Summary
The central protease of eukaryotes, the 26S proteasome, has a 20S proteolytic core particle (CP) and an attached 19S regulatory particle (RP). The RP is further subdivided into lid and base subcomplexes. Little is known about RP assembly. Here we show that four conserved assembly factors govern biogenesis of the yeast RP base. Nas2 forms a complex with the Rpt4 and Rpt5 ATPases and enhances 26S proteasome formation in vivo and in vitro. Other RP subcomplexes contain Hsm3, which is related to mammalian proteasome subunit S5b. Hsm3 also contributes to base assembly. Larger Hsm3-containing complexes include two additional proteins, Nas6 and Rpn14, which function as assembly chaperones as well. Specific deletion combinations affecting these four factors cause severe perturbations to RP assembly. Our results demonstrate that proteasomal RP biogenesis requires multiple, functionally overlapping chaperones and suggest a model in which subunits form specific subcomplexes that then assemble into the base.
Introduction
In eukaryotes, short-lived proteins are degraded primarily by the ubiquitin-proteasome system (Hochstrasser, 1996) (Heinemeyer et al., 2004). Defects in the system are linked to a variety of human diseases, and proteasomal inhibitors are used to treat several cancers (Goldberg, 2007) (Schwartz and Ciechanover, 2009). Most proteasome substrates are first modified by polyubiquitin chains, allowing recognition by the proteasome and degradation of the substrate. The 26S proteasome consists of a proteolytically active 20S proteasome core particle (CP) bound at one or both ends by a 19S regulatory particle (RP) (Schmidt et al., 2005). In addition to substrate binding, the RP is responsible for substrate unfolding, substrate translocation into the proteolytic chamber of the CP, and release of ubiquitin from the substrate.
High-resolution structural information is available for the CP, and it has been subject to extensive biochemical and genetic analysis (reviewed in Marques et al., 2009). The CP forms a cylinder of four stacked heptameric rings. Two structurally related classes of subunits make up the rings. The outer rings have α-type subunits and the inner rings β-type subunits, with the protease sites formed by specific β subunits.
The structure and activities of the RP are less well understood. The lid and base each contain at least nine different subunits, with additional polypeptides, such as Rpn10, associated more loosely or only under specific conditions (Fig. 1A) (Schmidt et al., 2005). The base includes six different AAA+ ATPase subunits, Rpt1-Rpt6, as well as the two largest subunits, Rpn1 and Rpn2. Recent structural analysis suggests that Rpn1 and Rpn2 form toroids, with Rpn1 stacked on Rpn2 and Rpn2 in direct contact with the CP α-ring (Rosenzweig et al., 2008). The ATPases are thought to form a hexameric ring that encircles the Rpn1-Rpn2 stack and also contacts the α ring (Ferrell et al., 2000) (Hartmann-Petersen et al., 2001).
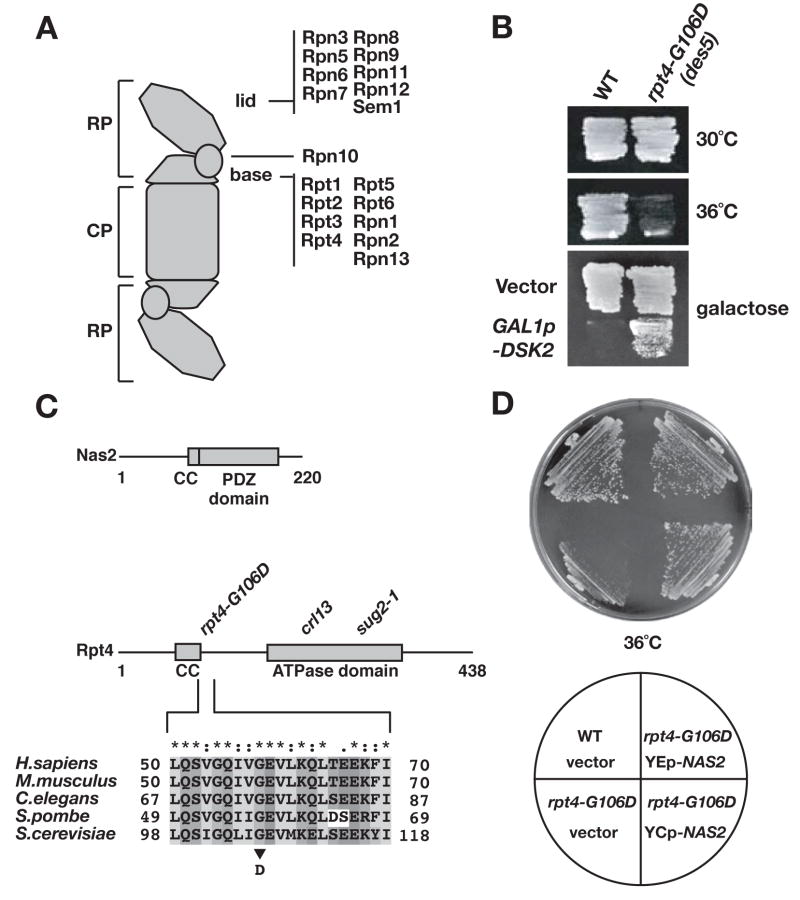
Fig. 1.
NAS2 is a dosage suppressor of the rpt4-G106D ATPase mutant.
A. Illustration of the doubly capped form of the 26S proteasome. The composition of the yeast 20S proteasome core particle (CP) and the 19S regulatory particle (RP) is given.
B. The des5 mutation (rpt4-G106D) suppresses the lethality caused by galactose-dependent overexpression of the Dsk2 polyubiquitin-binding protein (bottom panel) but causes temperature-sensitive growth.
C. Domain organization of the Nas2 and Rpt4 proteins and the position of the rpt4-G106D mutation (and two other tested mutations). CC, putative coiled coil. Rpt4-G106 is at the end of the CC.
D. Low-copy (YCp-NAS2) and high-copy (YEp-NAS2) plasmids expressing NAS2 suppress the temperature-sensitive growth of the rpt4-G106D mutant.
Rpn10 as well as the base subunit Rpn13 are proteasomal polyubiquitin receptors (Husnjak et al., 2008). Additional proteins function as mobile receptors that bridge polyubiquitin chain recognition and proteasome targeting. In yeast, the related Rad23, Dsk2, and Ddi1 proteins use their ubiquitin-like domains to dock onto Rpn1 and their UBA domains to bind substrate polyubiquitin chains (reviewed in Hurley et al., 2006). There is considerable redundancy for substrate binding among these receptors, and genetic data indicate additional receptors must exist (Diaz-Martinez et al., 2006) (Husnjak et al., 2008).
Although much is known about how the proteasome recognizes and degrades its substrates, investigations have just begun into how this highly abundant complex of ~2,500 kD and at least 33 different subunits is assembled in the first place. Understanding proteasome assembly should provide general insight into strategies of multisubunit protein assembly in vivo. It will also be crucial for finding ways to inhibit assembly as part of emerging therapies that target the proteasome (Goldberg, 2007). Analysis of CP assembly is far more advanced than for the RP (Marques et al., 2009) (Murata et al., 2009). Eukaryotic CP assembly initiates with formation of an α ring followed by ordered addition of β subunits to the α-ring heteroheptamer (Hirano et al., 2005) (Li et al., 2007). Joining of two half-proteasomes triggers autocatalytic processing of active-subunit propeptides and CP maturation (Chen and Hochstrasser, 1996). Moreover, a least three phylogenetically conserved CP-specific assembly chaperones facilitate proteasome biogenesis, and one of these is known to control CP composition as well (Kusmierczyk et al., 2008).
Whether de novo proteasomal RP assembly also requires dedicated assembly chaperones has been uncertain (Heinemeyer et al., 2004). Recent evidence suggests that the CP can enhance RP base biogenesis in the cell and might therefore be an RP assembly factor (Kusmierczyk et al., 2008). Additional proteins are known to associate with the proteasome, but the functional significance of most of these associations is unclear (Guerrero et al., 2008). Here we show that assembly of the RP base in yeast is orchestrated by at least four distinct assembly chaperones. One of these factors, Nas2, was identified as a dosage suppressor of an rpt4 mutant. The remaining three, Hsm3, Nas6, and Rpn14, were identified biochemically in subcomplexes with specific base subunits. None of them associates detectably with the mature 26S proteasome. Genetic and biochemical data suggest that Nas2 (orthologous to human p27/Bridge-1) and Hsm3 (human S5b) overlap more closely in function, as do Nas6 (human gankyrin) and Rpn14 (human PAAF1). These factors are conserved from yeast to human and were until now either unconnected to the proteasome or thought to be subunits or inhibitors of the proteasome. Shortly before submission of this paper, Le Tallec et al. (2009) also reported that Hsm3 has properties of an RP assembly factor. Our data unify this previously disconnected set of factors, which all function specifically in the assembly of the RP base and are not components of the mature 26S proteasome. The results lead to a model in which the RP base assembles from a set of discrete chaperone-associated base subunit complexes; once assembled, the base binds to the lid and all chaperones are released prior to or during RP-CP association.
Results
Identification of NAS2 as a dosage suppressor of rpt4-G106D
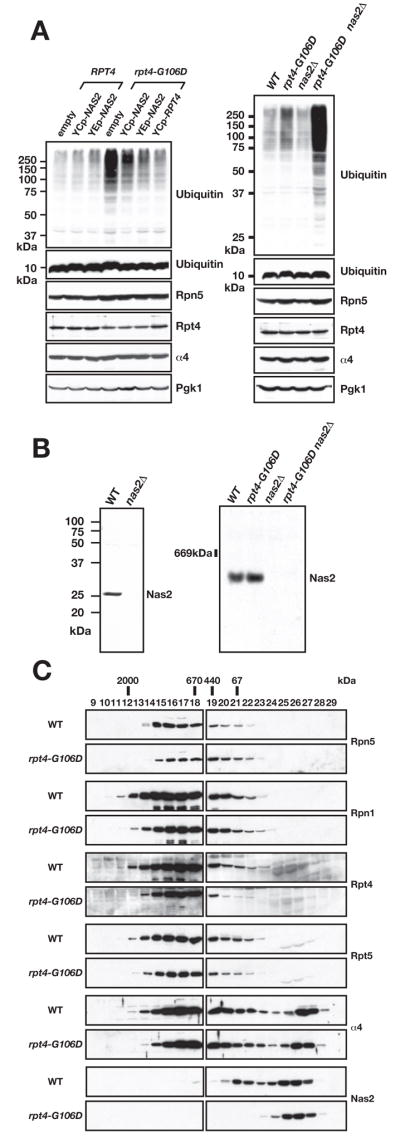
High levels of the Dsk2 polyubiquitin receptor are toxic to S. cerevisiae (Fig. 1B, bottom), and a previous genetic screen identified a series of suppressors of this toxicity (Funakoshi et al., 2002; 2004). All of the characterized suppressors were found to be recessive proteasome mutants. One previously uncharacterized mutant, designated des5, was found to be temperature-sensitive for growth, but multiple attempts to clone the affected gene failed, instead repeatedly yielding the unlinked NAS2 gene (Fig. 1C). We therefore identified the gene mutated in the des5 strain by chromosomal mapping and linkage analysis (see Suppl. Data). The des5 strain had a mutation in RPT4, which encodes one of the six ATPases of the proteasomal regulatory particle (RP) (Fig. 1A). The mutant allele encodes a protein with a single amino acid change, G106D, mapping to a conserved region of Rpt4 at the end of a predicted coiled coil (Fig. 1C). Nas2 is also predicted to have a coiled-coil domain. Both low-copy and high-copy plasmids bearing NAS2 were strong suppressors of the rpt4-G106D mutant (Fig. 1D).
Previous analysis of nas2Δ mutants had not uncovered any abnormalities (Watanabe et al., 1998) (Russell et al., 1999). Nas2 had been assessed for a potential role in proteasome function because of its ~35% sequence identity to mammalian p27/Bridge-1, which is a component of the “modulator” complex that also contains the Rpt4 and Rpt5 proteasomal ATPases (DeMartino et al., 1996) (Watanabe et al., 1998). Addition of purified mammalian modulator to purified RP and CP complexes leads to enhanced association of RP complexes with the CP (Adams et al., 1998). The mechanism of how this enhanced 26S formation occurs through the modulator has remained obscure.
We first attempted to link Nas2 to the in vivo function of the proteasome. Proteasomal defects lead to accumulation of polyubiquitinated proteins that can be detected by anti-ubiquitin immunoblotting. The rpt4-G106D mutant showed a pronounced build-up of such species, and these were suppressed by NAS2 when introduced into the mutant on a low-copy, or to a greater extent, a high-copy plasmid (Fig. 2A, left). Conversely, nas2Δ, which by itself caused no increase in bulk polyubiquitin conjugates, exacerbated the defect due to rpt4-G106D (Fig. 2A, right). Growth assays yielded parallel results, showing that nas2Δ did not cause any detectable growth defects, but it displayed a synthetic sick interaction with rpt4-106D (Suppl. Fig. S1A). Similarly, when we assayed levels of specific model substrates of the proteasome, nas2Δ had no effect, but rpt4-G106D mutants had increased amounts of these substrates, consistent with a proteolytic defect (Suppl. Fig. S1B). Levels were further increased in the rpt4-G106D nas2Δ double mutant. Based on growth assays, high-copy NAS2 could also weakly suppress another mutant rpt4 allele, called crl13 or sug2-13, but could not suppress an rpt4 allele (sug2-1) that is specifically defective in the transcriptional activation function of the RP but not in general proteolysis (Suppl. Fig. S2A and not shown) (Russell et al., 1996). Loss of Nas2 did not worsen the defects due to mutations in two other RP ATPases, Rpt1 (cim5-1) or Rpt6 (cim3-1), nor did high-copy NAS2 suppress these defects (Suppl. Fig. S2B, C).
Fig. 2.
Nas2 affects proteasome function but is not part of the 26S proteasome.
A. Anti-ubiquitin immunoblots showing polyubiquitin conjugate accumulation in rpt4-G106D cells. Left, effect of low-copy (YCp) and high-copy (YEp) expression of NAS2 on rpt4-G106D. Right, effect of nas2Δ on ubiquitin-conjugate profiles. Levels of proteasome components in the strains are shown below. Pgk1 serves as a loading control.
B. Anti-Nas2 immunoblot analysis of yeast proteins separated by SDS-PAGE (left) or by nondenaturing PAGE (right).
C. Gel filtration analysis of Nas2 and proteasomal proteins from WT and rpt4-G106D cells. Equivalent amounts of protein were loaded from each extract onto a Superose-6 column, and fractions were subjected to immunoblotting with antibodies to the indicated proteins.
Together, these results implicate Nas2 in the in vivo function of the proteasome, which previously had only been suggested from biochemical studies with human p27. Moreover, the genetic data indicate that Nas2 activity is linked to the Rpt4 subunit of the RP.
RP assembly defects in rpt4-G106D are enhanced by loss of Nas2
An earlier study did not find Nas2 in 26S proteasomes (Russell et al., 1999), but because the protein was epitope-tagged, it might have been defective for proteasome association. Affinity-purified antibodies to Nas2 reacted specifically with a single species of the expected size in SDS gel separations (Fig. 2B, left). By nondenaturing PAGE, a single band was observed, which migrated more quickly than assembled proteasomes (Fig. 2B, right; see Fig. 3D for size comparison). Similarly, using Superose-6 gel filtration, we found Nas2 in two peaks in wild-type (WT) cells: one peak that is probably monomeric Nas2 and a larger species that appeared to be less than 400 kDa (Fig. 2C). No reactivity was seen in earlier fractions from the column where mature proteasomal species migrate. We conclude that yeast Nas2 is not stably associated with the mature proteasome or RP. Notably, the larger Nas2 peak disappeared in extracts from rpt4-G106D cells, indicating that this species is dependent on functional Rpt4.
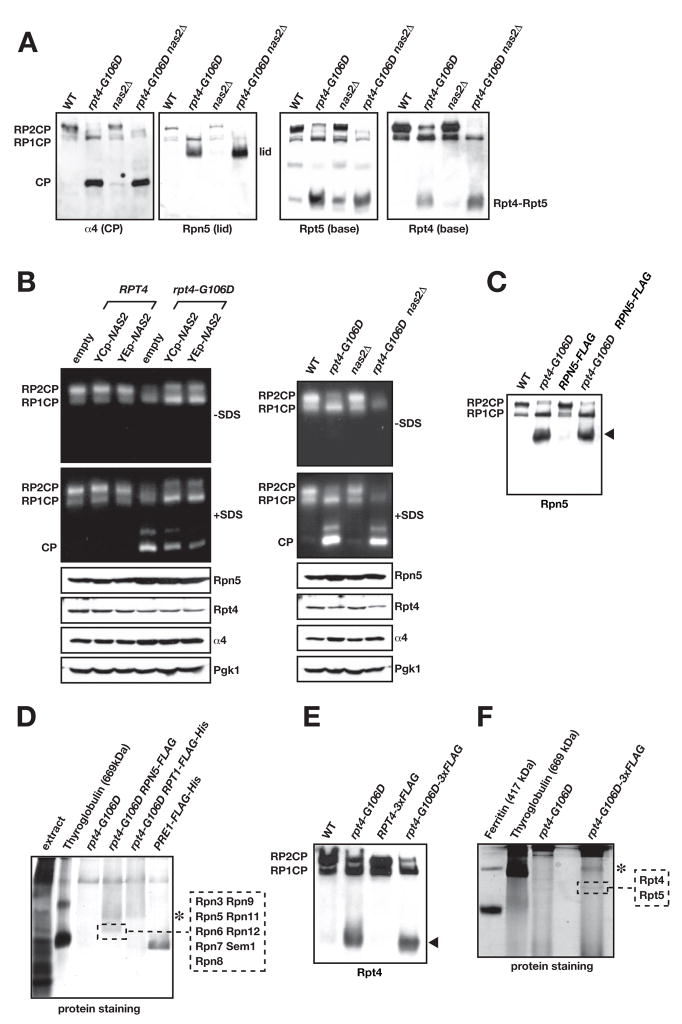
Fig. 3.
The rpt4-G106D and nas2Δ mutants have proteasome assembly defects.
A. Immunoblot analysis of nondenaturing PAGE-separated proteins from yeast whole-cell extracts. The faint central band in the anti-Rpt5 blot is of unknown composition.
B. Nondenaturing gel separation of proteins from yeast extracts followed by gel overlay with the fluorogenic substrate Suc-LLVY-AMC to assay proteasomal activity. Left panel, effect of low-copy (YCp) and high-copy (YEp) expression of NAS2 on functional proteasome formation. Right panel, effect of nas2Δ on assembly. Addition of SDS to the reaction buffer activates the otherwise latent CP and Blm10-CP (unlabeled band above free CP).
C. A C-terminal Flag tag on Rpn5 does not disrupt the Rpn5-containing RP subcomplex in rpt4-G106D cells (arrowhead).
D. Nondenaturing gel separation of Flag-affinity purified proteins from rpt4-G106D RPN5-FLAG cells. MS/MS analysis of the indicated band from the GelCode Blue-stained gel demonstrated that it was the 9-subunit RP lid. Asterisk, a band that is likely the free RP but was not analyzed. Unlike panel C, gel separation included a stacking gel and was run longer.
E. A triplicated Flag tag on Rpt4 does not disrupt the Rpt4-containing RP subcomplex in rpt4-G106D cells (arrowhead).
F. Nondenaturing gel separation of Flag-affinity purified proteins from rpt4-G106D RPT4-3xFLAG cells. MS/MS analysis of the indicated band from the GelCode Blue-stained gel yielded Rpt4 and Rpt5 from the RP base. Asterisk, as in panel D. PAGE was done with a 5.5% PA separating gel instead of the 4% used in other panels, and a stacking gel was used.
Since Nas2 is not a stable component of the proteasome, we hypothesized that it might function in proteasome assembly. Anti-α4 (CP) immunoblotting of nondenaturing gel-separated whole cell yeast lysates revealed a very small but reproducible increase in free CP in the nas2Δ mutant relative to WT (Fig. 3A), consistent with a deficiency in RP relative to CP levels. A much more pronounced increase in free CP, with a concomitant reduction of doubly capped 26S proteasomes (RP2CP), was seen in an rpt4-G106D strain by blotting with antibodies against the CP (α4), RP base (Rpt4, Rpt5), and RP lid (Rpn5) (Fig. 3A). These changes were also observed by a fluorogenic peptide substrate overlay assay (Fig. 3B). The deficiency in 26S proteasome complexes could be partially suppressed by introduction of extra copies of the NAS2 gene (Fig. 3B, left) and was exacerbated in the rpt4-G106D nas2Δ double mutant (right). These changes in the relative levels of free CP compared to full RP2CP and RPCP particles indicate that Nas2 contributes to RP (or RP-CP) assembly or stability.
The anti-Rpn5 (lid) and anti-Rpt4/Rpt5 (base) immunoblots in Fig. 3A also revealed protein complexes from the rpt4-G106D strains that were either absent or barely detectable in WT cells. To purify and determine the molecular composition of these complexes, chromosomal RPN5 or rpt4-G106D were modified with sequences for the Flag epitope. These tags did not interfere with accumulation of the novel RP subcomplexes (Fig. 3C, E, arrowheads). Extracts from large yeast cultures were then bound to an anti-Flag affinity column, and proteins were eluted with excess Flag peptide and resolved on nondenaturing gels that were stained for protein (Fig. 3D, F). The indicated complexes (and the same gel regions from purifications from untagged strains) were excised, cleaved with trypsin, and subjected to mass spectrometric (LC-MS/MS) sequencing. Based on these analyses, the Rpn5-containing complex was identified as the complete 9-subunit RP lid (Fig. 3D; Suppl. Table S1), and the Rpt4-containing complex had Rpt5 as its only other detectable constituent (Fig. 3F; Suppl. Table S2). (We note that our conditions for nondenaturing PAGE appear to break apart Nas2 and Rpt4-Rpt5, so what we refer to as the “Rpt4-Rpt5 complex” when analyzed by nondenaturing PAGE may have originally also contained Nas2 (see below)).
The assignment of lid identity was consistent with the reactivity of this species on native gels with antibodies to a lid subunit but not to base or CP subunits (Fig. 3A). It was further supported by comparing the migration on native gels of Rpn5-containing complexes from rpt4-G106D cells to those from rpn2-ΔN, a base mutant known to accumulate free lid (Isono et al., 2007) (Suppl. Fig. S3A; also see Fig. 6B). The identity of the Rpt4-Rpt5 complex was supported by immunoblotting experiments showing that the complex purified via the Flag-tagged rpt4-G106D protein reacted with antibodies to Rpt5 but not to Rpt1 or Rpn1 (Suppl. Fig. S3B). Therefore, the rpt4-G106D RP base mutation causes excess accumulation of free, fully assembled RP lid plus at least one subcomplex of the base (Rpt4-Rpt5), consistent with a defect in RP base assembly.
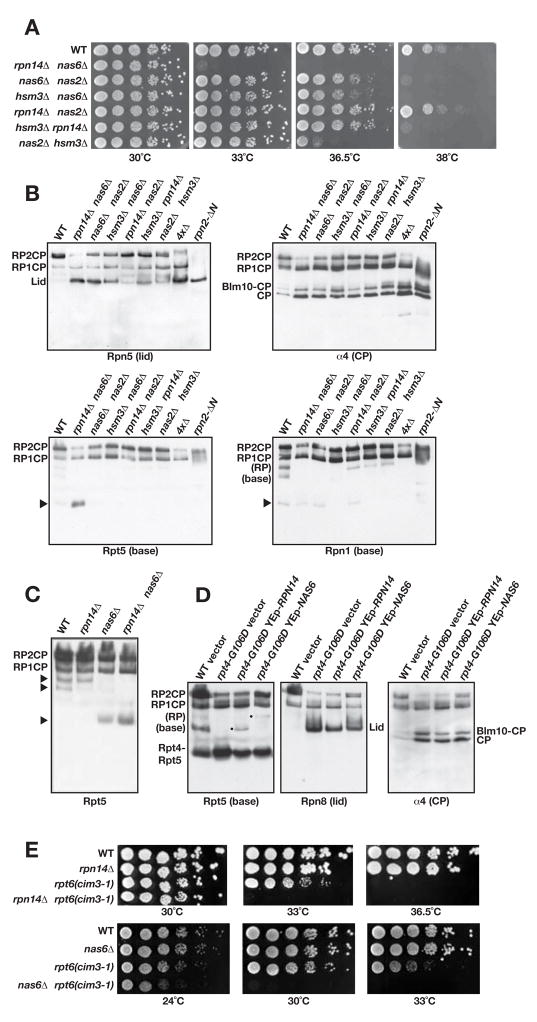
Fig. 6.
Nas6 and Rpn14 are also important for RP base assembly.
A. Serial dilutions of the indicated double mutants were spotted onto YPD plates and incubated at the indicated temperatures for 2 d.
B. Immunoblot analysis of nondenaturing gel-separated proteins from the indicated mutants. The 4xΔ strain is a nas2Δ nas6Δ hsm3Δ rpn14Δ quadruple mutant.
C. Comparison of strains by nondenaturing gel/anti-Rpt5 immunoblotting. The free RP and free base are labeled by the upper two arrowheads based on the MS/MS analysis done in Fig. 5F and immunoblot analyses. Lower arrowhead, likely Rpt4-Rpt5 complex (based on co-migration on native gels with species observed in rpt4-G106D).
D. High-copy RPN14 stimulates RP base formation whereas NAS6 stimulates full RP assembly in rpt4-G106D cells. Native gel immunoblotting was done as in panel B.
E. Growth of the strains combining mutations in Nas6 or Rpn14 and base subunit Rpt6 (cim3-1) was evaluated as in panel A.
Since the mammalian modulator complex consists of the orthologs of Rpt4, Rpt5, and Nas2, we next asked if the yeast Rpt4-Rpt5 complex, which is of very low abundance but detectable in WT cells, associates in vivo with Nas2. A fully functional Flag-tagged Nas2 expressed from its normal chromosomal locus was able to coprecipitate both Rpt4 and Rpt5 in WT cells but not the CP (α4), the lid (Rpn5, Rpn8), or other subunits of the base (Rpn1) (Fig. 4A). The three proteins, when overexpressed, can be purified as a stoichiometric complex from yeast as well (Fig. 4B). These results suggest that yeast contain a complex similar to the mammalian modulator and that Nas2 dissociates from Rpt4-Rpt5 once these proteins are incorporated into the RP. Interestingly, even though the Rpt4-Rpt5 complex was more abundant in rpt4-G106D cells, we failed to detect either subunit in the Nas2-Flag coprecipitations (Fig. 4A), which is consistent with the loss in this mutant of the early Nas2 peak seen by gel filtration (Fig. 2C). This suggests that the rpt4-G106D mutation, while not affecting Rpt4 binding to Rpt5, impairs Rpt4-Rpt5 binding to Nas2.
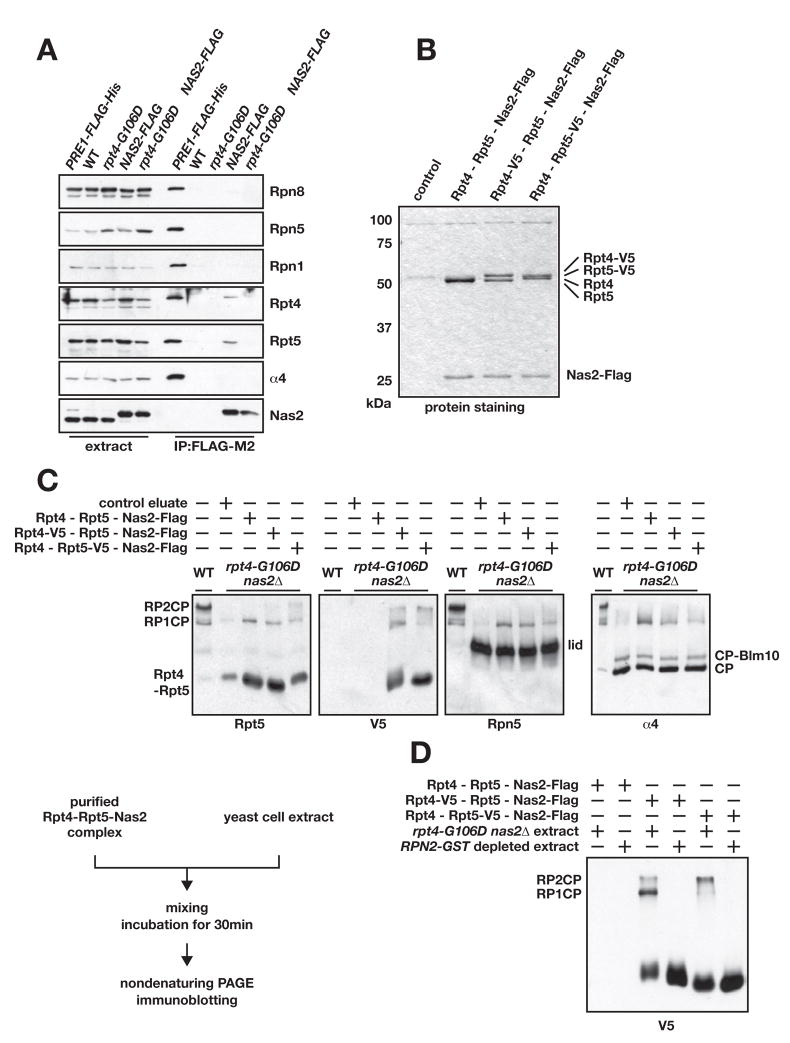
Fig. 4.
Nas2 forms a complex with Rpt4 and Rpt5, which stimulates 26S proteasome formation in vitro.
A. Co-purification of endogenous Rpt4 and Rpt5 with chromosomally expressed Nas2-Flag. Eluted proteins were tested for the indicated proteasomal subunits by immunoblotting. Pre1-Flag (CP β4 subunit) served as a positive purification control.
B. Affinity-purified Nas2-Flag-Rpt4-Rpt5 complexes resolved on an SDS gel and stained with Coommassie Blue. Control, purification from a strain without Flag-tagged Nas2.
C. Nas2-Rpt4-Rpt5 complexes stimulate 26S proteasome formation in vitro. Control eluate is derived from cells lacking the Flag tag on Nas2. Assembly reactions were done at 24°C. Diagram of the experimental scheme is shown.
D. Stimulation of assembly of 26S proteasomes by purified Nas2-Rpt4-Rpt5 requires proteasome components from the target yeast extract. 26S proteasomes and RP-derived components were pre-cleared from RPN2-GST cell extracts by binding to glutathione-Sepharose.
A Nas2-Rpt4-Rpt5 complex facilitates proteasome assembly in vitro
If a Nas2-Rpt4-Rpt5 complex acts as a functional intermediate in RP base assembly, it should be possible to transfer Rpt4-Rpt5 from such purified complexes to proteasomal precursors in vitro, leading to stimulation of 26S proteasome formation. We purified the Nas2-Flag-Rpt4-Rpt5 complexes from yeast strains that overexpressed these three polypeptides and also had chromosomal RPN2 tagged with a GST coding sequence. Yeast lysates were first depleted of assembled proteasomes and RPs by passing them over a glutathione-Sepharose resin; Nas2-Flag-containing complexes in the depleted lysates were then bound to an anti-Flag column and eluted with Flag peptide. This yielded Nas2-Rpt4-Rpt5 complexes of high purity (Fig. 4B).
We then made whole-cell lysates in low salt conditions from rpt4-G106D nas2Δ cells, which accumulate excess free CP and RP lid and various RP base intermediates (Fig. 3 and below). Purified Nas2-Rpt4-Rpt5 complexes were added to the lysates along with ATP and incubated for 30 minutes. Reaction products were resolved on nondenaturing gels and analyzed by immunoblotting (Fig. 4C). Three different preparations of Nas2-Rpt4-Rpt5 complexes all stimulated formation of 26S proteasomes (RPCP and RP2CP). Using V5-epitope tags present on either Rpt4 or Rpt5, we could detect incorporation of these ATPase subunits from the purified Nas2-Rpt4-Rpt5 complexes into 26S particles. Importantly, RPN2-GST cell extracts depleted of 26S proteasome components failed to assemble 26S complexes in the reconstitution assay, indicating that the 26S particles seen in Fig. 4C were not derived from contaminants of the purified Nas2-Rpt4-Rpt5 complexes (Fig. 4D). These results suggest that Nas2-Rpt4-Rpt5 complexes can indeed serve as intermediates in the assembly of proteasomes.
Additional RP subparticles identified in RP assembly mutants
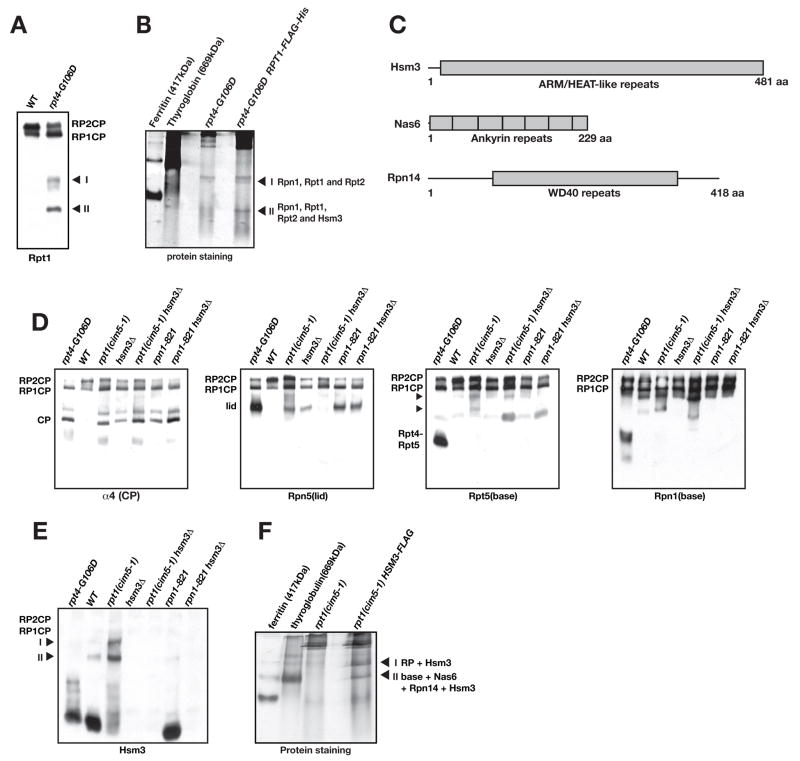
When we compared proteasome profiles from WT and rpt4-G106D cells by native gel immunoblotting with an anti-Rpt1 antibody, two novel species appeared in the mutant whole-cell extracts that were much less abundant in WT extracts (Fig. 5A, complexes I, II). These species were still detectable when Rpt1 was fused to a Flag-His6 tag (not shown); therefore, this tag was used for affinity purification of the two complexes and MS/MS identification of their components. Three base subunits, Rpn1, Rpt1, and Rpt2, comprised the less abundant complex I, whereas these same subunits, together with a novel protein, Hsm3, comprised complex II (Fig. 5B; Suppl. Table S4).
Fig. 5.
Hsm3 enhances RP assembly in vivo.
A. Two novel complexes that include the Rpt1 ATPase accumulate in rpt4-G106D cells (labeled I and II in the immunoblot).
B. Flag affinity-purified complexes from rpt4-G106D RPT1-FLAG-His6 ated by native gel PAGE, stained with GelCode Blue, excised, and analyzed by MS/MS. The proteins identified in each band are listed.
C. Domain organization of Hsm3, Nas6, and Rpn14 proteins.
D. RP assembly defects in hsm3Δ and base subunit mutants. Analysis was done as in Fig. 3A. Arrowheads, two Rpt5-containing complexes that accumulate in rpt1ts (cim5-1).
E. Two slow-migrating complexes from cim5-1 (and at a lower level, WT) cells contain Hsm3 (arrowheads labeled I and II).
F. Purification of complexes I and II from cim5-1 HSM3-FLAG cells with analysis as in panel B. The protein complexes identified by MS/MS from each band are listed.
Hsm3 was previously linked to DNA mismatch repair, but its molecular function had not been explored (Fedorova et al., 2000). A proteomic survey of proteins that could be crosslinked in vivo to the yeast 26S proteasome identified 471 proteins, one of which was Hsm3 (Guerrero et al., 2008), and a very recent report also links Hsm3 to RP assembly (Le Tallec et al., 2009). Hsm3 has a large central region with similarity to armadillo (ARM) or HEAT repeats (Fig. 5C). These related motifs form α-solenoid modules that contribute to curved or helical structures (Rosenzweig et al., 2008). Notably, our sequence analysis of Hsm3 indicated weak but significant similarity to the human S5b proteasome subunit (Gorbea et al., 2000) (Suppl. Fig. S4). Although regarded as an RP subunit, S5b is not detected in most MS/MS analyses of proteasome-associated proteins. S5b might associate only with RP-related complexes that are not part of full 26S proteasomes. It had been thought that S5b was specific to mammals. However, the idea that Hsm3 is in fact the yeast ortholog of S5b is strongly supported by the finding that human S5b forms part of a heterotetramer with human Rpt1, Rpt2, and Rpn1 when these proteins are co-expressed in vitro (Gorbea et al., 2000). These correspond exactly to the three base subunits that we isolated in a complex with Hsm3 in yeast (Fig. 5B).
Hsm3 is also an RP base assembly factor
Cells deleted for the HSM3 gene had no apparent growth deficit when grown on rich medium at several temperatures, but a growth defect was observed when hsm3Δ cells were grown on medium containing the amino-acid analog canavanine, especially at temperatures above 30°C (Suppl. Fig. S5A). When hsm3Δ was combined with other mutations in the proteasome, more severe growth defects were detected; enhanced growth defects were also evident for the nas2Δ hsm3Δ double mutant (Suppl. Fig. S5B and see Fig. 6A). Analysis of proteasomes by native PAGE/immunoblotting revealed a modest deficiency in RP2CP complexes in the hsm3Δ mutant and small increases in both free lid and free CP (Fig. 5D). These data are consistent with a defect in RP base assembly in hsm3Δ cells. A mutation in the Rpt1 ATPase, cim5-1, also caused an apparent defect in RP assembly, with two prominent particles migrating below the RPCP complex (arrowheads in Fig. 5D, anti-Rpt5). The pattern of particle accumulation in cim5-1 cells was altered when Hsm3 was also deleted. Interestingly, the growth defect of cim5-1 cells (and that of the rpn1-821 mutant) was suppressed by high-copy HSM3 expression but not by extra copies of NAS2 (Suppl. Fig. S5C, D).
In light of the possible link between Hsm3 and RP base assembly, we used an affinity-purified antibody to Hsm3 to test potential Hsm3 binding to the proteasome or proteasomal intermediates. In WT cells, most Hsm3 is associated with a fast-migrating species of undetermined composition (Fig. 5E). A small fraction is present in two slower-migrating bands (marked as I and II). Staining of these latter bands was much more intense in the cim5-1 mutant, and they migrated similarly to the two Rpt5-containing species highlighted in Fig. 5D. By tagging the chromosomal copy of HSM3 with a Flag epitope sequence, we could affinity-purify species I and II and determine their composition by MS/MS (Fig. 5F). The sequenced peptides (Suppl. Tables S5 and S6) indicated that band I was the full RP plus Hsm3 while band II was a complex of the full base, Hsm3, and two additional proteins, Nas6 and Rpn14.
Nas6 and Rpn14 have overlapping roles in RP assembly
Since neither Nas6 nor Rpn14 has been found in purified 26S proteasomes and both appear to be present at substoichiometric levels in RP-like particles, we considered the possibility that they also are RP assembly chaperones. Deletion of either factor alone had no obvious effects on growth or viability (Suppl. Fig. S5A and not shown). However, combining the two deletions caused a striking temperature-sensitive growth phenotype. At 30°C the double mutant grew comparably to WT, but at 33°C, minimal growth was observed (Fig. 6A). Combining nas6Δ, rpn14Δ, nas2Δ, and hsm3Δ in all possible double mutant combinations revealed varying degrees of temperature sensitivity, with nas6Δ rpn14Δ> nas2Δ hsm3Δ > nas2Δ nas6Δ ≅ hsm3Δ nas6Δ> hsm3Δ rpn14Δ> rpn14Δ nas2Δ ≅ WT (Fig. 6A). The same trends were seen in strains with triple and quadruple deletions of these genes, with the 4xΔ (quadruple) mutant being the most sensitive of all (Suppl. Fig. S6A). These data suggest unique but partially overlapping functions for these four proteins, and they imply particularly close links between Nas6 and Rpn14 on the one hand and Nas2 and Hsm3 on the other.
The synthetic growth interactions between deletions of the four putative RP-base assembly factors were paralleled by their effects on proteasome assembly as assayed by native gel immunoblotting (Fig. 6B and Suppl. Fig. S6B). Among the double mutants, anti-Rpn5 immunoblotting revealed the greatest accumulation of free lid, and lowest level of RP2CP, in the nas6Δ rpn14Δ mutant, congruent with its strong temperature-sensitive growth defect. The nas6Δ rpn14Δ strain also showed the greatest reduction in RP2CP particles, and a strong build-up of free CP (and Blm10-CP), based on anti-α4 (and anti-Blm10) blotting (Fig. 6B and not shown). All of these results are consistent with impairment of RP base assembly.
Blotting with antibodies to base subunits (Rpn1, Rpt5) revealed that distinct proteasomal subcomplexes accumulated in the various double mutants (Fig. 6B). The nas6Δ rpn14Δ mutant accumulated high levels of an Rpt5-containing species (Fig. 6B, arrowhead) that based on mobility could be equivalent to the Rpt4-Rpt5 complex seen in rpt4-G106D cells (Figs. 3 and 5D; also seen in nas6Δ, Fig. 6C). Consistent with this, loss of either Nas6 or Rpn14 exacerbated the rpt4-G106D growth defect (Suppl. Fig. S6D), and overexpression of either protein partially suppressed rpt4-G106D (not shown). The nas6Δ rpn14Δ strain, and to a slightly lesser extent the nas6Δ, nas6Δ hsm3Δ and nas6Δ nas2Δ mutants, also accumulated less free RP and free base than WT (Fig. 6B, C). The loss of free RP and concomitant increase in free lid whenever Nas6 is lost is striking and suggests that Nas6 has a role in lid-base joining. In support of this idea, high-copy NAS6 stimulated formation of an RP-like species in rpt4-G106D cells, whereas high-copy RPN14 enhanced levels of free base but not the RP (Fig. 6D). Native gel immunoblot analysis indicated that Nas6 associates with both free base and RP-like complexes, whereas Rpn14 associated with free base but was not detected in the RP (Suppl. Fig. S7A, B).
We also detected a faster migrating Rpn1-containing band (arrowhead in Fig. 6B) that is visible in WT cell extracts and a number of the chaperone mutants and may represent a species similar or identical to the Rpt1-Rpt2-Rpn1-Hsm3 complex that accumulates in rpt4-G106D cells (Fig. 5). This species is eliminated or altered in mobility when Hsm3 is deleted (Fig. 6B). The implication of these results is that blocking full RP base formation leads to a build-up of various base subcomplexes.
PAAF1, the mammalian ortholog of Rpn14, binds most tightly to the Rpt6 ATPase (Park et al., 2005). Consistent with this, we found strong enhancement of the growth defect of an rpt6ts (cim3-1) mutant when RPN14 was deleted (Fig. 6E). This was first evident at 33°C. Conversely, overexpression of RPN14 suppressed the cim3-1 growth defect (Suppl. Fig. S6C). Because nas6Δ interacts strongly genetically with rpn14Δ, we checked for nas6Δ cim3-1 interactions and found an even greater growth defect, which is evident already at 24°C (Fig. 6E). High-copy NAS6 also suppressed cim3-1 (Suppl. Fig. S6C). Collectively, these data strongly suggest that Nas6 and Rpn14 are key factors in RP assembly that have linked but distinct functions.
Discussion
The present results demonstrate that cells have at least four specific factors that are necessary for efficient RP base assembly in vivo. Their functions partially overlap, so the single gene knockouts have only mild defects. But when deleted in particular combinations, striking RP assembly abnormalities result. These factors are all characterized by well-known protein-protein interaction modules, such as coiled-coil and PDZ domains (Nas2), ARM/HEAT repeats (Hsm3), WD40 repeats (Rpn14), and ankyrin repeats (Nas6). These domains are expected to function in RP subunit binding; this is known to be true for Nas6 (Nakamura et al., 2007). Our data show that efficient in vivo proteasomal regulatory particle biogenesis depends on an unexpectedly large array of extrinsic factors and that these evolutionarily conserved proteins all function, at least in part, as RP base assembly chaperones.
Nas2, Hsm3, Nas6, and Rpn14 are RP assembly factors
Analysis of the mammalian Nas2 ortholog, p27, showed that it could be purified as a complex with an apparent size of ~300 kDa that included equal amounts of p27, Rpt4 and Rpt5 (DeMartino et al., 1996). Addition of this “modulator” to purified RP (called PA700) and CP complexes enhanced capping of the CP with RP complexes. Our in vitro assembly data are broadly comparable with these results. We suggest that in both yeast and mammalian cells, Nas2/p27 and the modulator stimulate RP base assembly from incomplete RP base precursors.
Yeast Nas2 and mammalian p27 have never been detected in fully assembled base, RP or 26S proteasome complexes, so it is possible that Nas2 binding is mutually exclusive with another component of the RP base. The rpt4-G106D coiled coil (CC)-disrupting mutation does not interfere with Rpt4-Rpt5 interaction but impairs binding to Nas2. However, rpt4-G106D cells have a much stronger assembly defect than do nas2Δ cells. The simplest hypothesis to explain these data is that the Rpt4 CC associates directly with the CC in Nas2 as well as another RP subunit(s), although other interactions between Rpt4-Rpt5 and Nas2 are also possible.
Yeast Hsm3, which is not part of mature 26S proteasomes, is related by its sequence and biochemical interactions to mammalian S5b, a protein previously thought to be part of 26S proteasomes (Gorbea et al., 2000). However, the isolation of S5b and biochemical analysis of its protein-protein interactions were done using purified RP complexes, so either the purified mammalian RPs included RP precursors that exclusively contained the S5b protein, or S5b can also associate with mature RP when the RP is not bound to the CP. Consistent with these ideas, our MS/MS and anti-Hsm3 native gel immunoblot analyses showed Hsm3 in a complex that appeared to include the complete RP, while no Hsm3 was detected in the RPCP or RP2CP species.
Congruent with what had been shown with in vitro synthesized human RP proteins, yeast Hsm3 forms a stable complex in vivo with the Rpt1, Rpt2, and Rpn1 subunits (Fig. 5B). This heteromer was detectable in WT cells but was much more abundant in the rpt4-G106D strain. Potentially, the rpt4 mutation interferes with the binding of Rpt4-Rpt5(-Nas2) to the Rpt1-Rpt2-Rpn1-Hsm3 complex, limiting formation of a larger RP-base precursor that has both subcomplexes. Rpt4 and Rpt5 are not believed to be direct ring neighbors with either Rpt1 or Rpt2 (Hartmann-Petersen et al., 2001), but a recent study suggests that Rpn1 fills the center of the ATPase ring (Rosenzweig et al., 2008), so Rpn1 might make contacts with multiple ATPases (see Fig. 7). Nas2 may facilitate this initial interaction but could get displaced from Rpt4-Rpt5 by other base subunits. Alternative routes of base assembly are also possible (see next section).
Fig. 7.
Model for the assembly of the proteasome regulatory particle and the role of RP base assembly chaperones. The inferred point of each chaperone’s release is shown at the step after the latest complex in which it was detected. See text for details.
Just prior to the submission of our work, an independent study was published on the potential role of yeast Hsm3 in RP assembly (Le Tallec et al., 2009). The overlapping data on Hsm3 from the two studies are largely in agreement. Le Tallec et al. did not analyze any of the other RP assembly factors identified here, nor did they determine the exact complexes in which Hsm3 resides. Our analysis of assembly defects were all done under conditions where cells were fully viable, whereas the other study required extended cell growth under conditions that are lethal to hsm3Δ.
Nas6 and Rpn14 had previously been associated with proteasome function, primarily in mammals (Whitby and Hill, 2007). Yeast Nas6 and its mammalian ortholog gankyrin bind directly to the Rpt3 ATPase; gankyrin does not bind detectably to the full 26S proteasome (Dawson et al., 2002). Interestingly, gankyrin also appears to function as an assembly platform for specific kinases and ubiquitin ligases, which is important for the regulation of the p53 and Rb tumor suppressors. From the crystal structure of the Nas6-Rpt3 complex, the ankyrin repeats that comprise most of Nas6 form a curved structure whose concave surface forms an extensive interface with Rpt3 (Nakamura et al., 2007). Modeling Rpt3 as part of a full ATPase ring suggests that Nas6 binds to the outer surface of the ring and would be compatible with binding to a full ring (Whitby and Hill, 2007). This is consistent with our MS/MS analysis, which identified Nas6 in a base precursor containing all 6 ATPases (and all 3 base Rpn subunits). Moreover, anti-Nas6 immunoblotting showed that Nas6 also associates with a full RP-like complex. We were only able to purify small amounts of these complexes (Fig. 5F), so Nas6 was likely missed in the MS/MS analysis of tryptic fragments (the same explanation is likely for our failure to detect Sem1, the smallest RP subunit; Suppl. Fig. S7B).
Previously, only minimal information was available for Rpn14 or its human ortholog, PAAF1. Deletion of yeast RPN14 causes an extremely mild proteolytic defect against certain substrates (Seong et al., 2007). In contrast, human PAAF1 has been proposed to act as an inhibitor of RP-CP interaction (Park et al., 2005). This inhibition contributes to the recruitment of the RP, without the CP, to HIV-1 promoters by the viral Tat protein, which stimulates transcriptional elongation (Lassot et al., 2007). PAAF1 binds most strongly to Rpt6 but shows some association with the remaining five proteasomal ATPases as well (Park et al., 2005). If Rpn14/PAAF1, which has a WD40/β-propeller domain, makes contacts with multiple ATPases, it is possible that it binds the surface of the ATPase ring that would contact the CP α-ring. This could account for the inhibition of RP-CP association when PAAF1 is overexpressed in tissue culture cells (Park et al., 2005).
The other three assembly factors described in our study are also composed substantially or almost entirely of structurally well-characterized protein interaction motifs. In Nas6 binding to the C-terminal domain of Rpt3, over 2250 Å2 of surface area are buried between Nas6 and Rpt3, which led to the suggestion that Nas6 is an intrinsic subunit of the proteasome (Nakamura et al., 2007). The fact that it is not, despite this very large interface with complementary charge patches, suggests that Nas6 release might require additional factors, such as molecular chaperones or regulated conformation changes in the ATPase ring. An earlier study indicated that Nas6 is only associated with the RP in the absence of ATP (Verma et al., 2000), and Hsp70 and Hsp90 family members contribute to proteasome assembly, although their exact functions remain uncertain (Murata et al., 2009).
The central region of Hsm3 is composed largely of ARM or HEAT repeats; based on known structures of proteins made of such repeats, the Hsm3 protein is likely to form a curved structure analogous to that formed by the ankyrin repeats of Nas6. We therefore suggest that Hsm3 will also bind to the outer surface of the ATPase ring. The specific genetic interaction between an rpt1ts mutation (cim5-1) and hsm3Δ, as well as the ability of high-copy HSM3 (but not NAS2) to suppress the cim5-1 growth defect, suggests that Hsm3 might bind the Rpt1 C-terminal domain, similar to the Nas6-Rpt3 complex.
Nas2 does not bear any obvious repeat structure but residues 96-191 of the 220-residue protein are predicted to comprise a PDZ domain. Most PDZ domains bind C-terminal peptide motifs in their target proteins (Nourry et al., 2003). It is possible that Nas2 binds a C-terminal peptide of one of the base subunits. Rpt5 terminates with the sequence FYA-COOH, which would fit the consensus for a class II PDZ target (X-Hb-X-Hb-COOH, where Hb is a hydrophobic residue; Nourry et al., 2003). Interestingly, this C-terminal segment of Rpt5 also conforms to the “HbYX motif” recently shown to insert into a pocket between subunits in the CP α-ring (Rabl et al., 2008) (Gillette et al., 2008). Nas2 might prevent premature association of the Rpt4-Rpt5 complex with the CP in addition to facilitating its incorporation into RP base precursors.
Model for RP assembly
We propose a general model for RP assembly, as outlined in Fig. 7. Although there may be a precise order of addition of subunits or subunit complexes into the RP base, we cannot deduce this from the data in hand. The general roles we envision for the assembly chaperones are: (1) stabilizing specific RP base subcomplexes; (2) facilitating incorporation of these subcomplexes into higher-order RP precursors; and/or (3) preventing subunit associations that slow assembly or lead to off-pathway intermediates. For the subcomplexes shown at the beginning of the pathway in Fig. 7, we have provided direct evidence for in vivo formation of Rpt4-Rpt5-Nas2 and Rpt1-Rpt2-Rpn1-Hsm3. These are both detected in WT cells but accumulate to higher levels in specific mutants. An Rpt3-Nas6 complex has been documented in biochemical and structural studies (Nakamura et al., 2007). The simplest RP subcomplex that we had seen containing the Rpt3, Rpt6, Rpn2, and Rpn13 base subunits was a full base precursor, but it is possible that these subunits also participate in other, more transient precursors.
In the preceding section, we suggested that the Rpt4-Rpt5-Nas2 and Rpt1-Rpt2-Rpn1-Hsm3 complexes might assemble cooperatively into an RP base precursor, consistent with the partially overlapping functions of Nas2 and Hsm3 inferred from analysis of multiple mutant combinations. Simultaneous loss of the other two chaperones, Nas6 and Rpn14, has even more striking effects on RP biogenesis. Moreover, high-copy RPN14 and NAS6 suppress rpt6 (cim3-1) growth defects, and overexpression of both genes in the same cells further enhances this suppression (Suppl. Fig. S6C and not shown). This argues for significant functional overlap between these two chaperones. Nevertheless, the individual mutations do have distinct effects. The rpn14Δ strain shows a selective loss of free base whereas nas6Δ cells lose a large fraction of both the free base and free RP (Fig. 6C). These data suggest that while both deletions lead to an RP base assembly defect, nas6Δ also may cause a defect in base-lid joining. Consistent with this, high-copy NAS6, but not RPN14, can drive formation of the full RP from free base and lid in certain rpt mutants (Fig. 6D and not shown).
It is interesting that Rpt3 and Rpt6, the two ATPases bound by Nas6 and Rpn14, respectively, are not direct neighbors in the ATPase ring (Fig. 7). However, in vitro binding and yeast two-hybrid analyses indicate that these two subunits can associate (Russell et al., 1996) (Richmond et al., 1997). Therefore, it is possible that an important function for the Nas6 and Rpn14 chaperones is to limit this potential off-pathway interaction. This could operate indirectly. For example, Rpn14 binding to Rpt6 might facilitate Rpt6 association with Rpt1-Rpt2-Rpn1-Hsm3, and Nas6 binding to Rpt3 might enhance interaction with Rpt4-Rpt5(-Nas2), with either interaction thereby limiting Rpt3-Rpt6 binding. Thus, only after loss of both Nas6 and Rpn14 will a strong assembly defect be detected. Experiments to test these mechanistic hypotheses are underway.
Conservation of RP assembly mechanisms
All of the assembly factors described here are conserved across a broad range of eukaryotic species. This is also true for CP assembly chaperones. Therefore, it is highly likely that the assembly mechanisms of the RP and 26S proteasome are very similar in most or all eukaryotes. At present we know of three dedicated CP assembly factors (Murata et al., 2009) and now four RP base assembly factors. Nothing is known yet about whether additional dedicated assembly chaperones will be needed for efficient RP lid biogenesis or lid-base joining in vivo, although this seems probable.
Proteasomal active site inhibitors are in clinical use for treating several cancers and are in trials for managing additional disorders (Goldberg, 2007). Impairing proteasome assembly represents a potential new strategy for inhibiting proteasome function for clinical purposes. This might prove particularly useful when patients develop resistance to active-site inhibitors. Research on the mechanisms of proteasome biogenesis and the roles of the emerging array of assembly chaperones will be essential for the development of effective assembly inhibitors.
Experimental Procedures
Yeast strains and media
All yeast manipulations were carried out according to standard protocols (Guthrie and Fink, 2002). Strains used in this study are listed in the Supplementary Data. For plating serial dilutions of cells, yeast cultures were grown overnight in rich medium (YPD) or minimal medium (SD) and diluted to an OD600 of 0.2 in water. Six-fold dilutions were prepared in water and spotted onto various media. Plasmids (Suppl. Data) were made by standard methods (Guthrie and Fink, 2002).
Immunoblot analysis
SDS-PAGE and immunoblot analysis of proteins were carried out according to standard procedures (Li et al., 2007). For immunoblotting, gel-separated protein samples were transferred to PVDF membranes (Millipore). Membranes were incubated with antibodies to ubiquitin (M.H. lab), Rpn5 or Rpn8 (D. Finley), Rpt4 (T. Kodadek (rabbit) or W. Tansey (mouse)), α4 (D.H. Wolf), Pgk1 or V5 (Invitrogen), Rpt5 (BIOMOL), Rpn1 or Rpt1 (W. Tansey), Blm10 (C. Enenkel) or affinity-purified antibodies to Nas2, Nas6, or Hsm3. Proteins were visualized by ECL or ECL-Advance (for mouse anti-Rpt4 antibody) (GE Healthcare).
Nondenaturing gel analyses of yeast extracts
Yeast cell extracts were prepared essentially as described (Kusmierczyk et al., 2008). Briefly, mid-to-late log phase cells (OD600 1.0–2.0) were washed with ice-cold water and frozen in liquid nitrogen. The frozen cells were ground with mortar and pestle, and the resultant cell powder was thawed in 26S buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 10% glycerol, 1 mM ATP). Extracts were centrifuged for 10 min at 15,000 × g to remove cell debris. After determining concentrations with the Bio-Rad Protein Assay, 24–25 μg of protein per sample were used for native PAGE followed by substrate overlay or immunoblotting; 3–6 μg of total protein were used for immunoblotting after SDS-PAGE. Nondenaturing PAGE was performed as described (Kusmierczyk et al., 2008).
MS/MS analysis of protein complexes
Purified proteasomes and proteasome subparticles were separated by 4–6% native PAGE and visualized with GelCode Blue as described (Li et al., 2007). Protein bands were excised and sent to Midwest Bio Services LLC for compositional analysis by Nano-LC-MS/MS. Samples were digested in-gel with trypsin, and the resulting peptide mixture was analyzed by LC-MS/MS.
Additional methods are described in the Suppl. Data.
Supplementary Material
Acknowledgments
We thank C. Enenkel, D. Finley, T. Kodadek, W. Tansey, and D.H. Wolf for antibodies, and A. Kusmierczyk and M. Kunjappu for comments on the manuscript. M.F. was supported in part by a JSPS Postdoctoral Fellowship from Japan. This work was supported initially by NIH grant GM046904 and more recently by GM083050 to M.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams GM, Crotchett B, Slaughter CA, DeMartino GN, Gogol EP. Formation of proteasome-PA700 complexes directly correlates with activation of peptidase activity. Biochem. 1998;37:12927–12932. doi: 10.1021/bi981482i. [DOI] [PubMed] [Google Scholar]
- Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- Dawson S, Apcher S, Mee M, Higashitsuji H, Baker R, Uhle S, Dubiel W, Fujita J, Mayer RJ. Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J Biol Chem. 2002;277:10893–10902. doi: 10.1074/jbc.M107313200. [DOI] [PubMed] [Google Scholar]
- DeMartino GN, Proske RJ, Moomaw CR, Strong AA, Song X, Hisamatsu H, Tanaka K, Slaughter C. Identification, purification, and characterization of a PA700-dependent activator of the proteasome. J Biol Chem. 1996;271:3112–3118. doi: 10.1074/jbc.271.6.3112. [DOI] [PubMed] [Google Scholar]
- Diaz-Martinez LA, Kang Y, Walters KJ, Clarke DJ. Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div. 2006;1:28. doi: 10.1186/1747-1028-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova IV, Kovaltzova SV, Korolev VG. The yeast HSM3 gene is involved in DNA mismatch repair in slowly dividing cells. Genetics. 2000;154:495–496. doi: 10.1093/genetics/154.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell K, Wilkinson CR, Dubiel W, Gordon C. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem Sci. 2000;25:83–88. doi: 10.1016/s0968-0004(99)01529-7. [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Li X, Velichutina I, Hochstrasser M, Kobayashi H. Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J Cell Sci. 2004;117:6447–6454. doi: 10.1242/jcs.01575. [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci USA. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- Gorbea C, Taillandier D, Rechsteiner M. Mapping Subunit Contacts in the Regulatory Complex of the 26S Proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J Biol Chem. 2000;275:875–882. doi: 10.1074/jbc.275.2.875. [DOI] [PubMed] [Google Scholar]
- Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci U S A. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular and Cell Biology. 350–351. San Diego: Academic Press; 2002. [Google Scholar]
- Hartmann-Petersen R, Tanaka K, Hendil KB. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch Biochem Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Ramos PC, Dohmen RJ. The ultimate nanoscale mincer: assembly, structure and active sites of the 20S proteasome core. Cell Mol Life Sci. 2004;61:1562–1578. doi: 10.1007/s00018-004-4130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Ann Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, Ueda T, Kikuchi Y, Tanaka K, Nakano A, Toh-e A. The Assembly Pathway of the 19S Regulatory Particle of the Yeast 26S Proteasome. Mol Biol Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- Lassot I, Latreille D, Rousset E, Sourisseau M, Linares LK, Chable-Bessia C, Coux O, Benkirane M, Kiernan RE. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol Cell. 2007;25:369–383. doi: 10.1016/j.molcel.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. Embo J. 2007;26:2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109 doi: 10.1021/cr8004857. in press. [DOI] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Umehara T, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem Biophys Res Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Sci STKE. 2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- Park Y, Hwang YP, Lee JS, Seo SH, Yoon SK, Yoon JB. Proteasomal ATPase-associated factor 1 negatively regulates proteasome activity by interacting with proteasomal ATPases. Mol Cell Biol. 2005;25:3842–3853. doi: 10.1128/MCB.25.9.3842-3853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond C, Gorbea C, Rechsteiner M. Specific interactions between ATPase subunits of the 26 S protease. J Biol Chem. 1997;272:13403–13411. doi: 10.1074/jbc.272.20.13403. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Sathyanarayana UG, Johnston SA. Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26 S proteasome. J Biol Chem. 1996;271:32810–32817. doi: 10.1074/jbc.271.51.32810. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Steger KA, Johnston SA. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J Biol Chem. 1999;274:21943–21952. doi: 10.1074/jbc.274.31.21943. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- Schwartz AL, Ciechanover A. Targeting Proteins for Destruction by the Ubiquitin System: Implications for Human Pathobiology. Annu Rev Pharmacol Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- Seong KM, Baek JH, Yu MH, Kim J. Rpn13p and Rpn14p are involved in the recognition of ubiquitinated Gcn4p by the 26S proteasome. FEBS Lett. 2007;581:2567–2573. doi: 10.1016/j.febslet.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe TK, Saito A, Suzuki M, Fujiwara T, Takahashi E, Slaughter CA, DeMartino GN, Hendil KB, Chung CH, Tanahashi N, Tanaka K. cDNA cloning and characterization of a human proteasomal modulator subunit, p27 (PSMD9) Genomics. 1998;50:241–250. doi: 10.1006/geno.1998.5301. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Hill CP. A versatile platform for inactivation and destruction. Structure. 2007;15:137–138. doi: 10.1016/j.str.2007.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.