Abstract
First-line therapy for patients with glioblastoma multiforme includes treatment with radiation and temozolomide (TMZ), an oral DNA alkylating chemotherapy. Sensitivity of glioma cells to TMZ is dependent on the level of cellular O6-methylguanine-DNA methyltransferase (MGMT) repair activity. Several common coding- region polymorphisms in the MGMT gene (L84F and the linked pair I143V/K178R) modify functional characteristics of MGMT and cancer risk. To determine whether these polymorphic changes influence the ability of MGMT to protect glioma cells from TMZ, we stably overexpressed enhanced green fluorescent protein (eGFP)-tagged MGMT constructs in U87MG glioma cells. We confirmed that the wild-type (WT) eGFP-MGMT protein is properly localized within the nucleus and found that L84F, I143V/K178R, and L84F/I143V/K178R eGFP-MGMT variants exhibited nuclear localization patterns indistinguishable from WT. Using MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H- tetrazolium bromide] proliferation and clonogenic survival assays, we confirmed that WT cells expressing eGFP-MGMT are resistant to TMZ treatment compared with control U87MG cells, and that each of the polymorphic eGFP-MGMT variants confers similar resistance to TMZ. However, upon exposure to O6-benzylguanine (O6-BG), a synthetic MGMT inhibitor, the L84F and L84F/I143V/K178R variants were degraded more rapidly than WT or I143V/K178R in a proteasome-dependent manner. Despite the increased O6-BG–stimulated protein turnover caused by the L84F alteration, cells expressing L84F eGFP-MGMT did not exhibit altered sensitivity to the combination of O6-BG and TMZ compared with WT cells. In conclusion, we demonstrated that the L84F polymorphic variant has altered protein turnover without modifying sensitivity of U87MG cells to TMZ or combined TMZ and O6-BG. These findings may provide a clue to determining the clinical significance of MGMT coding-region polymorphisms.
Keywords: glioma, O6-benzylguanine (O6-BG), O6-methylguanine-DNA methyltransferase (MGMT), polymorphism, temozolomide, U87MG
Glioblastoma multiforme (GBM) is a devastating form of brain cancer in which median patient survival remains at approximately 1 year. Currently, first-line therapy for all GBM patients after surgery consists of the combination of temozolomide (TMZ) and regional fractionated radiation followed by TMZ alone.1 The cytotoxicity of TMZ, an oral DNA alkylating agent, results in part from DNA strand breaks caused by the addition of alkyl groups to the O6-position of guanine. Endogenous O6-methylguanine-DNA methyltransferase (MGMT) provides a major mechanism of TMZ resistance by repairing alkylated guanines, and the level of MGMT activity is correlated with cellular resistance to alkylating therapeutic agents.2 Following repair of a single DNA adduct, each MGMT molecule is stoichiometrically inactivated and degraded. Application of MGMT inhibitors such as O6-benzylguanine (O6-BG), an MGMT pseudosubstrate, or dose-intensive TMZ regimens are being explored clinically to increase the efficacy of TMZ by depleting tumors of MGMT.3–5 The role of common coding-region polymorphisms in altering either the ability of MGMT to protect glioma cells from TMZ or the susceptibility to cellular depletion has not been well studied.
The biology of the common coding-region polymorphic variants of MGMT has recently been extensively reviewed.6,7 The three most common coding-region polymorphisms consist of the substitution of phenylalanine for leucine at amino acid position 84 (L84F) and a 100% linked pair of polymorphisms causing an isoleucine-to-valine substitution at 143 and a lysine-to-arginine substitution at 178 (I143V/K178R). These are found in the general population with approximate allelic frequencies of 12%–27% (L84F) and 11%–28% (I143V/K178R). In a study of patients with gliomas of all grades, more than 20% were found to be carriers of the L84F polymorphism.8 Interestingly, L84F was detected more frequently in de novo GBMs than in other gliomas, suggesting that this variant may be involved in the pathogenesis of GBM.9
Inspection of the primary amino acid sequence of MGMT shows that I143V is adjacent to the active site of MGMT, and L84F is adjacent to H85, one of the four residues that coordinates zinc binding10 (Fig. 1). Although these polymorphisms are located near active or regulatory regions of MGMT, initial in vitro studies of variants generated in bacteria have demonstrated no direct affect on the demethylating activity of MGMT.6 In contrast, many epidemiological studies indicate that these polymorphisms are associated with an increased risk of developing a variety of cancers, including bladder, breast, prostate, and lung.6 Accumulating evidence indicates that these polymorphic alterations may alter more subtle functional characteristics of MGMT. For example, in vitro inactivation studies on variant MGMT species generated in bacteria demonstrate that the I143V/K178R variant is resistant to inactivation by a variety of MGMT inhibitors, including O6-BG, with the effect due solely to the I143V alteration.11,12 Interestingly, lymphocytes isolated from healthy volunteers harboring either the L84F or I143V variant alleles had increased levels of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK)-induced chromosomal aberrations compared with wild type (WT), suggesting suboptimal repair activity of both the L84F and I143V variants.13 The same group demonstrated that lymphocytes isolated from smokers with the L84F variation, but not I143V, were associated with increased frequencies of hypoxanthine phosphoribosyltransferase (HPRT) mutations.14 Only one study to date has examined the relationship between these polymorphic variants and response to dacarbazine (a TMZ analogue).15 In this retrospective melanoma study, no significant correlations were observed, although a tendency toward a lower response rate was seen for those patients harboring the I143V/K178R polymorphism.
Fig. 1.
Schematic diagram of the eGFP (enhanced green fluorescent protein)-MGMT (O6-methylguanine-DNA methyltransferase) fusion protein amino acid sequence. The eGFP tag was linked to the N-terminus of MGMT by a five-amino-acid linker (SGLRS). The coding-region polymorphisms examined in this study are indicated for L84F (within the diamond) and for the linked pair I143V/K178R (within the circles). The residues involved in zinc-binding are underlined, and the active site residues are indicated within the rectangle.
Detailed studies to elucidate the functional effects of these polymorphic MGMT variants are required to understand the epidemiological associations between the common MGMT polymorphisms and risk of developing various cancers. Such functional studies are also valuable to predict whether the polymorphic variants affect the efficacy or toxicity of alkylating agents such as TMZ against tumor cells. In this study, we analyzed the localization and stability of the common polymorphic MGMT variants expressed in U87MG glioma cells and evaluated the effect of these alterations on the sensitivity of these cells to the cytotoxic effects of TMZ alone or combined with O6-BG.
Materials and Methods
Cell Culture
293T cells were maintained in Iscove’s modified Dulbecco’s medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Hyclone) and 100 U/ml penicillin and 100 μg/ml streptomycin (Lonza, Walkersville, MD, USA). U87MG cells were maintained in Dulbecco’s modified Eagle’s medium/F12 (Mediatech, Herndon, VA, USA) supplemented with 10% fetal bovine serum and 100 U/ml penicillin and 100 μg/ml streptomycin. All cells were grown in a 5% CO2 humidified atmosphere. 293T and U87MG cells were obtained from Dr. P. Mischel (University of California, Los Angeles, CA, USA).
Generation of WT N-Terminus Enhanced Green Fluorescent Protein–Tagged MGMT Fusion Construct
To generate the enhanced green fluorescent protein (eGFP)-MGMT fusion construct, DNA fragments encoding MGMT and eGFP were sequentially subcloned into the pLPCX retroviral expression vector (Clontech, Mountain View, CA, USA). The pLPCX-eGFP-MGMT construct was designed to lack a stop codon following eGFP and contained a five-amino-acid “linker” segment separating eGFP and MGMT. eGFP was on the 5′ end of MGMT (amino terminus of translated product) (Fig. 1). The coding region of MGMT was amplified from human MGMT cDNA (ATCC, Manassas, VA, USA) using adaptor primers (forward primer with the HindIII site underlined and the BsrGI sites italicized: 5′-GAT CAT AAG CTT TGT ACA AGT CCG GAC TCA GGT CTA TGG ACA AGG ATT GTG AAA TGA AAC GC-3′; reverse primer with the BglII site underlined and the ClaI site italicized: 5′-GAT CAT ATC GAT AGA TCT TCA GTT TCG GCC AGC AGG CGG-3′) and inserted into a HindIII- and ClaI-linearized pLPCX vector. The coding region of eGFP was amplified from the pEGFP-N1 (Clontech) vector using adaptor primers (forward primer with the HindIII site underlined: 5′-GAT CAT AAG CTT TCG CCA CCA TGG TGA GCA AGG GCG A-3′; reverse primer with the ClaI site underlined: 5′-GAT CAT ATC GAT TTA CTT GTA CAG CTC GTC CAT GCC GAG-3′) and inserted into the HindIII- and ClaI-linearized pLPCX. Next, pLPCX-eGFP was digested with BsrGI to excise the eGFP gene, which was then inserted into the BsrGI-linearized pLPCX-MGMT.
Generation of Polymorphic Variant eGFP-MGMT Fusion Constructs
The combinations of the L84F, I143V, and K178R MGMT alterations were generated in pLPCX-eGFP-MGMT using the Quikchange methodology (Strata-gene, La Jolla, CA, USA) with the following primer sequences: 5′ L84F (5′-CCC GTG CCG GCT TTT CAC CAT CCC G-3′), 3′ L84F (5′-CGG GAT GGT GAA AAG CCG GCA CGG G-3′), 5′ I143V (5′-CCT GTC CCC ATC CTC GTC CCG TGC CAC AGA GTG-3′), 3′ I143V (5′-CAC TCT GTG GCA CGG GAC GAG GAT GGG GAC AGG-3′), 5′ K178R (5′-GGC CAC CGG TTG GGG AGG CCA GGC TTG GGA GGG-3′), and 3′ K178R (5′-CCC TCC CAA GCC TGG CCT CCC CAA CCG GTG GCC-3′). Entire MGMT coding-region sequences of all constructs (including WT) were verified at the UCLA Sequencing Core Facility.
Retroviral Expression of eGFP-MGMT Fusion Constructs in U87MG Cells
This retroviral expression system has been previously described.16 Briefly, 293T cells were cotransfected with 10 μg of the designated pPLCX construct containing eGFP-MGMT, 5 μg Hit-60, and 5 μg vesicular stomatitis virus glycoprotein (VSV-G) using HEPES-buffered saline and 150 mM CaCl2. For infection of U87MG cells, conditioned medium collected from transfected 293T cells (containing recombinant virus particles) was supplemented with 5 μg/ml Polybrene (Sigma, St. Louis, MO, USA) and added to target U87MG cells. Infected U87MG cells were selected with 0.7 μg/ml puromycin (Calbiochem, La Jolla, CA, USA). For each construct, multiple clonal cell lines were derived from the selected populations by limited dilution. Of these, two representative clones of each construct were selected for functional studies.
Western Blot Analysis of eGFP-MGMT Fusion Proteins
Cells were lysed with RIPA buffer containing 0.5% sodium dodecyl sulfate (SDS; Teknova, Hollister, CA, USA), 1% NP-40 (U.S. Biological, Swampscott, MA, USA), and 0.25% sodium deoxycholate (Sigma) supplemented with Complete Mini Protease Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN, USA), 1 μM sodium vanadate (Fisher, Fair Lawn, NJ, USA), 1 μM sodium fluoride (Fisher), and 1 μM phenylmethanesulfonyl fluoride (Sigma). Cells were scraped and passed through a 27-5/8–gauge needle five times. The resulting lysates were spun down at maximum speed in a microcentrifuge, and the supernatants were collected. The BCA (bicinchoninic acid) Protein Assay Kit (Pierce, Rockford, IL, USA) was used to determine the total protein concentration of the resultant lysates. Total protein from U87MG eGFP-MGMT lysates (4 μg) or from U87MG eGFP lysates (1 μg) was loaded into precast Tris-HEPES-SDS gels (Pierce). Protein was transferred to nitrocellulose paper using a Transblot SD Semi-Dry Transfer Cell apparatus (Biorad, Hercules, CA, USA). Immunoblotting was performed by incubation in primary antibodies diluted in 1% milk in Tween/Tris-buffered saline using a 1:1,000 dilution of anti-GFP horseradish peroxidase (HRP; Santa Cruz Biotechnology, Santa Cruz, CA, USA; polyclonal sc-8334) or a 1:1,000 dilution of anti-MGMT (Chemicon, Temecula, CA, USA; monoclonal 16200) followed by incubation with a 1:10,000 dilution of secondary antibody conjugated to HRP (Biomeda, Foster City, CA, USA). Protein bands were visualized using SuperSignal West Pico Chemiluminescent Substrate (Pierce) and exposed to film.
Quantitation of Cellular eGFP-MGMT Expression
Fluorescence plate-reader–based quantification of relative eGFP-MGMT expression level was performed as previously described.17 Cell lysates (25 μl) were transferred to a clear 96-well plate, and fluorescence signal was quantified with a Wallac Victor2 plate reader using 485-nm excitation and 535-nm emission filters. Background signal from uninfected U87MG cells was subtracted from each reading. Experiments involved triplicate measurements, and three independent experiments were performed on each of the two representative clones per construct. Results were recorded as fluorescence units per microgram of total protein by dividing the fluorescence signal by the amount of total protein and are reported as means ± SEMs.
MTT Proliferation Assay
MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assays were performed as described with some modifications.18 Briefly, 2,000 cells were seeded in 96-well plates. After an overnight incubation, the specified concentration of TMZ (NCI/NIH Developmental Therapeutics Program, Rockville, MD, USA) and/or O6-BG (Sigma) in complete growth medium was added to the cells. Dimethyl sulfoxide (DMSO) solvent was added to control wells. Without replacing medium, 4 days after the addition of TMZ and/or O6-BG, the cells were treated with 0.5 mg/ml MTT (Sigma). After 4 h of incubation at 37ºC, the MTT was removed and the intracellular formazan crystals were solubilized with DMSO. Absorbance was quantified with a Wallac Victor2 plate reader at 535 nm and 660 nm (subtracted as reference). The results are expressed as a percentage of the absorbance measured for the untreated cultures of each corresponding clone. Three independent experiments were performed in multiples of six on two clones per construct (two experiments on one clone and one experiment on the other for each variant).
Clonogenic Cell Survival Assay
Clonogenic assays were performed as described with some modifications.19 Briefly, 500 cells were seeded in 6-cm tissue culture dishes. Following an overnight incubation, the cells were treated with the specified concentration of TMZ and/or O6-BG in complete growth medium. Both compounds were initially dissolved in DMSO, and equivalent volumes of DMSO solvent were added to untreated dishes. Without replacing medium, 12 days after the addition of treatments, the cells were fixed and stained with 0.25% crystal violet in methanol. Colonies containing more than 30 cells were manually counted. The results are reported as a percentage of the colonies in untreated cultures of each corresponding clone. Three independent experiments were performed in duplicate on two clones per construct, and data are expressed as means ± SEMs.
Cycloheximide Degradation Assay
Degradation (protein turnover) experiments were performed as previously described, with some modifications.5 Briefly, 300,000 cells were seeded in 3.5-cm tissue culture dishes and grown overnight. Cells were treated with 50 μM cycloheximide (CHX; MP Biomedicals, Aurora, OH, USA) and, in some experiments, 5 μM bortezomib (Millenium Pharmaceuticals, Cambridge, MA, USA) or 200 μM leupeptin hemisulfate (Fisher). Because bortezomib is supplied in mannitol, control treatments included the appropriate concentration of mannitol. One h later, 25 μM O6-BG or an equivalent volume of its DMSO solvent was added. At indicated times following O6-BG application, the medium was removed and centrifuged at 6,000 rpm. The resulting pellets and the cells that adhered to the dishes were lysed with RIPA buffer supplemented as described above. Lysate (88% of the volume) was transferred to wells on a 96-well plate, and fluorescence was quantified with a Wallac Victor2 plate reader using 485-nm excitation and 535-nm emission filters. Three independent experiments were performed in duplicate on two clones for each eGFP-MGMT variant or eGFP control. Results are normalized to control treatment (e.g., CHX + DMSO) at the indicated time point for each cell line, except as noted. Data are expressed as means ± SEMs.
Data Analysis
Differences were analyzed with the Student t-test. p-values < 0.05 were considered significant.
Results
Stable Expression of WT and Polymorphic Variant eGFP-MGMT Constructs in U87MG Glioma Cell Lines
To examine the subcellular localization and function of WT and MGMT coding-region polymorphic variants in glioma cells, we generated eGFP-tagged WT and mutant MGMT cDNA fusion constructs and stably expressed them in human U87MG glioma cells using the pLPCX retroviral expression system. The eGFP-tagged MGMT constructs consisted of eGFP joined to the N-terminus of MGMT by a five-amino-acid “linker” segment (Fig. 1). The identical eGFP-MGMT fusion protein containing a P144K mutation has previously been shown to be translated, properly localized, and functionally active.20 We selected the U87MG glioma cell line to conduct these studies because of their low endogenous MGMT expression and high sensitivity to TMZ.21,22
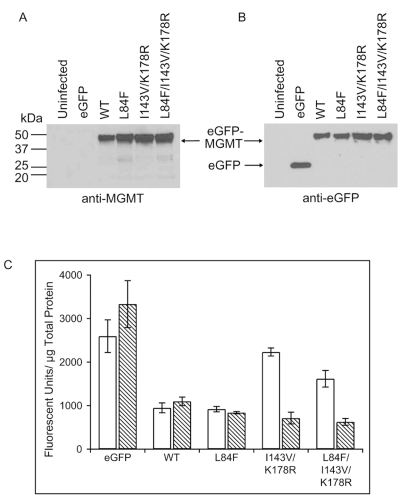
Total cell lysates of representative stable clonal cell lines expressing eGFP or various eGFP-MGMT constructs were subjected to Western blot analysis to confirm expression using both anti-MGMT and anti-GFP antibodies. Using a monoclonal anti-MGMT antibody, a prominent band representing the full-length eGFP-MGMT fusion protein at the predicted molecular weight of eGFP-MGMT (51 kDa) was detected for the cell lines expressing WT, L84F, I143V/K178R, and L84F/I143V/K178R along with faint apparent degradation bands (Fig. 2A). As expected, the 51-kDa band was not detected in uninfected U87MG cells or in U87MG cells expressing eGFP (Fig. 2A). Confirming the lack of appreciable endogenous MGMT expression in U87MG cells, no band was detected at the predicted molecular weight for MGMT (22 kDa) in any of the cell lines tested. Probing with an anti-GFP polyclonal antibody, a prominent band at 51 kDa in cells expressing the various eGFP-MGMT fusion constructs was again detected, whereas cells expressing eGFP demonstrated only the expected band at 29 kDa (Fig. 2B).
Fig. 2.
Western blot analysis of wild-type (WT) and polymorphic eGFP (enhanced green fluorescent protein)-MGMT (O6-methylguanine-DNA methyltransferase) fusion protein expression and quantitation of relative expression levels of stable clonal cell lines. Stable clonal U87MG cell lines expressing the eGFP-MGMT constructs or eGFP only were generated using the retroviral pLPCX expression vector. (A) Equal amounts (eGFP, 1 μg; all others, 4 μg) of lysates prepared from control U87MG cells (uninfected) or representative clones of U87MG cells stably expressing eGFP only (eGFP), WT, or mutant eGFP-MGMT were probed with an anti-MGMT monoclonal antibody. An approximately 51-kDa band representing the full-length fusion protein was produced by WT and mutant cell lines but not uninfected cells or cells expressing only eGFP. No endogenous MGMT was detected at 22 kDa. (B) The same lysates were also probed with an anti-GFP polyclonal antibody. The full-length fusion protein (~51-kDa band) was again detected for WT and mutant eGFP-MGMT–expressing cell lines. No band was detected in control cells, and the predicted 29-kDa eGFP band was detected in the eGFP-only cell line. (C) Quantitation of cellular eGFP, WT, or mutant eGFP-MGMT fusion proteins was performed by measuring eGFP fluorescence on a microplate reader and normalizing for total protein concentration determined in parallel. Relative fluorescence units per microgram of protein from total lysates were given for two independent clones isolated for each construct. Data shown were obtained from three experiments on each clonal cell line and represent means ± SEMs (n = 3).
To determine the relative eGFP-MGMT expression levels of each of the clonal cell lines used in subsequent functional studies, we quantified eGFP-MGMT expression levels for two representative clonal cell lines per construct by measuring eGFP fluorescence on a plate reader and normalizing for the amount of total cellular protein measured in parallel. The WT and L84F pairs of clones were fairly evenly matched in expression, whereas the I143V/K178R and L84F/I143V/K178R pairs differed by roughly 3-fold (Fig. 2C). Overall, differences in expression level in these pairs did not significantly affect interpretation of results of subsequent functional studies.
Polymorphic Variant eGFP-MGMT Fusion Proteins Are Properly Localized to the Nucleus
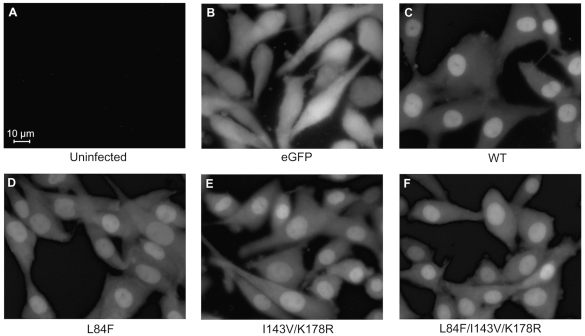
To determine the subcellular localization of the various eGFP-MGMT proteins, we performed fluorescence microscopy on live cells to visualize the distribution of eGFP fluorescence. We found that the WT eGFP-MGMT fusion protein was predominantly located in the nucleus with low-level diffuse signal in the cytoplasm (Fig. 3C), confirming that the amino-terminal eGFP-tagged MGMT maintained nuclear localization, as found in previous studies.20 The L84F, I143V/K178R, and L84F/I143V/K178R eGFP-MGMT fusion proteins were also localized to the nucleus in a pattern indistinguishable from that of WT (Fig. 3D–F), indicating that the polymorphisms individually or in combination do not affect the predominant, steady-state nuclear localization of MGMT.
Fig. 3.
Fluorescence microscopic analysis of subcellular localization of the eGFP (enhanced green fluorescent protein)-MGMT (O6-methylguanine-DNA methyltransferase) fusion proteins demonstrates a predominantly nuclear signal that is similar for wild-type (WT) and polymorphic eGFP-MGMT constructs. Clonal cell populations of live cells were used, and images were captured with a fluorescence microscope using a 40× objective. These data are representative of photographs on at least two clonal cell lines for each cell type. (A) Uninfected U87MG cells demonstrate the absence of fluorescence signal. (B) eGFP signal is distributed throughout the cytoplasm. (C–F) WT and mutant eGFP-MGMT fusion proteins localize primarily in the nucleus.
Overexpression of WT and Variant eGFP-MGMT Constructs in U87MG Cells Confers Similar Resistance to TMZ Cytotoxicity
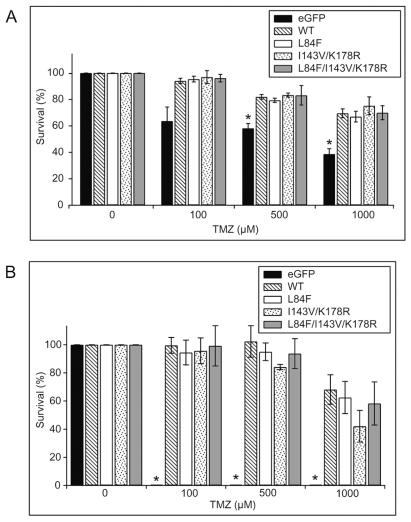
To evaluate whether L84F, I143V/K178R, or L84F/I143V/K178R mutations alter the ability of MGMT to protect U87MG cells from the cytotoxic effects of TMZ, we first confirmed whether expression of WT eGFP-MGMT increases resistance of U87MG cells to TMZ using both MTT proliferation and clonogenic survival assays. The results of the MTT assays indicated that cells expressing WT eGFP-MGMT were significantly (p < 0.05) more resistant to the 500- and 1,000-μM TMZ concentrations than were control U87MG cells stably expressing only eGFP (Fig. 4A). Similarly, the results of the clonogenic assays showed that cell lines expressing WT eGFP-MGMT were almost completely resistant to the effects of 100 and 500 μM TMZ and were only modestly sensitive to the effects of 1,000 μM TMZ (Fig. 4B), whereas the colony-forming ability of eGFP-expressing cell lines was completely abolished by treatment with 100–1,000 μM TMZ (Fig. 4B). These results confirm that amino-terminal eGFP-tagged MGMT retains repair activity and correspond with previous studies demonstrating that MGMT overexpression protects tumor cells, including glioma cells, from TMZ cytotoxicity.19,23–25
Fig. 4.
Expression of wild-type (WT) and polymorphic eGFP (enhanced green fluorescent protein)-MGMT (O6-methylguanine-DNA methyltransferase) constructs confers resistance to temozolomide (TMZ). (A) MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] proliferation assays were performed by seeding cells for 1 day, treating cells with the indicated concentrations of TMZ for 4 days without medium change, and then counting the cells. Survival is expressed as the percentage of untreated cells (0 μM TMZ [dimethyl sulfoxide (DMSO) only]) for the same corresponding cell line after subtracting background absorbance from all values. A total of at least three separate experiments were each performed in multiples of six, on two clones for each eGFP-MGMT variant or eGFP control, and data are expressed as means ± SEMs (n = 3). *Significant difference between eGFP and WT (p < 0.05) at 500- and 1,000-μM TMZ concentrations; no significant differences were observed between WT and the various eGFP-MGMT variants. (B) Clonogenic assays were performed by seeding 60-mm dishes with 500 cells for 1 day and treating cells with the indicated concentrations of TMZ for 14 days without medium change. Resultant colonies of at least 30 cells were counted. Survival is expressed as the percentage of untreated cells (0 μM TMZ [DMSO only]) for the same corresponding cell line. A total of three assays were performed in duplicate on two clones for each construct, and data are expressed as means ± SEMs (n = 3). At all concentrations of TMZ tested, eGFP-expressing U87MG cells were unable to form colonies compared with WT cells (*p < 0.05). No significant differences were observed between WT and the various eGFP-MGMT variants.
To determine whether introduction of polymorphic variants altered the protective ability of eGFP-MGMT against TMZ, we performed MTT and clonogenic assays on the various cell lines expressing the polymorphic variants. MTT analysis demonstrated that L84F, I143V/K178R, and L84F/I143V/K178R eGFP-MGMT conferred equal levels of protection against TMZ compared with WT eGFP-MGMT (Fig. 4A). Cells expressing WT and variant eGFP-MGMT also displayed comparable resistance against TMZ in clonogenic assays (Fig. 4B). These results indicate that the MGMT variants do not differ from WT in their ability to protect the cell against the cytotoxic effects of TMZ. Interestingly, inspection of individual experiments revealed no significant differences between the I143V/K178R and L84F/I143V/K178R pairs of clones despite the 3-fold difference in expression level (data not shown).
O6-BG but Not TMZ Induces Degradation of L84F Compared with WT eGFP-MGMT
With the observation that polymorphic MGMT alteration did not alter the ability of MGMT to protect U87MG cells from TMZ, we investigated whether the polymorphic variations alter the stability of the MGMT protein. To do so, we performed CHX protein turnover experiments using a fluorescence plate reader to measure eGFP-MGMT levels remaining at various time points. We first evaluated basal degradation of the eGFP-MGMT variants by measuring eGFP-MGMT remaining after incubation in CHX only. We detected a negligible level of basal degradation of all polymorphic variants over the 8-h period of the experiment; more than 80% of signal remained after 8 h (Fig. 5A).
Fig. 5.
O6-BG (benzylguanine) induces increased protein turnover of L84F eGFP (enhanced green fluorescent protein)-MGMT (O6-methylguanine-DNA methyltransferase) compared with wild type (WT). (A) Basal degradation of WT and the eGFP-MGMT variants is low. Cells were pretreated with cycloheximide (CHX) for 1 h to inhibit protein synthesis. At 0-, 4-, and 8-h time points, cells were lysed and eGFP fluorescence was quantified with a plate reader. Fluorescence remaining was calculated by normalizing to time 0 for each corresponding cell line. All experiments were performed in duplicate. For each cell type, a total of three independent experiments were performed on two clones, and results are expressed as means ± SEMs (n = 3). There was no appreciable difference between eGFP control cells and any of the eGFP-MGMT constructs. Results are normalized to time 0 for each cell line. (B) L84F eGFP-MGMT demonstrate increased turnover in the presence of O6-BG treatment compared with WT eGFP-MGMT. Cells were treated with CHX 1 h before treatment with 25 μM O6-BG. At 0, 4, and 8 h after the addition of O6-BG, cells were lysed, and eGFP fluorescence was quantified with a plate reader. In this case, fluorescence remaining was normalized to the basal degradation rate by dividing by the CHX-only treatment of the same cell line at the corresponding time point. For each cell type, a total of three independent experiments were performed on two clones, and results are expressed as means ± SEMs (n = 3). ‡Significant difference between WT and I143V/K178R versus eGFP control cells (p < 0.05). *Significant difference between L84F and L84F/I143V/K178R versus WT (p < 0.05). (C) Temozolomide (TMZ) does not induce increased turnover of either WT- or L84F-expressing cells. Cells were treated with CHX 1 h before treatment with either 25 μM O6-BG or 1,000 μM TMZ. After 8 h, cells were lysed and eGFP fluorescence was quantified with a plate reader. Fluorescence remaining normalized as in B to the basal degradation rate at 8 h is shown. For each cell type, a total of three independent experiments were performed on two clones, and results are expressed as means ± SEMs (n = 3). There was no difference between TMZ versus CHX-only degradation for either WT or L84F constructs. The degradation induced by O6-BG was significantly increased compared with TMZ for both WT and L84F constructs (*p < 0.05). (D) Bortezomib but not leupeptin reverses the effects of O6-BG treatment in both WT and L84F-expressing cells. Cells were treated with CHX and 5 μM bortezomib or 200 μM leupeptin 1 h prior to the addition of O6-BG. After 8 h, cells were lysed, and fluorescence was quantified with a plate reader. Results are normalized to basal degradation as in B. The values above 100% observed for WT and L84F cells treated with bortezomib indicated inhibition of basal degradation in addition to O6-BG–induced degradation. One representative experiment is shown.
Because O6-BG, a synthetic inhibitor of MGMT, potently induces rapid degradation of MGMT,26 we examined the stability of the various constructs upon addition of O6-BG. We found that treatment of cells with 25 μM O6-BG results in significantly (p < 0.05) greater degradation of L84F (50% signal remaining compared with untreated at 8 h) and L84F/I143V/K178R compared with WT and I143V/K178R (80% signal compared with untreated at 8 h; Fig. 5B), indicating that O6-BG induces increased degradation of L84F but not I143V/K178R MGMT. These results are normalized to show the degradation induced by O6-BG only. The rapid turnover of the L84F/I143V/K178R protein indicated that the presence of L84F resulted in increased degradation in the context of I143V/K178R. In contrast, the addition of 1,000 μM TMZ did not stimulate significant degradation of WT or L84F eGFP-MGMT constructs at 8 h (Fig. 5C). To confirm that the difference was not specific to the 25 μM O6-BG concentration, we titrated the concentration of O6-BG and found similar responses at O6-BG concentrations ranging between 25 and 100 μM (data not shown).
O6-BG–Stimulated Degradation of WT and L84F Is Sensitive to Proteasomal but Not Lysosomal Inhibition
Since MGMT degradation is a proteasomally dependent process,27,28 we sought to confirm whether the enhanced degradation of L84F is also dependent on the proteasome by treating the cell cultures with bortezomib, a clinically available dipeptide that reversibly inhibits the chymotryptic enzyme activity of the proteasome.30 We titrated the concentration of bortezomib between 0.1 and 5 μM and observed increasing inhibition of degradation, with 5 μM resulting in complete inhibition (data not shown). Using 5 μM bortezomib, we found that O6-BG–induced degradation of both WT and L84F was completely inhibited (Fig. 5D). Because of the normalization, the values above 100% represented inhibition of basal degradation by bortezomib in addition to that induced by O6-BG. These results indicate that both enhanced degradation of L84F stimulated by O6-BG and basal degradation are predominantly proteasomally dependent processes.
To determine whether O6-BG–induced degradation of WT and L84F is also dependent on the lysosome, we treated the cell lines with leupeptin, a lysosomal inhibitor. In contrast to the results obtained for bortezomib, the addition of 200 μM leupeptin did not inhibit O6-BG–induced degradation of either WT or L84F eGFP-MGMT (Fig 5D). The addition of higher concentrations of leupeptin (200–800 μM) also had no effect on the O6-BG–stimulated degradation of L84F (data not shown). These results indicate that degradation of MGMT is not a lysosomally dependent process.
Cells Expressing L84F Do Not Exhibit Altered Sensitivity to TMZ Combined with O6-BG
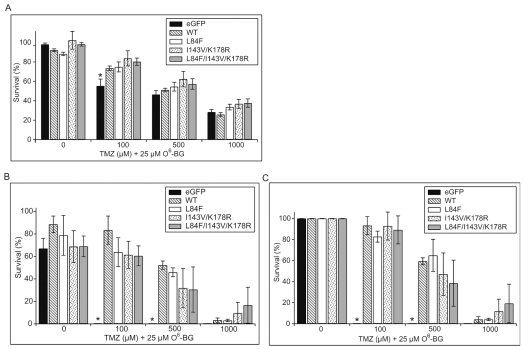
Based on our findings of increased susceptibility of the L84F variant to depletion by O6-BG, we hypothesized that cells expressing the L84F variant would be more sensitive to the combination of O6-BG and TMZ. To explore whether cells expressing the L84F MGMT variant have altered sensitivity to the cytotoxic effects of the combination of O6-BG and TMZ, we performed MTT and clonogenic assays on U87MG cells expressing the various eGFP-MGMT constructs. In these experiments, we used the same concentration of O6-BG (25 μM) as in the degradation experiments since this concentration was sufficient to induce differential degradation between WT and L84F. In MTT proliferation assays, treatment with 25 μM O6-BG alone had little effect on the proliferation of cells expressing eGFP or any of the eGFP-MGMT variants (Fig. 6A). In clonogenic assays, treatment with 25 μM O6-BG alone resulted in a small decrease in the colony-forming abilities of all the cell lines tested (Fig. 6B).
Fig. 6.
Cotreatment with O6-benzylguanine (O6-BG) sensitizes cells expressing wild-type (WT) and variant eGFP (enhanced green fluorescent protein)-MGMT (O6-methylguanine-DNA methyltransferase) constructs to temozolomide (TMZ) in a dose-dependent manner. (A) MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assays were performed as described for Fig. 4A, except that cells were treated with 25 μM O6-BG in addition to the indicated concentration of TMZ. WT and variant eGFP-MGMT all conferred a similar degree of resistance to the combination of TMZ and O6-BG. Survival is expressed as percentage of untreated cells (0 μM TMZ, 0 μM O6-BG [dimethyl sulfoxide (DMSO) only]) for the same corresponding cell line after subtracting background absorbance from all values. A total of at least three separate experiments were performed in multiples of six each on two clones for each eGFP-MGMT variant or eGFP control, and data are expressed as means ± SEMs (n = 3). There was no significant effect of O6-BG by itself on survival of the various cell lines. Resistance to TMZ was maintained when cotreated with 100 μM for eGFP-MGMT–containing cell lines versus eGFP control cells (*p < 0.05). However, when cell lines were cotreated with 500 and 1,000 μM TMZ, there was no significant difference between eGFP-MGMT–containing cell lines and eGFP control cells, or between WT and the variant constructs. (B and C) Clonogenic assays were performed as described for Fig. 4B except that cells were treated with 25 μM O6-BG in addition to the indicated concentrations of TMZ. (B) Survival is expressed as percentage of untreated cells (0 μM TMZ, 0 μM O6-BG [DMSO only]) for the same corresponding cell line. A total of at least three separate experiments were performed in duplicate on two clones for each eGFP-MGMT variant or eGFP control, and data are expressed as mean ± SEM (n = 3). This shows that O6-BG alone has modest toxicity. (C) To visualize the isolated effects of TMZ, we renormalized the same data from A and expressed survival as percentage of the 0 μM TMZ/25 μM O6-BG–treated value for the same corresponding cell line after subtracting background absorbance from all values. Resistance to TMZ was maintained when cotreated with O6-BG and 100 μM and 500 μM for eGFP-MGMT–containing cell lines versus eGFP control cells (*p < 0.05). However, when cell lines were cotreated with O6-BG and 1,000 μM TMZ, there was no significant difference between eGFP-MGMT–containing cell lines versus eGFP control cells or between WT and the variant constructs.
From the MTT assays, we observed that O6-BG renders WT and variant eGFP-MGMT–expressing cell lines as sensitive to 500- and 1,000-μM TMZ concentrations as control cells expressing eGFP (Fig. 6A). When the clonogenic assay results were normalized to remove the effects of O6-BG alone, we observed that O6-BG renders WT and variant eGFP-MGMT–expressing cell lines as sensitive to 1,000-μM TMZ concentrations as control cells expressing eGFP (Fig. 6C) However, both MTT and clonogenic assays indicated that the combination of O6-BG and TMZ sensitizes each of the WT and variant eGFP-MGMT cells equally (Fig. 6A–C). This indistinguishable sensitivity of the WT and variant cell lines to the combination of TMZ and O6-BG suggests that the enhanced turnover of the L84F mutant induced by O6-BG does not affect its ability to protect cells from TMZ cytotoxicity.
Discussion
We established a U87MG glioma cell culture system based on overexpression of N-terminus eGFP-tagged MGMT constructs to study the effects of polymorphic alterations on TMZ-induced glioma cell cytotoxicity. We confirmed that these constructs were properly targeted and functionally active in protecting cells from TMZ cytotoxicity. Furthermore, our model system confirms that the MGMT depletional strategy using O6-BG can transform an MGMT-expressing cell insensitive to TMZ into a cell that is sensitive to TMZ. In this cell culture model system, all of the polymorphic variants studied behaved similarly to WT in MTT proliferation and clonogenic survival assays. While the most obvious interpretation of this data is that introduction of these amino acid variations does not result in alteration of MGMT’s ability to repair TMZ-induced DNA alkylation, the possibility exists that subtle alterations are not detectable in our model system because of relatively high over-expression of the constructs. This is supported by results (data not shown) of our MTT and clonogenic experiments on I143V/K178R and L84F/I143V/K178R pairs of clones with roughly 3-fold differences in expression levels (Fig. 2C), in which increased resistance to TMZ in cell lines with higher expression was not observed. This may indicate that expression level of MGMT offers increased protection against TMZ cytotoxicity up to but not beyond a certain threshold. Another possibility is that the eGFP tag somehow prevents observation of a functional change due to these polymorphic variants. Lastly, phenotypic differences of these polymorphic variants may manifest only in cell or tissue types other than gliomas.
Our major finding is that the L84F alteration shows increased degradation of MGMT in the context of O6-BG exposure compared with WT. An equivalent effect was observed when L84F was introduced in the context of the I143V/K178R MGMT variation, but no effect was seen upon the introduction of I143V/K178R alone. In the three-dimensional structure of MGMT, the L84F residue is relatively close to the active site, raising the possibility that this alteration affects the binding of O6-BG to MGMT, resulting in enhanced degradation.7 Based on our finding that the differential degradation is sensitive to the proteasomal inhibitor bortezomib, it is also possible that the L84F variation affects the conformation of the benzylated form of MGMT. It is well established that upon DNA repair or inactivation by pseudosubstrates, MGMT undergoes a conformational change that targets it for proteasomal degradation.6 The L84F alteration may alter this conformational change so that either the ubiquitin acceptor site is more accessible or the binding of an E3 ligase is more favorable. Three-dimensional analysis of the benzylated form of L84F MGMT may demonstrate the actual structural perturbation, and generation of a series of L84 mutants may be useful to determine amino acid side-chain characteristics tolerated at that position. It will be important to determine whether tamoxifen and nitrous oxide, both recently shown to induce proteasomal degradation of MGMT, can also induce differential degradation of L84F versus WT MGMT.26,30 Such data may aid in understanding how these agents affect MGMT degradation.
Despite this difference in O6-BG–induced degradation between L84F-containing variants and WT MGMT, all of the MGMT polymorphic variants studied had nearly equivalent ability to protect U87MG cells from the cytotoxic effects of TMZ or combined TMZ/O6-BG. While the failure to discover phenotypic variation could result from inherent pitfalls of our model system, the simplest interpretation of our data is that increased clearance of alkylated MGMT does not affect repair ability of MGMT. If this is indeed the case, a clue to how increased clearance of an alkylated form of a polymorphic variant of MGMT may have a biological consequence is suggested by evidence supporting the role of alkylated MGMT in suppressing estrogen receptor-α (ERα-dependent transcription via interaction with ERα).31 This possibility has been raised by epidemiological studies showing an association between the L84F polymorphism and increased breast cancer risk.32 The increased turnover rate of the alkylated (benzylated) L84F MGMT may result in less suppression of ERα, resulting in increased proliferation.
In conclusion, our results indicate that the L84F variant of MGMT is degraded faster in the presence of O6-BG, but the potential functional consequences of this difference remain to be elucidated. It may still be important to explore the roles of MGMT polymorphisms as predictive biomarkers for response to TMZ (in either conventional or depletional regimens) and how they may influence response in relation to concurrent MGMT promoter methylation.33 Alternatively, it may be necessary to examine whether patients harboring these variants may be more susceptible to the myelotoxic effects of TMZ-based therapies independent of tumor efficacy.
Acknowledgments
This work was made possible by support from the Cancer Center of Santa Barbara and the Art of the Brain Foundation. A portion of this work was presented in abstract form at the Society for Neuro-Oncology 2007 annual meeting.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Liu L, Gerson SL. Targeted modulation of MGMT: clinical implications. Clin Cancer Res. 2006;12(2):328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- 3.Quinn JA, Desjardins A, Weingart J, et al. Phase I trial of temozolomide plus O6-benzylguanine for patients with recurrent or progressive malignant glioma. J Clin Oncol. 2005;23(28):7178–7187. doi: 10.1200/JCO.2005.06.502. [DOI] [PubMed] [Google Scholar]
- 4.Brandes AA, Tosoni A, Cavallo G, et al. Temozolomide 3 weeks on and 1 week off as first-line therapy for recurrent glioblastoma: phase II study from Gruppo Italiano Cooperativo di Neuro-oncologia (GICNO) Br J Cancer. 2006;95(9):1155–1160. doi: 10.1038/sj.bjc.6603376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23(5):823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 6.Pegg AE, Fang Q, Loktionova NA. Human variants of O6-alkylguanine-DNA alkyltransferase. DNA Repair (Amst) 2007;6(8):1071–1078. doi: 10.1016/j.dnarep.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugni JM, Han J, Tsai MS, Hunter DJ, Samson LD. Genetic association and functional studies of major polymorphic variants of MGMT. DNA Repair (Amst) 2007;6(8):1116–1126. doi: 10.1016/j.dnarep.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 8.Wiencke JK, Aldape K, McMillan A, et al. Molecular features of adult glioma associated with patient race/ethnicity, age, and a polymorphism in O6-methylguanine-DNA-methyltransferase. Cancer Epidemiol Biomarkers Prev. 2005;14(7):1774–1783. doi: 10.1158/1055-9965.EPI-05-0089. [DOI] [PubMed] [Google Scholar]
- 9.Inoue R, Isono M, Abe M, Abe T, Kobayashi H. A genotype of the polymorphic DNA repair gene MGMT is associated with de novo glioblastoma. Neurol Res. 2003;25(8):875–879. doi: 10.1179/016164103771954005. [DOI] [PubMed] [Google Scholar]
- 10.Schwarzl SM, Smith JC, Kaina B, Efferth T. Molecular modeling of O6-methylguanine-DNA methyltransferase mutant proteins encoded by single nucleotide polymorphisms. Int J Mol Med. 2005;16(4):553–557. [PubMed] [Google Scholar]
- 11.Fang Q, Loktionova NA, Moschel RC, Javanmard S, Pauly GT, Pegg AE. Differential inactivation of polymorphic variants of human O6-alkylguanine-DNA alkyltransferase. Biochem Pharmacol. 2008;75(3):618–626. doi: 10.1016/j.bcp.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margison GP, Heighway J, Pearson S, et al. Quantitative trait locus analysis reveals two intragenic sites that influence O 6-alkylguanine-DNA alkyltransferase activity in peripheral blood mononuclear cell. Carcinogenesis. 2005;26(8):1473–1480. doi: 10.1093/carcin/bgi087. [DOI] [PubMed] [Google Scholar]
- 13.Hill CE, Wickliffe JK, Wolfe KJ, Kinslow CJ, Lopez MS, Abdel-Rahman SZ. The L84F and the I143V polymorphisms in the O6-methylguanine-DNA-methyltransferase (MGMT) gene increase human sensitivity to the genotoxic effects of the tobacco-specific nitrosamine carcinogen NNK. Pharmacogenet Genomics. 2005;15(8):571–578. doi: 10.1097/01.fpc.0000167332.38528.a5. [DOI] [PubMed] [Google Scholar]
- 14.Hill CE, Wickliffe JK, Guerin AT, et al. The L84F polymorphism in the O6-methylguanine-DNA-methyltransferase (MGMT) gene is associated with increased hypoxanthine phosphoribosyltransferase (HPRT) mutant frequency in lymphocytes of tobacco smokers. Pharmacogenet Genomics. 2007;17(9):743–753. doi: 10.1097/FPC.0b013e3281111eb1. [DOI] [PubMed] [Google Scholar]
- 15.Ma S, Egyhazi S, Ueno T, et al. O6-Methylguanine-DNA-methyltransferase expression and gene polymorphisms in relation to chemotherapeutic response in metastatic melanoma. Br J Cancer. 2003;89(8):1517–1523. doi: 10.1038/sj.bjc.6601270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang MY, Lu KV, Zhu S, et al. Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res. 2006;66(16):7864–7869. doi: 10.1158/0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]
- 17.Lai A, Le D-N, Paznekas WA, Gifford WD, Jabs EW, Charles AC. Oculodentodigital dysplasia connexin43 mutations result in non-functional connexin hemichannels and gap junctions in C6 glioma cells. J Cell Sci. 2006;119(3):532–541. doi: 10.1242/jcs.02770. [DOI] [PubMed] [Google Scholar]
- 18.Figul M, Soling A, Dong HJ, Chou TC, Rainov NG. Combined effects of temozolomide and the ribonucleotide reductase inhibitors didox and trimidox in malignant brain tumor cells. Cancer Chemother Pharmacol. 2003;52(1):41–46. doi: 10.1007/s00280-003-0611-2. [DOI] [PubMed] [Google Scholar]
- 19.Hermisson M, Klumpp A, Wick W, et al. O6-Methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J Neurochem. 2006;96(3):766–776. doi: 10.1111/j.1471-4159.2005.03583.x. [DOI] [PubMed] [Google Scholar]
- 20.Choi U, DeRavin SS, Yamashita K, et al. Nuclear-localizing O6-benzylguanine-resistant GFP-MGMT fusion protein as a novel in vivo selection marker. Exp Hematol. 2004;32(8):709–719. doi: 10.1016/j.exphem.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Ueda S, Mineta T, Nakahara Y, Okamoto H, Shiraishi T, Tabuchi K. Induction of the DNA repair gene O6-methylguanine-DNA methyltransferase by dexamethasone in glioblastomas. J Neurosurg. 2004;101(4):659–663. doi: 10.3171/jns.2004.101.4.0659. [DOI] [PubMed] [Google Scholar]
- 22.Combs SE, Schulz-Ertner D, Roth W, Herold-Mende C, Debus J, Weber KJ. In vitro responsiveness of glioma cell lines to multimodality treatment with radiotherapy, temozolomide, and epidermal growth factor receptor inhibition with cetuximab. Int J Radiat Oncol Biol Phys. 2007;68(3):873–882. doi: 10.1016/j.ijrobp.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Cai S, Xu Y, Cooper RJ, et al. Mitochondrial targeting of human O6-methylguanine DNA methyltransferase protects against cell killing by chemotherapeutic alkylating agents. Cancer Res. 2005;65(8):3319–3327. doi: 10.1158/0008-5472.CAN-04-3335. [DOI] [PubMed] [Google Scholar]
- 24.Passagne I, Evrard A, Depeille P, Cuq P, Cupissol D, Vian L. O(6)-Methylguanine DNA-methyltransferase (MGMT) overexpression in melanoma cells induces resistance to nitrosoureas and temozolomide but sensitizes to mitomycin C. Toxicol Appl Pharmacol. 2006;211(2):97–105. doi: 10.1016/j.taap.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Roos WP, Batista LF, Naumann SC, et al. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26(2):186–197. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- 26.Liu L, Xu-Welliver M, Kanugula S, Pegg AE. Inactivation and degradation of O6-alkylguanine-DNA alkyltransferase after reaction with nitric oxide. Cancer Res. 2002;62(11):3037–3043. [PubMed] [Google Scholar]
- 27.Srivenugopal KS, Yuan XH, Friedman HS, Ali-Osman F. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chloroethyl)-1-nitrosourea. Biochemistry. 1996;35(4):1328–1334. doi: 10.1021/bi9518205. [DOI] [PubMed] [Google Scholar]
- 28.Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O(6)-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23(5):823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 29.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23(21):4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 30.Kuo CC, Liu JF, Shiah HS, Ma LC, Chang JY. Tamoxifen accelerates proteasomal degradation of O6-methylguanine DNA methyltransferase in human cancer cells. Int J Cancer. 2007;121(10):2293–2300. doi: 10.1002/ijc.22927. [DOI] [PubMed] [Google Scholar]
- 31.Teo AKC, Oh HK, Ali RB, Li BFL. The modified human DNA repair enzyme O6-methylguanine-DNA methyltransferase is a negative regulator of estrogen receptor-mediated transcription upon alkylation DNA damage. Mol Cell Biol. 2001;21(20):7105–7114. doi: 10.1128/MCB.21.20.7105-7114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han J, Tranah GJ, Hankinson SE, Samson LD, Hunter DJ. Polymorphisms in O6-methylguanine DNA methyltransferase and breast cancer risk. Pharmacogenet Genomics. 2006;16(7):469–474. doi: 10.1097/01.fpc.0000215065.21718.4c. [DOI] [PubMed] [Google Scholar]
- 33.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]