Abstract
Purpose
Separase, an endopeptidase, plays a pivotal role in chromosomal segregation by separating sister chromatids during the metaphase to anaphase transition. Using a mouse mammary tumor model we have recently demonstrated that overexpression of Separase induces aneuploidy and tumorigenesis (Zhang et al., 2008, PNAS 105:13033). In the present study, we have investigated the expression level of Separase across a wide range of human tumors.
Experimental Design
To examine the expression levels and localization of Separase in human tumors, we have performed immunofluorescence microscopy using human Separase antibody and tumor tissue arrays from osteosarcoma, colorectal, breast, and prostate cancers with appropriate normal controls.
Results
We show that Separase is significantly overexpressed in osteosarcoma, breast and prostate tumor specimens. There is a strong correlation of tumor status with the localization of Separase into the nucleus throughout all stages of the cell cycle. Unlike the normal control tissues, where Separase localization is exclusively cytoplasmic in non dividing cells, human tumor samples show significantly higher number of resting cells with a strong nuclear Separase staining. Additionally, overexpression of Separase transcript strongly correlates with high incidence of relapse, metastasis and lower 5 year overall survival rate in breast and prostate cancer patients.
Conclusion
These results further strengthen our hypothesis that Separase might be an oncogene, whose overexpression induces tumorigenesis, and indicates that Separase overexpression and aberrant nuclear localization are common in many tumor types and may predict outcome in some human cancers.
Keywords: Separase, Oncogene, Osteosarcoma, Breast cancer, Prostate cancer, Colon cancer
TRANSLATIONAL RELEVANCE
This study demonstrates that Separase, a chromosomal segregation protein is significantly overexpressed in multiple human solid tumors including breast, prostate and osteosarcoma. It is not only the total cellular level of Separase protein, but the constitutive localization of Separase in the nucleus that constitutes the strongest contributor to distinguish tumor from normal tissue. Cells with nuclear Separase localization represent the majority of the total tumor mass. By re-evaluating the available published data, we also found a strong positive correlation of Separase mRNA expression with tumor grade and a strong negative correlation with disease free and overall survival. These studies not only strengthen the hypothesis that Separase overexpression plays an important role in mammary carcinogenesis, but also suggest that it may be a more common feature of human malignancies. Our study strongly suggests that Separase expression analysis can be considered as a factor to predict metastasis and patient outcome for primary tumor analysis.
Introduction
An evolutionarily conserved protein complex called cohesin holds sister chromatids together to allow accurate separation of sister chromatids into two daughter cells. At the onset of anaphase, Separase, an endopeptidase, is activated and cleaves the cohesin subunit Rad21 (also called SCC1 or MCD1) which releases sister chromatid cohesion. Separase activity is tightly regulated via several mechanisms (for details see (1–3)) to ensure accurate and precise activation of cohesin Rad21 cleavage during the metaphase to anaphase transition (2–4). Separase is activated after its inhibitory chaperone securin is degraded by APC-mediated phosphorylation and ubiquitin-mediated degradation (1, 5–8). Additionally, phosphorylation of Separase on Ser1126 and Thr1326 residues is a second mechanism to inhibit Separase activity (9, 10). Therefore, Securin null cells are viable and appear to have a nearly normal cell cycle (11–13). However, premature separation of sister chromatids e.g. by premature activation of Separase or by insufficient inhibition of overexpressed Separase, is thought to result in aneuploidy (14).
Knockout of the Separase gene results in embryonic lethality in mice (13, 15). SiRNA mediated knockdown of Separase results in genomic instability (8, 16), also seen in Separase deficient mouse embryonic fibroblasts. No severe haploinsufficiency in Separase heterozygous mouse has been observed (Gouqing Ge and Debananda Pati, unpublished observation), suggesting that moderately lower level of Separase is sufficient for normal cell cycle progression in vivo. On the contrary, there are several lines of evidence that overexpression of Separase can lead to premature sister chromatid separation, anaphase bridges and lagging chromosomes (17, 18).
Aneuploidy is a hallmark of human cancers (19) and is especially high in osteosarcoma, breast, and prostate cancers (20, 21). Although there have been many proposed hypotheses, there is no general agreement as to why aneuploidy is so highly prevalent in cancer cells and whether it contributes to tumor progression (22–24). Recently we have demonstrated that overexpression of Separase induces aneuploidy and mammary tumorigenesis in mice (18). In the present study we have investigated the expression levels and intracellular localization of Separase in a variety of human tumors to probe the hypothesis that overexpression and/or mislocalization of Separase is associated with human cancers. Here, we document a striking correlation of Separase overexpression in human cancers. Furthermore, aberrant nuclear localization of Separase was detected in breast, prostate and bone tumors, which is not seen in the normal control cells.
Materials and Methods
Tissue Arrays
Arrays for colorectal and breast carcinoma were obtained from Cooperative Human Tissue Network (CHTN) version CHTN2003CRCprog and CHTNBrCaProg1, respectively. Additionally we used array TMA-040 from Protein Biotechnologies. From US Biomax we obtained arrays for Osteosarcoma BS26011 and Prostate samples BC19019 and BC19111. Scoring was performed by taking pictures of 3 randomly selected areas from any given tissue spot, which than were analyzed by two different investigators, with the second being uninformed of the origin of the sample and the first scoring result. In each field 100 cells where counted and evaluated for the magnitude of Separase expression determining the propensity (on a scale of 0–5) and the intensity (on a scale of 0–3) scores for a total of 300 cells per tissue spot. If there was a discrepancy in the scorings between the two, a third investigator made the judgment call or the sample was excluded from the study.
Tumor Specimens
A set of anonymized human breast tumors with matched normal breast tissues were obtained from the tissue repository of the M.D. Anderson Cancer Center. All tumors were harvested by a breast pathologist and quality controlled to ensure the nature and the neoplastic cell composition and histology in each specimen. Each tumor specimen contained at least 85% tumor cells. All tissues in this study were obtained after IRB approved informed consents. Information of the samples provided by the tissue bank is included in Supplemental Table 1C.
For each of the 10 tumors and 5 normal tissue specimens obtained from the MD Anderson repository, tissue was embedded in O.C.T (Sakura Tissue-Tek®) medium and refrozen at −0°C before cryosectioning and immunofluorescence staining.
Separase Overexpressed Cell lines
Human Separase was overexpressed constitutively in human cervical cancer cell line HeLa, and conditionally in the diploid, nontumorigeneic FSK3 mammary epithelial cells. In brief, HeLa cells were transfected with a CMV-driven, neomycin resistant HA-tagged-hSeparase plasmid and selected for stable integration by treating the cells in the presence of G418 (Geneticin) (Invitrogen, Carlsbad, CA). Empty vector transfected clones were served as controls. The hSeparase protein expression of transfectants was detected by Western blot analysis using HA-epitope antibody, and subsequently verified with commercially available Separase antiserum (Abnova, Taipei, Taiwan). The clone selection was based on the expression level of Separase protein, compared to the empty vector controls. Details of the Tet-inducible FSK3 Separase cells have been previously described (18).
Immunofluorescence (IF) microscopy
Human tissue arrays and paraffin embedded samples where baked for 2h at 60°C and deparaffinize in Xylene for 2x, 10 min respectively. Following stepwise rehydration in 100%, 95%, 80%, 75%, 30% Etanol for 10 min each and 2× 5 min in deionized water, antigen retrieval was performed in a Pressure Cooker at 121°C for 20 min in buffer (14.5 ml 0.1M citric acid monohydrate + 61.5 ml 0.1M sodium citrate in 750 ml ddH2O at pH 6.0). Nonspecific binding was blocked with 10% normal goat serum (NGS) in PBS at RT for 3h prior to incubation with primary anti Separase Ab (Abnova ESPL1-6H6) and normal mouse IgG as control Ab over night at 4°C in 10% NGS/PBS in a humidified chamber. After rinsing with PBS on a shaker for 4× 15min, incubation with secondary Ab (goat anti-mouse IgG-Rhodamine conjugate) was performed in 10% NGS/PBS in a humidified chamber at RT for 1.5h. To detect the proliferation status of the cells in the tissue array, a set of slides were counter stained with Ki67 antibody (ab833, Abcam, Cambridge, MA) followed by a secondary goat-anti-rabbit-FITC antibody (Molecular Probes, F-2765) for detection. After PBS rinse on a shaker 3× 10 min, slides where mounted using Vectashield mount medium with DAPI and kept at 4°C if not immediately imaged on a Nikon eclipse E800 microscope. The images were processed using background subtraction to remove shading due to non-uniform illumination and inhomogeneous staining effects, and color compensation to minimize the effects of spectral bleed through among the three color channels (red, green, and blue). The algorithms are described in detail elsewhere (25, 26).
Statistical Analysis
For each cancer subtype, the associations between the localization of Separase expression (nuclear vs. cytoplasm), the magnitude of the expression signal, and the disease status (tumor vs. normal) were modeled using a logistic regression framework. Forward selection model construction approach (27), in which covariates are incorporated into the regression model in the order of statistical significance, was used to construct the final logistic regression model and assess the strength of association between each of the considered covariates and disease status. ANOVA framework was used to establish the statistical significance for the associations between covariates and disease status.
The protein expression levels of Separase in human breast tumor specimens was compared statistically to the matched normal tissues using a set of paired tests including paired t-test, ranksum and signed rank test. Ranksum and signed rank tests are more robust to departures from normality and do not have restrictive assumptions. The IF staining data for Separase expression in these matched normal vs. tumor samples and the matched controls vs. disease from the tissue arrays were used for the paired t-test analysis. For the purposes of statistical analysis, the localization of Separase expression was separated into distinct binary variables indicating absence/presence of Separase in the nucleus and absence/presence of Separase in the cytoplasm. For each sample the average expression levels and localization in the 100 cells evaluated in each of the 3 randomly selected microscopy spots was calculated. The magnitude of expression, evaluated using either propensity and intensity scores, or using the combined total score, was also included in the statistical model (28, 29). Due to the high degree of correlation between propensity and intensity scores in the observed data, combining the two into the total score did not have a significant impact in the inferences drawn from the logistic regression model.
Results
To examine the expression levels and localization of Separase in human tumors, we have performed immunofluorescence microscopy using human Separase antibody and tumor tissue arrays from osteosarcoma, colorectal, breast, and prostate cancers with appropriate normal controls. Separase expression was scored according to standard pathology scoring with a propensity score (PS) ranging from 0 to 5 and an intensity score (IS) from 0 to 3 (28, 29). The combined addition of PS and IS was used as total score (TS) for logistic regression analysis in a stepwise forward model (27). Furthermore we also scored the expression depending upon Separase localization to cytoplasm, nucleus, or both compartments. Table 1 summarizes the dataset specifications with total number of cases and controls that were used for the analysis passing tissue quality control and scoring confidence. For a summary of the raw scoring data see Supplemental Table 1A–C. We first looked at the tumor types in different organs individually and where possible used a paired t-test for tumor and matched normal control tissue from the same patient.
Table 1.
Dataset specifications of tissue type and numbers of controls and cancer cases investigated.
| Tumor | Samples | Cases | Controls |
|---|---|---|---|
| Breast | 121 | 103 | 18 |
| Osteocarcoma | 59 | 55 | 4 |
| Prostate | 68 | 59 | 9 |
| Colon | 68 | 42 | 26 |
| Total | 316 | 259 | 57 |
Number of cases and controls that where included in the statistical analysis of Separase expression correlation with tumor status.
Breast cancer
The paired t-test for ductal carcinomas where we had matched normal tissue from the same patient (tissue repository) shows a highly significant correlation of high Separase expression in tumors for the propensity score (t=−4.36, p-value=0.0024), and an even higher correlation for the intensity score (t=−4.91, p-value=0.0012). Figure 1 shows representative samples of Separase staining in normal and breast cancer samples. Using a logistic regression model with forward selection on all normal versus all breast tumors (tissue repository samples and arrays), the factor of nuclear expression is found to be the most significant one with t=3.4228 and p=0.00075. Hence, overexpression of Separase in the nucleus is the strongest positive contributing factor for tumors, followed by intensity and propensity scores. In a full covariant model, the nuclear localization was the only statistically significant contributor at p=0.0020. Propensity and intensity are most likely not independent contributors and hence did not reach significance in this model. However, if total score (propensity + intensity) is used, the full covariant model predicts high Separase expression as the strongest contributor for tumors at t=6.503 (p=9.69×10−10) and the nuclear localization as the second strongest at t=5.172 (p=6.90×10−7).
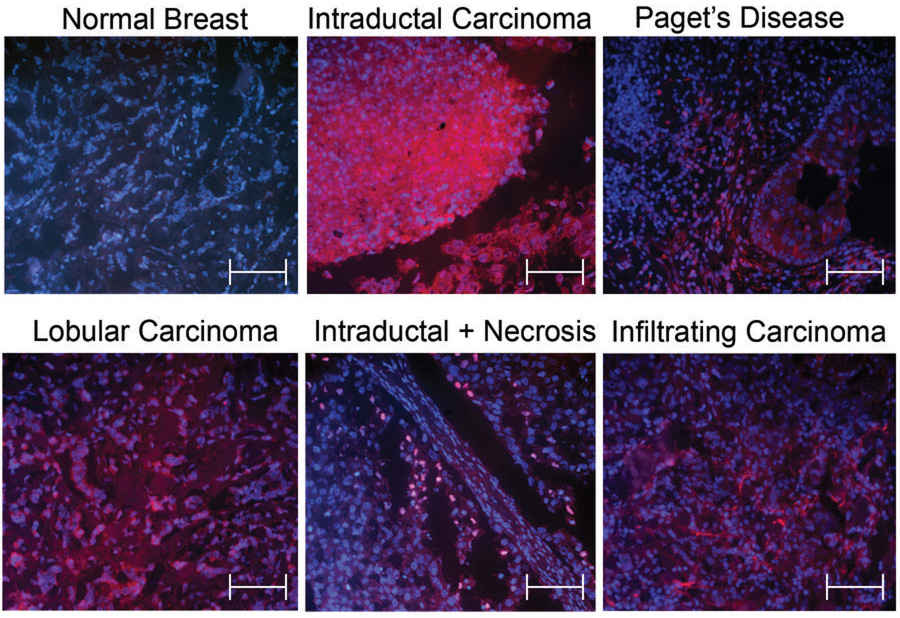
Figure 1.
Representative Immunofluorescence pictures of Separase staining comparing normal mammary gland with intraductal carcinomas with and without necrosis, Paget’s disease, lobular adenocarcinoma, and infiltrating ductal carcinoma. Separase is shown in red (Rhodamine conjugated secondary goat-anti mouse antibody), and DNA in blue (DAPI staining). Scale bar represent 100µm.
Prostate
We analyzed a total of 58 samples including 9 normal controls, 37 hyperplasias and 22 tumors. Since it is debatable if prostate tissue adjacent to a prostate adenocarcinoma actually is normal or not (30–32), we examined two types of control tissue including 3 samples from adjacent “normal” tissues and 6 samples from healthy prostates. The 3 adjacent normal tissue samples had slightly elevated Separase protein expression and a substantial amount of interphase cells with nuclear Separase localization. These findings where not seen in the 6 normal healthy controls. Therefore, the nuclear localization as predictor for prostate cancer did not reach significance with the 3 adjacent controls having nuclear Separase expression. If we exclude these 3 samples the nuclear localization is again the strongest predictor of the malignant phenotype.
Comparing the 22 prostate carcinomas with all 9 controls, the intensity of Separase expression is the strongest predictor for tumors at t=9.968 (p=7.10×10−11); followed by the propensity score at t=3.689 (p=0.00092). Figure 3 shows representative samples of Separase staining in normal and prostate adenocarcinoma samples. We observed detectable levels of Separase expression in less than 5% of the normal prostate cells. If we assume a higher proliferation index in hyperplastic and tumor samples, this still does not account for the number of strong Separase expressing cells, since we observed this in 30–100% of cells in different tumor specimens.
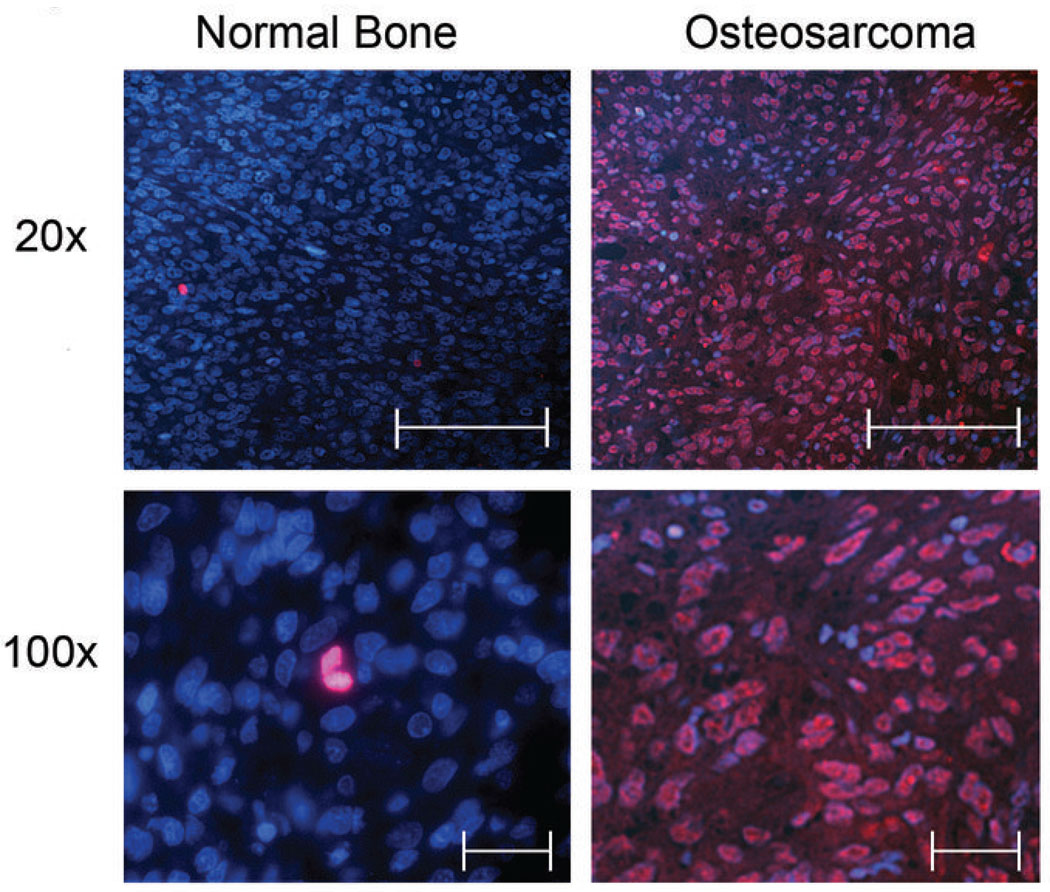
Figure 3.
Representative Immunefluorescence pictures of normal bone (left penal) and Osteosarcoma samples (right hand penal) at the indicated magnifications. Separase expression is represented in red, and DNA in blue (DAPI staining). 20× scale bars represent 150µm and 100× scale bars represent 30µm.
Based on linear discriminant analysis, 35 out of the 37 hyperplastic samples were closer to tumor samples than normal controls (with p-values 0.00243 to 1.71*10−8), implying that hyperplasia more closely resembles a stage closer to prostate cancer with regard to the high Separase expression and its nuclear localization. This is another indication, that the increase in Separase expression is one of the early steps in malignant transformation.
Osteosarcoma and colorectal carcinoma
Analysis of 63 osteosarcoma specimens and 4 normal controls on the CHTN2003CRCprog array indicated that nuclear overexpression of Separase was the strongest predictor of tumor tissue with a t-value of 10.506 at p=2.66×10−15. The intensity of Separase expression with t=3.262 (p=0.0018) as well as the propensity score with t=3.006 (p=0.0038) were also statistically significant contributors. Figure 3 shows two of the normal controls and two representative examples of Osteosarcoma.
However, in colorectal cancers Separase expression was found to be very high in both normal non-neoplastic and neoplastic colon. We did not detect any significant difference in either propensity, or intensity scores between colon cancer subtypes including adenoma, metastatic cancer, and primary carcinoma. Nuclear localization also did not reach significance to distinguish tumor from normal colon.
Separase localization and proliferation
To address the question, if stronger Separase staining correlates merely with increased proliferation, we counter stained tissue array samples and cultured cells with the proliferation marker Ki67 (green fluorescence) and Separase (red fluorescence). The majority of osteosarcoma and breast cancer samples on the tissue arrays clearly show that high Separase expression is constitutively nuclear regardless of the proliferative status of the cells (see Figure 4). In contrast, normal controls have very low or undetectable Separase expression except for the proliferating cells; hence a comparison of cytoplasmic versus nuclear localization is not feasible. The only normal breast sample that has relatively high Separase expression in resting cells shows clear exclusion of the Separase from the nucleus in the majority of interphase cells (see Figure 4). We were unable to test the prostate tumor samples for Ki67 counter staining due to the unavailability of additional array. Additional studies using a stable Separase overexpressing Hela clone and a Tet-inducible diploid, nontumorigenic mouse mammary epithelial cell line indicated that Separase nuclear localization correlates with its overexpression irrespective of the proliferative status (see Supplemental Figure 4). The induction of Separase expression in the FSK cell lines clearly shows a shift of Separase localization from exclusively cytoplasmic in uninduced cells to an evenly cytoplasmic and nuclear staining after 3 days of Doxocycline induced Separase expression (Supplemental Figure 4), suggesting that overexpression of Separase may contribute to its aberrant localization to nucleus.
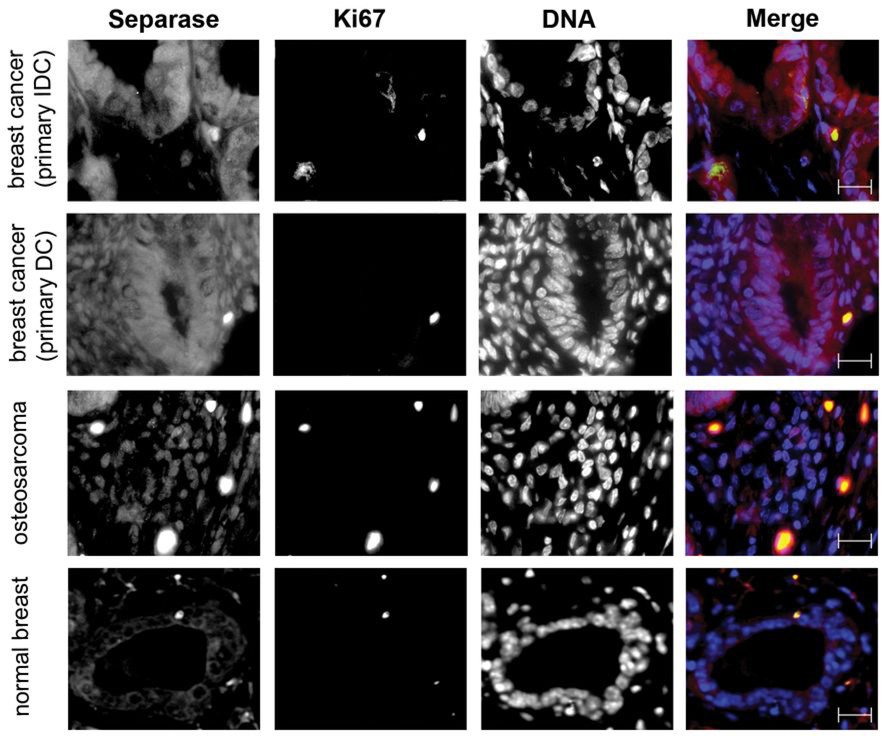
Figure 4.
Immunefluorescence pictures of tissue array samples co-stained with Ki67 (green) and Separase (red). DNA is represented in blue (DAPI staining). Top two rows depict two different types of breast cancer specimens (IDC, Interductal carcinoma; DC, Ductal Carcinoma), and the third row a representative osteosarcoma sample. Row 4 contains a normal breast specimen with detectable Separase expression showing the majority of Separase excluded from the nucleus. The majority of normal breast samples have undetectable or very low Separase staining. Black and white single channel and the merged pictures are shown for better appreciation of Separase localization to cytoplasm and nucleus in the breast cancer and osteosarcoma sample. Scale bars represent 25µm.
Summary
For all the investigated tumor types except colon carcinoma, not only high expression levels, but also the aberrant nuclear localization of Separase was the strongest predictor of tumor status. Both propensity and intensity score showed a significantly higher Separase protein expression in tumors compared with normal tissue controls. The correlation between Separase overexpression in tumor also holds good using total score. Table 2 summarizes the logistic regression data for all cancer types using a stepwise forward model. Our data clearly show very strong nuclear localization and increased expression of Separase in osteosarcoma, breast, and prostate tumors.
Table 2.
Summary of logistic regression analysis using stepwise forward model selection to identify parameters that predict disease status including all investigated samples.
| Parameters | AIC | Model coefficients | p-value |
|---|---|---|---|
| Breast Cancer status | |||
| nu | 33.15 | 0.129 | 0.00075 |
| nu + TS | 32.96 | 0.123 + 0.026 | 0.0018 + 0.075 |
| Osteocarcoma status | |||
| nu | −58.11 | 0.333 | 2.66*10−15 |
| nu + TS | −69.34 | 0.283 + 0.036 | 1.15*10−12 + 0.00040 |
| nu + TS + cy | −72.28 | 0.254 + 0.048 − 0.048 | 2.73*10−10 + 3.82*10−5 + 0.0320 |
| Prostate Cancer status | |||
| TS | 5.38 | 0.191 | 1.53*10−9 |
| TS + cy | −3.06 | 0.176 – 0.138 | 6.72*10−10 + .00023 |
| Combined all cancers | |||
| nu | 61.85 | 0.196 | 1.89*10−12 |
| nu + TS | 30.93 | 0.154 + 0.063 | 2.07*10−7 + 1.39*10−8 |
| nu + TS + cy | 27.12 | 0.132 + 0.071 – 0.068 | 6.9*10−7 + 9.69*10−10 + 0.0174 |
Associations between disease status and expression of separase: expression in the nucleus (nu), expression in cytoplasm (cy), and the degree of expression measured by combining propensity and intensity scores (TS). Consistent with the stepwise forward model selection paradigm, only the statistically significant contributors (covariates) are included in the model. Lower Akaike information criterion (AIC) values indicate a better predictive value. Statistical analysis was carried out using S-Plus software (TIBCO Software Inc., Palo Alto, CA)
Discussion
We recently reported that induced overexpression of Separase in a mouse mammary model causes aneuploidy and tumor formation in vivo (18). Using Western blot analysis of a limited number of breast tumor specimens we also showed that Separase protein is significantly overexpressed in human breast tumors as compared to the matched normal controls (18). There are several published studies analyzing the mRNA expression levels from breast cancers vs. normal controls and metastasis (21, 33–35). We reevaluated these data available on the Oncomine database (www.oncomine.org) for Separase mRNA levels with respect to tumor status and patient outcome data. Separase transcript level is highly correlated with tumor status (36–38). Separase mRNA levels are found to be consistently higher in tumors compared with normal controls at highly significant p-values. None of these studies identified Separase as one of the top 70 gene (39) or 64 gene (34) signature that would predict breast cancer patient outcome when measured in the primary tumor in these studies. Notably, there are only 3 genes in common between these two studies, indicating that the top 70 or 64 genes selected by the cut-off criteria might not be the most important genes, but the most intensely altered expressions in these patient cohorts. It is interesting to note that high Separase expression in both studies correlates with a higher incidence of recurrence (t-test: −7.415 at p-value: 2.8*10−11) and lower 5 year survival rate (t-test: −4.594 at p-value: 1*10−5) see supplemental Figure 1. Supplemental Figure 2 shows the correlation between high Separase mRNA levels and breast tumors if compared with normal breast (35).
Based on our analysis of Separase mRNA expression data from different tumor grades in the Oncomine data base, we also found a correlation between high Separase expression and high grade tumors. Additionally, the Uppsala study (33) also negatively correlates Separase expression with disease free and overall survival (t-test: −4.413 at p-value: 2*10−5) indicating that there might be a correlation in all breast cancer patients, even though it is not one of the top 70 genes with the highest alteration in expression levels.
At least one published study (31) contains mRNA expression data for Separase in prostate cancers positively correlating it with tumor status. In metastatic prostate tumors Separase expression is found to be particularly high with a correlation factor of 0.805 at p-value: 3.210−5 (see supplemental Figure 3). These results further strengthen our observation that overexpression of Separase correlates with malignant transformation and should be evaluated as a novel biomarker for detection and prediction.
In our study we have here demonstrated that Separase is significantly overexpressed in multiple human solid tumors including breast, prostate and osteosarcoma. Interestingly, it is not only the total cellular level of Separase protein, but also its aberrant nuclear localization that constitutes the strongest statistical contributor to distinguish tumor from normal tissue. These cells with nuclear Separase localization represent the majority of the total tumor mass. The mechanistic significance of nuclear Separase localization is unclear; but there are several possible explanations. First, it is possible that the normal mechanism of active nuclear exclusion of Separase (40) may be overwhelmed by Separase overexpression. Second, export of Separase from the nucleus of proliferating tumor cells may be inefficient owing to Separase overexpression. Third, high Separase level and its nuclear localization may poise the cells for division. Finally, it is known that cohesin is recruited to damaged sites along chromosomes during repair, and it is removed by Separase following DNA repair (16, 41). Hence, nuclear retention of Separase in proliferating tumor cells could result in premature removal of cohesin, a process normally occurring only after the repair process is complete. Premature cohesion removal would enhance mutation defects in the tumor DNA-damage response.
Separase might be important for DNA damage repair (16). How overexpression and nuclear localization is connected or contributes to tumor formation/progression is not yet understood. In mouse mammary epithelia cells, transcriptional regulation of Separase expression is regulated by estrogen and progesterone, and Separase expression is further facilitated by loss of p53 (17). The observation that Separase overexpression and loss or mutation of p53 strongly correlate in breast cancers (33, 42) might not be coincidental. These findings strengthen the hypothesis that misregulation of sister chromatid cohesion and segregation and the resultant aneuploidy could be a strong driving force for tumorigenesis and/or tumor progression.
Recently a number of studies have focused on characterizing tumor transcriptomes of gliomas (43), ovarian (44, 45), breast (21, 35, 39), bladder (46), and prostate cancers (31). While analyzing these published datasets, we found a strong positive correlation of Separase mRNA expression with tumor grade and a strong negative correlation with disease free and overall survival. Furthermore, studies show that overexpression of Separase strongly correlates with mutations in p53 (33, 42) as well as BRCA1 (34) in breast cancer patients. These studies not only strengthen our hypothesis that Separase overexpression plays an important role in mammary carcinogenesis (17), but also suggest that it may be a more common feature of human malignancies. Aberrant nuclear localization of the Separase in the tumor may not be the sole contributing factor since the IS (intensity score), PS (propensity score) or combined total scores are also highly significant in the forward selection model; indicating that Separase protein expression strongly correlates with tumor progression. Therefore, we suggest that Separase expression analysis can be considered as a factor to predict metastasis and patient outcome for primary tumor analysis.
Supplementary Material
Figure 2.
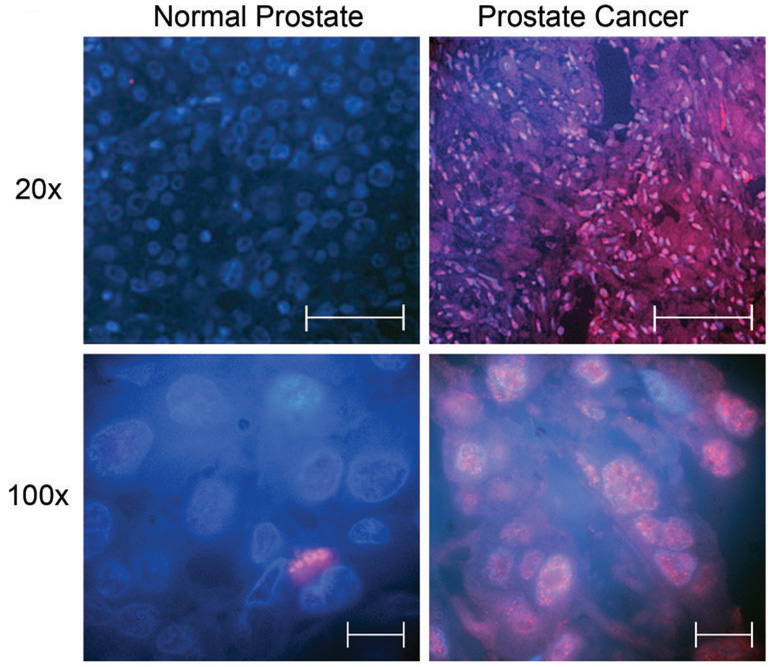
Representative Immunefluorescence pictures of two normal prostate samples (left penal) one at 20× (Scale bar represents 60µm) and one at 100× (Scale bar represents 10µm) in comparison with two prostate cancer samples (right hand penal) at the same magnifications. Separase is represented in red, and DNA by DAPI staining in blue.
Acknowledgements
We thank Drs Daniel Medina, Terzah M. Horton, Pulivarthi H. Rao, and Sarai Vega for critical reading of this manuscript, and Adel K. El-Naggar (MD Anderson Cancer Center) for providing the breast samples. This study was supported by Award Number 1RO1 CA109330 from the National Cancer Institute to D. Pati.
References
- 1.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13(16):2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 2.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293(5533):1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 3.Haering CH, Nasmyth K. Building and breaking bridges between sister chromatids. Bioessays. 2003;25(12):1178–1191. doi: 10.1002/bies.10361. [DOI] [PubMed] [Google Scholar]
- 4.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103(3):375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 5.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93(6):1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10(24):3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 7.Hornig NC, Knowles PP, McDonald NQ, Uhlmann F. The dual mechanism of separase regulation by securin. Curr Biol. 2002;12(12):973–982. doi: 10.1016/s0960-9822(02)00847-3. [DOI] [PubMed] [Google Scholar]
- 8.Waizenegger I, Gimenez-Abian JF, Wernic D, Peters JM. Regulation of human separase by securin binding and autocleavage. Curr Biol. 2002;12(16):1368–1378. doi: 10.1016/s0960-9822(02)01073-4. [DOI] [PubMed] [Google Scholar]
- 9.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107(6):715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Andreu-Vieyra CV, York JP, et al. Inhibitory phosphorylation of separase is essential for genome stability and viability of murine embryonic germ cells. PLoS Biol. 2008;6(1):e15. doi: 10.1371/journal.pbio.0060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfleghaar K, Heubes S, Cox J, Stemmann O, Speicher MR. Securin is not required for chromosomal stability in human cells. PLoS Biol. 2005;3(12):e416. doi: 10.1371/journal.pbio.0030416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pemberton HN, Franklyn JA, Boelaert K, et al. Separase, securin and Rad21 in neural cell growth. J Cell Physiol. 2007;213(1):45–53. doi: 10.1002/jcp.21086. [DOI] [PubMed] [Google Scholar]
- 13.Kumada K, Yao R, Kawaguchi T, et al. The selective continued linkage of centromeres from mitosis to interphase in the absence of mammalian separase. J Cell Biol. 2006;172(6):835–846. doi: 10.1083/jcb.200511126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9(3):204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 15.Wirth KG, Wutz G, Kudo NR, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol. 2006;172(6):847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagao K, Adachi Y, Yanagida M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature. 2004;430(7003):1044–1048. doi: 10.1038/nature02803. [DOI] [PubMed] [Google Scholar]
- 17.Pati D, Haddad BR, Haegele A, et al. Hormone-induced chromosomal instability in p53-null mammary epithelium. Cancer Res. 2004;64(16):5608–5616. doi: 10.1158/0008-5472.CAN-03-0629. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, Ge G, Meyer R, et al. Overexpression of Separase in mouse mammary epithelial cells induces aneuploidy and tumorigenesis. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0801610105. in press 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci U S A. 1998;95(23):13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearney TJ, Price EA, Lee S, Silberman AW. Tumor aneuploidy in young patients with colorectal cancer. Cancer. 1993;72(1):42–45. doi: 10.1002/1097-0142(19930701)72:1<42::aid-cncr2820720110>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 21.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 22.Duesberg P. Are centrosomes or aneuploidy the key to cancer? Science. 1999;284(5423):2091–2092. doi: 10.1126/science.284.5423.2089f. [DOI] [PubMed] [Google Scholar]
- 23.Zimonjic D, Brooks MW, Popescu N, Weinberg RA, Hahn WC. Derivation of human tumor cells in vitro without widespread genomic instability. Cancer Res. 2001;61(24):8838–8844. [PubMed] [Google Scholar]
- 24.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18(6):658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Merchant F. Fluorescence Microscopy. In: Wu QJ, Merchant FA, Castleman KR, editors. Microscope Image Processing. 1st Edition. Academic Press; 2008. pp. 247–290. [Google Scholar]
- 26.Merchant FC, KR . Computer Assisted Microscopy. In: Bovik AC, editor. Handbook of Image and Video Processing. 2nd Edition. San Diego, California: Academic Press; 2005. pp. 1311–1340. [Google Scholar]
- 27.Agresti A. Categorical Data Analysis. Second edition. Hoboken, New Jersey: John Wiley & Sons, Inc; 2002. (ISBN: 0-471-36093-7). [Google Scholar]
- 28.Conger A. Integration and generalisation of Kappas for multiple raters. Psychol Bull. 1980;88:322–328. [Google Scholar]
- 29.van Diest PJ, van Dam P, Henzen-Logmans SC, et al. A scoring system for immunohistochemical staining: consensus report of the task force for basic research of the EORTC-GCCG. European Organization for Research and Treatment of Cancer-Gynaecological Cancer Cooperative Group. J Clin Pathol. 1997;50(10):801–804. doi: 10.1136/jcp.50.10.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Bergh A, Damber JE. Chronic inflammation in benign prostate hyperplasia is associated with focal upregulation of cyclooxygenase-2, Bcl-2, and cell proliferation in the glandular epithelium. Prostate. 2004;61(1):60–72. doi: 10.1002/pros.20061. [DOI] [PubMed] [Google Scholar]
- 31.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8(5):393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Barclay WW, Woodruff RD, Hall MC, Cramer SD. A system for studying epithelial-stromal interactions reveals distinct inductive abilities of stromal cells from benign prostatic hyperplasia and prostate cancer. Endocrinology. 2005;146(1):13–18. doi: 10.1210/en.2004-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 34.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Ginestier C, Cervera N, Finetti P, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12(15):4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 37.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 38.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 39.van't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y, Yu H, Zou H. Nuclear exclusion of separase prevents cohesin cleavage in interphase cells. Cell Cycle. 2006;5(21):2537–2542. doi: 10.4161/cc.5.21.3407. [DOI] [PubMed] [Google Scholar]
- 41.Adachi Y, Kokubu A, Ebe M, Nagao K, Yanagida M. Cut1/separase-dependent roles of multiple phosphorylation of fission yeast cohesion subunit Rad21 in post-replicative damage repair and mitosis. Cell Cycle. 2008;7(6):765–776. doi: 10.4161/cc.7.6.5530. [DOI] [PubMed] [Google Scholar]
- 42.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shai R, Shi T, Kremen TJ, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22(31):4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 44.Lancaster JM, Dressman HK, Whitaker RS, et al. Gene expression patterns that characterize advanced stage serous ovarian cancers. J Soc Gynecol Investig. 2004;11(1):51–59. doi: 10.1016/j.jsgi.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66(3):1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 46.Stransky N, Vallot C, Reyal F, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38(12):1386–1396. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.