = S0 (t), where S1 (t) is the survivorship of (long-lived) mice belonging to experimental treatments at time t, and S0 (t) is the survivorship of mice belonging to corresponding control treatments at time t (Collett, 2003). Hazard ratios were calculated using the Cox proportional hazards (PH) model and are defined as the ratio of age-specific mortality in control treatments to age-specific mortality in (long-lived) experimental treatments. Both deceleration factors and hazard ratios were estimated using maximum likelihood. A description of each genetic manipulation, including genetic background of mice and sample sizes, is provided in Table 1.
| Comparison | Deceleration Factor (95% CI) | Hazard Ratio (95% CI) |
|---|---|---|
| Prop1(df/df) | 1.48 (1.34, 1.63) | 7.82 (3.98, 15.4) |
| Pit1(dw/dw) | 1.39 (1.29, 1.50) | 21.2 (7.0, 64.4) |
| PappA(−/−) | 1.32 (1.22, 1.43) | 8.77 (3.34, 23.0) |
| Clk1(+/−)(S2) | 1.32 (1.22, 1.44) | 11.9 (2.2, 64.9) |
| Irs2(+/−)(M)a | 1.29 (1.22, 1.35) | 515 (25, 10500) |
| p66(−/−) | 1.25 (1.17, 1.34) | 5.90 (1.98, 17.6) |
| Igf1r(+/−)(F) | 1.24 (1.05, 1.46) | 2.86 (1.36, 6.03) |
| Ghr(−/−) | 1.23 (1.15, 1.32) | 4.69 (2.75, 7.98) |
| αMUPA | 1.18 (1.10, 1.28) | 2.92 (1.70, 5.01) |
| bIrs2(+/−)b | 1.17 (1.12, 1.24) | 3.55 (2.49, 5.05) |
| Ghrhr(lit/lit) | 1.17 (1.10, 1.24) | 3.28 (1.90, 5.68) |
| MCAT | 1.16 (1.10, 1.22) | 2.42 (1.72, 3.42) |
| Clk1(+/−)(S1) | 1.16 (1.09, 1.24) | 4.91 (2.06, 11.7) |
| fIr(−/−) | 1.14 (1.07, 1.21) | 2.00 (1.47, 2.72) |
| Irs2(+/−)(F)c | 1.14 (1.10, 1.17) | 24.0 (5.99, 95.8) |
| Klothod | 1.13 (1.05, 1.22) | 2.15 (1.45, 3.21) |
| bIrs2(−/−)e | 1.13 (1.06, 1.20) | 5.09 (2.76, 9.39) |
| TRX-Tgf | 1.12 (0.98, 1.28) | 1.39 (0.75, 2.60) |
| p66(+/−) | 1.11 (1.03, 1.19) | 2.41 (0.84, 6.93) |
| Hcrt-UCP2g | 1.08 (1.01, 1.16) | 1.57 (1.07, 2.32) |
| Surf1(−/−)h | 1.07 (1.02, 1.13) | 3.54 (1.77, 7.05) |
| Gpx4(+/−) | 1.03 (0.98, 1.08) | 1.22 (0.82, 1.82) |
Maternal ID was modeled as a covariate.
Date of birth was modeled as a covariate.
Paternal ID was modeled as a covariate.
Gender was modeled as a covariate.
Gender, paternal ID and date of birth were modeled as covariates.
Gender was modeled as a covariate.
Gender and paternal ID were modeled as covariates.
Date of birth was modeled as a covariate.
Figure 2.
AFT model deceleration factor estimates. The deceleration factor represents the parameter c in the relation S1 (ct) = S0 (t), where S1 (t) is the survivorship of the experimental cohort at time t and S0 (t) represents survivorship of the control cohort at time t (Equation 1). The value of 100(c − 1) provides an estimate of the percent treatment difference in lifespan (experimental versus control) for any survival time quantile. For each comparison (see Table 1), filled symbols indicate the estimated deceleration factor value and bars represent a 95% confidence interval. Some deceleration factor estimates have been adjusted for covariates such as parental IDs, date of birth or gender (see Table 2).
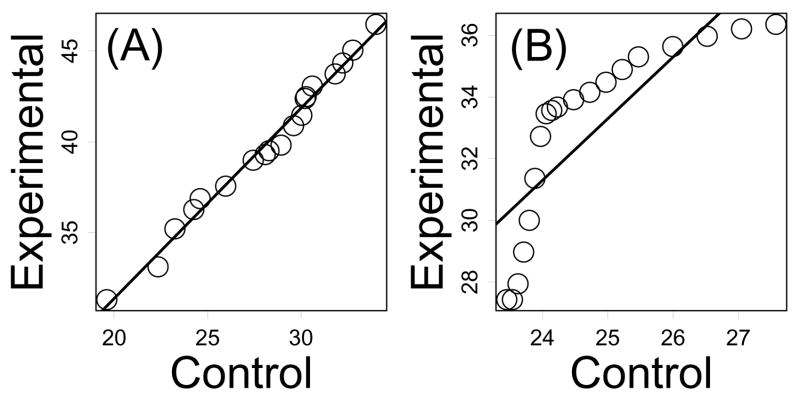
Diagnostic analysis indicated that AFT models adequately described treatment effects on survival time. This was indicated by QQ plots, which in most cases, revealed a linear relationship between lifespan quantiles in control and experimental treatments, reflecting consistent treatment effects across early, middle and late stages of the lifespan (Figure 3 and Supplemental Data File 1). Moreover, for 18 of 22 comparisons, there was no significant evidence that genetic manipulations altered the Weibull shape parameter of survival time distributions, which is consistent with the location-shift treatment effect assumed by the AFT model (exceptions were bIrs2(−/−), bIrs2(+/−), fIr(−/−) and Klotho). There were six comparisons for which the AFT model was questionable, based upon QQ plots, residual analysis and comparisons between observed and fitted survival curves (Irs2(+/−)(M), bIrs2(−/−), bIrs2(+/−), Igf1r(+/−)(F), Clk(+/−)(S2), TRX-TG). In each case, a similar trend was present, in which the treatment effect was strong at early ages, but weakened at more advanced ages (see Supplemental Data File 1). This treatment effect is more complex than that assumed to exist under the AFT model, and could reflect legitimate age-dependent treatment effects, or potentially, the absence of important (unmeasured) covariate variables. It should be noted that, even in these six cases, deceleration factor estimates were still informative, and represented an average between the strong and weak treatment effects early and late in life (Table 2).
Figure 3.
Quantile-Quantile plots. Survival time quantiles calculated from control cohort survival times are plotted against corresponding survival time quantiles calculated from experimental cohort survival times. Part (A) shows a QQ plot for the Pit1(dw/dw) comparison and part (B) shows a QQ plot for the Clk1(+/−)(S2) comparison. The solid line represents a least-square regression line. Part (A) indicates that the AFT model appropriately describes the treatment effect for the Pit1(dw/dw) comparison, since points approximate a straight line. Part (B) suggests that the AFT model may not be appropriate for the Clk1(+/−)(S2) comparison, since points do not approximate a straight line. QQ plots for all 22 Table 1 comparisons are shown in Supplemental Data File 1.
Case-deletion diagnostic analysis was performed to evaluate whether outlying observations had strong influence on deceleration factor estimates (Supplemental Data File 2). A common pattern observed in several experiments was the presence of an especially long-lived individual from the control cohort that decreased the deceleration factor estimate (Prop1(df/df), Clk1(+/−)(S2), PappA(−/−), αMUPA, bIrs2(+/−), Igf1r(+/−)(F), Ghr(−/−), Ghrhr(lit/lit), Klotho, Hcrt-UCP2, Gpx4(+/−)). There were only two cases, however, in which the influence was strong enough to have a substantial impact on the estimated deceleration factor. For the Prop1(df/df) comparison, a deceleration factor of ĉ = 1.48 was obtained using the complete dataset, and this value increased to ĉ = 1.54 (95% CI: 1.41, 1.69) if the outlying observation was eliminated. Likewise, for the Clk1(+/−)(S2) comparison, a deceleration factor of ĉ = 1.32 (95% CI: 1.22, 1.44) was obtained using the complete dataset, and this value increased to ĉ = 1.41 (95% CI: 1.30, 1.52) if the outlying observation was eliminated. The elimination of outlying observations is always a delicate issue in statistical modeling, and should generally depend upon whether there is reason to believe that outliers were driven by experimental artifacts, which is not known for the experiments under consideration.
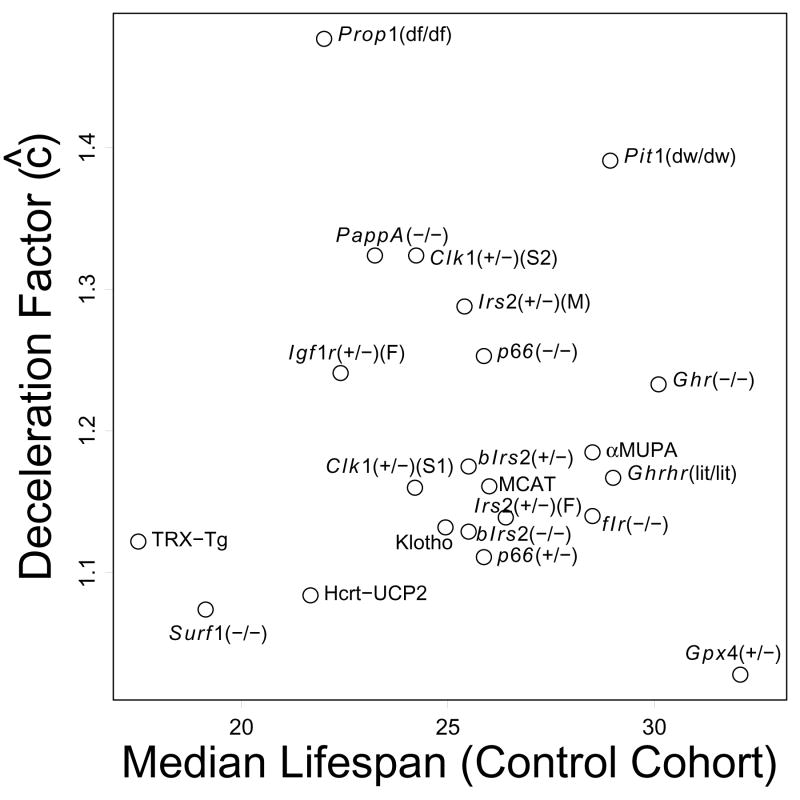
There was little evidence to indicate that large treatment effects were commonly driven by premature mortality within control cohorts, such as that arising from pathogen infection. Overall, in fact, there was a positive relationship between control cohort longevity and deceleration factor estimates (Figure 4). Nevertheless, some comparisons did involve relatively strong or weak longevity within control cohorts, which should be considered when evaluating treatment effects for certain comparisons (e.g., Prop1(df/df), αMUPA, Ghrhr(lit/lit), Gpx4(+/−)) (Figure 4).
Figure 4.
Deceleration factors estimates and control cohort lifespan. The plot shows a weak positive association between deceleration factor estimates and median lifespan estimates from control cohorts. Each point represents one of the 22 comparisons listed in Table 1.
3.3 Cox Proportional Hazard (PH) model
Hazard ratios varied considerably among comparisons, and in general, there was only a limited relationship between hazard ratio and deceleration factor estimates (r = 0.22, rs = 0.71) (see Table 2). In some cases, exceedingly large hazard ratios were obtained. For example, the hazard ratio estimated for the Irs2(+/−)(M) comparison suggested that age-specific mortality rates were 515 times greater in the control cohort as compared to the experimental cohort (95% CI: 25, 10500). While such a large estimate could represent a departure from modeling assumptions, this observation suggests that, in comparison to the AFT model approach, the PH model hazard ratios are less stable and more sensitive to deviations from model assumptions.
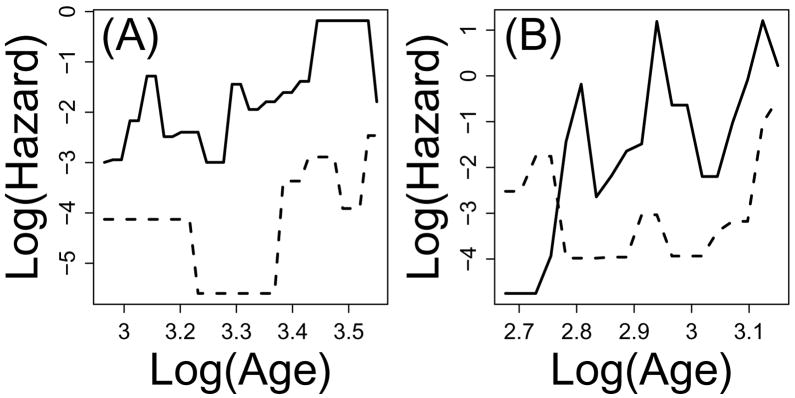
The proportional hazards assumption was questionable for half of the comparisons, as indicated by crossing of log-transformed hazard functions of control and experimental treatments (see Figure 5) (see p66(−/−), αMUPA, bIrs2(+/−), MCAT, Clk1(+/−)(S1), Klotho, bIrs2(−/−), TRX-Tg, Hcrt-UCP2, Surf1(−/−), Igf1r(+/−)(F), Ghr(−/−), Gpx4(+/−) in Supplemental Data File 3). The proportional hazards assumption was most clearly violated for the bIrs2(+/−) and bIrs2(−/−) comparisons. This was supported based upon graphical evidence and also statistical tests of the proportional hazards assumption. For each of three statistical tests, there was significant evidence to suggest that hazards for bIrs2(+/−) and bIrs2(−/−) were non-proportional (P < 0.01) (Table 3). The proportional hazards assumption was also doubtful for the Surf1(−/−) comparison, based upon graphical evidence and two of three statistical tests (Table 3). For one of three statistical tests, hazards were non-proportional with respect to the Prop1(df/df), fIr(−/−), TRX-Tg and Gpx4(+/−) comparisons, although in some cases this inference was not supported based upon inspection of the log-cumulative hazard plot (Supplemental Data File 3).
Figure 5.
Log-cumulative hazard plots. Part (A) shows a log-cumulative hazard plot for the Prop1(df/df) comparison, while part (B) shows the log-cumulative hazard plot for the Surf1(−/−) comparison. In both (A) and (B), the dotted line represents the logarithm of the estimated hazard function for the experimental treatment, while the solid line represents the logarithm of the estimated hazard function for the control treatment. Part (A) shows that, for the Prop1(df/df) comparison, the difference between log-hazard functions of control and experimental treatments is roughly consistent over time (as assumed by the PH model). Part (B) shows that, for the Surf1(−/−) comparison, the difference between log-hazard functions of control and experimental treatments varies over time, which suggests that the standard PH model may not be appropriate. Log-cumulative hazard plots for each of the 22 Table 1 comparisons are shown in Supplemental Data File 3.
Table 3.
Statistical tests of proportional hazards. The table lists p-values from each of three tests, where significant p-values indicate non-proportional hazards (contrary to the PH model assumption). For comparisons in which covariates besides genotype were included in models, results correspond to global tests and indicate non-proportionality for at least one of the multiple covariates. Log-cumulative hazard plots are shown in Supplemental Data File 3 as a graphical tool for assessing proportionality of hazards between experimental and control treatments.
| Comparison | Weighted Residuals | Score Test | Smooth Test |
|---|---|---|---|
| Prop1(df/df) | 0.350 | 0.043 | 0.220 |
| Pit1(dw/dw) | 0.620 | 0.740 | 0.610 |
| PappA(−/−) | 0.750 | 0.400 | 0.590 |
| Clk1(+/−)(S2) | 0.840 | 0.240 | 0.430 |
| Irs2(+/−)(M) | 0.960 | 0.570 | 0.570 |
| p66(−/−) | 0.210 | 0.094 | 0.093 |
| Igf1r(+/−)(F) | 0.980 | 0.690 | 0.730 |
| Ghr(−/−) | 0.470 | 0.580 | 0.560 |
| αMUPA | 0.130 | 0.250 | 0.130 |
| bIrs2(+/−) | 0.000 | 0.000 | 0.000 |
| Ghrhr(lit/lit) | 0.280 | 0.075 | 0.280 |
| MCAT | 0.550 | 0.220 | 0.830 |
| Clk1(+/−)(S1) | 0.590 | 0.750 | 0.640 |
| fIr(−/−) | 0.055 | 0.014 | 0.140 |
| Irs2(+/−)(F) | 0.910 | 0.340 | 0.340 |
| Klotho | 0.450 | 0.220 | 0.770 |
| bIrs2(−/−) | 0.008 | 0.010 | 0.006 |
| TRX-Tg | 0.330 | 0.330 | 0.006 |
| p66(+/−) | 0.170 | 0.280 | 0.380 |
| Hcrt-UCP2 | 0.200 | 0.230 | 0.230 |
| Surf1(−/−) | 0.008 | 0.440 | 0.000 |
| Gpx4(+/−) | 0.081 | 0.008 | 0.150 |
3.4 Maximum Lifespan and Quantile Regression
Treatment effects on maximum lifespan were evaluated using the Score statistic and contingency table approach advocated by Wang et al. (2004), as well as asymptotic rank inversion confidence intervals on coefficient estimates from a quantile regression model (Koenker, 1994) (see Methods). Based on the Score statistic, treatment effects on maximum lifespan were non-significant for seven comparisons (Irs2(+/−)(M), p66(−/−), Clk1(+/−)(S2), Klotho, TRX-Tg, p66(+/−), Hcrt-UCP2) (Table 4). In most cases, this non-significance could reflect weak statistical power resulting from low sample size. Among the seven non-significant results, for example, there were only two cases in which sample sizes for both experimental and control treatments were moderately large (n > 20 per treatment) (Klotho and Hcrt-UCP2). In both of these cases, moreover, significant treatment effects were found based upon rank inversion confidence intervals from a quantile regression model, which accounted for the potential influence of covariates (Table 4). There was one comparison for which the treatment effect on maximum lifespan was significant using the Score statistic (Surf1(−/−); P = 0.003), but not with respect to rank inversion confidence intervals (95% CI: 0.99, 1.03) (Table 4). This difference could reflect the influence of date of birth on the maximum lifespan treatment difference, since the latter statistical approach accounted for this covariate.
Table 4.
Maximum lifespan. The Score Test p-values evaluate whether treatments differ significantly in the number of mice that survive to the 90th percentile survival time (calculated from both treatments combined) (see Wang et al., 2004). The treatment effect column lists the ratio between the 90th percentile survival time in experimental treatment and the 90th percentile survival time in the control treatment. Asymptotic 95% confidence intervals on treatment effect estimates were calculated using rank inversion (Koenker, 1994), with adjustment for one or several covariate variables in some cases (e.g., gender, date of birth, etc.) (see Table 2). Confidence intervals are only calculated for comparisons in which sample sizes are moderately large in both experimental and control treatments (n > 20) (see Table 1).
| Comparison | P-Value (Score Test) | Treatment Effect (95% CI) |
|---|---|---|
| Prop1(df/df) | 0.042 | 1.37 (1.21, 1.60) |
| Pit1(dw/dw) | 0.010 | 1.38 (1.29, 1.50) |
| PappA(−/−) | 0.040 | 1.31 (1.23, 1.39) |
| Clk1(+/−)(S2) | 0.406 | 1.30 (—, — ) |
| Irs2(+/−)(M) | 0.105 | 1.30 ( —, — ) |
| p66(−/−) | 0.211 | 1.26 (—, — ) |
| Igf1r(+/−)(F) | 0.008 | 1.15 (—, — ) |
| Ghr(−/−) | 0.010 | 1.21 (1.15, 1.27) |
| αMUPA | 0.012 | 1.16 (1.09, 1.23) |
| bIrs2(+/−) | < 0.001 | 1.10 (1.05, 1.29) |
| Ghrhr(lit/lit) | 0.038 | 1.14 (1.04, 1.24) |
| MCAT | < 0.001 | 1.12 (1.01, 1.20) |
| Clk1(+/−)(S1) | 0.049 | 1.12 (—,—) |
| fIr(−/−) | 0.011 | 1.08 (1.01, 1.17) |
| Irs2(+/−)(F) | 0.049 | 1.15 (—, — ) |
| Klotho | 0.051 | 1.13 (1.06, 1.20) |
| bIrs2(−/−) | 0.035 | 1.13 (1.06, 1.20) |
| TRX-Tg | 0.750 | 1.00 ( —, —) |
| p66(+/−) | 0.056 | 1.14 (—, — ) |
| Hcrt-UCP2 | 0.376 | 1.07 (1.01, 1.11) |
| Surf1(−/−) | 0.003 | 1.00 (0.99, 1.05) |
| Gpx4(+/−) | 0.671 | 0.99 (0.97, 1.03) |
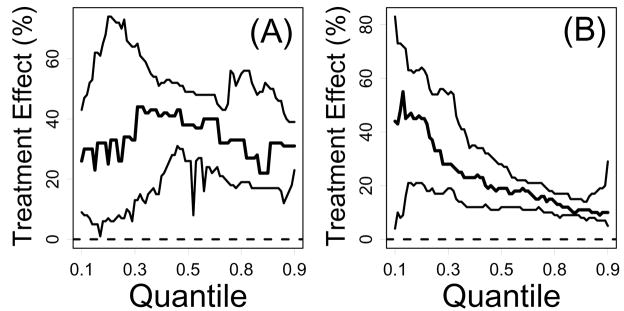
Quantile regression was also used to provide a detailed profile of how treatment effects varied across the lifespan (Koenker and Geling, 2001). The approach is illustrated by Figure 6 for one case in which treatment effects were similar across time (PappA(−/−); Figure 6A) and a second case for which treatment effects were time-dependent (bIrs2(+/−); Figure 6B). In the first case, the quantile regression approach confirms results from the AFT model analysis, and shows that the PappA(−/−) mutation increases lifespan by approximately 30%, with a consistent treatment effect across the lifespan (Figure 6A). In the second case, the deviation from the AFT model assumption is apparent, and it is clear that treatment effects are strong early in the lifespan, but weaken later in the lifespan, with an overall average effect that is summarized well by the estimated AFT model deceleration factor ( ĉ = 1.17, 95% CI: 1.12, 1.24) (Figure 6B). Similar quantile regression analyses for all Table 1 comparisons are shown in Supplemental Data File 4.
Figure 6.
Quantile regression estimation of treatment effects. Quantile regression was used to estimate treatment effects across a range of survival time quantiles (τ = 0.10,…,0.90). For a given quantile τ (horizontal axis), the vertical axis represents the percent increase in survivorship associated with an experimental treatment (Koenker and Geling, 2001). In part (A), results for the PappA(−/−) treatment are shown, and in part (B), results for the bIrs2(+/−) treatment are shown. In each plot, the middle line represents the calculated effect of experimental treatments at each survival time quantile, while the upper and lower lines outline a 95% confidence region (Koenker and Geling, 2001).
4. Discussion
The number of genetic manipulations known to extend mouse lifespan can be expected to grow considerably in the coming decades. In the S. cerevisiae and C. elegans model systems, genomic screens suggest that lifespan is increased by manipulating the activity of 0.1 – 3% of genes (Kennedy, 2008). These results raise the possibility that there may be over 500 single-gene manipulations that significantly extend longevity in the laboratory mouse. Mouse survivorship data is subject to experimental noise and there is the potential that uncontrolled covariates will influence treatment comparisons. Indeed, two separate laboratories can carry out a similar survivorship experiment, and yet arrive at dissimilar conclusions (Taguchi et al., 2007; Selman et al., 2008a; Selman et al., 2008b). The statistical analysis of experimental survivorship data will thus be critical for prioritizing among known and yet to be discovered models of mouse longevity. This study showed that AFT models are well-suited to evaluating the effects of genetic manipulations on survivorship, since most manipulations have a multiplicative effect on survivorship, with similar treatment effects at early, middle and late ages. Analysis of 16 datasets revealed that AFT model deceleration factors are an informative indicator of treatment effect size, and are robust to departures from modeling assumptions that are characteristic of mouse survival data. Quantile regression methods naturally complement the AFT approach, and for large experiments, are useful for detailed evaluation of treatment effects and survivorship patterns at late ages. These statistical methods, based upon the survival curve rather than the hazard function, have not been widely used by previous investigators, but provide valuable tools for evaluating the effects of interventions on survivorship in the laboratory mouse.
The AFT model deceleration factor has been viewed as an indicator of how a treatment alters the “biological clock” of subjects, since it quantifies the direct effect of an experimental treatment on survival time (Nardi and Schemper, 2003). This interpretation gives a strong advantage to the AFT model within the context of experimental aging research. The Cox PH model is a more commonly used alternative to the AFT approach, and unlike the AFT model, is based on the hazard function (Cox, 1972). This contrasts, however, with the tendency of most researchers to summarize treatment effects in terms of survival time (e.g., % increase in mean or median lifespan). Further, among research reports describing new long-lived mouse models, survival curve estimates are displayed much more frequently than estimated hazard functions (e.g., Miskin and Masos, 1997; Migllaccio et al., 1999; Blüher et al., 2003; Holzenberger et al., 2003; Schriner et al., 2005; Conti et al., 2006; Conover and Bale, 2007; Taguchi et al., 2007; Yan et al., 2007). Another advantage of the AFT model is that it can often be used when hazards are non-proportional between experimental treatments (Orbe et al., 2002; Patel et al., 2006). Investigators commonly fail to evaluate proportionality of hazards when fitting PH models (Altman et al., 1985), and this can lead to loss of statistical power and inaccurate conclusions regarding treatment effects (Hess, 1994; Abrahamowicz et al., 1996). In this study, hazards between experimental treatments were clearly non-proportional for at least three comparisons (bIrs2(+/−), bIrs2 (−/−), TRX-Tg), and proportionality was questionable for several others (p66(−/−), αMUPA, MCAT, Clk1(+/−)(S1), Igf1r(+/−)(F), Klotho, Hcrt-UCP2, Surf1(−/−)). In such cases, the addition of age-dependent covariates can sometimes be used to adequately fit PH models, but such remedial measures may not be required using the AFT approach.
AFT model deceleration factors appear to be more robust than PH model hazard ratios. Among all comparisons examined, AFT model deceleration factors were within a narrow range (1.03 < ĉ < 1.48), and even when treatment effects were time-dependent, sensible deceleration factor estimates were obtained that corresponded well to the percent treatment difference in median lifespan. In contrast, PH model hazard ratios varied considerably among comparisons (1.22 < HR < 515), and in some cases led to counter-intuitive conclusions. For instance, based upon hazard ratios, the treatment effect of the Irs2(+/−) mutation in females (HR = 24.0) is several times larger than the effect of the Prop1(df/df) mutation (HR = 7.82). It may be that the fitted PH model was in some way not optimized for evaluating the effect of the Irs2(+/−) mutation, and that certain remedial measures would have provided a more informative hazard ratio (e.g., altering the functional form of covariates, adding time-dependent covariates, removing outliers, stratification). However, this example illustrates that, for mouse survivorship experiments, AFT model deceleration factors often provide a more intuitive effect size measure than hazard ratio estimates, which may reflect the fact that deceleration factors are less sensitive to the model deviations apt to occur in data from mouse survivorship studies. Another consideration is that, for most mouse survivorship experiments, treatment effects will be much larger than those in standard epidemiological analyses. For large treatment effects, some analyses have suggested that AFT parameter estimates are, asymptotically, more efficient than those of the PH model (Oakes, 1977; Cox and Oakes, 1984).
The purpose of this report is not to dismiss the Cox PH model as a potentially useful tool for the analysis of mouse survivorship data. Analyzing treatment effects based on the hazard function may sometimes reveal trends not evident from analysis of survival curves (Royston and Parmar, 2002), and indeed, many aging researchers have based ideas and concepts on the rate of age-specific mortality (e.g., Pletcher et al., 2000; Swindell and Bouzat, 2006; Lenaerts et al., 2007). Moreover, if survival times follow a Weibull distribution, the Cox PH model can be re-parameterized as a Weibull AFT model, and AFT model deceleration factors should correspond to log-transformed hazard ratios (Collett, 2003). Additionally, with the PH model, it is not necessary to specify a particular distribution for the baseline hazard function. In contrast, for the AFT model, the investigator must choose a particular survival time distribution, although this is usually easily done based upon the Akaike’s Information Criterion (Akaike, 1974), and a recent AFT modeling approach has been proposed that avoids specification of the survival time distribution (Orbe et al., 2002). Overall, a reasonable course of action for investigators may be to apply both the PH and AFT models and evaluate carefully which method is most appropriate for the particular dataset under consideration. If both modeling approaches are appropriate, reporting results from two methods would demonstrate that conclusions are robust and consistent between alternative techniques.
Treatment effects on maximum lifespan are the most important consideration when evaluating the relevance of mouse longevity models to aging research (Wang et al., 2004; Flurkey et al., 2007). If AFT modeling is appropriate, the effects of an experimental treatment on survivorship are similar at early, middle and late stages of the lifespan. In such cases, the estimated deceleration factor reflects the treatment effect at all ages (including late ages), and is therefore informative with regard to maximum lifespan. Results from this analysis show that, in fact, most genetic manipulations do have multiplicative effects on survivorship that do not depend strongly on time (Supplemental Data Files 1 and 4). This is, however, an assumption that should be evaluated on a case-by-case basis for individual survivorship experiments. In this analysis, there were six genetic manipulations for which treatment effects were clearly stronger early in life compared to late in life (Irs2(+/−)(M), bIrs2(−/−), bIrs2(+/−), Igf1r(+/−)(F), Clk1(+/−)(S1), TRX-TG). For these comparisons, AFT model deceleration factors provide an “averaged” estimate of the treatment effect across the lifespan, but overestimate treatment effects at late ages. For such cases, quantile regression modeling with rank inversion confidence intervals was more informative than either AFT or PH model results (Koenker, 1994; Koenker and Geling, 2001). This quantile regression approach is most appropriate for comparisons with large sample sizes in both experimental and control treatments (n > 50 per treatment), although for smaller experiments, treatment effects at specific quantiles can be investigated using the contingency table approach described by Wang et al. (2004).
The present analysis provides an informative and quantitative comparison among a number of genetic manipulations in terms of their positive influence on mouse survivorship. An interesting aspect of this side-by-side comparison is the decline in survivorship effects among mutations that inhibit increasingly downstream elements of the growth hormone/insulin-like growth factor I (GH/IGF-I) signaling pathway (see: Prop1(df/df), Pit1(dw/dw), Ghrhr(lit/lit), Ghr(−/−), PappA(−/−), Igf1r(+/−)(F), Irs2(+/−)(M), Irs2(+/−)(F), bIrs2(+/−), bIrs2(−/−)). The Prop1(df/df) and Pit1(dw/dw) mutations, for example, inhibit GH/IGF-I signaling and have the strongest overall effects on survivorship (ĉ ≥1.39 ), while the Ghrhr(lit/lit), Ghr(−/−), PappA(−/−), Igf1r(+/−) and Irs2 mutations inhibit the same pathway, but each has a weaker effect on survivorship (1.13≤ ĉ ≤1.32 ). One possibility is that prolactin or thyroid stimulating hormone deficiencies in Prop1(df/df) and Pit1(dw/dw) mice contribute to increased survivorship, apart from the effects of inhibited GH/IGF-I signaling. These endocrine deficiencies are only characteristic of the Prop1(df/df) and Pit1(dw/dw) mice, and both of these long-lived models stand apart from all others in terms of the magnitude by which survivorship is increased (Figure 2). Vergara et al. (2004) found that administration of thyroxine did, in fact, diminish the lifespan of Pit1(dw/dw) dwarf mice relative to controls. Therefore, although the GH/IGF-I pathway is clearly important for longevity determination in the laboratory mouse, secondary endocrine effects associated with Prop1(df/df) and Pit1(dw/dw) mice may also warrant investigation.
There has been remarkable progress since Brown-Borg et al. (1996)’s original finding that mouse lifespan is significantly extended by a single gene mutation, and important directions remain for future experimental work. For example, genetic background can have a substantial impact on survivorship effects (Spencer et al., 2003; Toivonen et al., 2007), but only few studies have evaluated survivorship of long-lived mutants on multiple genetic backgrounds (Coschigano et al., 2003; Liu et al., 2005). Additionally, some genetic manipulations appear to increase survivorship through independent mechanisms (e.g., PappA(−/−) and MCAT), but it is unknown whether certain combinations of mutations have additive effects on survivorship. The longevity studies required to address these issues require considerable time and expense, but the statistical analysis of resulting data requires only a modest time investment. No one statistical approach will perform optimally for every dataset and it is usually profitable to experiment with several methods. The results presented here, however, argue that the AFT model should be utilized more widely in aging research, along with quantile regression modeling as a complementary follow-up approach. These methods are based upon the survival curve, generate appealing and robust summary statistics with confidence intervals, and can be used to calculate treatment effects adjusted for variables not controlled experimentally. These approaches therefore provide useful tools that can be help maximize the insight obtained from experimental studies of mouse survivorship.
Supplementary Material
Acknowledgments
This work was supported by NIA training grant T32-AG000114 and the University of Michigan Department of Pathology. Two anonymous reviewers provided helpful comments and suggestions for this manuscript. Additionally, the author thanks a number of researchers that contributed the experimental survivorship data analyzed in this report, including Andrzej Bartke, Michael S. Bonkowski, Cheryl A. Conover, Bruno Conti, Marco Giorgio, Siegfried Hekimi, Makoto Kuro-o, Richard A. Miller, Piegiuseppe Pelicci, Peter S. Rabinovitch, Qitao Ran, Arlan Richardson and Junji Yodoi.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamowicz M, MacKenzie T, Esdaile JM. Time-dependent hazard ratio: modeling and hypothesis testing with application in lupus nephritis. J Amer Statist Assoc. 1996;91:1432–1439. [Google Scholar]
- Akaike A. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716–723. [Google Scholar]
- Altman DG, De Stavola BL, Love SB, Stepniewska KA. Review of survival analyses published in cancer journals. British Journal of Cancer. 1985;72:511–518. doi: 10.1038/bjc.1995.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrodale I, Roberts F. Solution of an overdetermined system of equations in the ℓ1 norm. Communications of the ACM. 1974;17:319–320. [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Bonkowski MS, Rocha JS, Masternak MM, Al Regaiey KA, Bartke A. Targeted disruption of growth hormone receptor interferes with the beneficial actions of caloric restriction. Proc Natl Acad Sci. 2006;103:7901–7905. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the aging process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Collett D. Modelling Survival Data in Medical Research. 2. CRC Press; Boca Raton: 2003. [Google Scholar]
- Conover CA, Bale LK. Loss of pregnancy-associated plasma protein A extends lifespan in mice. Aging Cell. 2007;6:727–729. doi: 10.1111/j.1474-9726.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- Conti B, Sanchez-Alavez M, Winsky-Sommerer R, Concetta Morale M, Lucero J, Brownell S, Fabre V, Huitron-Resendiz S, Henriksen S, Zorrilla EP, de Lecea L, Bartfai T. Transgenic mice with a reduced core body temperature have an increased life span. Science. 2006;314:825–828. doi: 10.1126/science.1132191. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Holland AN, Riders ME, List EO, Flyvbjerg A, Kopchick JJ. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology. 2003;144:3799–3810. doi: 10.1210/en.2003-0374. [DOI] [PubMed] [Google Scholar]
- Cox DR. Regression models and life-tables. J Roy Statist Soc Ser B. 1972;34:187–220. [Google Scholar]
- Cox DR, Oakes D. Analysis of Survival Data. Chapman & Hall; London: 1984. [Google Scholar]
- Dell’Agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux I, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, Currer JM, Harrison DE. The Mouse in Aging Research. In: Fox JG, Davisson MT, Quimby FW, Barthold SW, Newcomer CE, Smith AL, editors. The Mouse in Biomedical Research. 2. Elsevier; Burlington, MA: 2007. pp. 637–672. [Google Scholar]
- Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- Hess KR. Assessing time-by-covariate interactions in proportional hazards regression models using cubic spline functions. Statist Med. 1994;13:1045–1062. doi: 10.1002/sim.4780131007. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Géloën A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Hutton JL, Monaghan PF. Choice of parametric accelerated life and proportional hazards models for survival data: asymptotic results. Lifetime Data Analysis. 2002;8:375–393. doi: 10.1023/a:1020570922072. [DOI] [PubMed] [Google Scholar]
- Keene O. Alternatives to the hazard ratio in summarizing efficacy in time-to-event studies: an example from influenza trials. Statist Med. 2002;21:3687–3700. doi: 10.1002/sim.1312. [DOI] [PubMed] [Google Scholar]
- Koenker R. In: Mandl P, Hušková M, editors. Confidence intervals for regression quantiles; Asymptotic statistics: proceedings of the 5th Prague Symposium; Heidleberg: Physica-Verlag; 1994. pp. 349–359. [Google Scholar]
- Koenker R. Quantile Regression. Cambridge U. Press; Cambridge: 2005. [Google Scholar]
- Koenker R, Geling R. Reappraising medfly longevity: a quantile regression approach. J Am Stat Assoc. 2001;96:458–468. [Google Scholar]
- Krauss D. Identifying nonproportional covariates in the cox model. Comm Statist Theory Methods. 2008;37:617–625. [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomur I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro-o M. Suppression of aging in mice by the hormone klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ren J. Influence of cardiac-specific overexpression of insulin-like growth factor 1 on lifespan and aging-associated changes in cardiac intracellular Ca2+ homeostasis, protein damage and apoptotic protein expression. Aging Cell. 2007;6:799–806. doi: 10.1111/j.1474-9726.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- Lenaerts I, van Eygen S, van Fleteren J. Adult-limited dietary restriction slows gompertzian aging in Caenorhabditis elegans. Ann NY Acad Sci. 2007;1100:442–448. doi: 10.1196/annals.1395.049. [DOI] [PubMed] [Google Scholar]
- Liang H, Masoro EJ, Nelson JF, Strong R, McMahan CA, Richardson A. Genetic mouse models of extended lifespan. Exp Gerontol. 2003;38:1353–1364. doi: 10.1016/j.exger.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Lin DY, Wei LJ, Ying Z. Checking the cox model with cumulative sums of martingale-based residuals. Biometrika. 1993;80:557–572. [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and lifespan in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Floyd RA, Flurkey K, Hensley KL, Javors MA, Leeuwenburgh C, Nelson JF, Ongini E, Nadon NL, Warner HR, Strong R. An aging interventions testing program: study design and interim report. Aging Cell. 2007;6:565–575. doi: 10.1111/j.1474-9726.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- Miskin R, Masos T. Transgenic mice overexpressing urokinase-type plasminogen activator in the brain exhibit reduced food consumption, body weight and size, and increased longevity. J Gerontol A Biol Sci Med Sci. 1997;52:B118–B124. doi: 10.1093/gerona/52a.2.b118. [DOI] [PubMed] [Google Scholar]
- Mitsui A, Hamuro J, Nakamura H, Kondo N, Hirabayashi Y, Ishizaki-Koizumi S, Hirakawa T, Inoue T, Yodoi J. Overexpression of human thioredoxin in transgenic mice controls oxidative stress and life span. Antioxid Redox Signal. 2002;4:693–696. doi: 10.1089/15230860260220201. [DOI] [PubMed] [Google Scholar]
- Nardi A, Schemper M. Comparing cox and parametric models in clinical studies. Statist Med. 2003;22:3597–3610. doi: 10.1002/sim.1592. [DOI] [PubMed] [Google Scholar]
- Oakes D. The asymptotic information in censored survival data. Biometrika. 1977;64:441–448. [Google Scholar]
- Orbe J, Ferreira E, Núñez-Antón V. Comparing proportional hazards and accelerated failure time models for survival analysis. Statist Med. 2002;21:3493–3510. doi: 10.1002/sim.1251. [DOI] [PubMed] [Google Scholar]
- Patel K, Kay R, Rowell L. Comparing proportional hazards and accelerated failure time models: an application in influenza. Pharmaceut Statist. 2006;5:213–224. doi: 10.1002/pst.213. [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Khazaeli AA, Curtsinger JW. Why do life spans differ? Partitioning mean longevity differences in terms of age-specific mortality parameters. J Gerontol A Biol Sci Med Sci. 2000;55:B381–B389. doi: 10.1093/gerona/55.8.b381. [DOI] [PubMed] [Google Scholar]
- Pourhoseingholi MA, Hajizadeh E, Dehkordi BM, Safaee A, Abadi A, Zali MR. Comparing cox regression and parametric models for survival of patients with gastric carcinoma. Asian Pacific J Cancer Prev. 2007;8:412–416. [PubMed] [Google Scholar]
- Ran Q, Liang H, Ikeno Y, Qi W, Prolla TA, Jackson Roberts L, II, Wolf N, VanRemmen H, Richardson A. Reduction in glutathione peroxidase 4 increases life span through increased sensitivity to apoptosis. J Gerontol Biol Sci. 2007;62A:932–942. doi: 10.1093/gerona/62.9.932. [DOI] [PubMed] [Google Scholar]
- Redden DT, Fernández JR, Allison DB. A simple significance test for quantile regression. Statist Med. 2004;23:2587–2597. doi: 10.1002/sim.1839. [DOI] [PubMed] [Google Scholar]
- Royston P, Parmar MK. Flexible parametric proportional-hazards and proportional-odds models for censored survival data, with application to prognostic modeling and estimation of treatment effects. Statist Med. 2002;21:2175–2197. doi: 10.1002/sim.1203. [DOI] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, Wallace DC, Rabinovitch PS. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, Clements M, Ramadani F, Okkenhaug K, Schuster E, Blanc E, Piper MD, Al-Qassab H, Speakman JR, Carmignac D, Robinson ICA, Thornton JM, Gems D, Partridge L, Withers DJ. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008a;22:807–818. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- Selman C, Lingard S, Gems D, Partridge L, Withers DJ. Comment on “Brain Irs2 signaling coordinates life span and nutrient homeostasis”. Science. 2008b;320:1012. doi: 10.1126/science.1152366. [DOI] [PubMed] [Google Scholar]
- Spencer CC, Howell CE, Wright AR, Promislow DE. Testing an ‘aging gene’ in long-lived drosophila strains: increased longevity depends on sex and genetic background. Aging Cell. 2003;2:123–130. doi: 10.1046/j.1474-9728.2003.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Bouzat JL. Inbreeding depression for male survivorship parameters in Drosophila melanogaster: implications for senescence theory. Genetics. 2006;172:317–327. doi: 10.1534/genetics.105.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- Toivonen JM, Walker GA, Martinez-Diaz P, Bjedov I, Driege Y, Jacobs HT, Gems D, Partridge L. No influence of Indy on lifespan in Drosophila after correction for genetic and cytoplasmic background effects. PloS Genet. 2007;3:e95. doi: 10.1371/journal.pgen.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara M, Smith-Wheelock M, Harper JM, Sigler R, Miller RA. Hormone-treated snell dwarf mice regain fertility but remain long lived and disease resistant. J Gerontol A Biol Sci Med Sci. 2004;59:1244–1250. doi: 10.1093/gerona/59.12.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li Q, Redden DT, Weindruch R, Allison DB. Statistical methods for testing effects on maximum lifespan. Mech Age Develop. 2004;125:629–632. doi: 10.1016/j.mad.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Wei LJ. The accelerated failure time model: a useful alternative to the Cox regression model in survival analysis. Statist Med. 1992;11:1871–1879. doi: 10.1002/sim.4780111409. [DOI] [PubMed] [Google Scholar]
- Wu S, Li Q, Du M, Li SY, Ren J. Cardiac-specific overexpression of catalase prolongs lifepsan and attenuates ageing-induced cardiomyocyte contractile dysfunction and protein damage. Clin Exp Pharmacol Physiol. 2007;34:81–87. doi: 10.1111/j.1440-1681.2007.04540.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Vatner DE, O’Connor JP, Ivessa A, Ge H, Chen W, Hirotani S, Ishikawa Y, Sadoshima J, Vatner SF. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130:247–258. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.