Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are sarcomas with poor prognosis and limited treatment options. Evidence for a role of epidermal growth factor receptor (EGFR) and receptor tyrosine kinase erbB2 in MPNSTs led us to systematically study these potential therapeutic targets in a larger tumor panel (n = 37). Multiplex ligation-dependent probe amplification and fluorescence in situ hybridization analysis revealed increased EGFR dosage in 28% of MPNSTs. ERBB2 and three tumor suppressor genes (PTEN [phosphatase and tensin homolog deleted on chromosome 10], CDKN2A [cyclin-dependent kinase inhibitor 2A], and TP53 [tumor protein p53]) were frequently lost or reduced. Reduction of CDKN2A was linked to appearance of metastasis. Comparison of corresponding neurofibromas and MPNSTs revealed an increase in genetic lesions in MPNSTs. No somatic mutations were found within tyrosine-kinase–encoding exons of EGFR and ERBB2. However, at the protein level, expression of EGFR and erbB2 was frequently detected in MPNSTs. EGFR expression was significantly associated with increased EGFR gene dosage. The EGFR ligands transforming growth factor α and EGF were more strongly expressed in MPNSTs than in neurofibromas. The effects of the drugs erlotinib and trastuzumab, which target EGFR and erbB2, were determined on MPNST cell lines. In contrast to trastuzumab, erlotinib mediated dose-dependent inhibition of cell proliferation. EGF-induced EGFR phosphorylation was attenuated by erlotinib. Summarized, our data indicate that EGFR and erbB2 are potential targets in treatment of MPNST patients.
Keywords: EGFR, ERBB2, MPNST, targeted therapy, tumor suppressor gene
Approximately half of malignant peripheral nerve sheath tumors (MPNSTs) develop in the setting of neurofibromatosis type 1 (NF1), a hereditary tumor syndrome with an incidence of 1:3,500.1 NF1-associated MPNSTs generally arise from plexiform neurofibromas (pNFs). Loss of the tumor suppressor gene NF1 constitutes only a first step in tumorigenesis. During the course of malignant progression, further genetic and regulatory alterations, such as mutations in TP53 (tumor protein p53), CDKN2A (cyclin-dependent kinase inhibitor 2A), or PDGFRA (platelet-derived growth factor receptor α) or upregulation of MMP-13 (matrix metalloprotease 13), are acquired.2–6 Furthermore, EGFR (epidermal growth factor receptor) gene amplification and increased transcript levels have been detected.7,8
Currently, treatment options for MPNST patients are still unsatisfactory. Thus, a better knowledge of molecular alterations in MPNSTs is of major importance for therapeutic strategies that aim to target proteins specifically altered in tumor cells (targeted therapy).
Several observations point toward a role of the erbB family, which consists of four members (erbB1–4), in nerve sheath tumors. It has been shown that normal Schwann cells are EGFR (erbB1) negative, whereas neurofibromas and MPNSTs express EGFR.9 Evidence for a causal role of EGFR in nerve sheath tumor formation comes from transgenic mice expressing EGFR in Schwann and other glial cells. These animals developed neurofibromas and occasionally MPNSTs.10 EGFR and receptor tyrosine kinase erbB2 (HER2/Neu) were also expressed in sarcoma cell lines generated from an NF1 mouse model.11 Moreover, Schwann cell tumors from animals exposed to the carcinogen N-ethyl-N-nitrosourea harbor ERBB2 mutations.12,13
EGFR and erbB2 are of special therapeutic interest because drugs targeting these receptors, including erlotinib (Tarceva) and trastuzumab (Herceptin), are already available for cancer treatment. Erlotinib is a low-molecular-weight inhibitor that binds to the kinase domain of EGFR, thereby inhibiting signal transduction. Trastuzumab is a humanized antibody targeting the extracellular domain of erbB2.
Although there is cumulating evidence of a role for EGFR and erbB2 in nerve sheath tumors,14,15 previous studies did not systematically analyze these potential therapeutic targets in larger panels of human MPNSTs. We therefore studied genetic alterations and expression of erbB2 and EGFR in a set of 37 human MPNSTs and four MPNST cell lines. We also examined the effect of trastuzumab and erlotinib treatment on MPNST cell lines.
Materials and Methods
Tumor Tissue, DNA, and RNA Extraction
Tumor samples were collected from University Hospital Eppendorf (Hamburg, Germany), Robert-Rössle-Hospital (Berlin, Germany), Otto-von-Guericke-University (Magdeburg, Germany), and Charité-Universitätsmedizin Berlin (Berlin, Germany). MPNSTs of 37 patients were analyzed for genetic alterations and receptor expression. Four patients also contributed corresponding pNFs. Frozen tissue from eight neurofibromas was used for Western blotting. In addition, DNA and lysates from cell lines S462, ST88-14, NSF-1 (kindly provided by V.M. Riccardi from the Neurofibromatosis Institute, La Crescenta, CA, USA), and low-passage culture 31002 (<8 passages) were examined. The S462 cell line was established from MPNST 24472. Twenty-nine patients were diagnosed with NF1, and one patient with NF2; seven individuals were non-NF1 patients. Following initial diagnosis by local neuropathologists, all tumor samples were reviewed by the same pathologist. Histopathological examination was based on the modified Fédération Nationale des Centres de Lutte Contre le Cancer system.16 Tumor sections were examined histologically prior to extraction of DNA and proteins. Tumor areas were scraped from the slides for subsequent extraction. In cases of frozen tissue, DNA was extracted using Trizol reagent from Invitrogen (Karlsruhe, Germany). DNA extraction from paraffin-embedded material was carried out according to the QIAamp DNA Mini Kit protocol (Qiagen, Hilden, Germany). Adjacent pNF was available for MPNSTs 24626, 24772, 24776, and 24324. DNA from pNF and MPNST areas was separately extracted. Breast cancer sample 31842, which served as control for ERBB2 amplification, was kindly provided by Dr. Konrad Kölble (Department of Pathology, Charité-Universitätsmedizin Berlin). The investigations were carried out with the informed consent of the patients.
Single-Strand Conformational Polymorphism and Sequencing
Electrophoresis of PCR products (ranging from 170 to 270 bp) was performed on polyacrylamide gels. To enhance sensitivity, two different gel and running conditions were applied. Detailed information on primer sequences, amplification, and gel conditions is provided in Table 1. All PCR products showing mobility shifts were confirmed by independent PCRs and compared with PCR products of corresponding normal tissue. Aberrantly migrating bands were excised, and the DNA was extracted. After reamplification, PCR products were sequenced bidirectionally (model 3730; Applied Biosystems, Foster City, CA, USA). MPNST 24776 was analyzed for only ERBB2, and cell line ST88-14 only for EGFR. Sequences were compared to accession numbers AF288738 (EGFR) and AC079199 (ERBB2).
Table 1.
Primer sequences, amplification, and gel conditions
| Primer Name | Primer Sequence (5′–3′) | Product Size | Temp (°C)a |
Condition 1b |
Condition 2b |
||
|---|---|---|---|---|---|---|---|
| Gel | Run | Gel | Run | ||||
| ERBB2 Ex17f | AATCCCTGACCCTGGCTTC | 196 bp | 59.0 | 10% A | 3 W; 15 h | 14% A | 3 W; 18 h |
| ERBB2 Ex17r | CGGGCTGGGAGGACTTCA | ||||||
| ERBB2 Ex18f | ACCCACCACCCCCTCAC | 211 bp | 61.0 | 8% A + 10% Gly | 7 W; 15 h | 14% A | 3 W; 18 h |
| ERBB2 Ex18r | CGACCACACCCCTCCA | ||||||
| ERBB2 Ex19f | CCCACGCTCTTCTCACTCAT | 183 bp | 58.6 | 14% A | 3 W; 18 h | 8% A + 10% Gly | 7 W; 15 h |
| ERBB2 Ex19r | GGGTCCTTCCTGTCCTCCTA | ||||||
| ERBB2 Ex20f | CTCTCAGCGTACCCTTGTCC | 230 bp | 58.6 | 10% A | 3 W; 15 h | 14% A | 3 W; 18 h |
| ERBB2 Ex20r | CAAAGAGCCCAGGTGCATAC | ||||||
| ERBB2 Ex21f | TACATGGGTGCTTCCCATTC | 204 bp | 58.6 | 10% A | 3 W; 15 h | 14% A | 3 W; 18 h |
| ERBB2 Ex21r | TCTGCTCCTTGGTCCTTCAC | ||||||
| ERBB2 Ex22f | TAGCCCATGGGAGAACTCTG | 243 bp | 61.0 | 10% A | 3 W; 15 h | 14% A | 3 W; 18 h |
| ERBB2 Ex22r | AGCTCTCATCCTCCCTCCAG | ||||||
| ERBB2 Ex23f | ACTCCTGACCCTGTCTCTGC | 200 bp | 58.6 | 10% A | 3 W; 15 h | 14% A | 3 W; 18 h |
| ERBB2 Ex23r | AGGCAGCCAGCACAGCTC | ||||||
| ERBB2 Ex24f | ATGCTGACCTCCCTCCTG | 170 bp | 63.0 | 14% A + 5% Gly | 6 W; 18 h | 10% A | 3 W, 15 h |
| ERBB2 Ex24r | GAGGGTGCTCTTAGCCACAG | ||||||
| EGFR Ex18f | CATGGTGAGGGCTGAGGTGA | 202 bp | 61.0 | 14% A | 3 W, 18 h | 8% A + 10% Gly | 7 W; 15 h |
| EGFR Ex18r | AGCCCAGAGGCCTGTGCCA | ||||||
| EGFR Ex19f | CACAATTGCCAGTTAACGTC | 189 bp | 54.0 | 8% A + 10% Gly | 7 W; 15 h | 14% A | 3 W; 18 h |
| EGFR Ex19r | GCCTGAGGTTCAGAGCCAT | ||||||
| EGFR Ex20f | CTTCTGGCCACCATGCGAA | 270 bp | 56.0 | 14% A + 5% Gly | 6 W; 20 h | 14% A | 3 W, 18 h |
| EGFR Ex20r | ATCTCCCCTCCCCGTATCT | ||||||
| EGFR Ex21f | ATGATGATCTGTCCCTCACAG | 222 bp | 58.0 | 14% A | 3 W; 18 h | 18% A + 10% Gly | 7 W; 15 h |
| EGFR Ex21r | TGGCTGACCTAAAGCCACCT | ||||||
| EGFR Ex22f | TAGGTCCAGAGTGAGTTAAC | 205 bp | 57.6 | 10% A | 3 W; 15 h | 14% A | 3 W; 18 h |
| EGFR Ex22r | AGCCAGCTTGGCCTCAGTAC | ||||||
| EGFR Ex23f | GTTCATTCATGATCCCACTGCC | 221 bp | 60.9 | 10% A | 3 W; 15 h | 14% A | 3 W; 18 h |
| EGFR Ex23r | AGTGTGGACAGACCCACCAG | ||||||
| EGFR Ex24f | CAATGCCATCTTTATCATTTC | 191 bp | 54.8 | 10% A | 3 W; 15 h | 14% A | 3 W; 18 h |
| EGFR Ex24r | CAATGGAAGCACAGACTGC | ||||||
Abbreviations: ERBB2, receptor tyrosine-protein kinase; Ex, exon; f, forward; r, reverse; EGFR, epidermal growth factor receptor.
Primer annealing temperature.
Gel and running condition of single-strand conformational polymorphism analysis: A, acrylamide; Gly, glycerin.
Multiplex Ligation-Dependent Probe Amplification Performance and Analysis
The SALSA P105 Oligodendroglioma-2 kit (Vs. 03, lot 0804) containing 9 PTEN probes, 5 CDKN2A probes, 8 TP53 probes, 3 EGFR probes, 2 ERBB2 probes, and 16 control probes was employed (MRC Holland, Amsterdam, the Netherlands). Five control probes were omitted from the panel of probes used for internal normalization because of their localization to chromosomal segments that are frequently altered in MPNSTs (11q23.3, 3p22, 16q24.3, 22q11.21, and 7q31.2).
Samples suspected of harboring EGFR amplification were also analyzed with the new version of the P105 kit (Vs. 04, lot 0306), which contains 11 EGFR probes, to detect possible EGFRvIII mutants lacking exons 2–7 that are frequently found in glioblastomas (GBMs). We used 150 μg of template DNA for hybridization and performed PCR reactions in a volume of 25 μl for 30 cycles. The PCR products were analyzed on a semiautomated sequencer (ABI377, Applied Biosystems). We mixed 1 μl of the PCR product with 4 μl loading buffer and heat denatured it. We loaded 0.5 μl on acrylamide gels. The GeneScan 500 TAMRA size Standard (Applied Biosystems) served as molecular weight marker.
Samples were normalized by dividing each peak area by the combined peak areas of all peaks in a lane. This procedure generates proportions of individual peaks of the total peak area (relative peak value). The mean value of the 11 internal control probes was calculated, and all relative peak values were divided by the mean value of the controls. Tumor values were then divided by the values obtained from normal control DNA to calculate a ratio. Mean values of the probes binding a distinct gene were calculated. Samples exceeding twice the standard deviation of normal DNA were scored as genetically altered. Values > 1.9 were scored as gene amplification. Values between 1.9 and 1.31 were interpreted as increased gene dosage (borderline). Values between 1.3 and 0.8 were scored as normal gene dosage. Recently, thresholds of 1.2 and 0.8 were suggested for gains and losses with this kit.17 We interpreted values between 0.79 and 0.4 as reduced gene dosage, caused by loss of one allele. Values below 0.4 were interpreted as loss of both alleles. GBM 6236 and the breast cancer sample 31842 with known gene amplification served as positive controls for EGFR (31 copies) and ERBB2 (4 copies) amplification, respectively. Interpretation was sometimes difficult because probes that localized to different regions of a gene produced nonuniform results. If two or more probes produced signals below 0.8, we scored the gene dosage to be reduced. Few cases showed reduced gene dosage according to single probes. The results were accepted if they were reproducible and if adjacent probes yielded normal gene dosage.
Fluorescence In Situ Hybridization
Slices 3–4 μm thick from paraffin material and cytospins, fixed in 3:1 methanol:acetic acid, were used for fluorescence in situ hybridization (FISH). Samples were incubated according to the manufacturer’s recommendations in pretreatment solution (Abbott, Ludwigshafen, Germany) and then in protease or pepsin. The LSI EGFR SpectrumOrange/CEP 7 SpectrumGreen probes (Abbott) were used for overnight hybridization in a HYBrite denaturation/hybridization system (Abbott) according to the manufacturer’s protocol. Slides were slightly counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and analyzed using a fluorescence microscope with appropriate filters (Leica, Wetzlar, Germany). For each FISH analysis, 60 tumor cells were analyzed. Green and red signals were counted for each cell. Amplification was noted if the ratio of target signals to CEP 7 probe signals was ≥2 or if clusters (i.e., more than 15 or innumerable target signals) were seen.
Reverse Transcriptase PCR Analysis of EGFR, EGF, and Transforming Growth Factor α Gene TGFA
Real-time PCR was performed as reported previously.5 EGFR primers 5′-ATGCCCG-CATTAGCTCTTAG-3′ and 5′-GCAACTTCCCAAAATGTGCC-3′ generated a product of 98 bp. Desmin primers 5′-ACTCCCAGC-CCCTGGTATAG-3′ and 5′-AGGGTAAGGAGCCCA-GACAG-3′ served as reference and generated a product of 180 bp. Conventional reverse transcriptase (RT)-PCR was performed for the EGFR ligands using EGF primers (5′-CCTGCCTAGTCTGCGTCTTT-3′ and 5′-CACAATACCCAGAGCGAACA-3′) and transforming growth factor α (TGFA) primers (5′-GGATTGACA-CAGAAGGAACCA-3′ and 5′-GCCTTGACCCAT-TCAGAAAA-3′) in a volume of 12.5 μl. Optimal PCR cycles for semiquantitative analysis were determined to be 36 cycles for EGF and TGFA (155 bp and 150 bp) and 32 cycles for the reference gene RPS3 (205 bp). PCR fragments (5 μl) were separated on 2% agarose gels.
Western Blotting
Tissues and cell cultures were homogenized in ice-cold lysis buffer (1% Triton X-100, 100 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA) containing protease and phosphatase inhibitor cocktail. Homogenization was enhanced by sonification. Tumor lysates were heat denaturated and loaded onto 7.5% acrylamide gels for subsequent protein separation. MagicMark XP from Invitrogen was applied as a size standard. After transfer of proteins to nitrocellulose membranes (Invitrogen), the membranes were blocked in 3% nonfat dry milk with 0.05% Tween/Tris-buffered saline for 1 h and incubated overnight at 4°C with antibodies to cellular-erbB2 (diluted 1:600; A0485), EGFR (diluted 1:200; sc-03), phosphorylated EGFR (diluted 1:200; sc-12351), and phosphorylated tyrosine residues (diluted 1:10,000; sc-7020). All antibodies were purchased from Santa Cruz Biotechnology (Heidelberg, Germany), with the exception of c-erbB2 antibody (DakoCytomation, Hamburg, Germany). After washing, the membranes were incubated for 1 h with a secondary peroxidase-labeled antibody. Visualization was performed with enhanced chemiluminescence (ECL 1&2 Substrate or Advanced ECL 1&2 Substrate; Amersham Biosciences, Freiburg, Germany). Sensitivity of erbB2 detection was enhanced by biotin-conjugated second antibodies, followed by 1 h incubation with a 1:2,000 dilution of ExtrAvidin from Sigma (Munich, Germany). Rehybridization of membranes was performed with anti-β-actin antibody (diluted 1:6,000; AC-15) from Sigma.
Immunohistochemistry and Scoring
Detection of EGFR was performed with the Ventana Benchmark system (Ventana, Strasbourg, France). EGFR antibody (diluted 1:100; M3563) was obtained from DakoCytomation. Antigen retrieval was achieved by pretreatment of the tissue slices for 8 min with pronase. Visualization was performed with diaminobenzidine. Expression of erbB2 was analyzed by immunofluorescence using an erbB2 antibody (diluted 1:50; A0485 from DakoCytomation) and a second Cy3-conjugated antibody. Nuclei were counterstained with DAPI I from Abbott, and antigen retrieval was enhanced by heating. Negative controls without primary antibodies were carried out and did not produce signals. As positive controls, we used skin for EGFR expression and a breast cancer metastasis for erbB2 expression. Scoring was performed according to the percentage of positive cells: <5% was classified as negative (–), and 6%–100% was classified as positive: 6%–30% of positive cells were scored with +, 31%–60% with ++, and >60% with +++. A blinded repeated test produced similar results.
Cell Culture Assays
MPNST cell lines S462, ST88-14, NSF-1, and low-passage culture 31002 were maintained in Dulbecco’s modified Eagle’s medium (DMEM) plus Glutamax-I (1,000 mg/l glucose; Invitrogen) containing 10% fetal bovine serum (FBS) and 5 μg/ml gentamycin. During the drug assays, cells were maintained in DMEM containing 5% FBS. We seeded 3 × 103 cells (4 × 103 for ST88-14) in 300 μl medium into 24-well plates and allowed them to adhere overnight. Erlotinib and trastuzumab (kindly provided by Genentech, San Francisco, CA, USA) and imatinib (kindly provided by Novartis Pharma AG, Basel, Switzerland) were added in 100 μl medium to obtain the indicated concentrations. Negative controls contained vehicle only. We exchanged 300 μl medium containing respective drug concentrations on days 3 and 5. Cell proliferation was evaluated on days 4 and 7 posttreatment with the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Mannheim, Germany). The experiments were performed in duplicate and repeated three times. The fractional product concept was used to determine whether drug combinations yielded additive or synergistic effects. Erlotinib effects on EGFR phosphorylation were determined in cell culture dishes (diameter, 10 cm). Semiconfluent cells were serum starved for 24 h. Erlotinib was added to a final concentration of 5 μM and incubated for 1 h. Cells were then stimulated with EGF (100–50 ng/ml) for 10 min at 37°C, washed with phosphate-buffered saline, scraped, centrifuged, and resuspended in 70 μl lysis buffer.
Statistical Methods
SPSS version 12.0 (SPSS, Chicago, IL, USA) was used for statistical analysis. Survival rates were determined using the Kaplan-Meier method and the log rank test. Association of parameters was assessed with the Pearson correlation and Fisher exact test. A p-value of <0.05 was considered significant.
Results
Genetic Alterations of EGFR, ERBB2, and Tumor Suppressor Genes
Tyrosine-kinase–encoding domains (exon 18–24) of EGFR and ERBB2 were screened for sequence alterations by single-strand conformational polymorphism in samples from 34 patients (37 MPNSTs, 4 corresponding pNFs, and MPNST cell lines S462 and ST88-14). In addition, ERBB2 exon 17, encoding the transmembrane domain, was analyzed because point mutations have been described in this region in peripheral nerve sheath tumors of domesticated animals.18 Somatic mutations were not found, but polymorphisms were detected in exons 20, 21, and 23 of EGFR and in exon 17 of ERBB2 (Table 2). All EGFR variants were silent. The ERBB2 variant in codon 655 results in an exchange from isoleucine to valine. Comparison with the National Center for Biotechnology Information’s dbSNP database (www.ncbi.nlm.nih.gov/SNP) revealed no significant differences in polymorphism frequency in our series compared to that in the normal population (data not shown).
Table 2.
Genetic alterations and expression of epidermal growth factor receptor (EGFR) and receptor tyrosine-protein kinase (erbB2) in malignant peripheral nerve sheath tumors (MPNSTs) and MPNST cell lines
| MPNST ID | NF1a | Gradeb | EGFRampc,d | EGFR IHC/WBe | ERBB2ampc,d | erbB2 IHC/WBe | PTENd | CDKN2Ad | TP53d |
|---|---|---|---|---|---|---|---|---|---|
| 24256 | Y | 3 | 4.5 amp | + | R | +++ | R | L | n |
| 24326 | Y | 2 | nf | − | NA | +++ | |||
| 24626 | Y | 2 | R | − | n | +++ | R | L | n |
| 24534 | Y | 3 | nf | − | NA | ++ | |||
| 24668 | Y | 3 | nf | − | − | + | |||
| 24670 | Y | 3 | n | − | n | + | R | L | R |
| 24748 | Y | 2 | n | − | n | +++ | L | R | R |
| 24772 | Y | 2 | 1.36 amp | ++ | R | +++ | L | L | n |
| 24776 | Y | 1 | nf | − | NA | − | |||
| 24472g | Y | 3 | n | +/WB++ | R | −/WB++ | n | n | R |
| 24480 | Y | 2 | n | + | n | − | R | n | n |
| 24484 | Y | 3 | n | NA | n | ++ | n | n | n |
| 24476 | Y | 2 | n | − | n | − | n | n | n |
| 24784 | Y | 1 | n | −/WB++ | n | ++/WB++ | n | n | n |
| 21914 | Y | 2 | 7.5 amp exon 1 | −/WB+++ truncated | R | − | R | L | R |
| 21852 | Y | 2 | 1.45 amp | WB+ | n | WB+ | R | n | R |
| 24308 | Y | 3 | n | NA | R | NA | R | R | n |
| 24310 | Y | 2 | n | NA | n | NA | L | R | n |
| 24324 | Y | 1 | 6.9 amp | +++ | R | + | n | L | R |
| 24332 | Y | 2 | nf | − | NA | ++ | |||
| 24354 | Y | 1 | nf | − | NA | + | |||
| 24694 | Y | 2 | n | − | n | +++ | R | n | n |
| 26592 | Y | 2 | nf | − | NA | +++ | |||
| 28650 | Y | 2 | n | − | n | +++ | n | n | n |
| 28652 | Y | 1 | n | − | n | +++ | n | n | n |
| 27724 | Y | 3 | nf | − | NA | +++ | |||
| 26580 | N | 3 | n | NA | R | NA | n | L | R |
| 26582 | N | 3 | n | − | R | +++ | R | n | R |
| 26584 | N | 2 | R | (+) | n | ++ | Amp/R | L | L |
| 26586 | N | 2 | nf | − | NA | +++ | |||
| 26588 | N | 3 | 1.38 amp | − | n | NA | R | R | R |
| 168 | NF2 | NA | n | NA | n | NA | n | R | n |
| 524 | N | NA | 1.8 amp | NA | n | NA | R | R | n |
| 5050 | N | NA | R | NA | 1.33 amp | NA | n | n | n |
| 29250 | Y | 3 | n | WB+ | n | WB++ | n | L | n |
| 31472 | Y | 3 | 1.42 amp | +++ | n | ++ | 1.43 amp | L | n |
| 31474 | Y | 3 | n | +++ | n | ++ | n | L | R |
| S462g | Y | — | 1.47 amp | WB+++ | R | WB++ | n | n | R |
| ST88–14 | Y | — | n | WB+++ | R | WB+ | R | n | R |
| 31002 | N | — | 1.48 amp | WB++ | n | WB++ | R | n | n |
| NFS-1 | Y | — | 1.76 amp | WB+++ | n | WB+++ | R | L | n |
Abbreviations: PTEN, phosphatase and tensin homolog deleted on chromosome 10; CDKN2A, cyclin-dependent kinase inhibitor 2A; TP53, tumor protein p53; NA, not assessed (lack of material). Cell lines are listed in the lower part of the table.
Neurofibromatosis type 1 status of the patient: N, no; Y, yes; NF2, neurofibromatosis type 2.
Tumor grade according to the modified Fédération Nationale des Centres de Lutte Contre le Cancer system.
Amplification status (amp) according to multiplex ligation-dependent probe amplification is indicated by fold increase relative to the normal gene dose. ErbB2, receptor tyrosine-protein kinase.
R, monoallelic loss; L, biallelic loss; n, normal gene dose.
Immunohistochemistry (IHC)/Western blot quantification (WB): strong (+++), medium (++), weak (+).
Samples analyzed for EGFR dosage by real-time PCR.
Samples belonging to the same patient.
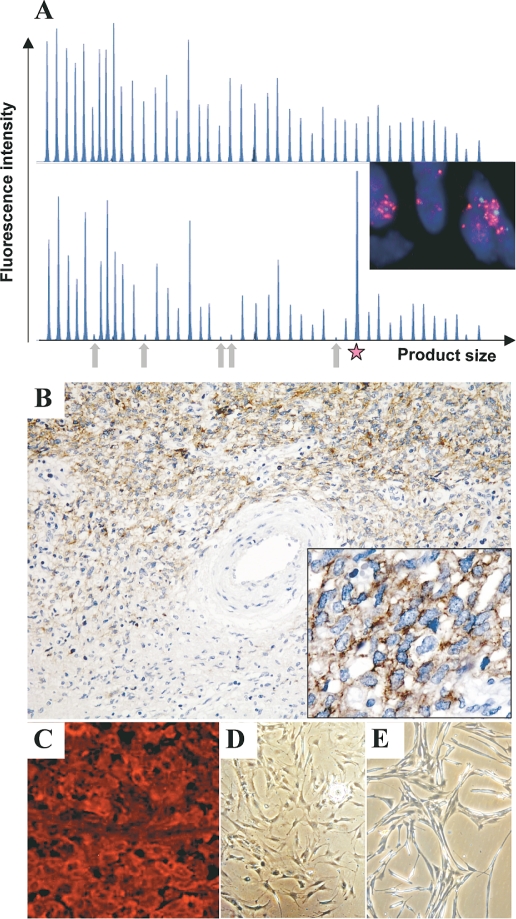
DNA from 31 patients was available for gene dosage analysis by multiplex ligation-dependent probe amplification (MLPA; Table 3). Initial analysis of EGFR dosage was performed by real-time PCR and produced results similar to those from MLPA. For nine samples, indicated in Table 3, only PCR data are available. Increased EGFR dosage was observed in 28% of MPNSTs. Most of these samples yielded borderline values between 1.3 and 1.9. However, three MPNSTs harbored EGFR gene dosages > 4.0. EGFR amplification in MPNST 21914 was repeatedly shown to be restricted to exon 1 (Fig. 1A). ERBB2 gene dosage was reduced in 32% of MPNSTs. Reduction in gene dosage was also observed for TP53 (39%) and for PTEN and CDKN2A (58% each). The PTEN pattern of MPNST 26584 was remarkable (and reproducible) because two probes recognizing exon 1 yielded values of 0.5, whereas probes binding to exons 2–9 generated values of 2.0.
Table 3.
Allelic variants of epidermal growth factor receptor (EGFR) and receptor tyrosine-protein kinase (erbB2) in malignant peripheral nerve sheath tumor patients
| Gene | Codon | Triplet | Amino acid | Allele frequency (f) |
|---|---|---|---|---|
| EGFR exon 20 | 787 | CAG>CAA | Silent | A787 = 0.63 |
| G787 = 0.37 | ||||
| EGFR exon 21 | 836 | CGC>CGT | Silent | C836 = 0.97 |
| T836 = 0.03 | ||||
| EGFR exon 23 | 903 | ACC>ACT | Silent | C903 = 0.80 |
| T903 = 0.20 | ||||
| ERBB2 exon 17 | 655 | ATC>GTC | I >V | Ile655 = 0.74 |
| Val655 = 0.26 |
Fig. 1.
Genetic alterations, receptor expression, and morphology of malignant peripheral nerve sheath tumors (MPNSTs) and MPNST cell cultures. (A) Electropherograms depicting gene dosage of MPNST 21914 (lower panel) and corresponding plexiform neurofibroma (pNF) 21912 (upper panel) generated with the SALSA P105 Oligodendroglioma-2 multiplex ligation-dependent probe amplification kit (Vs. 04). The arrows indicate signal reduction of the five CDKN2A probes in the MPNSTs. The star marks the increased signal of the EGFR exon 1 probe (inset). Fluorescence in situ hybridization analysis demonstrates cluster amplification of epidermal growth factor receptor gene EGFR (red signals; centromer: green signals). (B) Immunohistochemistry of EGFR on MPNST 24324. Note the strong EGFR expression in the cellular MPNSTs compared to the area with differentiation corresponding to pNF. Original magnification: ×200; right corner, ×400. (C) Receptor tyrosine-protein kinase erbB2 immunofluorescence of MPNST 24626. Original magnification: ×400. (D and E) Morphology of low-passage MPNST culture 31002 and MPNST cell line NSF-1, respectively.
Benign precursor pNFs were available from three patients and were compared to corresponding MPNSTs (Table 4). MPNST 21914 exhibited multiple genetic alterations, whereas the corresponding pNF had normal gene dosage within the analyzed genes (Fig. 1A). EGFR gene dosage was increased in MPNST 24324 by a factor of 6.9, whereas the corresponding pNF 28580 showed an increase by a factor of 2.2 relative to the normal dosage. MPNST 24626 had stronger alterations in, for example, CDKN2A than in two corresponding pNFs. pNF 24624 was pure neurofibroma, whereas pNF 28578 was scraped from a slice also containing MPNSTs. One patient contributed primary MPNST 21852 and corresponding relapse 22318 (resected 5 months later). The relapse had acquired a reduction of ERBB2 and CDKN2A.
Table 4.
Multiplex ligation-dependent probe amplification analysis on tumor pairs
| Tumor ID | EGFR | ERBB2 | PTEN | CDKN2A | TP53 |
|---|---|---|---|---|---|
| 21914 MPNST | 7.5 amp | R | R | L | R |
| 21912 pNF | n | n | n | n | n |
| 24324 MPNST | 6.9 amp (7.6)a | R | n | L | R |
| 28580 pNF | NA (2.2)a | NA | NA | NA | NA |
| 24626 MPNST | R | n | R | L | n |
| 28578 pNF I | n | n | R | R | n |
| 24624 pNF II | n | n | R | n | n |
| 21852 MPNST | 1.5 amp | n | R | n | R |
| 22138 relapse | 1.8 amp (2.4)a | R | R | R | R |
Abbreviations: EGFR, epidermal growth factor receptor; erbB2, receptor tyrosine-protein kinase; PTEN, phosphatase and tensin homolog deleted on chromosome 10; CDKN2A, cyclin-dependent kinase inhibitor 2A; TP53, tumor protein p53; MPNST, malignant peripheral nerve sheath tumor; amp, amplification status; R, monoallelic loss; L, biallelic loss; pNF, plexiform neurofibroma; n, normal gene dose; NA, not assessed.
EGFR dosage determined by real-time PCR.
In order to verify MLPA data with an independent method we applied FISH in selected cases with increased EGFR values. FISH analysis revealed chromosome 7 polysomy in all samples. Moreover, MPNSTs 21914 and 24256 also harbored EGFR gene amplification in most cells (Table 5 and Fig. 1A). MPNST 21852 and the corresponding recurrent MPNST 22318 had borderline amplifications (1.6 and 2.0) that matched well with MLPA data (1.5 and 1.8).
Table 5.
Fluorescence in situ hybridization analysis of malignant peripheral nerve sheath tumors with increased epidermal growth factor receptor (EGFR) dosage
| Tumor ID | % Ampa | % Polysomyb | Ratioc |
|---|---|---|---|
| 21914 | 90 | 40 | Clusterd |
| 21852 | <10 | 60 | 1.6 |
| 22138 | 50 | 55 | 2.0 |
| 24772 | <10 | 15 | 1.3 |
| S462 cells | 0 | 90 | 1.0 |
| 24256 | 80 | 30 | Clusterd |
Gene amplification was accepted when target gene/centromer was ≥2 and is given as percentage of cells with gene amplification.
Polysomy was accepted when centromer signals were three or more per nucleus and is given as percentage of cells with polysomy.
Ratio: EGFR signals/centromer signals.
Cluster indicates ≥15 signals per target and nucleus.
Expression of EGFR, erbB2, and EGFR Ligands
EGFR and erbB2 expression was determined by immunohistochemistry in MPNSTs from 28 patients (Table 2; Fig. 1, B and C). EGFR was detected in 29% (8 of 28) and erbB2 in 82% (23 of 28) of MPNSTs. Stronger EGFR expression in MPNST 24324 than in adjacent pNF (Fig. 1B) corresponds well with underlying EGFR amplification in these tumors (Table 4).
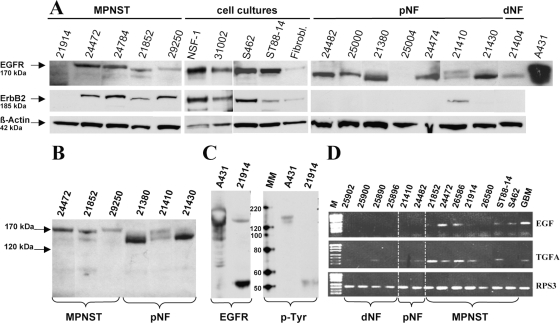
Western blot detected EGFR in all five MPNSTs, four MPNST cell cultures, and seven of eight neurofibromas (Fig. 2A). Interestingly, different EGFR isoforms were detected in MPNSTs (170 kDa) and in neurofibromas (150 kDa), as depicted in Fig. 2, A and B. MPNST 21914 was exceptional because the major signal was detected at 50 kDa (Fig. 2C). To determine whether this small EGFR isoform was active, we examined phosphorylation of tyrosine residues. EGF-stimulated A431 carcinoma cells served as positive control. Phosphorylated EGFR was detected at 170 kDa in A431 lysate and at 50 kDa in MPNST 21914. ErbB2 was present in four of five MPNSTs and all MPNST cultures but in only one of eight neurofibromas (Fig. 2A). The signals were detected at the expected size of 185 kDa. For comparison, EGFR and erbB2 were also examined in nontumorous cells (dermal fibroblasts), which showed weak expression of both receptors.
Fig. 2.
Detection of epidermal growth factor receptor (EGFR), EGFR ligands, and receptor tyrosine-protein kinase (erbB2). (A) Western blot showing EGFR and erbB2 expression in malignant peripheral nerve sheath tumors (MPNSTs), MPNST cell lines, fibroblasts (Fibrobl.), and neurofibromas (NF). Lysate of EGF-stimulated A431 carcinoma cells served as EGFR positive control. Detection of β-actin demonstrates equal loading. (B) Different EGFR isoforms in MPNSTs and plexiform neurofibromas (pNFs) separated on the same gel. (C) Detection of phosphorylated proteins using the p-Tyr antibody specific for phosphotyrosine residues. Two sets of A431 and 21914 lysates were run on the same gel and blotted. After cutting the membrane one set was incubated with an EGFR antibody, the other set was incubated with p-Tyr. MM, MagicMark XP size standard. (D) Expression of EGFR ligands EGF and transforming growth factor α (TGFA) was determined by reverse transcriptase PCR in neurofibromas, MPNSTs, and MPNST cell lines ST88-14 and S462. RPS3 served as housekeeping gene. Abbreviations: M, size standard PUC 19 DNA/MspI; GBM, glioblastoma cell line DBTRG-05 MG, serving as positive control; dNF, dermal neurofibromas.
EGFR ligand expression (EGF, TGFA) was performed with RT-PCR on five MPNSTs, two MPNST cell lines, and six neurofibromas (Fig. 2D). EGF was absent in neurofibromas and expressed in two of five MPNSTs and in both MPNST cell lines. Weak TGFA expression was detected in three of six neurofibromas. By contrast, MPNSTs showed stronger expression, with the exception of MPNST 26580 and cell line S462, which were negative.
Effect of Receptor Tyrosine Kinase Inhibitors on MPNST Cells
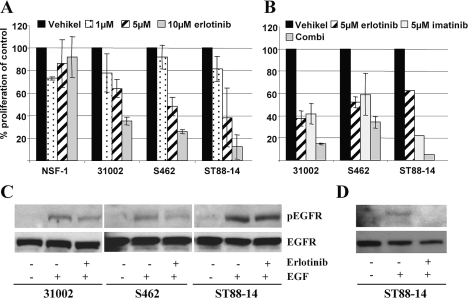
The effects of erlotinib and trastuzumab on MPNST cells were examined. Trastuzumab, tested in a concentration range of 10–100 μg/ml on S462 and ST88-14 cells, yielded a 20%–30% reduction of proliferation on S462 cells with concentrations of 10 μg/ml and 50 μg/ml (data not shown). A concentration of 100 μg/ml resulted in 20% inhibition, demonstrating that a dose-dependent effect was not achieved. ST88-14 cells were not affected by trastuzumab treatment.
Erlotinib concentrations of 1, 5, and 10 μM were tested on MPNST cell lines S462, ST88-14, NSF-1, and low-passage culture 31002 (Fig. 3A). Morphology of low-passage culture 31002 and cell line NSF-1 is shown in Fig. 1, D and E. Proliferation was measured on days 4 and 7 posttreatment. Effect of incubation times on cell growth was negligible (data not shown). The erlotinib concentration inhibiting cell growth by 50% (IC50) on day 7 posttreatment was 7 μM for 31002, 5 μM for S462, and 4 μM for ST88-14. NSF-1 cells were not affected by erlotinib treatment. The fractional product concept was applied to determine the effect of drug combination. This method can be used for drugs that act mutually nonexclusively. Fractions of unaffected, proliferating cells treated with single drugs were multiplied. The result is the calculated effect for additive-acting drugs. Experimental data were compared to the calculated value. If experimental and calculated data match, the effect is additive. Smaller experimental values indicate a synergistic effect, whereas larger values indicate an antagonistic mode of action.
Fig. 3.
Effect of erlotinib and imatinib on proliferation and epidermal growth factor receptor (EGFR) phosphorylation. (A) Effect of different erlotinib concentrations on cell proliferation in four malignant peripheral nerve sheath tumor lines after 7 days of incubation. (B) Single and combination (Combi) treatment with 5 μM of erlotinib and imatinib. Error bars represent the SEM of three independent experiments. ST88-14 in B was tested once in duplicate. (C) Effect of erlotinib on EGF-induced EGFR phosphorylation. The cells were cultivated 24 h without serum and then treated with vehicle, EGF (100 ng/ml), or erlotinib (5 μM) plus EGF. (D) Cells were stimulated with 50 ng/ml EGF. The phosphorylated form of EGFR (pEGFR) was detected by a phosphospecific EGFR antibody. Rehybridization of the membranes with an EGFR antibody shows equal loading.
Combinations of erlotinib and imatinib (5 μM each) approximated an additive effect on S462 and 31002 cells (Fig. 3B). The effect on ST88-14 appeared to be stronger than additive. While erlotinib and imatinib had similar effects on S462 and 31002 cells, imatinib was superior on ST88-14. To determine if growth inhibition of erlotinib is actually mediated by inhibition of EGFR signaling, the effect of 5 μM erlotinib on EGF-induced EGFR phosphorylation was determined. Reduction of EGFR phosphorylation was achieved in 31002 and S462 but not in ST88-14 cells (Fig. 3C). To clarify whether stimulation with 100 ng/μl EGF may override the effect of erlotinib, we tested 50 ng/μl EGF. Under these conditions, erlotinib inhibited EGFR phosphorylation (Fig. 3D).
Statistical Analysis
Molecular characteristics of the tumors as determined by MLPA, immunohistochemistry, and Western blotting were compared with each other and with clinical information of MPNST patients. Presence of metastasis was associated with reduced CDKN2A (p = 0.054, Fisher exact test; n = 22). Survival analysis was close to significance (p = 0.09, log rank). Notably, all patients with metastases (n = 5) had reduced CDKN2A. EGFR status and EGFR protein expression were significantly associated (p = 0.026, Fisher exact test; n = 26). When the four different staining and gene status levels were taken into account, the correlation was even more significant (p = 0.016, Pearson correlation). Tumor grade was linked to PTEN gene dosage (p = 0.017, Fisher exact test; n = 24). None of the grade 1 MPNSTs had affected PTEN, whereas 82% of grade 2 and 40% of grade 3 MPNSTs had reduced PTEN.
Discussion
We detected gene dosage alterations in the oncogenes EGFR and ERBB2 and in the tumor suppressor genes PTEN, CDKN2A, and TP53. Increased EGFR dosage was present in 28% of MPNSTs and resulted from chromosome 7 polysomy sometimes combined with EGFR amplification. Gain of chromosome 7 in MPNSTs has been reported previously.7,19,20 Most cell lines (three of four) showed slightly elevated EGFR values. Enrichment of cells with EGFR amplifications under culture conditions is possible but appears unlikely since EGFR amplifications have been reported to disappear under culture conditions.21
The frequent reduction of ERBB2 dosage may be explained by its colocalization with the NF1 gene on the long arm of chromosome 17. Frequent loss of chromosome 17 in MPNSTs has been shown.19 TP53, localizing to the short arm of chromosome 17, showed reduced gene dosage in 40% of MPNSTs (3% corresponding to biallelic inactivation) and might also be caused by chromosome 17 loss. However, 9 of 31 MPNSTs showed alterations in either ERBB2 or TP53, indicating that their loss is not always combined. Reduced CDKN2A dosage in 57% of MPNSTs (37% with values corresponding to homozygous deletions) agrees with a previous study showing CDKN2A deletions in 75% of MPNSTs (45% with homozygous deletions).7 Similar results were found by other groups.4,22 Here, we provide evidence for an association of reduced CDKN2A with metastasis. However, these results need confirmation with larger patient numbers. Nevertheless, the association of CDKN2A alteration and disease progression, including metastasis, has been reported for other cancers.23,24 We show for the first time reduced PTEN dosage, which was present in 57% of MPNSTs (10% with values corresponding to homozygous deletions). PTEN reduction corresponded mainly to monoallelic loss. It remains to be clarified whether the remaining allele is also inactivated. Up to now, only one study determined PTEN mutations in 12 MPNSTs but did not detect any.25 However, epigenetic regulation of PTEN has been reported for different tumors26–28 and could also take place in MPNSTs. In fact, our preliminary data point toward PTEN promoter methylation in the majority of MPNSTs but not in neurofibromas. A detailed analysis of PTEN alterations is currently being performed.
Further, PTEN haploinsufficiency may be sufficient to promote tumor progression.29 PTEN controls the Akt/mTOR (mammalian target of rapamycin) pathway by antagonizing phosphoinositide 3-kinase. MPNST cell lines were found to be sensitive to the mTOR inhibitor rapamycin.30 It is possible that PTEN loss contributes to the activation of this pathway. Therefore, it might be of interest to test whether MPNSTs with affected PTEN respond better to mTOR inhibitors than do those with unaffected PTEN.
EGFR and erbB2 were frequently expressed in MPNSTs, as shown by immunohistochemistry and Western blotting. ErbB2, detected in the vast majority of MPNSTs, was rarely expressed in neurofibromas, thereby pointing toward a progression-associated event. The proliferation-associated expression of erbB2 is also observed physiologically, with erbB2 being expressed in developing Schwann cells and downregulated in adulthood, but reexpressed in proliferating Schwann cells of traumatic neuromas or upon stimulation with growth factors.31,32 Although reduction of ERBB2 dosage was detected in 32% of MPNSTs, most of the affected cases still expressed erbB2. This result may appear contradictory, but haploinsufficiency does not necessarily lead to reduced expression. Given a scenario in which one allele of ERBB2 is accidentally co-deleted with NF1, regulatory processes could account for erbB2 expression, which might be important for tumor cell proliferation. EGFR was detected in MPNSTs and neurofibromas, but the isoforms differed. In MPNSTs, we detected bands at 170 kDa, which correspond to glycosylated EGFR. Neurofibromas mainly showed bands at 150 kDa, possibly representing an unglycosylated isoform of EGFR.33 Proper glycosylation is important for ligand binding, correct folding, and kinase activity.33 Therefore, it is tempting to speculate that glycosylated EGFR in MPNSTs is more potent in transmitting mitogenic signals upon ligand binding than is the unglycosylated form in neurofibromas. EGFR may also be upregulated during disease progression, as shown in Fig. 1B. In general, EGFR expression was associated with increased EGFR dosage, thereby providing evidence for the underlying mechanism. Furthermore, increased transcript levels of EGF and TGFα were detected in MPNSTs compared to neurofibromas. Thus, stronger expression of EGF ligands might contribute to tumor progression.
Our in vitro assays demonstrated a dose-dependent effect of erlotinib but not trastuzumab on MPNST cell proliferation. Trastuzumab is approved for treatment of erbB2-overexpressing breast cancers with underlying ERBB2 amplification. Although the exact mode of trastuzumab action is not fully understood, it is anticipated that trastuzumab is effective only in tumor cells with ERBB2 amplification surpassing a certain erbB2 expression threshold.34 A unique feature of erbB2 is its ability for spontaneous dimerization when overexpressed. Further, antibody-dependent cell-mediated cytotoxicity is thought to contribute to the effect mediated by trastuzumab in vivo.35 Absence of ERBB2 amplification in MPNST cell lines and lack of immune cells in our assay may explain failure of trastuzumab. Erlotinib, targeting the kinase domain of EGFR, inhibited most tested cell lines. NSF-1 cells were not affected, although they express EGFR. Independence from EGFR signaling may be explained by numerous other alterations. Modulating effects of tumor suppressor genes on therapy have been reported.36 Recently, an inhibitory effect of erlotinib on MPNST cell lines S462 and STS26T was reported.37 IC50 values in S462 cells were similar to ours, and an antiangiogenic effect was observed in an STS26T xenograft model. Taken together, IC50 values of our cell culture assays were similar to plasma concentrations in patients38 and suggest that erlotinib may be a candidate for therapeutic use in MPNST patients.
Combination therapies are likely to yield best results in highly malignant tumors with numerous genetic and epigenetic alterations. Therefore, we combined imatinib, previously shown to be effective on S462 cells,5 with erlotinib. ST88-14 cells were more sensitive to combination treatment than were the other cells. A possible explanation is the TP53 wild-type status of ST88-14, whereas S462 cells harbor mutant TP53 (unpublished observation, Nikola Holtkamp). A modulating effect of p53 status on sensitivity to the EGFR inhibitor cetuximab was observed in two hepatocellular cancer cell lines. Despite strong EGFR expression in both cell lines, the one with mutant p53 responded less to cetuximab than the one with wild-type p53.39 Thus, TP53 alterations may be considered for individualized therapies of MPNSTs.
In summary, our results provide a molecular rationale for clinical application of EGFR inhibitors. Further, erbB2 was expressed by most MPNSTs. In contrast to other erbB family members (EGFR, erbB3, erbB4), erbB2 lacks a ligand-binding domain. ErbB2 must therefore form heterodimers with other family members, all of which were detected in MPNSTs,15 to respond to growth factors. Application of pan-erbB inhibitors appears tempting to inhibit this cross talk (heterodimerization). Prior analysis of EGFR and erbB2 expression in tumors will probably help to choose patient subgroups most likely to benefit from treatment with specific inhibitors. Finally, a combination of drugs is likely to be most effective in combating MPNSTs.
Acknowledgments
We thank Petra Matylewski for excellent technical assistance, Horst Skarabis for his advice on statistical evaluation, and Michael Baier for helpful discussion. This work was supported by the U.S. Department of Defense Neurofibromatosis Research Program (NF050145).
References
- 1.Huson SM. Neurofibromatosis 1: a clinical and genetic overview. In: Huson SM, Hughes RAC, editors. The Neurofibromatoses. London: Chapman and Hall Medical; 1994. pp. 160–203. [Google Scholar]
- 2.Menon AG, Anderson KM, Riccardi VM, et al. Chromosome 17p deletions and p53 gene mutations associated with the formation of malignant neurofibrosarcomas in Recklinghausen neurofibromatosis. Proc Natl Acad Sci U S A. 1990;87:5435–5439. doi: 10.1073/pnas.87.14.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legius E, Dierick H, Wu R, et al. TP53 mutations are frequent in malignant NF1 tumors. Genes Chromosomes Cancer. 1994;10:250–255. doi: 10.1002/gcc.2870100405. [DOI] [PubMed] [Google Scholar]
- 4.Kourea HP, Orlow I, Scheithauer BW, Cordon-Cardo C, Woodruff JM. Deletions of the INK4A gene occur in malignant peripheral nerve sheath tumors but not in neurofibromas. Am J Pathol. 1999;155:1855–1860. doi: 10.1016/S0002-9440(10)65504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holtkamp N, Okuducu AF, Mucha J, et al. Mutation and expression of PDGFRA and KIT in malignant peripheral nerve sheath tumors, and its implications for imatinib sensitivity. Carcinogenesis. 2006;27:664–671. doi: 10.1093/carcin/bgi273. [DOI] [PubMed] [Google Scholar]
- 6.Holtkamp N, Mautner V, Friedrich R, et al. Differentially expressed genes in neurofibromatosis 1-associated neurofibromas and malignant peripheral nerve sheath tumors. Acta Neuropathol Berl. 2004;107:159–168. doi: 10.1007/s00401-003-0797-8. [DOI] [PubMed] [Google Scholar]
- 7.Perry A, Kunz SN, Fuller CE, et al. Differential NF1, p16, and EGFR patterns by interphase cytogenetics (FISH) in malignant peripheral nerve sheath tumor (MPNST) and morphologically similar spindle cell neoplasms. J Neuropathol Exp Neurol. 2002;61:702–709. doi: 10.1093/jnen/61.8.702. [DOI] [PubMed] [Google Scholar]
- 8.Holtkamp N, Reuss DE, Atallah I, et al. Subclassification of nerve sheath tumors by gene expression profiling. Brain Pathol. 2004;14:258–264. doi: 10.1111/j.1750-3639.2004.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeClue JE, Heffelfinger S, Benvenuto G, et al. Epidermal growth factor receptor expression in neurofibromatosis type 1-related tumors and NF1 animal models. J Clin Invest. 2000;105:1233–1241. doi: 10.1172/JCI7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling BC, Wu J, Miller SJ, et al. Role for the epidermal growth factor receptor in neurofibromatosis-related peripheral nerve tumorigenesis. Cancer Cell. 2005;7:65–75. doi: 10.1016/j.ccr.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Velasco-Miguel S, Vass WC, Parada LF, DeClue JE. Epidermal growth factor receptor signaling pathways are associated with tumorigenesis in the Nf1:p53 mouse tumor model. Cancer Res. 2002;62:4507–4513. [PubMed] [Google Scholar]
- 12.Kindler-Rohrborn A, Kolsch BU, Fischer C, Held S, Rajewsky MF. Ethylnitrosourea-induced development of malignant schwannomas in the rat: two distinct loci on chromosome of 10 involved in tumor susceptibility and oncogenesis. Cancer Res. 1999;59:1109–1114. [PubMed] [Google Scholar]
- 13.Nakamura T, Ushijima T, Ishizaka Y, et al. Neu proto-oncogene mutation is specific for the neurofibromas in a N-nitroso-N-ethylurea-induced hamster neurofibromatosis model but not for hamster melanomas and human Schwann cell tumors. Cancer Res. 1994;54:976–980. [PubMed] [Google Scholar]
- 14.Carroll SL, Stonecypher MS. Tumor suppressor mutations and growth factor signaling in the pathogenesis of NF1-associated peripheral nerve sheath tumors: II. The role of dysregulated growth factor signaling. J Neuropathol Exp Neurol. 2005;64:1–9. doi: 10.1093/jnen/64.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Stonecypher MS, Byer SJ, Grizzle WE, Carroll SL. Activation of the neuregulin-1/ErbB signaling pathway promotes the proliferation of neoplastic Schwann cells in human malignant peripheral nerve sheath tumors. Oncogene. 2005;24:5589–5605. doi: 10.1038/sj.onc.1208730. [DOI] [PubMed] [Google Scholar]
- 16.Coindre JM, Trojani M, Contesso G, et al. Reproducibility of a histopathologic grading system for adult soft tissue sarcoma. Cancer. 1986;58:306–309. doi: 10.1002/1097-0142(19860715)58:2<306::aid-cncr2820580216>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Jeuken J, Cornelissen S, Boots-Sprenger S, Gijsen S, Wesseling P. Multiplex ligation-dependent probe amplification: a diagnostic tool for simultaneous identification of different genetic markers in glial tumors. J Mol Diagn. 2006;8:433–443. doi: 10.2353/jmoldx.2006.060012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoica G, Tasca SI, Kim HT. Point mutation of neu oncogene in animal peripheral nerve sheath tumors. Vet Pathol. 2001;38:679–688. doi: 10.1354/vp.38-6-679. [DOI] [PubMed] [Google Scholar]
- 19.Bridge RS, Jr, Bridge JA, Neff JR, Naumann S, Althof P, Bruch LA. Recurrent chromosomal imbalances and structurally abnormal breakpoints within complex karyotypes of malignant peripheral nerve sheath tumour and malignant triton tumour: a cytogenetic and molecular cytogenetic study. J Clin Pathol. 2004;57:1172–1178. doi: 10.1136/jcp.2004.019026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plaat BE, Molenaar WM, Mastik MF, Hoekstra HJ, te Meerman GJ, van den Berg E. Computer-assisted cytogenetic analysis of 51 malignant peripheral-nerve-sheath tumors: sporadic vs. neurofibromatosis-type-1-associated malignant schwannomas. Int J Cancer. 1999;83:171–178. doi: 10.1002/(sici)1097-0215(19991008)83:2<171::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 21.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 22.Perrone F, Tabano S, Colombo F, et al. p15INK4b, p14ARF, and p16INK4a inactivation in sporadic and neurofibromatosis type 1-related malignant peripheral nerve sheath tumors. Clin Cancer Res. 2003;9:4132–4138. [PubMed] [Google Scholar]
- 23.Suzuki H, Sugimura H, Hashimoto K. p16INK4A in oral squamous cell carcinomas—a correlation with biological behaviors: immunohistochemical and FISH analysis. J Oral Maxillofac Surg. 2006;64:1617–1623. doi: 10.1016/j.joms.2005.11.097. [DOI] [PubMed] [Google Scholar]
- 24.Rodolfo M, Daniotti M, Vallacchi V. Genetic progression of metastatic melanoma. Cancer Lett. 2004;214:133–147. doi: 10.1016/j.canlet.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 25.Mawrin C, Kirches E, Boltze C, Dietzmann K, Roessner A, Schneider-Stock R. Immunohistochemical and molecular analysis of p53, RB, and PTEN in malignant peripheral nerve sheath tumors. Virchows Arch. 2002;440:610–615. doi: 10.1007/s00428-001-0550-4. [DOI] [PubMed] [Google Scholar]
- 26.Mirmohammadsadegh A, Marini A, Nambiar S, et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006;66:6546–6552. doi: 10.1158/0008-5472.CAN-06-0384. [DOI] [PubMed] [Google Scholar]
- 27.Whang YE, Wu X, Suzuki H, et al. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci U S A. 1998;95:5246–5250. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salvesen HB, MacDonald N, Ryan A, et al. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int J Cancer. 2001;91:22–26. doi: 10.1002/1097-0215(20010101)91:1<22::aid-ijc1002>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 29.Kwabi-Addo B, Giri D, Schmidt K, et al. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc Natl Acad Sci U S A. 2001;98:11563–11568. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen JA, Yachnis AT, Arai M, Davis JG, Scherer SS. Expression of the neu proto-oncogene by Schwann cells during peripheral nerve development and Wallerian degeneration. J Neurosci Res. 1992;31:622–634. doi: 10.1002/jnr.490310406. [DOI] [PubMed] [Google Scholar]
- 32.Schlegel J, Muenkel K, Trenkle T, Fauser G, Ruschoff J. Expression of the ERBB2/neu and neurofibromatosis type 1 gene products in reactive and neoplastic Schwann cell proliferation. Int J Oncol. 1998;13:1281–1284. doi: 10.3892/ijo.13.6.1281. [DOI] [PubMed] [Google Scholar]
- 33.Fernandes H, Cohen S, Bishayee S. Glycosylation-induced conformational modification positively regulates receptor-receptor association: a study with an aberrant epidermal growth factor receptor (EGFRvIII/DeltaEGFR) expressed in cancer cells. J Biol Chem. 2001;276:5375–5383. doi: 10.1074/jbc.M005599200. [DOI] [PubMed] [Google Scholar]
- 34.Burstein HJ. The distinctive nature of HER2-positive breast cancers. N Engl J Med. 2005;353:1652–1654. doi: 10.1056/NEJMp058197. [DOI] [PubMed] [Google Scholar]
- 35.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 36.Fujita T, Doihara H, Kawasaki K, et al. PTEN activity could be a predictive marker of trastuzumab efficacy in the treatment of ErbB2-overexpressing breast cancer. Br J Cancer. 2006;94:247–252. doi: 10.1038/sj.bjc.6602926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahller YY, Vaikunth SS, Currier MA, et al. Oncolytic HSV and erlotinib inhibit tumor growth and angiogenesis in a novel malignant peripheral nerve sheath tumor xenograft model. Mol Ther. 2007;15:279–286. doi: 10.1038/sj.mt.6300038. [DOI] [PubMed] [Google Scholar]
- 38.Hidalgo M, Siu LL, Nemunaitis J, et al. Phase I and pharmacologic study of OSI-774, an epidermal growth factor receptor tyrosine kinase inhibitor, in patients with advanced solid malignancies. J Clin Oncol. 2001;19:3267–3279. doi: 10.1200/JCO.2001.19.13.3267. [DOI] [PubMed] [Google Scholar]
- 39.Huether A, Hopfner M, Baradari V, Schuppan D, Scherubl H. EGFR blockade by cetuximab alone or as combination therapy for growth control of hepatocellular cancer. Biochem Pharmacol. 2005;70:1568–1578. doi: 10.1016/j.bcp.2005.09.007. [DOI] [PubMed] [Google Scholar]