Abstract
In the last two decades, both diffuse optical tomography (DOT) and blood oxygen level dependent (BOLD)-based functional magnetic resonance imaging (fMRI) methods have been developed as noninvasive tools for imaging evoked cerebral hemodynamic changes in studies of brain activity. Although these two technologies measure functional contrast from similar physiological sources, i.e., changes in hemoglobin levels, these two modalities are based on distinct physical and biophysical principles leading to both limitations and strengths to each method. In this work, we describe a unified linear model to combine the complimentary spatial, temporal, and spectroscopic resolutions of concurrently measured optical tomography and fMRI signals. Using numerical simulations, we demonstrate that concurrent optical and BOLD measurements can be used to create cross-calibrated estimates of absolute micromolar deoxyhemoglobin changes. We apply this new analysis tool to experimental data acquired simultaneously with both DOT and BOLD imaging during a motor task, demonstrate the ability to more robustly estimate hemoglobin changes in comparison to DOT alone, and show how this approach can provide cross-calibrated estimates of hemoglobin changes. Using this multimodal method, we estimate the calibration of the 3 tesla BOLD signal to be −0.55% ± 0.40% signal change per micromolar change of deoxyhemoglobin.
Keywords: functional magnetic resonance imaging, near-infrared spectroscopy, diffuse optical tomography, multimodality imaging, Bayesian modeling
1 Introduction
In recent years there has been an emergence of an assortment of imaging modalities for noninvasively studying the brain. Among these, functional magnetic resonance imaging (fMRI)1–4 and diffuse optical tomography (DOT)5–8 are two techniques that have been developed largely in parallel to study cerebral functional hemodynamic responses. While both of these technologies are being applied successfully to a wide range of similar neuroscience and clinical topics, there are intrinsic limitations to each method, which are imposed by the governing physics of each technology (reviewed in Refs. 9 and 10). For example, while fMRI techniques such as blood oxygen level dependent (BOLD) can provide a measurement of blood oxygen saturation changes with fairly high spatial resolution (typically 2 to 4 mm for functional studies), these signals are physiologically ambiguous, owing to the indirect relationship between changes in the transverse relaxation rate of hydrogen nuclei and physiological hemodynamic parameters (i.e., deoxyhemoglobin and blood oxygen saturation) (reviewed in Ref. 11). Although such ambiguity does not impede the use of BOLD for mapping the spatial patterns of evoked changes, this does limit the use of BOLD to directly relate physiological parameters between subjects without additional calibration methods. Calibration of the BOLD signal is possible by inducing isometabolic changes in cerebral blood flow using hypercapnia or similar vasoactive agents.12–18 However, these hypercapnic-calibration methods require the subject to inhale increased levels of carbon dioxide gas for prolonged periods of time (up to several minutes). This procedure is both technically challenging and subject to several possible sources of systematic error19 that may render the technique difficult to translate to clinical applications. While the use of hypercapnia-calibrated fMRI techniques to provide quantitative measurements of blood oxygen saturation changes has been important in applying MR techniques to study metabolism, an alternative to hypercapnia calibration is needed to make the estimation of functional CMRO2 changes more routine. As CMRO2 is more directly related to neural-metabolic coupling, these measurements could have significant impact in better understanding the connections between neural and hemodynamic function in health and disease (reviewed in Ref. 20).
Continuous wave (cw)-based DOT has several complementary features to fMRI methods, including the ability to record a spectroscopic measurement of both oxygenated (oxyhemoglobin, HbO2) and deoxygenated (deoxyhemoglobin, HbR) forms of hemoglobin. In comparison to fMRI, optical methods generally have very high temporal resolutions, with acquisition rates capable of more than several hundred hertz. This resolution is much faster than needed to capture the typical slow evoked responses and fast enough to prevent aliasing of systemic physiological signals, such as cardiac pulsation and other physiology, which can be a major source of noise in fMRI studies due to undersampling.21,22 A drawback of the DOT technology is its lower spatial resolution, which is intrinsically limited by the propagation of photons through highly scattering biological tissue (reviewed in Ref. 23) and by the typically low number of optical measurement pairs recorded. Although DOT has the theoretical potential to provide quantitatively accurate measurements of hemoglobin concentration changes in the brain, in practice this can seldom be achieved because of the partial-volume effects introduced by the low spatial resolution and depth sensitivity of this method. In addition, the tomographic reconstruction of hemoglobin changes from optical measurements is generally an underdetermined and ill-posed inverse problem.7 Tomographic images can be improved with a greater number of measurement combinations, including overlapping measurements to provide more uniform sensitivities;24,25 however, regularization schemes must still be used to constrain the image reconstructions of the underlying absorption changes. In general, the accuracy of these reconstructions depends on the method and amount of regularization applied. In recent years, a great deal of attention has been given to this topic (reviewed in Ref. 26); however, more work is still needed. One promising approach—the incorporation of prior knowledge of the spatial location of the hemodynamic change by either anatomical-based8,27–29 or functionally-based priors30— improves the quantitative ability of DOT by constraining the solutions to the image reconstruction problem, and thus minimizing the errors introduced by partial-volume effects. With respect to optical imaging of the brain, the use of functional MRI data as such a statistical prior for the location of brain activation area has been suggested to improve DOT reconstructions.31 While it is believed that the introduction of statistical priors from structural or functional MRI may improve the localization of the optical signal, the implementation of such methods still has several unresolved issues. In particular, regularized reconstructions require a choice for the proper weight of the prior, as recently reviewed in Gibson, Hebden, and Acridge.26 In one extreme, the use of a strict (hard) prior (e.g., Ref. 32) will produce images with identical spatial resolution as the original prior (e.g., the functional MR image). However, this constraint assumes that the value, location, and boundaries of the prior have negligible uncertainty. Although the signal quality of fMRI images has greatly improved in recent years due to advancements in pulse-sequence design, RF coil design, and a move to higher magnetic field strengths, background physiology, intertrial variability, and other subject-related factors are still non-negligible sources of error in these measurements and will contribute to uncertainty in a fMRI-based prior. On the other hand, the use of a statistical prior (e.g., Refs. 30 and 33), while favorable in respect to the inclusion of the statistical uncertainty about the prior MRI information, requires knowledge of the proper statistical weight for the constraint. The optimal choice of this weighting depends on the relative measurement noise in both the fMRI and optical signals, and requires a proper statistical model of measurement noise. Concurrent multimodal measurements are unique in that physiological noise (for example, intertrial variability of the evoked response) is simultaneously recorded by each modality, while measurement noise is usually independent between instruments. This property of concurrent measurements provides an opportunity to use mutual information within multimodal measurements to help define the optimal statistical weighting of each modality in a joint image reconstruction. This concept of a bottom-up data fusion model has been previously introduced for neural imaging methods such as multimodal electroencephalography (EEG) and magnetoencephalography (MEG),34,35 but has not yet been demonstrated for multimodal hemodynamic measurements or optical methods. In this work, we describe a new analysis method for fusion of simultaneously acquired DOT and BOLD data that provides a joint estimate of the underlying physiological contrast giving rise to the concurrent measurements from both modalities. This approach makes use of the statistical properties of concurrent measurements and the commonality of the underlying physiology and fluctuations giving rise to these measurements. We use a Bayesian framework to jointly estimate brain activation changes from MR and optical using a single image reconstruction step. This approach enables us estimate oxy- and deoxyhemoglobin changes in the brain, with better spatial accuracy than DOT image reconstructions alone through the incorporation of time-varying spatial information from BOLD observations. Because the fMRI information constrains the spatial extent of the reconstruction, this helps to correct partial volume errors associated with optical reconstructions alone. Likewise, the spectroscopic information of the optical data defines the deoxyhemoglobin calibration of the BOLD signal. We find that the resulting fusion images contain quantitative information about micromolar changes in hemoglobin based on the cross-calibration of these two modalities.
We first present numerical simulations to examine the quantitative accuracy of hemoglobin estimates by our data fusion methods. Next, we apply the model to experimental data recorded simultaneously with DOT and BOLD imaging during a 2-s duration finger-walking task in five subjects.
2 Theory
2.1 Notation
In the following descriptions, we use the notations superscript T for the transpose operator, ⊗ for the Kronecker tensor product, and I for the identity matrix. In addition, for modality specific operators, a subscript will be used to reference the modality.
2.2 General Model Description
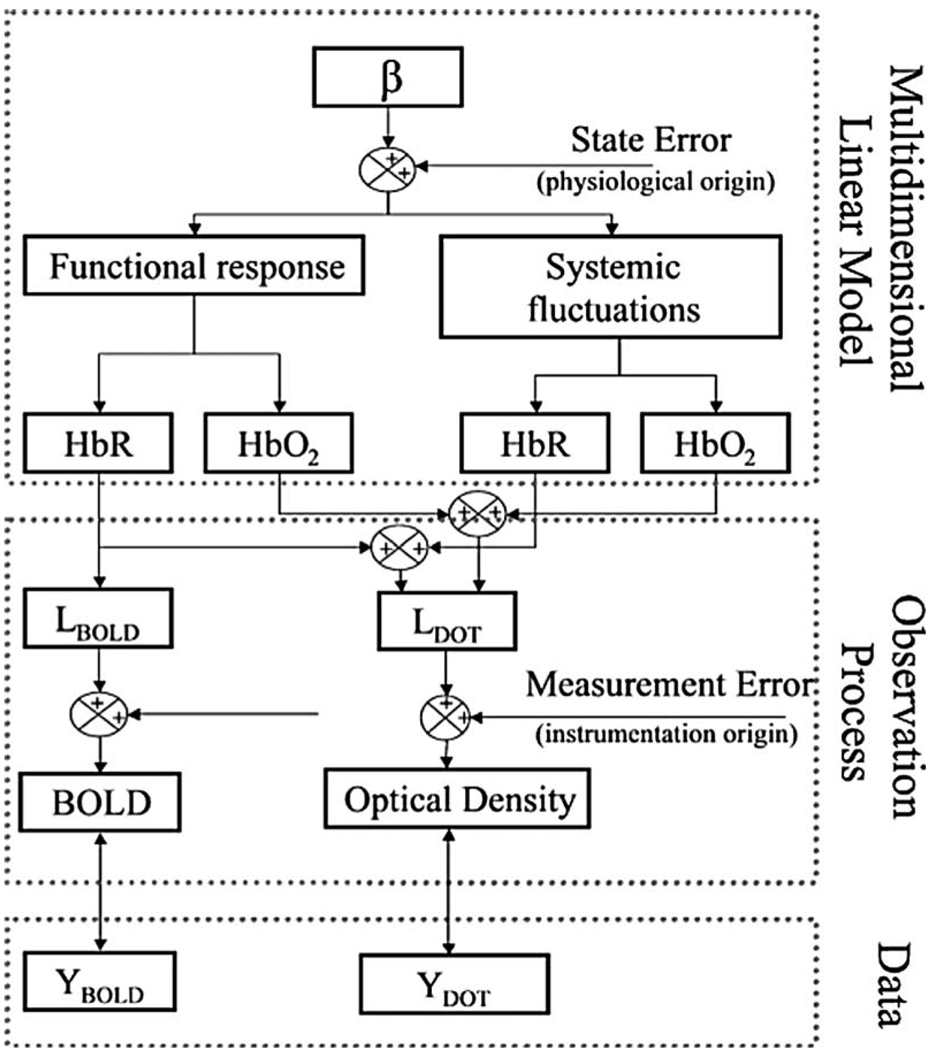
The functional contrast underlying both the BOLD and DOT signals derives from similar changes in hemoglobin concentrations and blood oxygen saturation. However, the details of the relationships between the measurements and underlying physiology are based on vastly different biophysical principles in the two modalities. These principles must be properly considered to incorporate both MRI and optical datasets into a single model. In Fig. 1, we present a schematic outline of our state model, which outlines how the measurement models for both MRI and optical are connected to a common model of physiological contrast. The framework for this model is described by three components, which are described in the following sections: 1. a multidimensional linear model by which a set of spatial-temporal basis functions is used to describe functional and systemic components of oxy- and deoxyhemoglobin concentration changes; 2. a set of measurement model equations describing the measurement biophysics for each modality and connecting the observations and underlying physiology; and 3. the least-squares minimization routine in which the experimental observations are fused to create a joint estimate of the underlying physiology.
Fig. 1. Schematic outline of model.
The multimodal fusion model is based on a state-space approach and consists of three components. The set of coefficients to be estimated (β) multiplies a convolution kernel of spatial and temporal basis functions in the multidimensional linear model to model the volumetric changes in oxy- and deoxyhemoglobin due to evoked activation and systemic fluctuations. Linear observation models connect the underlying changes in these hemodynamic variables to expected measurements by both DOT and BOLD technologies. The states are finally estimated by minimizing the error with respect to the experimental data using a Bayesian formulation of the linear inversion operation.
2.3 Multidimensional Linear Model
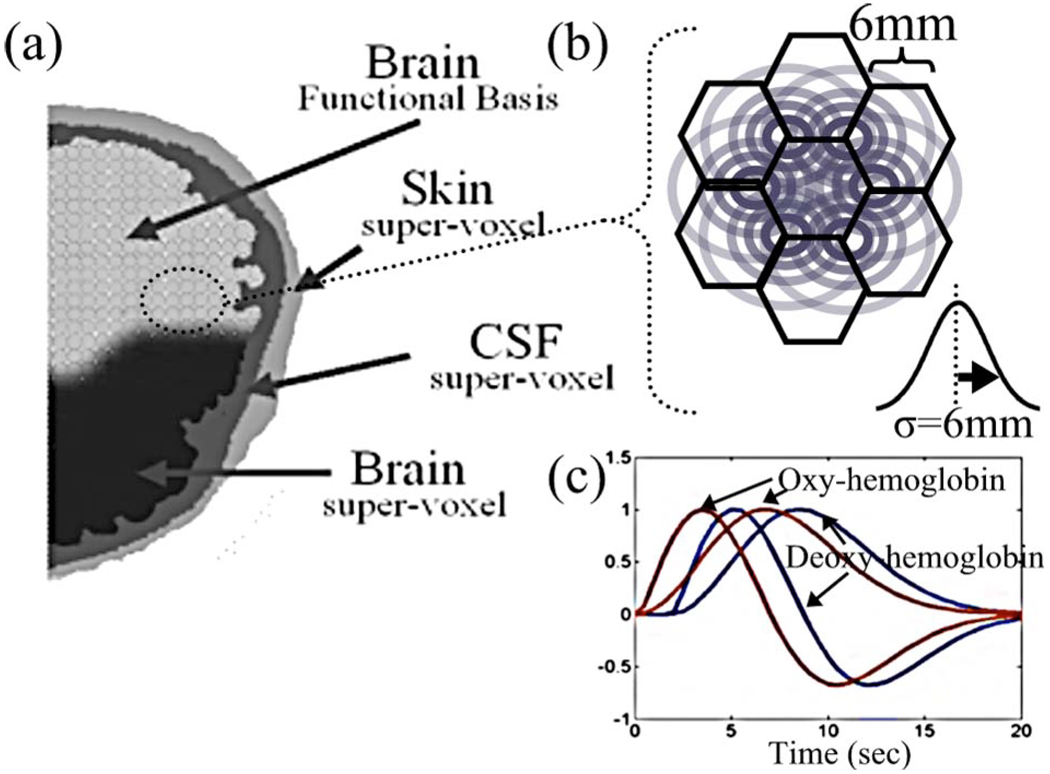
To describe the underlying physiological changes within the brain, we assume that changes in oxy- and deoxyhemoglobin can be described as linear combinations of a set of spatial and temporal basis functions designated to capture the functional and physiological hemodynamic fluctuations. In principle, this approach is similar to the general linear model (GLM) which has been previously introduced for fMRI36,37 and optical38,39 analysis, but has been modified here to include both spatial and temporal basis supports. Motivated by the anatomy of the brain and head and physiology of hemodynamic fluctuations, we introduced four distinct pairings of spatial and temporal basis functions in our model. These groups were; 1. the functional brain elements (denoted as subscript “functional” or F), 2. a global brain physiology basis (subscript “brain global” or B), 3. the cerebral spinal fluid (CSF) layer surrounding the brain (subscript CSF or C), and 4. the superficial skin layer (subscript “skin” or S). These four groups of spatial basis functions are diagrammed in Fig. 2.
Fig. 2. Basis functions in linear model.
A set of spatial and temporal basis functions are used to regularize the model estimates of evoked and systemic hemodynamic changes. To model the different aspects of the physiology, four sets of basis sets are used. In the volume corresponding to either gray or white brain matter, a gamma-variant function (and derivative) was used as a temporal basis (subplot C). These allow the modeling of the differing temporal dynamics of both oxy- and deoxyhemoglobin changes. A spatial basis of overlapping Gaussian spheres is used to reduce the dimensionality of the state space and replace a smoothing operation on the data (subplot B). To model the systemic contributions, the volume corresponding to skin, skull, and cerebral spinal fluid (CSF) layers are grouped into basis functions and given sine and cosine temporal functions to model their dynamics.
The first group of canonic functions representing the individual functional brain elements was derived from a spatial basis set composed of overlapping Gaussian spheres positioned on a hexagonal grid (as shown in Fig. 2). This basis is equivalent to a spatial smoothing kernel and is used in place of a separate smoothing operation applied to the data, as is usually typical of fMRI analysis. This basis also serves to reduce the unknown degrees of freedom of the model and makes the model more computationally tractable. We used a 6-mm standard deviation Gaussian kernel, which generated between 200 and 250 independent spatial functions (depending on the exact anatomy of the subject’s brain). These basis functions are restricted to those voxels that had been identified as either gray or white cortical matter by a tissue segmentation algorithm using the MRI anatomical images (see Methods in Sec. 3). The Gaussian spatial kernels were truncated at the anatomical boundaries between the brain and the CSF layers to impose a cortical constraint on the reconstruction of functional activation. The basis functions and reconstructions were limited to the contralateral hemisphere (opposite to hand movement) where optical measurements were recorded. The matrices representing each of the four types of spatial basis sets (denoted G in our model) have matrix dimensions of the number of independent basis functions (e.g., 200 to 250 for the functionally associated regions) by the number of voxels in the volume used for the optical and MRI forward models. When analyzing the experimental data, there are 28,672 columns in each of these matrixes (the fMRI images had 64 × 64 in-plane resolution and seven axial slices). To model the temporal dynamics of evoked functional changes within each of these Gaussian bases, we used a linear combination of a two-parameter (σ and τ) modified gamma function and its derivative (dispersion function), as given by the equations
and
| (1) |
The canonic temporal support vectors (t1 and t2) are generated from the evaluation of these respective continuous-time functions [Eq. (1)] at the discrete time points spanning the hemodynamic response (0 to 20 s at 2 Hz). This basis support is similar to the temporal basis often used in GLM analysis (e.g., the analysis of functional neuroimages (AFNI)40 or statistical parametric mapping (SPM)41 software packages). Since the temporal dynamics of the oxy- and deoxyhemoglobin responses are known to differ, we used separate timing parameters (σ and τ) for each hemoglobin species and denote these with subscript HbO2 and HbR, respectively. These timing parameters were empirically estimated by a nonlinear fit to the group average of the region-of-interest average of the DOT time courses as a preprocessing step of this analysis. The empirical values of τ and σ used in the model were (0.1 s ± 0.4 s, 6.7 s ± 0.3 s) and (1.8 s ± 0.4 s, 6.7 s ± 0.3 s) for oxy- and deoxyhemoglobin, respectively (standard errors estimated from the five individual subjects). We note that the inclusion of the second dispersion support in the linear model allows for sufficient flexibility in the temporal shape of the estimated response to model each of the individual subject’s data, as demonstrated in previous MRI studies (e.g., Ref. 42). For example, in this study, we found that a linear combination of these two temporal functions can account for most of the evoked responses in each of five subjects (R2|HbO2 = 0.81 ± 0.08 and R2|HbR = 0.86 ± 0.03; p<0.001 for all; for the average of five subjects ± StdErr).
The overall temporal model of the functional component of the hemodynamic signals can be expressed as the convolution of the experimental stimulus timing (U) and the functional impulse response functions [t1 and t2 in Eq. (1) for either oxy- or deoxyhemoglobin]:
| (2) |
where U is the binary vector describing the experimental paradigm (i.e., the timing of stimulus presentation) and spans the temporal duration of the experiment. The dimensions of the T matrix are the number temporal basis functions by the number of measurement time points.
In addition to the first pairing of spatial-temporal basis functions, which is used to model the evoked functional response, the global brain, CSF, and skin groups of temporal basis functions are used as systemic regressors of background physiology. These were included to model nuisance physiological contributions to the BOLD and/or DOT measurements by using larger (“super-voxel”) representations of the brain, CSF, and skin layers, as described in Fig. 2 and similar to the methods previously introduced to model systemic contributions to DOT signals.43 For each of these basis groups, we paired the spatial basis with a series of sine and cosine functions (1/20 to 1.0 Hz in 1/20-Hz steps) to describe the temporally oscillating systemic physiology. The skin-confined basis group models the systemic contributions to predominantly the optical signals and the brain-confined nuisance regressor models physiology common to both modalities.
In total, the set of all spatial-temporal bases is combined into a single convolution operator (GT), giving a total of eight unique basis groups (four groups for both HbO2 and HbR). This matrix is formed by the convolution of the individual spatial (G) and temporal (T) basis functions at each time instance for each of the eight groups. The matrix GT is given by the equation:
| (3) |
where the subscripts F, B, C, and S indicate the functional, global brain, CSF, and skin basis groups. The matrix GT has dimensions of twice the number of image reconstruction voxels multiplied by the number of discrete measurement time points (rows) by the total number of model unknowns (columns). The total number of model unknowns is equal to the number of spatial basis functions times the number of temporal basis functions and summed over the four tissue classifications and two hemoglobin species. For example, in our 6 min of experimental data, the GT matrix is approximately 41,000 × 3000 elements, which would be 200 gigabytes in size for 16-bit numerical precision. Fortunately, in practice, this full matrix never needs to be stored in memory, because in the inverse problem we only need the inner product of two such matrices (i.e., GTTGT), and this can be built up on a per time-point basis by making use of the block structure of this matrix and simple matrix operations. The details of this procedure are not discussed in this work.
The basis function matrix (GT) describes the spatial-temporal supports for the image reconstruction. Following the standard notation used previously to describe the general linear model in fMRI analysis (i.e., Ref. 44), the spatial and temporal dynamics of the underlying hemoglobin changes are described by a linear sum of the spatial-temporal basis function weighted by a vector of unknown coefficients denoted β. The vector containing the modeled hemoglobin changes (both systemic and evoked) at each volume element and time point is thus given by the equation:
| (4) |
where k is the total number of imaging parameters in the model and is equal to the number of time points multiplied by the number of volume elements. This matrix is a vectorized form of the reconstructed image (including time dimension) and is reshaped to a volumetric matrix in final analysis before displaying the images/movies.
2.4 Observation Models
The second component of our fusion model (as shown in Fig. 1) incorporates the measurement processes that relate the underlying physiological changes [oxy- and deoxyhemoglobin given by Eq. (4)] to the observations of each modality. The measurement equations for DOT and fMRI describe the biophysics of each instrument measurement. In this work, we have approximated this biophysics using linear approximations to these measurement equations. The validity of these linear simplifications has been discussed by a number of researchers that have examined the consistency of these DOT and fMRI modalities with these hypothesized underlying biophysical theories by correlating the temporal (reviewed in Ref. 10) and spatial45–47 components of measurements. These works have suggested that linearity between the change in deoxyhemoglobin and the BOLD signal can be justified. Note that although our current model has been formulated using these linear approximations, nonlinear extensions of both the optical and fMRI models are theoretically possible using similar methodologies (although the size of the model and computational memory requirements may currently limit this implementation in practice).
2.4.1 Blood oxygen level dependent measurement model
The BOLD signal has a complex origin arising from both intra- and extravascular tissue.4,48,49 These signals depend on not only the user acquisition parameters, such as echo time, magnetic field strength, and imaging echo type, but on features of the subject anatomy as well, such as vascular architecture and the orientation between blood vessels and the imaging fields.50 In this work, we simplify the BOLD measurement model by considering only the extravascular (EV) signal contribution. The EV signal is believed to compose the majority component of the BOLD signal at the 3 tesla (T) field strength.49 The EV signal is linear to changes in the concentration of deoxyhemoglobin per volume tissue and is given by the equation4,48,51
| (5) |
where Vo is the frequency offset in hertz of water at the outer surface of a magnetized vessel (=80.6 s−1 at 3 T52). Eo is the resting oxygen extraction fraction, and Vo is the baseline blood volume to tissue fraction. TE is the echo time used in the MR pulse sequence (30 ms in this study). [HbR0] is the baseline deoxyhemoglobin concentration. In general, the coefficients of this equation are not known with the certainty required for quantitative imaging, and moreover, may vary between subjects according to the baseline state or spatial region.53 For this reason, empirically calibrated fMRI techniques have been proposed to remove the influences of these parameters by using ratios of the blood flow and oxygen saturation (i.e., BOLD) signals.12
To account for these unknown calibration parameters in the BOLD measurement equation, these factors are lumped into a single parameter (α≡−4.3⋅υo⋅Vo⋅Eo⋅TE/[HbR0]). In our method, we use the information in the optical data to help determine the BOLD calibration factor, as we describe in Methods in Sec. 3. In our measurement model, the BOLD signal is a projection of the deoxyhemoglobin component of the model plus an additive noise term (υBOLD) by the equation
| (6) |
2.4.2 Diffuse optical tomography measurement model
In the DOT technique, near-infrared light is introduced into the head and propagates through the dense scattering layers of the scalp and skull into the brain. A small fraction of the light introduced eventually exits the head and is recorded a distance away from the source position. Hemodynamic changes in oxy- and deoxyhemoglobin concentrations affect the absorption properties of the brain and thus result in changes in the intensity of light as it migrates along a diffuse trajectory through the head. Using multiple measurements taken by an array of light source and detector positions spaced several centimeters apart (shown in Fig. 3), the DOT method can be used to spatially resolve these absorption changes and relate these to changes in oxy- and deoxyhemoglobin. The DOT measurement equation is described by the spatial profile of the light propagation through the head, which determines the sensitivity of these measurements. To derive the distribution of photons in a medium with a complex distribution of absorption and reduced scattering coefficients [μa(r) and , respectively], such as the human head, the photon density must be modeled by empirical means using computerized simulations. In Monte-Carlo-based modeling, the distribution of these photons is statistically modeled based on the probability of an absorption or scattering event at each region of space as described by μa(r) and . 54,55
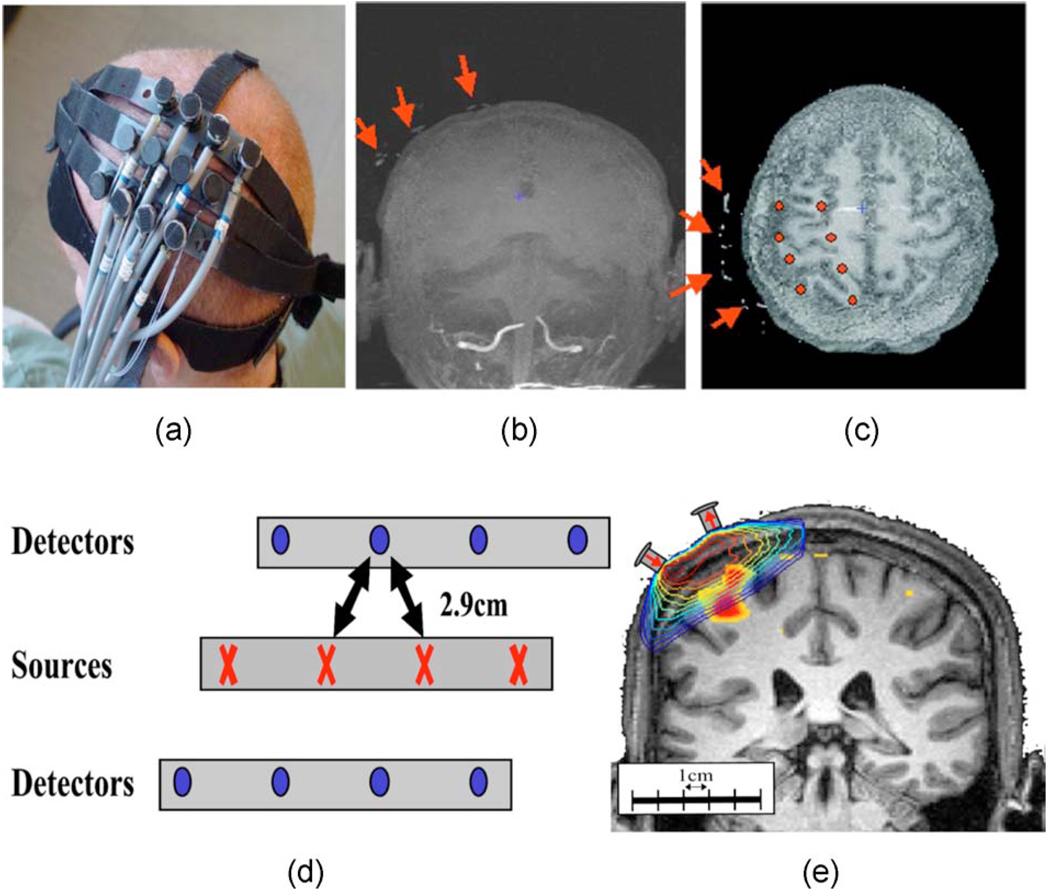
Fig. 3. Diffuse optical tomography.
Diffuse optical tomography uses spectroscopic measurements of optical absorption changes to record hemoglobin concentration changes. (a) The optical probe was placed over the subject’s primary motor area. (d) The probe contained four source and eight detector positions spaced 2.9 cm apart. (b) and (c) Fiducial markers on the probe were visible in the anatomical MR images. (e) The position of the probe and a segmented layer model for each subject were used to generate the optical sensitivity profiles using Monte Carlo methods.
For brain activation, the changes in the optical properties are generally small (on the order of a few percent), and thus, a linear approximation is believed to be sufficient for predicting the changes in optical measurements produced by localized changes in the optical properties (reviewed in Ref. 23). The spectroscopic forward model for such optical measurements is of the form56–58
| (7) |
where Y is the vector of measured optical signal changes for each source-detector pair and Aλ is a matrix describing the linear projection from absorption changes from within the volume to optical signal changes between each measurement pair. λ indicates wavelength dependence (690- and 830-nm wavelengths were used in this study). Each row of the matrix A describes the light propagation between a particular optical source and detector pair (shown in Fig. 3), which describes the spatial sensitivity for that measurement. The Aλ matrix is a projection operator, which integrates absorption changes over the volume to predict the measurements between sources and detectors. It has been shown that this linear operator is approximated by the adjunct product between the photon density distributions for each given source and each given detector involved in each optical measurement.7 In Eq. (6), the projection operators are combined with the spectral extinction coefficients (ελ) to give a direct forward operator between the concentration changes in HbO2 and HbR (per volume element) and the measured optical density changes at multiple wavelengths. In our model, υλ is an additive measurement noise term specific to each measurement channel and wavelength.
2.5 Data Fusion Model
Rather than treating the DOT and BOLD measurements independently or first computing an MR-based spatial map to be used as a reconstruction prior, our model tries to preserve the mutual information in the multimodal data by simultaneously considering measurements from both instruments. We concatenate the two measurement equations [Eqs. (6) and (7)] into a single joint-observation operator
| (8) |
The joint set of multimodal data can be expressed as the multiplication of the underlying model of oxy- and deoxyhemoglobin changes by the linear observation operator. Combining Eqs.(4) and (8), the complete model equation can be written as
| (9) |
where In is an identity matrix with size equal to the number of measured time points (n). The total model considers both time and space simultaneously (as defined by the spatial and temporal basis matrix GT). This final model can be written as a single matrix equation,
| (10) |
where Y has dimensions of total number of measurements (number of optical plus MRI measurements for all time points) and can be written by concatenating the set of measurements for all time points (t ∈ [1 n]) as
| (11) |
Similarly, the measurement model matrix L must be extended by replicating this matrix along the diagonal for each of the measurement time points. In our experimental data, both optical and MRI data have the same sample rate (2 Hz) and thus all discrete observation points use the same joint measurement equation, which includes both optical and fMRI measurement models. However, in general, the fMRI acquisition rate will be considerably slower than that of the DOT, and this can be accounted for in our model by time indexing the measurement operator (L) to use observations for one or both of the appropriate modalities at each observation point, depending on the samples acquired at that time. Thus, this model can be used to interpolate the reconstructed image using the higher temporal resolution DOT data, while maintaining a solution that is less frequently spatially constrained by fMRI measurements.
2.6 Bayesian Inversion Routine
To estimate the coefficients for the spatial-temporal basis sets describing changes in HbO2 and HbR, the linear model Eq. [(10)] is solved using a Bayesian formulation of the linear inverse operator59
| (12) |
In Eq. (12), R−1 and Γ2Q−1 represent the inverses of the covariance for the observation noise and state noise models, respectively. We have chosen this formulation over the Tikhonov regularization (Moore-Penrose generalized inverse), which is more commonly used in DOT, because the Bayesian formulation separates the regularization parameters into both an observation noise (R−1) and a state noise (Γ2Q−1) preconditioning term. A similar regularized inverse scheme was recently described by Yalavarthy et al.60 This weighted least-squares method provides a better framework to introduce differential observation noise for each modality. From a statistical standpoint, Γ2Q−1 is a covariance prior on the states β. This definition offers some intuitive guidance on how to tune the inversion, since we expect physiological fluctuations to be approximately on the order of micromolar magnitudes. In practice, however, the multidimensional linear model is only an approximation of the actual physical system, and the assumed Gaussian distributed prior on β is imperfect. These approximations effectively mean that the Bayesian inverse is a regularization scheme that must be tuned, and we have introduced a scalar parameter Γ for that purpose. In addition, we define the variance of the observation (measurement) noise model (υ) as a diagonal matrix formed from the variance [var()] of each measurement, i.e.,
| (13) |
Thus, the measurement noise covariance can be estimated from experimental data and is used to reflect our confidence in the measurements taken from the two different modalities as a statistical prior, as described in Methods in Sec. 3. In this model, we have assumed that both the process and the measurement error for each modality are zero-mean random variables.
3 Methods
3.1 Experimental Methods
The experimental protocol used in this study has been previously described.61 The data used in this analysis have been previously reported in that paper for comparison of the temporal characteristics of optical methods and fMRI. Five healthy, right-handed subjects (4 male, 1 female) were imaged in this experiment. The task consisted of a two-second duration finger-walking on the right (dominant) hand. The Institutional Review Board at Massachusetts General Hospital approved these procedures.
3.2 Diffuse Optical Tomography Acquisition and Preprocessing
We used a multichannel continuous-wave optical imager (CW4, TechEn Incorporated, Milford, Massachusetts) to obtain the measurements as previously described.62 The DOT imager has 18 lasers—nine lasers at 690 nm (18 mW) and nine at 830 nm (7 mW)—and 16 detectors of which only four source positions and eight detectors were used here. The laser wavelengths were 690 and 830 nm (18 and 7 mW, respectively).
The DOT probe was made from flexible plastic strips with plastic caps inserted in it to hold the ends of the 10-m-source/detector fiber optic bundles. The probe consisted of two rows of four detector fibers, and one row of four source fibers arranged in a rectangular grid pattern and spaced 2.9 cm between nearest neighbor source-detector pairs (shown in Fig. 3). This plastic probe was then secured to the subject’s head centered over the contralateral primary motor cortex (M1) via Velcro straps and foam padding. The 10-m fibers were run through the magnet bore to the back of the scanner and through a port into the control room, where they were connected to the DOT instrument.
Following collection and separation of source-detector pairs, the timing of the DOT data was synchronized to the MRI images (as described in Ref. 61). The data were down-sampled using a Nyquist filter to the same 2-Hz sample frequency as the fMRI. The raw light fluence measurements were converted to changes in optical density by the negative log of the normalized incident light intensity {ΔODλ(t)=−log[Iλ(t)/Iλ(0)]}.
Monte Carlo methods were used to generate the optical sensitivity profiles describing the DOT measurement equation [Eq. (6)].45,55,63 Anatomical MP-RAGE images acquired during the session were segmented into a five-layered head model for each of the subjects,45 which was used for these Monte Carlo simulations and also to define the spatial basis functions used in the linear model. From the vitamin E fiducial markers used to mark the optical probe, the locations of the DOT optodes were located. The photon migration paths were sampled from the simulated trajectories of 108 photons at both 690- and 830-nm wavelengths. These simulations were used to calculate the linear optical forward matrices (Aλ) and substituted into Eq. (7) (see Fig. 3). To create the spectroscopic forward operator, the hemoglobin extinction coefficients were used from the Oregon Medical Laser Center (http://omlc.ogi.edu/).
3.3 Functional Magnetic Resonance Imaging Acquisition and Preprocessing
BOLD-fMRI measurements were performed using a 3 tesla Allegra scanner (Siemens Medical Systems, Erlangen Germany). fMRI data were collected using a gradient echo planar imaging (EPI) sequence [TR/TE/α=500 ms/30ms/90 deg] with seven 5-mm oblique orientation slices (1-mm spacing) and 3.75-mm in-plane spatial resolution. Structural scans were performed using a T1-weighted MPRAGE sequence (1 × 1 × 1.33-mm resolution, TR/TI/TE/α = 2530 ms/1100 ms/3.25 ms/7 deg]. The BOLD time series was preprocessed using a motion-correction algorithm64 and mean normalized before being used in the model.
3.4 Numerical Simulations
We used simulation studies to test the quantitative accuracy of this method. This enables us to examine the ability to quantify hemoglobin changes, since no empirical “gold-standard” method exists that can provide quantitative measurements with which to validate our experimental results. Simulated inclusions were placed shallow (~18 mm) and deep (~25 mm) from the surface of the head (shown in Fig. 4). The hemodynamic response was simulated with a maximum peak value of 8 µM and −2.5 µM for oxy- and deoxyhemoglobin concentration changes, respectively. Varied levels of measurement noise were added to the simulated measurements to create varied levels of contrast-to-noise ratios.
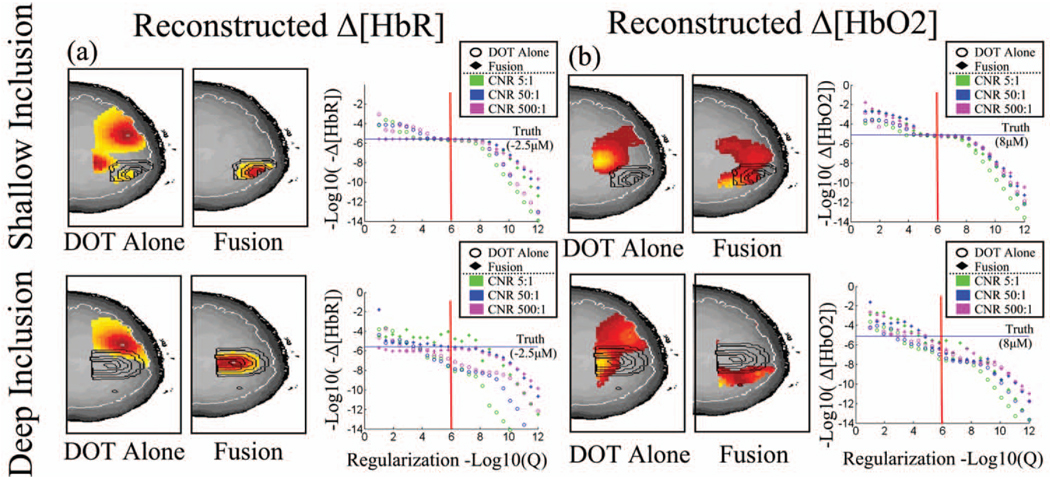
Fig. 4. Characterization of model quantitative accuracy.
The quantitative accuracy of the model was examined with observations simulated from a shallow (18 mm, toprow) and deep (25 mm, bottomrow) inclusion. The simulated inclusions are shown contoured (black) in each image (axial projections). The boundaries of the cortex are outlined in white. Measurement noise was added to achieve a 5:1, 50:1, and 500:1 contrast-to-noise ratio (CNR). Hemoglobin changes were reconstructed using the fusion model and DOT data alone for comparison. Reconstructions were performed at various regularization amounts to examine the dependence of the recovered magnitude of the response on the regularization parameter. The images are shown for the reconstruction at 1/Γ = 1µM (indicated by the vertical red line) for the CNR=50:1 case.
3.5 Optical Calibration of the Blood Oxygen Level Dependent Signal
The BOLD measurement model depends on an unknown calibration factor(α), which depicts the several baseline and structural unknown factors within Eq. (5). This calibration factor is determined empirically by comparing the spatial and temporal profiles of the DOT and BOLD data as described in Ref. 45. Using the linear forward operator describing the DOT measurement model [Eq. (7)], we project the model estimate of the BOLD signal from its natural volume space representation into the optical measurement (i.e., source-detector) space. By substituting Eq. (6) into Eq. (7), the expected optical signals due to a change in deoxyhemoglobin can be modeled from the model estimated BOLD signal, as discussed in Ref. 45.
| (14) |
These BOLD derived multiwavelength optical density changes can be used to estimate the change in deoxyhemoglobin predicted by the spatial location and magnitude of the BOLD signal (Δ[HbR]BOLD) for each source-detector pair using the modified Beer-Lambert law,65,66 and compared to the measured optical data to find the calibration factor (α) using the equation
| (15) |
In the modified Beer-Lambert equation [top in Eq. (15)], L is the linear distance between the position of a source and detector, and DPF is the differential path-length factor, which is a dimensionless coefficient defined as the effective path length through the head divided by the source-detector separation.63,65,66 This is calculated from the Monte Carlo simulations. Since this projection uses the optical forward model, it accounts for the DOT partial volume errors in the calibration of the BOLD signal. A single calibration factor (α) was used, which is consistent with findings of the spatial correlation of response amplitudes between the two modalities.45,46 To estimate this parameter in the model, we employ an iteration routine between state and parameter estimation. Alpha (α) is first estimated via Eq. (15) using the raw optical and MRI data, and then the states (β) are estimated using Eq. (12). The estimated optical and BOLD signals are recovered from the forward model using the estimated value of beta, and the alpha is re-estimated from the modeled BOLD and optical signals using the bottom of Eq. (15). This beta and alpha estimation process was repeated for 20 iterations. Alpha was found to converge below a +/−10% variation after a few iterations (approximately five iterations).
3.6 Estimation of Noise Covariance
To estimate the covariance of the measurement noise (R), we calculated the variance of the residual for each measurement by iteratively solving the state estimate and estimating R to be the variance of the residual of the measurements. The initial seed of R was calculated from linear regression of the data with the temporal basis.
An identity matrix was used for the state covariance matrix Q. The regularization tuning parameter (Γ) was adjusted in the analysis, as noted in Sec. 4. A single regularization parameter was used to scale the variance of both the oxy- and deoxyhemoglobin states.
4 Results
4.1 Simulation Studies
To investigate the improvements in the reconstruction accuracy, forward simulations of synthetic data and inverse reconstructions were first performed as described in Sec. 3. The simulated observation vectors were reconstructed using the DOT only and the multimodal fusion (DOT and BOLD) data in the reconstructions. Reconstructed axial slices are shown in Fig. 4 for the shallow and deep simulated inclusions. Using only the DOT data, the image reconstruction of both oxy- and deoxyhemoglobin depend strongly on the regularization parameter used (Γ) In Fig. 4, reconstructions are represented at regularization values of 1/Г=(1 µM). To examine the dependence of the reconstruction accuracy on the regularization parameter, we looked at the reconstructed response amplitude and goodness-of-fit of the model for varied levels of regularization. This was repeated for both the shallow and deep inclusions and at several levels of simulated instrument noise (5:1, 50:1, or 500:1 contrast-to-noise ratio). The results, shown in Fig. 4, demonstrate the robustness of the fusion model to the choice of this regularization for deoxyhemoglobin reconstructions. Since the estimate of deoxyhemoglobin change is overdetermined in the presence of both optical and BOLD measurements, accurate estimates of the response are recovered at minimal regularization. When over-regularization is applied, the response is underestimated, as expected. In contrast, reconstructions using only the DOT data show high dependence on the regularization amount. This result is consistent with prior expectations of the behavior of the DOT inverse problem with our measurement geometry. The fusion reconstruction of oxyhemoglobin changes is also dependent on the regularization applied. This is due to the fact that the reconstruction of oxyhemoglobin changes is only indirectly informed by the fMRI data through the off-diagonal terms in the spectroscopic optical forward model [Eq. (7)]. Since only the DOT measurements contain direct information about oxyhemoglobin, this component of the inverse problem is still ill posed and requires significant regularization. However, the oxyhemoglobin reconstructions are still modestly improved by the fusion reconstruction over the DOT-only reconstruction.
4.2 Experimental Studies
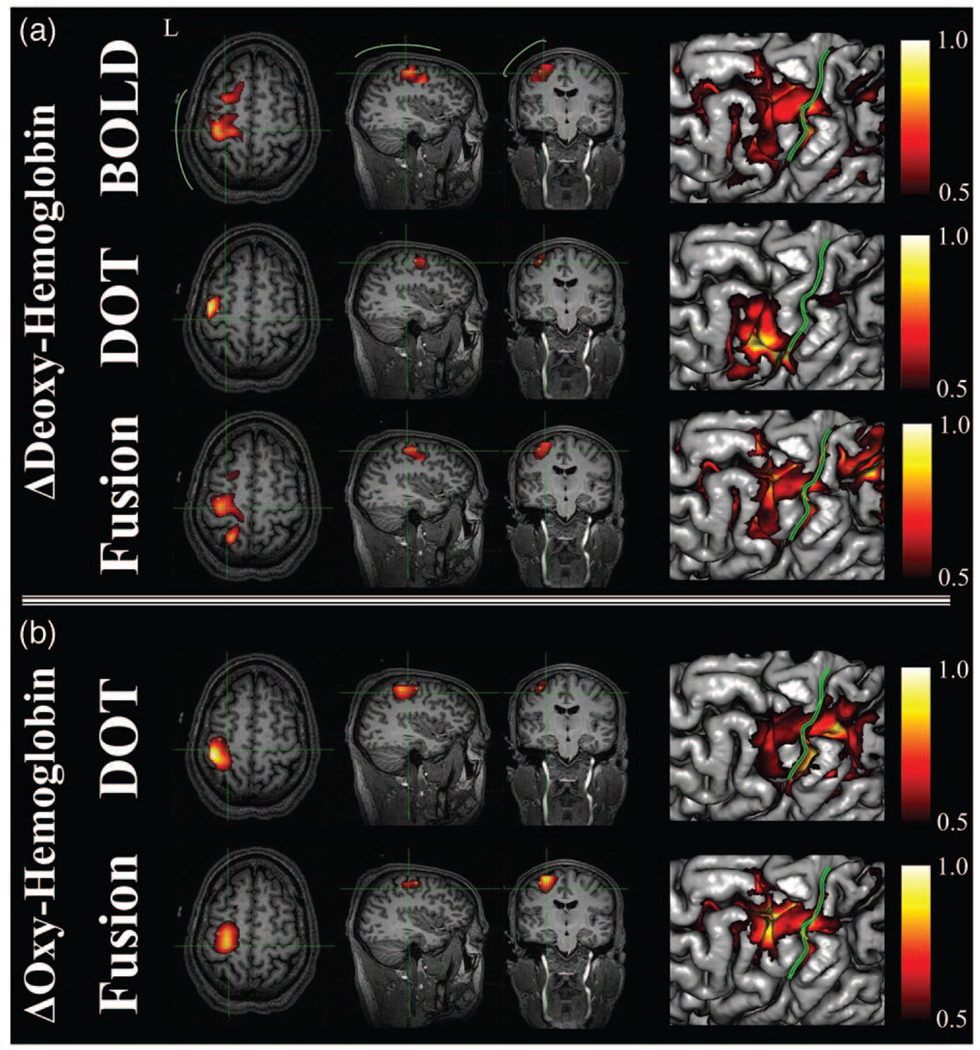
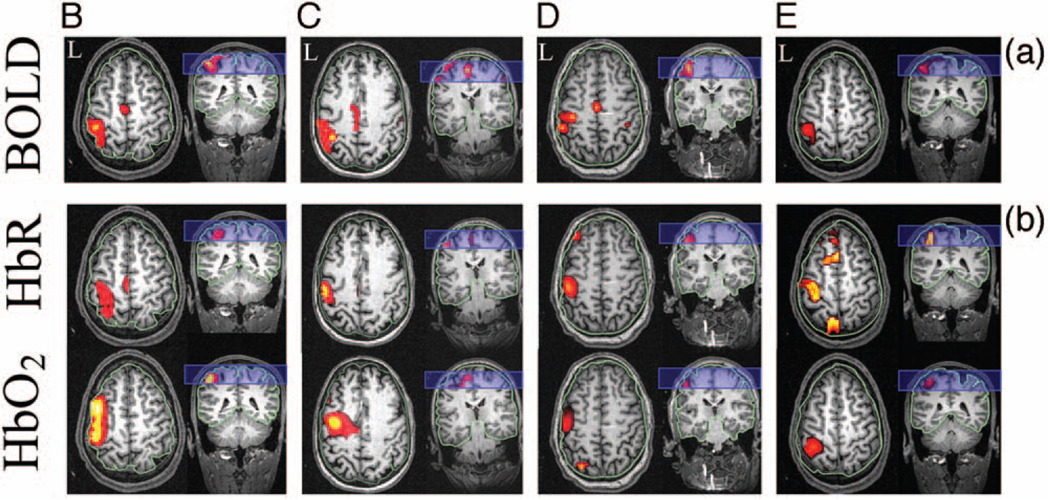
Images of evoked hemoglobin changes were reconstructed from the BOLD alone, DOT alone, and multimodal (fusion) experimental datasets for all five subjects, as shown in Fig. 5 and Fig.6. In Fig. 5, we show an in-depth look at the results for a single subject (subject A), and show the reconstructions of deoxyhemoglobin (upper rows) and oxyhemoglobin (bottom rows). The right column of images in Fig. 5 shows the surface rendering of the activation regions with the central sulcus marked for clarification. In Fig. 6, we show the reconstructed results for the data-fusion method for subjects B through E and comparison to the BOLD model. In agreement with previous analysis of this data,61 we found localized functional activation in the motor-cortex (M1; precentral gyrus) and primary sensory (S1; post-central gyrus) regions for each of the individual five subjects for the fMRI and fusion reconstructions. The peak amplitudes of the estimated evoked oxy- and deoxyhemoglobin responses are given in Table 1. In general, the multimodal fusion and BOLD images were similar, although a few notable differences were observed. These differences are most likely the result of differences in the regularization in the two models. A regularization amount (1/Γ) of 10 µM was used in the DOT and fusion reconstructions, and 10% in the BOLD images, which may account for these differences. In addition, the fusion images are more sensitive than single modality methods to both temporal and spatial registration errors between the two modalities, which can include intrinsic differences due to differential vascular sensitivity. This can introduce disinformation between modalities, which will result in the loss of confidence of an activation event and lower functional effects statistics (e.g., more likely to reject a null hypothesis).
Fig. 5. Direct hemoglobin reconstructions from empirical data.
This figure shows (b) the model reconstructions using the DOT alone, BOLD alone, and (a) fusion datasets for deoxy- and oxyhemoglobin changes. Data are represented for subject A. All responses have been normalized for comparison. Reconstructions were performed at a regularization level of 1/Γ = 10µM for the DOT and fusion models and 1/Γ = 10% for the BOLD model. Functional changes are shown masked below the half-max response amplitude. In (a) the approximate location of the optical probe is shown in green. The right-most images show the maximum intensity projections of the activation patterns onto the brain pial surface [surfaces generated using Freesurfer (The Massachusetts General Hospital; see http//surfer.nmr.mgh.harvard.edu)] and displayed using NeuroLens [Université de Montréal; (see http://www.neurolens.org)]. The central sulcus is outlined (green line) in each surface image.
Fig. 6. Comparison of fusion and MRI image reconstructions.
In this figure, we show the BOLD (a) and fusion reconstructions (b) for subjects B through E (left to right). Subject A was presented in Fig. 5. Regularization was applied as described for Fig. 5. All images are shown normalized to the maximum response and masked below the half-max amplitude. Axial and coronal projections are shown. The outline of the boundary between the brain and superficial layers is shown in green. The blue rectangle indicates the location of the imaging volume from the fMRI.
Table 1.
Comparison of reconstructed hemoglobin amplitudes. The maximum (minimum) response amplitude was calculated for a region of interest in each of the DOT alone and fusion reconstructions for the five subjects. Regions of interest were manually defined in the motor cortex area. The BOLD calibration factor (α) was calculated as described in the text. The values shown are for the reconstructions at a regularization 1/Γ = 10 µM.
| DOT-only | Data fusion | BOLD calibration (α) | |||
|---|---|---|---|---|---|
| Subject | ΔHbO2 | ΔHbR | ΔHbO2 | ΔHbR | |
| A | 5.5 µM | −2.9µM | 6.1µM | −1.2µM | −0.41 %-BOLD/µM |
| B | 0.3 µM | −1.8µM | 3.9µM | −1.5µM | −0.52 %-BOLD/µM |
| C | 3.1 µM | −1.4µM | 4.4µM | −0.7µM | −1.24 %-BOLD/µM |
| D | 4.9 µM | −1.0µM | 4.5µM | −0.3µM | −0.19 %-BOLD/µM |
| E | 7.3 µM | −3.µM | 1.6µM | −0.5µM | −0.39 %-BOLD/µM |
| Group | 4.2 µM | −2.0µM | 4.1µM | −0.9µM | −0.55 %-BOLD/µM |
In the model reconstructions that only used the DOT data, we found that the amplitudes of the estimated hemoglobin changes were dependent on the regularization applied, which was consistent with the simulation results and prior expectations on the behavior of the ill-posed inversion of the optical forward model and minimum norm estimator. The reconstructions shown in Fig. 5 and Fig.6 and values given in Table 1 were obtained at a regularization value of 1/Γ=10 µM, which provided the best reconstruction from the optical data alone when qualitatively compared to the fusion or BOLD reconstructions. We have chosen to present images using this regularization point to give the best possible representation of optical-only reconstructions for comparison to the data fusion method. With this choice of regularization, the optical-only results were qualitatively close to the fMRI images (as demonstrated in Fig. 5 for subject A), although distinct biases in the spatial locations were notable. Even with the cortical constraint of our model, we noted that our optical-only reconstructions tended to be biased toward the locations of optodes, which, in most cases, displaced the location of the DOT reconstructions relative to the BOLD alone or fusion derived estimates. In contrast to the deoxyhemoglobin reconstructions, the improvements of the fusion model to estimate of oxyhemoglobin changes over the DOT alone model were less dramatic. However, we found that the fusion estimates of oxyhemoglobin were less susceptible to biases toward the optode positions, although they were still superficially biased toward the head’s surface. The optical probe used in this study was based on a simple nearest-neighbor geometry. Recent work has shown that optical-only reconstructions could be further improved using tomographic probe designs, including the use of overlapping measurements (e.g., Refs. 24 and 25). These methods would be expected to further improve the accuracy of the reconstructions performed here for both optical-only and fusion methods.
In addition to the functionally evoked responses, our model also includes regressors for the background physiological oscillations modeled as global effects using large spatial basis supports. The inclusion of these regressors accounted for an average of an additional 8.5%±4.4% of the total model variance based on partial R2 analysis of the model (average of five subjects±StdErr; range 3.3%[subject B] to 26.3%[subject E]). As recently demonstrate by Diamond et al.43 using optical measurements and Glover, Li, and Ress67 using fMRI measurements, the use of direct measurements of systemic physiology by pulse-oximtery or noninvasive blood pressure methods as additional model regressors may provide further reduction of these cerebral physiological signals and should be examined in future work.
4.3 Optical Calibration of Blood Oxygenation Level Dependent
In Table 1, we show the recovered values of the optically calibrated BOLD scaling factor (α) for each of the five subjects (mean: −0.55%-BOLD/µM±0.40%-BOLD/µM). The high variance in the group estimate may be the result of differences in the baseline volume or oxygen extraction levels between subjects (particularly subject C). In addition, the values of the BOLD model parameters given in Eq. (5) are based on additional assumptions about the size and orientation of blood vessels (i.e., Ref. 50) and may also contribute to the differences observed between the five subjects. We can compare our measured values of alpha with theoretical estimates derived from Eq. (4) and data from previous literature. Assuming a baseline blood volume fraction of 3 to 5%,68,69 a total hemoglobin concentration of 60 to 100 µM,70 and an oxygen extraction fraction of 30 to 40%,69,71 the expected value for this calibration factor can be estimated to be between −0.3 to −0.9%-BOLD/µM--HbR for the MRI acquisition parameters used in this study. This calibration factor has also been determined experimentally by the hypercapnia method of BOLD calibration described by Davis et al.12 The hypercapnia calibration method determines the maximal BOLD change possible if all deoxyhemoglobin were displaced from the region (reviewed in Ref. 19). Thus, according to our approximation of a primarily extravascular water contribution to the BOLD signal, as given by Eq. (5), this hypercapnia calibration factor (usually denoted M) can be related to our alpha parameter by multiplying by the baseline deoxyhemoglobin levels (i.e., M≈α × [HbR0]). The empirical value of M has been reported between 7 to 25% as tabulated in Ref. 19, which corresponds to a value of alpha between −0.4 and −1.4%-BOLD/µM using the ranges of baseline total hemoglobin and oxygen extraction cited before. Indeed, both the empirical and theoretical estimates of this calibration factor are consistent with our values calculated for the five subjects, as shown in Table 1. In future work, it will be necessary to validate the optical-calibration approach against other calibration methods like the hypercapnic method.
5 Discussion
Concurrent multimodal measurements are observations of common underlying changes in underlying functional contrast. Our fusion model combines this mutual information from optical and fMRI modalities into a joint estimate of underlying hemoglobin changes. This approach provides a framework to combine the advantages of the high spatial and temporal resolutions contained between both modalities. In this work, we have developed a model that incorporates the biophysical principles that describe the relationships between the optical and fMRI measurements and underlying cerebral physiology. Using a single image reconstruction step, we can obtain direct estimates of hemoglobin changes that are simultaneously consistent with all sets of observations. This allows us to use the high spatial resolution of fMRI as a spatial prior to improve the optical reconstruction, while at the same time, to use high temporal and spectroscopic information from the DOT as priors on the reconstruction of the BOLD functional changes. The fusion of the higher spatial information from fMRI measurements and the spectroscopic information of the optical technique provided cross-calibrated estimates of hemoglobin changes. The Bayesian framework used in this model allows us to optimally incorporate data from different sensors based on knowledge of the statistical errors in each measurement type. In comparison to methods using fMRI as a statistical prior for the optical reconstruction, our approach incorporates multimodal information based on the statistical properties of concurrent measurements. We believe that this approach provides a framework to more efficiently consider concurrent multimodal datasets and to utilize the mutual information in the signals to improve the accuracy of estimates of the functional response. The advantages of this approach have been discussed for data fusion of magnetoencephalography and electroencephalography data using similar mutual information models (e.g., Refs. 34 and 35). We believe that future extensions of our method will offer similar utilities for hemodynamic imaging.
5.1 Comparison of Reconstruction Methods
In the comparisons between the optical only, BOLD only, and fusion reconstructions of the simulated data, we found that fusion methods produced the most accurate estimates of hemoglobin changes both in terms of spatial localization and quantitative accuracy. In particular, we found that for the fusion reconstructions of deoxyhemoglobin, the amplitude of these changes was relatively independent of the magnitude of the regularization applied and were fairly robust to errors in underestimation of the regularization parameter (Γ) as demonstrated in Fig. 4. In the fusion model, enough information is available to make the deoxyhemoglobin reconstruction problem overdetermined. Even at a minimal regularization level, the quantitatively accurate magnitudes of the deoxyhemoglobin responses were still recovered for both the deep and shallow deoxyhemoglobin inclusions. As expected, an over-regularization of the linear model resulted in underestimation of the hemodynamic response in all models. In contrast to deoxyhemoglobin, which is directly informed by the BOLD model, we found that reconstructions of oxyhemoglobin changes in the fusion were still dependent on the regularization. This is because oxyhemoglobin is only indirectly informed by the BOLD signal and still represents an underdetermined problem.
In comparison, we found that our DOT alone model was generally more superficially biased than the fusion model at recovering the spatial profile of both targets, and that the amplitude was very sensitive to the amount of regularization applied. This bias will result in an underestimation of the response amplitudes, since the optical sensitivity profiles fall exponentially with increasing depth. This finding is consistent with prior expectations concerning the accuracy of DOT reconstructions. Although the spatial basis functions used in this work ensured a cortical constraint to functional activations, the reconstructed amplitudes were underestimated by up to several orders of magnitude, in particular for the deep simulated inclusion, and the magnitude of the DOT estimated hemoglobin changes were highly dependent on the regularization parameters used.
The DOT alone and fusion reconstructions of the empirical data were consistent with the findings from the simulations. The spatial locations of the fusion reconstructions were more consistent with the profiles from the BOLD alone. Although the magnitudes of the changes were comparable in our final images, the DOT reconstructions were highly dependent on the regularization applied, which was chosen here to provide the best possible DOT reconstructions based on the BOLD result. Thus, this may be misleading to the quality of the reconstructions that can be obtained routinely by DOT alone.
5.2 Optically Calibrated Blood Oxygen Level Dependent Signals
In addition to the Bayesian fusion model that we have introduced in this work, we have also presented a method for using the spectroscopic information of the optical data to provide insight into the BOLD signal, allowing for optically calibrated BOLD imaging. In this work, we have assumed that a single calibration factor can be applied to calibrate the BOLD signal. In both the simulation and experimental results, we found that the estimate of this factor converged after only a few iterations of the model. In the simulation results, we found that we could recover this calibration factor. In the experimental results, we found approximate agreement between our estimates of this factor and the expectation from theoretical work approximated at 3T and the empirical hypercapnia-based BOLD calibration.
5.3 Model Limitations and Future Extensions
While our data fusion method greatly improves the reconstruction and quantitative accuracy of deoxyhemoglobin changes, the improvements to oxyhemoglobin quantification were more modest. In principle, the state covariance matrix (Q) used in the Bayesian pseudoinverse model [Eq. (12)] can be used to introduce a statistical prior between deoxy- and oxyhemoglobin maps with the inclusion of off-diagonal elements connecting the two matrix quadrants for oxy- and deoxyhemoglobin. However, in the work described here, we have purposefully not included these off-diagonal terms based on recent work that has suggested that underlying vascular structures may displace the locations of the two chromophores due to differential arterial versus venous weightings.45–47,72 In support of this, several recent fMRI-based studies have also shown spatial displacements between the BOLD signal and MR measures of blood volume73 and blood flow,74 which corroborate a spatial displacement of arterial and venous-weighted measurements. In future studies, we suggest that the incorporation of an MR measure of blood volume changes73,75,76 into this data fusion model may be more appropriate to constrain total-hemoglobin changes and consequently the quantification of oxyhemoglobin.
An assumption made in this model was that we only examined the extravascular contribution to the BOLD signal. This assumption ignores the contribution from the water molecules within the blood vessels that interact directly with the deoxyhemoglobin heme group.1,4,48,77 This assumption is supported by recent work by Lu and van Zijl determined that the extravascular signal composes approximately 67% of the intrinsic relaxation at 3 tesla.49 Further evidence of the dominance of the extravascular signal has also been demonstrated in the empirical comparison of optical and BOLD signals, which have shown a strong temporal correlation between the BOLD signal and the optical measure of deoxyhemoglobin (reviewed by Ref. 10). Thus, the source of BOLD functional contrast at 3 tesla is expected to be predominantly extravascular, which we believe justifies the assumption in this model in this current work. However, at lower field strengths, alternative MR acquisition protocols, or for more quantitatively accurate results, the intravascular component may become necessary. The inclusion of the intervascular component of the fMRI signal creates a nonlinearity in the BOLD measurement equation, which additionally depends on changes in the venous blood volume that determine oxygen saturation changes (i.e., Ref. 48). The future extension of this model to incorporate this term can be achieved by the replacement of the linear (extravascular BOLD) measurement model in Eq. (5) with a more detailed state-space model of the vascular physiology (i.e., Refs. 78–81). In future work, vascular models such as the Balloon77 or Windkessel82 models will enable the incorporation of more modalities into the model, such as blood flow.83 This nonlinear extension of this bottom-up model could allow the direct estimation of cerebral metabolism changes and provide a means to fuse multimodal data from a broader scope of methods. Recent work by Riera et al.79 proposed a similar state-space model for the time-series analysis of simultaneous BOLD and EEG, which could directly extend the work preformed here.
6 Conclusions
Concurrent multimodal measurements have unique statistical properties, and while an increasing number of publications have implored multimodal acquisition methods, further advancements need to be made to improve specific analysis techniques optimized for this unique form of data. In this work, we describe a fusion model that allows the direct reconstruction of hemoglobin changes from simultaneous optical and fMRI data. This model combines the advantages of both optical and fMRI methods, and allows the estimation of hemoglobin changes with improved temporal, spatial, and quantitative accuracy. Using concurrent optical and fMRI measurements, we estimate the calibration of the 3T BOLD signal to be −0.55% ± 0.40% signal change per micromolar change of deoxyhemoglobin. This is the first demonstration of a multimodal-based calibration of the BOLD signal.
Acknowledgments
Huppert was funded by the Howard Hughes Medical Institute predoctorial fellowship program. This work was supported by the National Institutes of Health (R01-EB002482, RO1-EB001954, R01-EB006385, T32-CA09502, and P41-RR14075). The authors would like to thank Christiana Andre, Danny Joseph, Div Bolar, and Joe Mandeville, Rick Hoge, and Maria Franceschini for their help in collecting this data. The authors would also like to thank Ville Kolehmainen, Jari Kaipio, and Simon Arridge for useful discussions of this work.
Contributor Information
Theodore J. Huppert, The Massachusetts General Hospital, Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, Massachusetts 02129 and University of Pittsburgh, Pittsburgh, Pennsylvania 15213
Solomon G. Diamond, The Massachusetts General Hospital, Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, Massachusetts 02129 and Dartmouth College, Thayer School of Engineering, Hanover, New Hampshire 03755
David A. Boas, The Massachusetts General Hospital, Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, Massachusetts 02129 and Harvard-MIT, Division of Health Sciences and Technology, Cambridge, MA 02139
References
- 1.Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. U.S.A. 1990;87(24):9868–9872. doi: 10.1073/pnas.87.24.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belliveau JW, Kennedy DN, Jr, McKinstry RC, Buchbinder BR, Weisskoff RM, Cohen MS, Vevea JM, Brady TJ, Rosen BR. Functional mapping of the human visual cortex by magnetic resonance imaging. Science. 1991;254(5032):716–719. doi: 10.1126/science.1948051. [DOI] [PubMed] [Google Scholar]
- 3.Belliveau JW, Kwong KK, Kennedy DN, Baker JR, Stern CE, Benson R, Chesler DA, Weisskoff RM, Cohen MS, Tootell RBH, Fox PT, Brady TJ, Rosen BR. Magnetic resonance imaging mapping of brain function. Human visual cortex. Invest. Radiol. 1992;27 Suppl 2:S59. doi: 10.1097/00004424-199212002-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogawa S, Menon RS, Tank DW, Kim SG, Merkle H, Ellermann JM, Ugurbil K. Functional brain mapping by blood oxygenation level-dependent contrast magnetic resonance imaging. A comparison of signal characteristics with a biophysical model. Biophys. J. 1993;64(3):803–812. doi: 10.1016/S0006-3495(93)81441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delpy DT, Cope MC, Cady EB, Wyatt JS, Hamilton PA, Hope PL, Wray S, Reynolds EO. Cerebral monitoring in newborn infants by magnetic resonance and near infrared spectroscopy. Scand. J. Clin. Lab Invest Suppl. 1987;188:9–17. [PubMed] [Google Scholar]
- 6.Villringer A, Chance B. Non-invasive optical spectroscopy and imaging of human brain function. Trends Neurosci. 1997;20(10):435–442. doi: 10.1016/s0166-2236(97)01132-6. [DOI] [PubMed] [Google Scholar]
- 7.Arridge SR. Optical tomography in medical imaging. Inverse Probl. 1999;15:14–93. [Google Scholar]
- 8.Baŕbour RL, Graber HL, Chang J, Baŕbour SS, Koo PC, Aronson R. MRI-guided optical tomography: prospectives and computation for a new imaging method. IEEE Comput. Sci. Eng. 1995;2:63–77. [Google Scholar]
- 9.Dunn JF, Zaim-Wadghiri Y, Pogue BW, Kida I. BOLD MRI vs. NIR spectrophotometry. Will the best technique come forward? Adv. Exp. Med. Biol. 1998;454:103–113. [PubMed] [Google Scholar]
- 10.Steinbrink J, Villringer A, Kempf F, Haux D, Boden S, Obrig H. Illuminating the BOLD signal: combined fMRI-fNIRS studies. Magn. Reson. Imaging. 2006;24(4):495–505. doi: 10.1016/j.mri.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 11.Norris DG. Principles of magnetic resonance assessment of brain function. J. Magn. Reson Imaging. 2006;23(6):794–807. doi: 10.1002/jmri.20587. [DOI] [PubMed] [Google Scholar]
- 12.Davis TL, Kwong KK, Weisskoff RM, Rosen BR. Calibrated functional MRI: mapping the dynamics of oxidative metabolism. Proc. Natl. Acad. Sci. U.S.A. 1998;95(4):1834–1839. doi: 10.1073/pnas.95.4.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoge RD, Atkinson J, Gill B, Crelier GR, Marrett S, Pike GB. Linear coupling between cerebral blood flow and oxygen consumption in activated human cortex. Proc. Natl. Acad. Sci. U.S.A. 1999;96(16):9403–9408. doi: 10.1073/pnas.96.16.9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kida I, Kennan RP, Rothman DL, Behar KL, Hyder F. High-resolution CMR(O2) mapping in rat cortex: a multiparametric approach to calibration of BOLD image contrast at 7 Tesla. J. Cereb. Blood Flow Metab. 2000;20(5):847–860. doi: 10.1097/00004647-200005000-00012. [DOI] [PubMed] [Google Scholar]
- 15.Hyder F, Kida I, Behar KL, Kennan RP, Maciejewski PK, Rothman DL. Quantitative functional imaging of the brain: towards mapping neuronal activity by BOLD fMRI. NMR Biomed. 2001;14(7–8):413–431. doi: 10.1002/nbm.733. [DOI] [PubMed] [Google Scholar]
- 16.Kannurpatti SS, Biswal BB, Hudetz AG. Regional dynamics of the fMRI-BOLD signal response to hypoxia-hypercapnia in the rat brain. J. Magn. Reson Imaging. 2003;17(6):641–647. doi: 10.1002/jmri.10311. [DOI] [PubMed] [Google Scholar]
- 17.Cohen ER, Rostrup E, Sidaros K, Lund TE, Paulson OB, Ugurbil K, Kim SG. Hypercapnic normalization of BOLD fMRI: comparison across field strengths and pulse sequences. Neuroimage. 2004;23(2):613–624. doi: 10.1016/j.neuroimage.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Fujita N, Matsumoto K, Tanaka H, Watanabe Y, Murase K. Quantitative study of changes in oxidative metabolism during visual stimulation using absolute relaxation rates. NMR Biomed. 2006;19(1):60–68. doi: 10.1002/nbm.1001. [DOI] [PubMed] [Google Scholar]
- 19.Chiarelli PA, Bulte DP, Piechnik S, Jezzard P. Sources of systematic bias in hypercapnia-calibrated functional MRI estimation of oxygen metabolism. Neuroimage. 2007;34(1):35–43. doi: 10.1016/j.neuroimage.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Raichle ME. Neuroscience. The brain’s dark energy. Science. 2006;314(5803):1249–1250. [PubMed] [Google Scholar]
- 21.Wang S, Luo L, Liang X, Gui Z, Chen C. Estimation and removal of physiological noise from undersampled multi-slice fMRI data in image space. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2005;2:1371–1373. doi: 10.1109/IEMBS.2005.1616683. [DOI] [PubMed] [Google Scholar]
- 22.Frank LR, Buxton RB, Wong EC. Estimation of respiration-induced noise fluctuations from undersampled multislice fMRI data. Magn. Reson. Med. 2001;45(4):635–644. doi: 10.1002/mrm.1086. [DOI] [PubMed] [Google Scholar]
- 23.Boas DA, Dale AM, Franceschini MA. Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy. Neuroimage. 2004;23 Suppl 1:S275–S288. doi: 10.1016/j.neuroimage.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Joseph DK, Huppert TJ, Franceschini MA, Boas DA. Design and validation of a time division multiplexed continuous wave diffuse optical tomography (DOT)system optimized for brain function imaging. Appl. Opt. 2006;45(31):814208151. doi: 10.1364/ao.45.008142. [DOI] [PubMed] [Google Scholar]
- 25.Zeff BW, White BR, Dehghani H, Schlaggar BL, Culver JP. Retinotopic mapping of adult human visual cortex with high-density diffuse optical tomography. Proc. Natl. Acad. Sci. U.S.A. 2007;104(29):12169–12174. doi: 10.1073/pnas.0611266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson AP, Hebden JC, Arridge SR. Recent advances in diffuse optical imaging. Phys. Med. Biol. 2005;50(4):R1–R43. doi: 10.1088/0031-9155/50/4/r01. [DOI] [PubMed] [Google Scholar]
- 27.Pogue BW, Paulsen KD. High-resolution near-infrared to-mographic imaging of the rat cranium by use of a priori magnetic resonance imaging structural information. Opt. Lett. 1998;23:1716–1718. doi: 10.1364/ol.23.001716. [DOI] [PubMed] [Google Scholar]
- 28.Boas DA, Dale AM. Simulation study of magnetic resonance imaging-guided cortically constrained diffuse optical tomography of human brain function. Appl. Opt. 2005;44(10):1957–1968. doi: 10.1364/ao.44.001957. [DOI] [PubMed] [Google Scholar]
- 29.Guven M, Yazici B, Intes X, Chance B. Diffuse optical tomography with a priori anatomical information. Phys. Med. Biol. 2005;50(12):2837–2858. doi: 10.1088/0031-9155/50/12/008. [DOI] [PubMed] [Google Scholar]
- 30.Intes X, Maloux C, Guven M, Yazici B, Chance B. Diffuse optical tomography with physiological and spatial a priori constraints. Phys. Med. Biol. 2004;49(12):N155–N163. doi: 10.1088/0031-9155/49/12/n01. [DOI] [PubMed] [Google Scholar]
- 31.Toronov V, Webb A, Choi JH, Wolf M, Michalos A, Gratton E, Hueber D. Investigation of human brain hemodynamics by simultaneous near-infrared spectroscopy and functional magnetic resonance imaging. Med. Phys. 2001;28(4):521–527. doi: 10.1118/1.1354627. [DOI] [PubMed] [Google Scholar]
- 32.Ntziachristos V, Yodh AG, Schnall MD, Chance B. MRI-guided diffuse optical spectroscopy of malignant and benign breast lesions. Neoplasia. 2002;4(4):347–354. doi: 10.1038/sj.neo.7900244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li A, Boverman G, Zhang Y, Brooks D, Miller EL, Kilmer ME, Zhang Q, Hillman EM, Boas DA. Optimal linear inverse solution with multiple priors in diffuse optical tomography. Appl. Opt. 2005;44(10):1948–1956. doi: 10.1364/ao.44.001948. [DOI] [PubMed] [Google Scholar]
- 34.Dale AM, Halgren E. Spatiotemporal mapping of brain activity by integration of multiple imaging modalities. Curr. Opin. Neurobiol. 2001;11(2):202–208. doi: 10.1016/s0959-4388(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 35.Baillet S, Garnero L, Marin G, Hugonin JP. Combined MEG and EEG source imaging by minimization of mutual information. IEEE Trans. Biomed. Eng. 1999;46(5):522–534. doi: 10.1109/10.759053. [DOI] [PubMed] [Google Scholar]
- 36.Friston KJ, Frith CD, Turner R, Frackowiak RS. Characterizing evoked hemodynamics with fMRI. Neuroimage. 1995;2(2):157–165. doi: 10.1006/nimg.1995.1018. [DOI] [PubMed] [Google Scholar]
- 37.Worsley KJ, Friston KJ. Analysis of fMRI time-series revisited—again. Neuroimage. 1995;2(3):173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Brooks DH, Boas DA. A haemodynamic response function model in spatio-temporal diffuse optical tomography. Phys. Med. Biol. 2005;50(19):4625–4644. doi: 10.1088/0031-9155/50/19/014. [DOI] [PubMed] [Google Scholar]
- 39.Cohen-Adad J, Chapuisat S, Doyon J, Rossignol S, Lina JM, Benali H, Lesage F. Activation detection in diffuse optical imaging by means of the general linear model. Med. Image Anal. 2007;11(6):616–629. doi: 10.1016/j.media.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 41.Henson RNA. Human brain function. In: Frackowiak RSJ, Friston KJ, Frith C, Dolan R, Friston KJ, Price CJ, Zeki S, Ashburner J, Penny WD, editors. Human Brain Function. San Diego, CA: Academic Press; 2003. [Google Scholar]
- 42.Chen H, Yao D, Liu Z. A comparison of Gamma and Gaussian dynamic convolution models of the fMRI BOLD response. Magn. Reson. Imaging. 2005;23(1):83–88. doi: 10.1016/j.mri.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Diamond SG, Huppert TJ, Kolehmainen V, Franceschini MA, Kaipio JP, Arridge SR, Boas DA. Physiological system identification with the Kalman filter in diffuse optical tomography. Med. Image Comput. Comput. Assist. Interv. Intl. Conf. Mod. Image Comput. Comput. Assist. Interv. 2005;8(Pt 2):649–656. doi: 10.1007/11566489_80. [DOI] [PubMed] [Google Scholar]
- 44.Friston KJ, Holmes AP, Poline JB, Grasby PJ, Williams SC, Frackowiak RS, Turner R. Analysis of fMRI time-series revisited. Neuroimage. 1995;2(1):45–53. doi: 10.1006/nimg.1995.1007. [DOI] [PubMed] [Google Scholar]
- 45.Huppert T, Hoge RD, Dale AM, Franceschini MA, Boas DA. A quantitative spatial comparison of diffuse optical imaging with BOLD- and ASL-based fMRI. J. Biomed. Opt. 2006;11(6):064018. doi: 10.1117/1.2400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sassaroli A, de BFB, Tong Y, Renshaw PF, Fantini S. Spatially weighted BOLD signal for comparison of functional magnetic resonance imaging and near-infrared imaging of the brain. Neuroimage. 2006;33(2):505–514. doi: 10.1016/j.neuroimage.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Toronov VY, Zhang X, Webb AG. A spatial and temporal comparison of hemodynamic signals measured using optical and functional magnetic resonance imaging during activation in the human primary visual cortex. Neuroimage. 2007;34(3):1136–1148. doi: 10.1016/j.neuroimage.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obata T, Liu TT, Miller KL, Luh WM, Wong EC, Frank LR, Buxton RB. Discrepancies between BOLD and flow dynamics in primary and supplementary motor areas: application of the balloon model to the interpretation of BOLD transients. Neuroimage. 2004;21(1):144–153. doi: 10.1016/j.neuroimage.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 49.Lu H, van Zijl PC. Experimental measurement of extravascular parenchymal BOLD effects and tissue oxygen extraction fractions using multi-echo VASO fMRI at 1.5 and 3.0 T. Magn. Reson. Med. 2005;53(4):808–816. doi: 10.1002/mrm.20379. [DOI] [PubMed] [Google Scholar]
- 50.Boxerman JL, Bandettini PA, Kwong KK, Baker JR, Davis TL, Rosen BR, Weisskoff RM. The intravascular contribution to fMRI signal change: Monte Carlo modeling and diffusion-weighted studies in vivo. Magn. Reson. Med. 1995;34(1):4–10. doi: 10.1002/mrm.1910340103. [DOI] [PubMed] [Google Scholar]
- 51.Yablonskiy DA, Haacke EM. Theory of NMR signal behavior in magnetically inhomogeneous tissues: the static dephasing regime. Magn. Reson. Med. 1994;32(6):749–763. doi: 10.1002/mrm.1910320610. [DOI] [PubMed] [Google Scholar]
- 52.Mildner T, Norris DG, Schwarzbauer C, Wiggins CJ. A qualitative test of the balloon model for BOLD-based MR signal changes at 3 T. Magn. Reson. Med. 2001;46(5):891–899. doi: 10.1002/mrm.1274. [DOI] [PubMed] [Google Scholar]
- 53.Chiarelli PA, Bulte DP, Piechnik S, Jezzard P. Sources of systematic bias in hypercapnia-calibrated functional MRI estimation of oxygen metabolism. Neuroimage. 2007;34(1):35–43. doi: 10.1016/j.neuroimage.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Jacques SL, Zheng L. MCML—Monte Carlo modeling of light transport in multi-layered tissues. Comput. Methods Programs Biomed. 1995;47(2):131–146. doi: 10.1016/0169-2607(95)01640-f. [DOI] [PubMed] [Google Scholar]
- 55.Boas D, Culver J, Stott J, Dunn A. Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head. Opt. Express. 2002;10:159–170. doi: 10.1364/oe.10.000159. [DOI] [PubMed] [Google Scholar]
- 56.Li A, Zhang Q, Culver JP, Miller EL, Boas DA. Reconstructing chromosphere concentration images directly by continuous-wave diffuse optical tomography. Opt. Lett. 2004;29(3):256–258. doi: 10.1364/ol.29.000256. [DOI] [PubMed] [Google Scholar]
- 57.Brooksby B, Srinivasan S, Jiang S, Dehghani H, Pogue BW, Paulsen KD, Weaver J, Kogel C, Poplack SP. Spectral priors improve near-infrared diffuse tomography more than spatial priors. Opt. Lett. 2005;30(15):1968–1970. doi: 10.1364/ol.30.001968. [DOI] [PubMed] [Google Scholar]
- 58.Srinivasan S, Pogue BW, Jiang S, Dehghani H, Paulsen KD. Spectrally constrained chromophore and scattering near-infrared tomography provides quantitative and robust reconstruction. Appl. Opt. 2005;44(10):1858–1869. doi: 10.1364/ao.44.001858. [DOI] [PubMed] [Google Scholar]
- 59.Devore JL. Probability and Statistics for Engineering and the Sciences. Belmont, CA: Wadsworth Inc; 1995. Simple linear regression and correlation; pp. 474–522. [Google Scholar]
- 60.Yalavarthy PK, Pogue BW, Dehghani H, Paulsen KD. Weight-matrix structured regularization provides optimal generalized least-squares estimate in diffuse optical tomography. Med. Phys. 2007;34(6):2085–2098. doi: 10.1118/1.2733803. [DOI] [PubMed] [Google Scholar]
- 61.Huppert TJ, Hoge RD, Diamond SG, Franceschini MA, Boas DA. A temporal comparison of BOLD, ASL, and NIRS hemodynamic responses to motor stimuli in adult humans. Neuroimage. 2006;29(2):368–382. doi: 10.1016/j.neuroimage.2005.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franceschini MA, Fantini S, Thompson JH, Culver JP, Boas DA. Hemodynamic evoked response of the sensorimotor cortex measured noninvasively with near-infrared optical imaging. Psychophysiology. 2003;40(4):548–560. doi: 10.1111/1469-8986.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strangman G, Franceschini MA, Boas DA. Factors affecting the accuracy of near-infrared spectroscopy concentration calculations for focal changes in oxygenation parameters. Neuroimage. 2003;18(4):865–879. doi: 10.1016/s1053-8119(03)00021-1. [DOI] [PubMed] [Google Scholar]
- 64.Cox RW, Jesmanowicz A. Real-time 3D image registration for functional MRI. Magn. Reson. Med. 1999;42(6):1014–1018. doi: 10.1002/(sici)1522-2594(199912)42:6<1014::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 65.Cope M, Delpy DT, Reynolds EO, Wray S, Wyatt J, van der Zee P. Methods of quantitating cerebral near infrared spectroscopy data. Adv. Exp. Med. Biol. 1988;222:183–189. doi: 10.1007/978-1-4615-9510-6_21. [DOI] [PubMed] [Google Scholar]
- 66.Delpy DT, Cope M, van der Zee P, Arridge S, Wray S, Wyatt J. Estimation of optical pathlength through tissue from direct time of flight measurement. Phys. Med. Biol. 1988;33(12):1433–1442. doi: 10.1088/0031-9155/33/12/008. [DOI] [PubMed] [Google Scholar]
- 67.Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RET-ROICOR. Magn. Reson. Med. 2000;44(1):162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 68.Ito H, Kanno I, Fukuda H. Human cerebral circulation: positron emission tomography studies. Ann. Nucl. Med. 2005;19(2):65–74. doi: 10.1007/BF03027383. [DOI] [PubMed] [Google Scholar]
- 69.Buxton RB, Uludag K, Dubowitz DJ, Liu TT. Modeling the hemodynamic response to brain activation. Neuroimage. 2004;23 Suppl 1:S220–S233. doi: 10.1016/j.neuroimage.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 70.Torricelli A, Pifferi A, Taroni P, Giambattistelli E, Cubeddu R. In vivo optical characterization of human tissues from 610 to 1010 nm by time-resolved reflectance spectroscopy. Phys. Med. Biol. 2001;46(8):2227–2237. doi: 10.1088/0031-9155/46/8/313. [DOI] [PubMed] [Google Scholar]
- 71.Herman P, Trubel HK, Hyder F. A multiparametric assessment of oxygen efflux from the brain. J. Cereb. Blood Flow Metab. 2006;26(1):79–91. doi: 10.1038/sj.jcbfm.9600165. [DOI] [PubMed] [Google Scholar]
- 72.Yamamoto T, Kato T. Paradoxical correlation between signal in functional magnetic resonance imaging and deoxygenated haemoglobin content in capillaries: a new theoretical explanation. Phys. Med. Biol. 2002;47(7):1121–1141. doi: 10.1088/0031-9155/47/7/309. [DOI] [PubMed] [Google Scholar]
- 73.Scouten A, Constable RT. Applications and limitations of whole-brain MAGIC VASO functional imaging. Magn. Reson. Med. 2007;58(2):306–315. doi: 10.1002/mrm.21273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Detre JA, Wang J. Technical aspects and utility of fMRI using BOLD and ASL. Clin. Neurophysiol. 2002;113(5):621–634. doi: 10.1016/s1388-2457(02)00038-x. [DOI] [PubMed] [Google Scholar]
- 75.Lu H, Golay X, Pekar JJ, Van Zijl PC. Functional magnetic resonance imaging based on changes in vascular space occupancy. Magn. Reson. Med. 2003;50(2):263–274. doi: 10.1002/mrm.10519. [DOI] [PubMed] [Google Scholar]
- 76.Lu H, Law M, Johnson G, Ge Y, van Zijl PC, Helpern JA. Novel approach to the measurement of absolute cerebral blood volume using vascular-space-occupancy magnetic resonance imaging. Magn. Reson. Med. 2005;54(6):1403–1411. doi: 10.1002/mrm.20705. [DOI] [PubMed] [Google Scholar]
- 77.Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: the balloon model. Magn. Reson. Med. 1998;39(6):855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- 78.Friston KJ. Bayesian estimation of dynamical systems: an application to fMRI. Neuroimage. 2002;16(2):513–530. doi: 10.1006/nimg.2001.1044. [DOI] [PubMed] [Google Scholar]
- 79.Riera J, Aubert E, Iwata K, Kawashima R, Wan X, Ozaki T. Fusing EEG and fMRI based on a bottom-up model: inferring activation and effective connectivity in neural masses. Philos. Trans. R. Soc. London, Ser. B. 2005;360(1457):1025–1041. doi: 10.1098/rstb.2005.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deneux T, Faugeras O. Using nonlinear models in fMRI data analysis: model selection and activation detection. Neuroimage. 2006;Vol. 32(4):1669–1689. doi: 10.1016/j.neuroimage.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 81.Huppert TJ, Allen MS, Benav H, Jones P, Boas DA. A multi-compartment vascular model for inferring arteriole dilation and cerebral metabolic changes during functional activation. J. Cereb. Blood Flow Metab. 2007;27:1262–1279. doi: 10.1038/sj.jcbfm.9600435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mandeville JB, Marota JJ, Ayata C, Zaharchuk G, Moskowitz MA, Rosen BR, Weisskoff RM. Evidence of a cerebrovascular postarteriole windkessel with delayed compliance. J. Cereb. Blood Flow Metab. 1999;19(6):679–689. doi: 10.1097/00004647-199906000-00012. [DOI] [PubMed] [Google Scholar]
- 83.Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magn. Reson. Med. 1992;23(1):37–45. doi: 10.1002/mrm.1910230106. [DOI] [PubMed] [Google Scholar]