Abstract
Vg1, a member of the transforming growth factor-β family involved in mesoderm induction, is translated subsequent to the localization of its mRNA to the vegetal pole of Xenopus oocytes. Whereas the localization of Vg1 mRNA is known to be directed by the 3′ untranslated region (UTR), the basis of its translational regulation is unknown. We show here that the 3′ UTR of Vg1 causes translational repression of two different reporter mRNAs in Xenopus oocytes. A 350-nucleotide region of the 3′ UTR, which is distinct from the localization element, is necessary and sufficient for mediating translational repression and specifically binds to a 38-kDa polypeptide. The translational repression activity is found throughout the oocyte and at all stages of oogenesis. These results suggest that factors colocalized with Vg1 mRNA at the vegetal pole relieve translational repression to allow expression of Vg1 protein.
Specification of cell fates in the developing embryo requires the coordinated spatial and temporal expression of multiple regulatory factors. One way in which this regulation is accomplished is by localizing mRNAs encoding important regulatory molecules to particular regions of the developing oocyte (1–5). The localization of mRNAs allows the developing oocyte and embryo to generate asymmetric distributions of proteins encoded by localized transcripts by virtue of the fact that the proteins are most highly enriched in those regions where their messages are found. These early mRNA-sorting events form the basis for determining cell fate and pattern formation in a number of organisms (6). However, in order for mRNA localization to achieve a high degree of fidelity, it is important to have mechanisms to prevent the premature translation of mRNAs before arriving at their final destination (7, 8).
Such a phenomenon of localization-dependent translation has been described for both the oskar and nanos transcripts in Drosophila melanogaster (9, 10). In these examples, the processes of localization and translational control are both controlled by sequences in their respective 3′ untranslated regions (UTRs). Moreover, the translational control elements (TCEs) lie within the regions of the 3′ UTR that mediate localization (10–13). These results have led to a model where the localization machinery competes for the same binding sites as the machinery involved in translational repression (7). According to this view, assembly of the localization complexes on the 3′ UTR relieves translational repression, thereby coupling proper localization with expression. Whether this paradigm is universally used during early developmental regulation of other transcripts remains unclear.
In Xenopus laevis, the transforming growth factor-β family member Vg1 is regulated both at the level of mRNA localization and translation during early development (5, 14–16). This maternally supplied transcript is localized in a microtubule-dependent manner to the vegetal pole during stages III and IV of oogenesis (17, 18). Only after the localization of Vg1 mRNA has been completed in stage IV can Vg1 protein expression be detected for the first time (14, 16). The temporal correlation between the completion of Vg1 mRNA localization and the onset of Vg1 translation suggests that these two events are coordinately regulated. In contrast to the mRNAs in Drosophila, relatively little is known about the mechanism of localization-dependent translational activation of Vg1 mRNA. The 3′ UTR of Vg1 contains a 340-nucleotide element that is necessary and sufficient to direct its vegetal pole localization (19). Recently, two RNA binding proteins, vera/VgRBP and VgRBP60, that bind specifically to the Vg1 localization element have been identified (20–22). However, the role of these proteins in localization and their relationship to translational regulation remain unknown.
In this study, we examine the translational control of the Vg1 mRNA. We find that the 3′ UTR of Vg1 mRNA mediates its translational repression. In contrast to nanos and oskar mRNAs, the TCE in the 3′ UTR of Vg1 lies outside of the previously defined localization element. The TCE is necessary and sufficient for repression and specifically binds to a 38-kDa protein. Surprisingly, the translational repression activity is present and equally distributed throughout the oocyte even in stages when the endogenous Vg1 mRNA is localized and actively being translated. Together, these results suggest that additional factors localized to the vegetal pole are responsible for relieving translational repression of Vg1 mRNA once it is correctly localized.
Materials and Methods
Construction of Plasmids.
The sp64-derived bovine prolactin expression construct, which contains an SP6 promoter and the 5′ UTR from Xenopus globin preceding the prolactin coding region, has been described (23). The Prl-Vg1 construct was engineered as follows; A plasmid containing the full Vg1 3′ UTR (19) was digested with BstEII, treated with Klenow fragment, and digested with EcoRI, and this fragment was ligated into the bovine prolactin expression construct that had been digested with BlpI, treated with Klenow fragment, and digested with EcoRI. The resulting plasmid was digested with EcoRI, and oligonucleotides encoding a poly(A)20 tail were inserted to generate Prl-Vg1. The control Prl construct was made by digesting Prl-Vg1 with SpeI and EcoRI, treating with Klenow fragment, and recircularizing the plasmid. Prl and Prl-Vg1 were linearized at either XbaI (located just after the A20 sequence; see Figs. 2 and 4) or EcoRI (located just before the A20 sequence; Fig. 1; see also Fig. 3; as indicated in the legends) before transcription. Transcript for Prl-NS was generated by linearizing the Prl construct at DraI, ≈1.2 kb beyond the stop codon in the vector, before transcription. β-Lac was made by replacing the prolactin coding region of Prl (excised with NcoI and BlpI) with a PCR product encoding full-length β-lactamase (β-lac). β-Lac-Vg1 was made by inserting the SpeI to XbaI segment of the Vg1 3′ UTR into the BlpI- and XbaI-digested β-lac construct. β-Lac-LE was made by inserting a PCR product encoding the 340-nucleotide minimal localization element of Vg1 into β-lac digested with BlpI and EcoRI. Construct C2 was made by digesting Prl-C2 with HindII and SpeI, treating with Klenow, and recircularizing the plasmid. C3 was made by digesting Prl-Vg1 with HindII and BsmI, treating with Klenow, and recircularizing the plasmid. C7 was made by ligating a PCR fragment encoding the Vg1 3′ UTR from the BsmI to NsiI sites into Prl-Vg1 digested with HindII and EcoRI. C8 was made by digesting Prl-C8 with HindII and SpeI, treating with Klenow, and recircularizing the plasmid. The resulting plasmids were linearized at EcoRI before transcription.
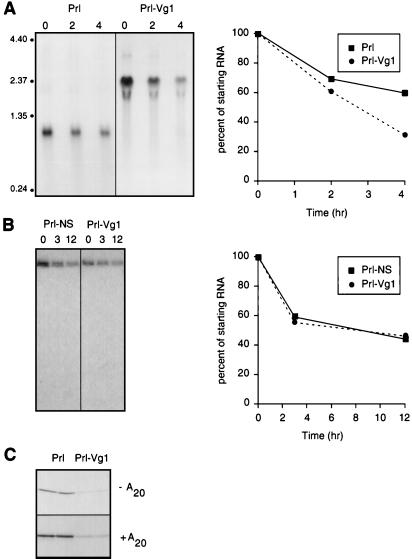
Figure 2.
Characterization of Vg1 3′ UTR-mediated translational repression. (A) [32P]UTP-labeled Prl and Prl-Vg1 transcripts were microinjected into stage VI labeled transcripts oocytes and incubated for the times (hr) indicated above each lane, and total oocyte RNA was isolated. The RNA was separated on a 1% formaldehyde-agarose gel and transferred to nitrocellulose membranes, and the labeled RNA was visualized by autoradiography. The positions of RNA standards are indicated to the left. Quantitation of the autoradiograph is shown on the right (●, Prl-Vg1; ■, Prl). (B) [32P]UTP-labeled transcripts encoding Prl-NS and Prl-Vg1 were analyzed for their relative stabilities over a 12-hr time period essentially as in A. Equal amounts of total oocyte RNA were analyzed by 5% acrylamide gel electrophoresis. The autoradiograph of the dried gel (Left) is shown alongside the quantitation of the radiolabeled RNA bands (●, Prl-Vg1; ■, Prl-NS). (C) Prl and Prl-Vg1 transcripts at 20 nM, with (+A20) or without (−A20) a poly(A)20 tail, were analyzed for translational repression as in Fig. 1. Duplicate samples are shown. The repression was ≈5.5-fold for both −A20 and +A20 transcripts.
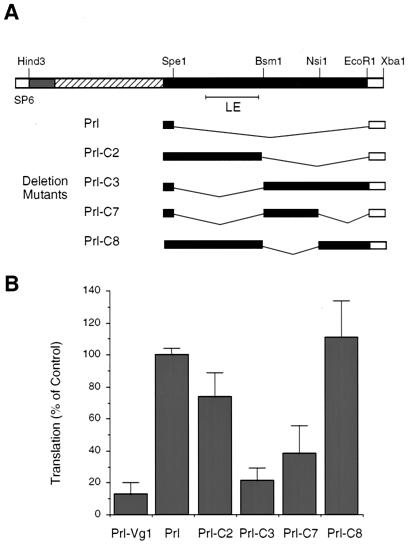
Figure 4.
The TCE is distinct from the mRNA localization element. (A) Diagram of the constructs used to map the TCE. The SP6 promoter, 5′ UTR (shaded), coding region (hatched), 3′ UTR (black), localization element (LE), poly(A)20 tail (white), and relevant restriction sites are indicated on the Prl-Vg1 construct. Prl, Prl-C2, Prl-C3, and Prl-C8 have deletions from SpeI to EcoRI, BsmI to EcoRI, SpeI to BsmI, and BsmI to NsiI, respectively, as diagrammed. Prl-C7 contains two deletions, from SpeI to BsmI and from NsiI to EcoRI. (B) The relative translational efficiencies of the constructs diagrammed in A were determined (at the RNA concentration of 20 nM) as described in Fig. 1. Values are the average of at least three measurements; standard errors are indicated by the error bars.
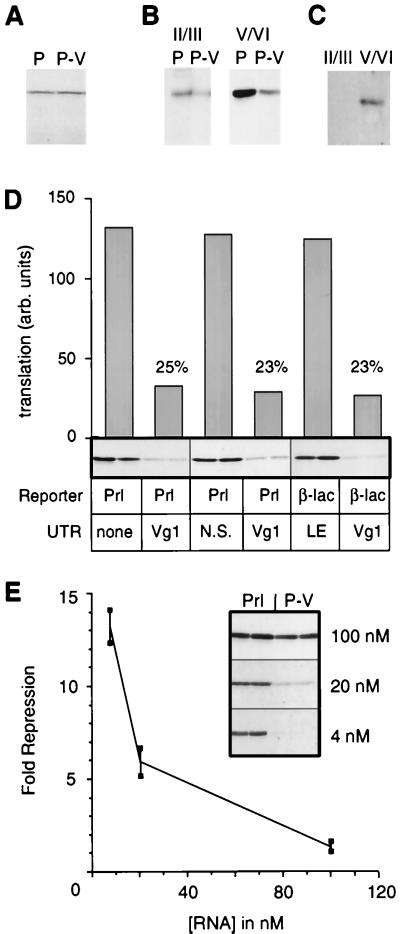
Figure 1.
Vg1 3′ UTR mediates translational repression of heterologous mRNA. (A) Plasmids encoding either prolactin (Prl, lane P) or prolactin followed by the 1.2-kb 3′ UTR from Vg1 (Prl-Vg1, lane P-V) were used to generate transcripts that were translated in a wheat germ extract containing [35S]methionine. Equal amounts of the translation reactions were separated by SDS/PAGE and the newly synthesized proteins were visualized by autoradiography. (B) Transcripts of Prl (P) and Prl-Vg1 (P-V) were injected into stage II/III (≈10 nl per oocyte) or stage V/VI (≈50 nl per oocyte) oocytes. After the labeling of newly synthesized proteins for 2 hr with 500 μCi/ml [35S]methionine, the relative amounts of radiolabeled prolactin were assessed by immunoprecipitation, SDS/PAGE, and autoradiography. (C) Western blot analysis for Vg1 expression in early-stage (II/III) and late-stage (V/VI) Xenopus oocytes. The proteins derived from the equivalent of 0.6 oocytes (early-staged sample) and 0.1 oocytes (late-staged sample) were separated by SDS/PAGE, transferred to nitrocellulose, and probed with anti-Vg1 antibodies. (D) Constructs encoding the indicated reporter (Prl, prolactin; β-lac, β-lactamase) fused to the indicated UTR (Vg1, the ≈1.2-kb Vg1 3′ UTR; N.S., 1.2 kb of nonspecific vector sequence; LE, the 340-nucleotide localization element of the Vg1 3′ UTR) were analyzed in duplicate for translational repression in stage VI oocytes as in B. The autoradiograph of the immunoprecipitated proteins as well as its quantitation are shown. The percentage of translational activity of each Vg1 containing transcript, relative to its appropriate control, is indicated. Note that the LE does not contain translational repression activity (see Fig. 4) and serves as a negative control for the β-lac-Vg1 construct. (E) Translational repression is saturable. Prl and Prl-Vg1 transcripts at an approximate concentration of 400 nM were diluted to 4, 20, and 100 nM before injection into stage VI oocytes (≈50 nl per oocyte). The oocytes were incubated for 2 hr at 18°C to allow the transcripts to diffuse throughout the oocyte. They were then labeled with [35S]methionine, and the synthesized prolactin was immunoprecipitated and analyzed by SDS/PAGE and autoradiography as in B. Shown are the autoradiograms (Inset) and quantitation of duplicate samples for Prl and Prl-Vg1 at the three different RNA concentrations.
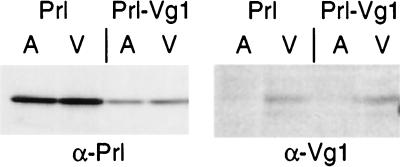
Figure 3.
Translational repression activity is distributed evenly throughout the oocyte. Prl and Prl-Vg1 transcripts at ≈20 nM were injected into stage VI oocytes (50 nl per oocyte), and the oocytes were incubated in MBSH for 12 hr at 18°C to ensure complete diffusion of the RNA within the oocytes. They were then labeled for 60 min with MBSH containing 1 μCi/ml [35S]methionine, fixed, and then manually dissected into animal (A) and vegetal (V) halves. Either prolactin or Vg1 was then immunoprecipitated from each half and analyzed by SDS/PAGE and autoradiography.
Cell-Free Transcription and Translation.
Cell-free transcription was performed by using SP6 RNA polymerase (New England Biolabs) as described (23). Quantitation of RNA yield in our standard transcription reactions (40°C for 60 min) was performed by measuring the incorporation of [32P]UTP and did not vary significantly from one experiment to another. Preparation of wheat germ extract and in vitro translation with this extract were performed as described (23).
Xenopus Oocyte Expression.
Oocytes were removed from Xenopus laevis frogs and maintained at 18°C in modified Barth's saline (MBSH; ref. 24). They were used within 5 days of harvest. Oocytes were staged according to published criteria, and transcripts were microinjected into the vegetal hemisphere as indicated in the figure legends. It is important to note that RNAs containing the Vg1 3′ UTR are localized only when injected into oocytes of stages I–III and cultured through at least stage IV. RNA injected into stage II/III oocytes, but not cultured, or injected into stage V/VI oocytes will not be localized (17). Thus, microinjected RNAs in the experiments described in this study are free to diffuse throughout the oocyte. Labeling was performed by incubation (at 18°C) in MBSH containing between 250–1,000 μCi/ml [35S]methionine (1 Ci = 37 GBq) for various times as indicated in the figure legends. Where unspecified, labeling was for 2 hr. After labeling, the oocytes were washed once in MBSH to remove free label and homogenized in 50 volumes of harvest buffer (1% SDS/0.1 M Tris⋅HCl, pH 8.9). The samples were heated to 100°C for 5 min and clarified by centrifugation at 12,000 × g for 2 min to remove any insoluble material, and the samples processed for immunoprecipitation with anti-prolactin (United States Biochemical), anti-Vg1, or anti-β-lac antibodies as described (25). The microinjected RNA concentration in the oocyte was estimated by using an injection volume of 50 nl and a stage VI oocyte volume of 1,000 nl (26) and by assuming significant diffusion of the RNA during the 2-hr incubation period before the labeling. Thus, in Fig. 1E, the RNA concentration within the oocyte is estimated to be 20-fold less than the injected concentration. Endogenous Vg1 mRNA concentration was estimated based on previous work that determined that it consisted of ≈0.05–0.1% of the 50–100 ng of poly(A) RNA per oocyte (15).
UV Crosslinking of RNA.
Collagenased oocytes were washed thoroughly and dounce homogenized in an equal volume of ice-cold homogenization buffer (0.25 M sucrose/25 mM Hepes, pH 7.4/50 mM KCl/1 mM MgAcetate/0.1 mM EDTA/2 mM PMSF/0.2 mM benzamidine/5 μg/ml aprotinin/10 μM leupeptin/1 mM DTT) and centrifuged at 10,000 × g for 10 min, the supernatant was recentrifuged at 200,000 × g for 40 min. This high speed supernatant was used at a final protein concentration of 0.75 mg/ml in the UV crosslinking reactions (see below). 32P-labeled RNA encoding the various constructs was synthesized as described (27). For UV crosslinking assays, transcription reactions containing bromo-UTP were assembled as described (27). In instances where the absolute amount of transcript was determined, [35S]UTP-labeled RNA was synthesized (27). The transcription reaction was then loaded on a Sepharose G-50 micro spin column (Amersham Pharmacia) to separate the free nucleotide from the RNA. The flow-through was phenol extracted and then ethanol precipitated overnight. An aliquot of the resuspended RNA was counted in a scintillation counter to determine the percent incorporation of radioactive nucleotide. The percentage of incorporation was used to estimate the moles of transcript synthesized. UV crosslinking reactions (20 μl ) consisting of 0.75 mg/ml extract (final concentration), transcript, and extract buffer were assembled on ice and then incubated for 30 min at room temperature. Heparin was then added to the reaction mix at a final concentration of 5 mg/ml. The crosslinking reactions were kept on ice and irradiated with a 312-nm UV light source placed 6 cm above the samples for 8 min. After irradiation, the reactions were treated with RNaseA at 1 mg/ml final concentration and incubated at 37°C for 15 min. The samples were then denatured at 65°C and resolved by SDS/PAGE.
Results
The Vg1 3′ UTR Mediates Translational Repression.
The observation that Vg1 translation begins coincident with the completion of Vg1 mRNA localization in stage IV suggests that these two events may be regulated coordinately (14, 16). One mechanism to achieve this coordination would be to regulate the repression machinery so that it is active during the early stages of oogenesis and then inactivated at stage IV, when Vg1 protein begins to be synthesized. Alternatively, the translational repression machinery could be actively maintained, with the Vg1 message being transported into a protected environment. We wished to address whether the Vg1 message is under direct translational control, and if so, what mechanisms might operate to coordinate its expression with localization.
Because translational control of mRNA is often mediated by sequences in the 3′ UTR, we wished to determine whether the 3′ UTR of Vg1 could also mediate translational regulation. To make this determination, we assessed the ability of the Vg1 3′ UTR to influence the translation of other coding regions. We prepared reporter constructs consisting of either the prolactin coding sequence with no 3′ UTR (Prl) or the prolactin coding sequence followed by the Vg1 3′ UTR (Prl-Vg1). Both Prl and Prl-Vg1 transcripts translated equally well in a wheat germ in vitro translation system (Fig. 1A). In contrast, the translational efficiency of the Prl-Vg1 mRNA was markedly reduced relative to that of Prl mRNA when injected into either early-stage (II/III) or late-stage (V/VI) Xenopus oocytes (Fig. 1B), despite the fact that endogenous Vg1 was expressed only in the late-stage oocytes (Fig. 1C). This repression was sequence specific, because a nonspecific 3′ UTR (consisting of the sp64 vector backbone, Prl-NS) did not produce this effect when fused to the prolactin coding sequence (Fig. 1D). Moreover, the effects of Vg1 3′ UTR seem to be reporter-independent, because fusion of the Vg1 3′ UTR to another reporter mRNA, β-lac, also caused translational repression (Fig. 1D).
Translational repression of Prl-Vg1 could be overcome by microinjecting increasing amounts of mRNA. An ≈10-fold reduction in 3′ UTR-specific translational repression was observed when the microinjected transcript concentration was increased from 4 to 100 nM (Fig. 1E), which suggests that the translational repression machinery is limiting. The range of RNA concentrations in which translational repression is detected is comparable to the physiologic concentration of the Vg1 message in the oocyte, and derepression only begins to saturate at mRNA concentrations that are ≈20-fold above the concentration of the endogenous Vg1 message (15). Together, these experiments demonstrate that the 3′ UTR of Vg1 is sufficient to confer translational repression in a heterologous context. Thus, it seems that within the context of the oocyte, the 3′ UTR of Vg1 mRNA is involved in its translational control.
The Vg1 3′ UTR Does Not Change mRNA Stability or Poly(A) Tail Length.
Several mechanisms of 3′ UTR mediated translational control have been described. These include altering mRNA stability (28), changing poly(A) tail length (29), or recruiting sequence-specific RNA-binding proteins that promote translational repression (30). To investigate whether differential mRNA stability is involved in Vg1 3′ UTR-mediated repression, Prl and Prl-Vg1 transcripts were microinjected into stage VI oocytes, and total RNA was isolated 0, 2, and 4 hr later. We were unable to observe a systematic or significant difference between the rates of RNA degradation (Fig. 2A), arguing against differential RNA stability as the cause of the 10-fold difference observed in their protein expression. Similarly, a comparison of the stability of Prl-Vg1 with Prl-NS showed no difference over a 12-hr time period (Fig. 2B).
To investigate the role of polyadenylation in Vg1 translational repression, we examined the effect of adding a synthetic poly(A)20 tail to Prl and Prl-Vg1. The translational efficiency of both transcripts was independent of the presence or absence of the poly(A) tail (Fig. 2C). The finding that deletions of more than 250 nucleotides at the 3′ end of the UTR (which are likely to disrupt elements used for cytoplasmic polyadenylation) still result in repression (see Fig. 4; described below), together with the absence of detectable size differences in transcript length over time (Fig. 2A), also argues against cytoplasmic polyadenylation as the mechanism of translational repression. The failure of mRNA stability or polyadenylation to explain the lower protein expression of Prl-Vg1 suggests that translational repression may operate via the recruitment of a protein complex to the 3′ UTR. This translational repression machinery is predicted to be present throughout oogenesis based on the results of Fig. 1B.
Translational Repression Activity Is Equal in Both the Animal and Vegetal Hemispheres.
The finding that injected Prl-Vg1 transcripts are translationally repressed in late-stage (V/VI) oocytes is somewhat surprising given that the endogenous Vg1 message is being actively translated at this time (Fig. 1C). This result raises the possibility that the endogenous, localized Vg1 message is in an environment that is free from repression. One way in which this regional translation might occur is through a spatial gradient of translational repression activity, with the lowest levels of repression being found in the vegetal pole. Alternatively, translational repression activity may be uniformly distributed, but factors that are colocalized with Vg1 at the vegetal pole may alleviate the repression. To distinguish between these possibilities, we compared the extent to which the Prl-Vg1 transcript is repressed in the animal versus vegetal hemispheres.
It is important to note that Prl-Vg1, despite containing the full Vg1 3′ UTR, will be localized only if injected into oocytes of stages I–III and cultured through at least stage IV. RNA injected into stage II/III oocytes, but not cultured or injected into stage V/VI oocytes, will not be localized (17). Thus, on injection into stage VI oocytes, Prl-Vg1 transcript is free to diffuse throughout the oocyte, allowing us to compare relative levels of translational repression in the animal versus vegetal halves. We found that repression of the Prl-Vg1 mRNA was equal in the animal and vegetal halves of stage V/VI oocytes (Fig. 3). Although the possibility of highly regional differences in translational repression activity cannot be excluded, this result argues against the existence of a global gradient of translational repression. Thus, it is possible that factors colocalizing at the vegetal pole with Vg1 relieve the message of its translational repression. This idea is similar to models proposed for nanos in Drosophila, where the localization machinery itself relieves the repression of transported messages (7).
Mapping of the Vg1 TCE.
To explore further the idea that localization and translation of Vg1 are coupled by having their respective complexes compete for common binding sites, we mapped the region of the Vg1 3′ UTR that is involved in translational repression. Deletion mutagenesis coupled with subsequent functional analyses for repression revealed a 348-nucleotide element, which we term the TCE, that is both necessary and sufficient for translational repression (Fig. 4). Interestingly, the TCE is distinct from the region of the 3′ UTR that had been determined to mediate localization (19). Thus, translational repression and localization of the Vg1 mRNA involve distinct RNA recognition elements and, hence, may be mediated by distinct factors.
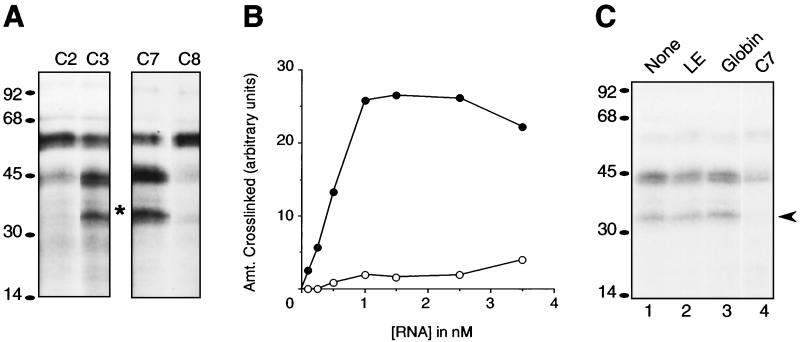
To identify proteins that might be involved in translational repression, 32P-labeled RNAs encoding different regions of the 3′ UTR were added to a cytosolic oocyte extract and then subjected to UV irradiation to crosslink proteins bound to the RNA. A 38-kDa protein bound and crosslinked to RNAs containing the TCE in a saturable manner but bound with very low affinity to RNAs lacking the TCE (Fig. 5 A and B). Two other prominent polypeptides of 45 kDa and 55 kDa were also observed. However, the 55-kDa polypeptide seemed to crosslink to all regions of the 3′ UTR and may represent a nonspecific RNA-binding protein (Fig. 5A). Consistent with this interpretation, immunoprecipitation experiments have identified the 55-kDa crosslinked polypeptide as FRGY2, a general RNA-binding protein (data not shown). The 45-kDa polypeptide appeared more specific to regions containing the TCE (Fig. 5A), but binding was competed only partially by the minimal TCE (Fig. 5C). In contrast, the binding of the 38-kDa protein to the TCE was specific and competed by a 50-fold molar excess of unlabeled TCE but not by transcripts encoding globin or the localization element (Fig. 5C). Thus, the 38-kDa protein and possibly the 45-kDa protein may be involved in mediating translational repression of Vg1 mRNA.
Figure 5.
Crosslinking of proteins specific to the TCE. (A) The constructs C2, C3, C7, and C8, which correspond to Prl-C2, Prl-C3, Prl-C7 and Prl-C8, respectively, but lack the 5′ UTR and coding regions, were used to synthesize 32P-labeled RNA. An amount of each RNA corresponding to equal incorporated counts was incubated with Xenopus oocyte extract and subjected to UV-mediated crosslinking. The labeled proteins were separated by SDS/PAGE and visualized by autoradiography. The asterisk indicates the position of a 38-kDa protein that appears to be specifically crosslinked to RNAs containing the TCE. (B) Effect of substrate RNA titration on crosslinking. Various concentrations of 32P-labeled C2 (○) and C3 (●) RNA were used for UV crosslinking as in A. The 38-kDa band was quantitated by densitometry (with local background subtraction) and plotted as a function of RNA concentration. (C) Binding of 38-kDa protein is competed by the TCE but not by the minimum localization element or globin. RNA corresponding to the globin coding region, localization element (LE), or TCE (C7) were synthesized and quantitated. A 50-fold excess of each was mixed with 32P-labeled C7 RNA (0.25 nM final concentration), and UV crosslinking was performed as in A. The arrowhead indicates the position of the 38-kDa crosslink. Lane 1 is the crosslinking reaction containing no competitor RNA.
Discussion
In this study, we have demonstrated that the Vg1 3′ UTR contains a TCE that is physically distinct from the Vg1 localization element and that specifically binds a 38-kDa protein. The translational repression mediated by this element is saturable and is present in both the animal and vegetal hemispheres of the oocyte. Furthermore, the repression activity is present during all stages of oogenesis, including those where the endogenous Vg1 message is being actively translated, and is not caused by changes in polyadenylation or message stability. Thus, Vg1 likely represents the first example of localization-dependent translation identified outside of D. melanogaster.
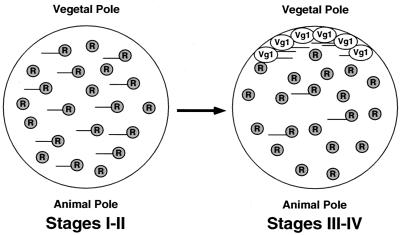
Our data suggest a model for the coordinate control of translation and localization (Fig. 6). In this model, a homogeneously distributed repressor maintains Vg1 mRNA in a translationally repressed state during stages I and II of oogenesis. During stages III and IV, the Vg1 transcripts are recruited into transport complexes and localized to the vegetal cortex where they are anchored to elements of the cytoskeleton. The final anchoring step may then cause the displacement of the repressor and allow the Vg1 protein to be synthesized. A similar model also has been proposed for nanos mRNA, which is translationally repressed until it localizes to the posterior pole of the Drosophila oocyte (7).
Figure 6.
Model for coordinate control of Vg1 mRNA localization and translation. During stages I–II of oogenesis, all Vg1 mRNA is complexed with the translational repression machinery (R) and is distributed homogeneously throughout the oocyte. During stages III–IV of oogenesis, Vg1 mRNA becomes localized to the vegetal pole where the repression machinery is displaced and Vg1 protein is synthesized. However, the translational repression machinery is still active throughout the oocyte during these stages and those Vg1 messages that have not been localized continue to be repressed.
Translational repression may serve to prevent Vg1 mRNA, which encodes a secretory protein, from being docked at sites of protein translocation at the endoplasmic reticulum (ER) during localization. This role for translational repression is consistent with recent observations that the localization element-binding protein vera/VgRBP is an ER-associated protein and that the Vg1 message colocalizes with a vegetal subcompartment of the ER during localization (31). Thus, translational repression may ensure that Vg1 mRNA is targeted only to those portions of the ER undergoing vegetal transport via vera/VgRBP. Prevention of premature or promiscuous interactions with the ER may also be important for gurken, a secreted transforming growth factor-α family member whose mRNA is localized to the anterodorsal region of the Drosophila oocyte. In this case, sequences in both the 5′ UTR and 3′ UTR are required for correct localization of the message, a situation that is unique among all known localized messages (32, 33). Assembly of a localization complex at the 5′ UTR would present an obstacle to translation and may be the mechanism by which translation and targeting of the message to the ER is prevented during localization. Thus, in contrast to mRNAs encoding cytosolic factors, it may be particularly important for messages encoding secreted proteins to be translationally repressed during localization.
The coupling of translational activation to proper mRNA localization to prevent the precocious expression of key regulatory proteins is a conserved theme in the development of both Xenopus and Drosophila. During Drosophila development, oskar and nanos mRNAs are not translated if their localization is disrupted, indicating the presence of a translational repressor whose action is alleviated by the proper localization of the transcript (8–10). In the case of nanos, a 120-kDa protein, Smaug, has been identified recently that is required for translational repression of nanos mRNA (34, 35). In the case of oskar, an 80-kDa RNA-binding protein, Bruno, has been shown to be required both for translational repression in vivo and proper pattern formation (10, 36). The candidate 38-kDa translational repressor of Vg1 is unlikely to be the Xenopus homologue of Smaug or Bruno because of the molecular mass differences between the proteins. Furthermore, there do not appear to be Bruno- or Smaug-binding sites in the TCE of Vg1 (data not shown). These findings suggest that distinct translational repressors operate on different transcripts or that the repression machinery may differ between organisms. Further work directed at identifying components of the repression machinery in both of these organisms should provide an answer to this question.
Acknowledgments
We thank V. Lingappa for support and guidance, members of the Vale Lab for critical reading of the manuscript, and C. Zimmerman and A. Calayag for excellent technical assistance. We also thank D. Melton for providing constructs and antibodies to Vg1 and M. Murray for providing antibodies to FRGY2. J.E.W. and R.S.H. were supported by Medical Scientist Training Program Grant GM 07618.
Abbreviations
- β-lac
β-lactamase
- ER
endoplasmic reticulum
- MBSH
modified Barth's saline
- TCE
translational control element
- UTR
untranslated region
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ephrussi A, Dickinson L K, Lehmann R. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 3.Kim-Ha J, Smith J L, Macdonald P M. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann R, Nusslein-Volhard C. Development (Cambridge, UK) 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 5.Melton D A. Nature (London) 1987;328:80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- 6.Bashirullah A, Cooperstock R L, Lipshitz H D. Annu Rev Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- 7.Bergsten S E, Gavis E R. Development (Cambridge, UK) 1999;126:659–669. doi: 10.1242/dev.126.4.659. [DOI] [PubMed] [Google Scholar]
- 8.Rongo C, Gavis E R, Lehmann R. Development (Cambridge, UK) 1995;121:2737–2746. doi: 10.1242/dev.121.9.2737. [DOI] [PubMed] [Google Scholar]
- 9.Gavis E R, Lehmann R. Nature (London) 1994;369:315–318. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- 10.Kim-Ha J, Kerr K, Macdonald P M. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 11.Gavis E R, Curtis D, Lehmann R. Dev Biol. 1996;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- 12.Gavis E R, Lunsford L, Bergsten S E, Lehmann R. Development (Cambridge, UK) 1996;122:2791–2800. doi: 10.1242/dev.122.9.2791. [DOI] [PubMed] [Google Scholar]
- 13.Kim-Ha J, Webster P J, Smith J L, Macdonald P M. Development (Cambridge, UK) 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- 14.Dale L, Matthews G, Tabe L, Colman A. EMBO J. 1989;8:1057–1065. doi: 10.1002/j.1460-2075.1989.tb03473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebagliati M R, Weeks D L, Harvey R P, Melton D A. Cell. 1985;42:769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- 16.Tannahill D, Melton D A. Development (Cambridge, UK) 1989;106:775–785. doi: 10.1242/dev.106.4.775. [DOI] [PubMed] [Google Scholar]
- 17.Yisraeli J K, Melton D A. Nature (London) 1988;336:592–595. doi: 10.1038/336592a0. [DOI] [PubMed] [Google Scholar]
- 18.Yisraeli J K, Sokol S, Melton D A. Development (Cambridge, UK) 1990;108:289–298. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- 19.Mowry K L, Melton D A. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- 20.Cote C A, Gautreau D, Denegre J M, Kress T L, Terry N A, Mowry K L. Mol Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 21.Deshler J O, Highett M I, Abramson T, Schnapp B J. Curr Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 22.Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz S P, Standart N, Yisraeli J K. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrews D W, Lauffer L, Walter P, Lingappa V R. J Cell Biol. 1989;108:797–810. doi: 10.1083/jcb.108.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heasman J, Holwill S, Wylie C C. Methods Cell Biol. 1991;36:213–230. doi: 10.1016/s0091-679x(08)60279-4. [DOI] [PubMed] [Google Scholar]
- 25.Hegde R S, Lingappa V R. Cell. 1996;85:217–228. doi: 10.1016/s0092-8674(00)81098-3. [DOI] [PubMed] [Google Scholar]
- 26.Smith L D, Xu W L, Varnold R L. Methods Cell Biol. 1991;36:45–60. doi: 10.1016/s0091-679x(08)60272-1. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz S P, Aisenthal L, Elisha Z, Oberman F, Yisraeli J K. Proc Natl Acad Sci USA. 1992;89:11895–11899. doi: 10.1073/pnas.89.24.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binder R, Horowitz J A, Basilion J P, Koeller D M, Klausner R D, Harford J B. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheets M D, Wu M, Wickens M. Nature (London) 1995;374:511–516. doi: 10.1038/374511a0. [DOI] [PubMed] [Google Scholar]
- 30.Ostareck-Lederer A, Ostareck D H, Standart N, Thiele B J. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deshler J O, Highett M I, Schnapp B J. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 32.Saunders C, Cohen R S. Mol Cell. 1999;3:43–54. doi: 10.1016/s1097-2765(00)80173-2. [DOI] [PubMed] [Google Scholar]
- 33.Thio G L, Ray R P, Barcelo G, Schupbach T. Dev Biol. 2000;221:435–446. doi: 10.1006/dbio.2000.9690. [DOI] [PubMed] [Google Scholar]
- 34.Dahanukar A, Walker J A, Wharton R P. Mol Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- 35.Smibert C A, Lie Y S, Shillinglaw W, Henzel W J, Macdonald P M. RNA. 1999;5:1535–1547. doi: 10.1017/s1355838299991392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webster P J, Liang L, Berg C A, Lasko P, Macdonald P M. Genes Dev. 1997;11:2510–2521. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]