Abstract
Background
Neuropeptides regulate a broad range of physiological and behavioral processes. Elucidation of neuropeptide function requires identifying the cells that respond to neuropeptide signals and determining the molecular, cellular, physiological, and behavioral consequences of activation of their cognate GPCRs in those cells. As a novel tool for answering these questions, we have developed genetically encoded neuropeptides covalently tethered to a glycosylphosphatidyl inositol (GPI) glycolipid anchor on the extracellular leaflet of the plasma membrane (“t-peptides”).
Results
We show that t-peptides cell-autonomously induce activation of their cognate GPCRs in cells that express both the t-peptide and its receptor. In the neural circuit controlling circadian rest-activity rhythms in Drosophila melanogaster, rhythmic secretion of the neuropeptide Pigment Dispersing Factor (PDF) and activation of its GPCR (PDFR) are important for intercellular communication of phase information and coordination of cellular oscillations of multiple circadian clock neurons. Broad expression of t-PDF in the circadian control circuit overcomes arrhythmicity induced by pdf01 null mutation, most likely due to activation of PDFR in PDFR-expressing clock neurons that do not themselves secrete PDF. More restricted cellular expression of t-PDF suggests that activation of PDFR accelerates cellular timekeeping in some clock neurons, while decelerating others.
Conclusions
The activation of PDFR in pdf01 null-mutant flies—and thus the absence of PDF-mediated intercellular transfer of phase information—induces strong rhythmicity in constant darkness, thus establishing a distinct role for PDF signaling in the circadian control circuit independent of the intercellular communication of temporal phase information. The t-peptide technology we have developed and validated should provide a useful tool for cellular dissection of bioactive peptide signaling in a variety of organisms and physiological contexts.
Introduction
In both flies and mammals, autonomous cellular clocks that underlie circadian cycles of rest and activity have been localized to particular clock neurons that are organized into circuits in the central nervous system [1, 2]. Clock neurons coordinate their phases with one another and communicate phase information to downstream neural targets via activity-dependent synaptic release of neurotransmitters and neuropeptides [for review, see 3, 4]. Intercellular communication via neuropeptides is essential in mediating circadian inputs, circadian outputs, and circadian synchronization, but the specific pharmacological and cellular mechanisms for such communication remains poorly understood.
The Drosophila circadian control circuit drives rhythmic locomotor activity and comprises six anatomically distinguishable bilateral groups: the small ventral, large ventral, and dorsal subgroups of the lateral group of neurons (sLNV, lLNV, and LND), and three subgroups of the dorsomedial group of neurons (DN1, DN2, and DN3) [for review, see 3]. These anatomical groupings have functional correlates. The sLNVs are considered to be “morning” (M) cells that drive the anticipatory increase in locomotor activity that occurs before lights-on in 12 hr:12 hr light:dark conditions (LD) and are also required for free-running rhythmicity in constant darkness (“DD”) [5–7]. In contrast, dorsal clock neurons—LNDs and DNs—include those that are considered “evening” (E) cells that drive the anticipatory increase in locomotor activity that occurs before lights-off in LD and are capable under certain circumstances of generating free-running rhythms in constant light (LL) [5, 7–10]. Furthermore, M cells and E cells transfer phase information to one another and they alternate setting the phase of locomotor activity depending on photoperiod, with M cells setting the phase of morning and evening peaks in short-day 10 hr:14 hr LD conditions and E cells setting the phase of both peaks in long-day 14 hr:10 hr LD [10, 11].
The sLNv M and lLNv neurons produce the neuropeptide PIGMENT DISPERSING FACTOR (PDF), which is thought to signal circadian phase to downstream neural elements, including non-PDF-expressing dorsal E clock neurons and, possibly, the direct locomotor control circuitry [6, 11–16]. pdf01 null mutation induces substantial arrhythmicity in DD and eliminates morning anticipation in LD [6]. The PDF receptor (PDFR) is a seven-transmembrane-domain GPCR that signals through adenylate cyclase/cAMP, is expressed in various clock and non-clock neurons, and is also required for robust free-running behavioral rhythmicity in DD [17–19]. Recent studies using an in vivo fluorescent reporter of cytoplasmic cAMP demonstrate that sLNVs, LNDs, and some DNs respond to bath applied PDF, and thus presumably possess PDFRs [20]. The specific functional role(s) of PDFR activation in particular subsets of PDFR-expressing clock neurons remains unknown. In addition, it remains an open question how PDF communicates phase information to E cells, and whether M cells might use PDF signals to gate the ability of E cells themselves to drive locomotor rhythms.
To address these questions, we have developed genetically encoded neuropeptides covalently tethered to a glycosylphosphatidyl inositol (GPI) glycolipid anchor on the extracellular leaflet of the plasma membrane. These GPI-tethered neuropeptides (“t-peptides”) induce activation of their cognate GPCRs with appropriate pharmacological specificity. t-peptides activate their GPCRs cell-autonomously, i.e., without activating their receptors on neighboring t-peptide non-expressing cells. This establishes the t-peptide system as a novel tool for the cellular dissection of neuropeptide signaling. We show that broad expression of t-PDF in the Drosophila circadian control circuit overcomes arrhythmicity in DD induced by pdf01 null mutation, most likely due to activation of PDFR in dorsal E cells. More restricted cellular expression of t-PDF suggests that activation of PDFR accelerates cellular timekeeping in some clock neurons, while decelerating others. These studies support the hypothesis that PDF signals from sLNV M cells gate the ability of dorsal E cells to drive locomotor rhythms, thus revealing a distinct role for PDF signaling in the circadian control circuit independent of the intercellular communication of temporal phase information.
Results
GPI-tethered peptides are pharmacologically specific, cell-autonomous activators of their cognate GPCRs in vitro
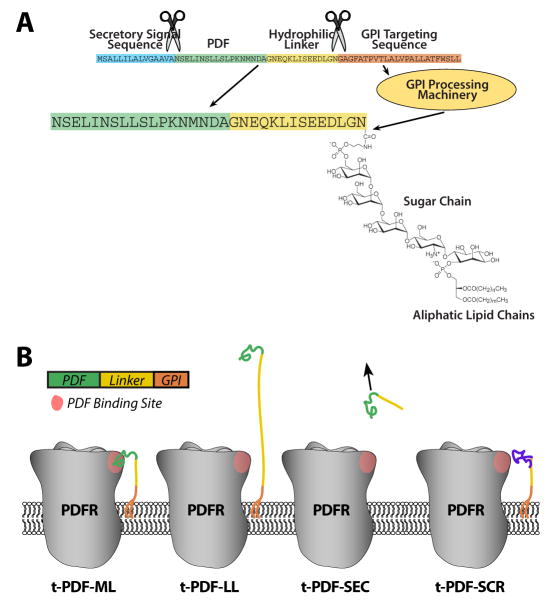
In order to probe the roles of neuropeptide GPCR activation in specific cellular contexts in intact organisms, we developed the t-peptide system. Each t-peptide comprises (from N to C terminus) a secretory signal sequence for targeting to the secretory pathway, a mature cleaved peptide sequence, a hydrophilic linker sequence with an embedded c-Myc epitope tag, and a GPI targeting sequence (Figure 1A) [based on the t-toxin system of 21]. t-PDF isoforms generated include t-PDF-ML and t-PDF-LL (possessing 14 and 40 amino acid linkers, respectively), t-PDF-SEC (lacking the GPI targeting sequence, and thus untethered), and t-PDF-SCR (with the amino acid sequence of the PDF peptide moiety scrambled) (Figure 1B).
Figure 1. Structure of GPI-tethered PDF isoforms.
(A) The medium-linker isoform of GPI-tethered PDF (t-PDF-ML) contains trypsin signal sequence (blue), mature cleaved PDF peptide sequence (green), hydrophilic linker comprising the c-Myc epitope tag flanked by single glycine-asparagine (GN) repeats (yellow), and the GPI targeting signal from lynx1 protoxin (orange). After processing in the secretory pathway, the secretory signal and GPI targeting sequences are cleaved, and the C terminus is covalently linked to GPI whose aliphatic lipid chains are intercalated in the extracellular leaflet of the plasma membrane. (B) Schematics depicting PDFR and the isoforms of t-PDF (not to scale), which are identical to t-PDF-ML except as follows: The linker of t-PDF-LL contains c-Myc epitope flanked by four N terminal GN repeats and eleven C terminal GN repeats, t-PDF-SEC contains no GPI targeting sequence, and the PDF sequence of t-PDF-SCR has been replaced by a scrambled sequence comprising the same amino acids as PDF.
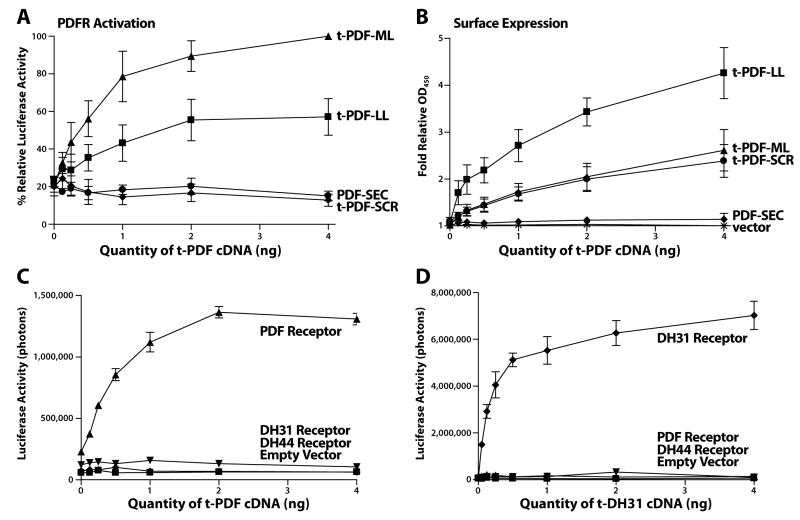
We co-expressed various t-peptides together with corresponding GPCRs in mammalian HEK293 tissue culture cells. Activation of GPCRs that signal through adenylate cyclase-mediated cAMP production is detected through co-transfection of a cAMP-sensitive CRE-luciferase reporter plasmid along with GPCR and t-peptide cDNAs. As seen in Figure 2A, co-expression of PDFR with increasing quantities of either t-PDF-ML or t-PDF-LL results in substantial dose-dependent steady-state cAMP increases 48 hours after transfection. (Figure 2A, B). The total activity increase induced by acute application of 1 μM saturating soluble PDF [see 17] to cells already co-expressing t-PDF and PDFR for 48 hours is not related to the pre-existing degree of activation of PDFR by co-expressed t-PDF (Supplemental Figure 1). Furthermore, when cells only expressing PDFR are mixed with cells only expressing t-PDF-ML, there is no receptor activation (Supplemental Figure 2). These results indicate that t-peptides are cell-autonomous activators of their cognate receptors and do not lead to substantial sustained desensitization.
Figure 2. t-PDF activates cloned PDF receptor when co-expressed in vitro in mammalian tissue culture cells.
Varying quantitites of cDNA encoding t-peptides are co-transfected into HEK 293 mammalian tissue culture cells with constant quantities of GPCR and cAMP-sensitive CRE-luciferase reporter cDNAs. Forty-eight hours after transfection, cells are either lysed for luciferase bioluminescence assay or kept intact and unpermeabilized for cell-surface anti-Myc ELISA assay. (A) t-PDF-ML and t-PDF-LL each dose-dependently increase steady-state intracellular cAMP, indicating activation of PDFR, with t-PDF-ML inducing greater increases than t-PDF-LL. t-PDF-SEC and t-PDF-SCR have no activity. (B) t-PDF-LL is expressed on the cell-surface at higher levels than t-PDF-ML and t-PDF-SCR, while t-PDF-SEC is undetectable. (C) t-PDF-ML only activates PDFR, and not the related receptors for the peptides DH31 or DH44. (D) t-DH31-ML, identical to t-PDF-ML except the PDF peptide sequence has been replaced with that for DH 31 (TVDFGLARGYSGTQEAKHRMGLAAANFAGGP), only activates DH31R, and not PDFR or DH44R. (Error bars are S.D; N = 3 repeats for all measurements).
While co-transfection with a given quantity of t-PDF-ML cDNA is more effective at activating PDFR than the same quantity of t-PDF-LL cDNA, detectable surface expression of t-PDF-ML is less than t-PDF-LL for the same quantity of cDNA (Figure 2A, B). This indicates a greater molar activity of t-PDF-ML than t-PDF-LL. The absence of activity of t-PDF-SEC (Figure 2A)—which is not detectable on the surface of the plasma membrane (Figure 2B)—indicates a molar activity of t-PDF with a C terminal linker liberated into the tissue culture medium that is too low to activate PDFR at all. This is not surprising, as native secreted PDF is C terminal amidated and the non-amidated form is approximately 300-fold less active in a bioassay of synthetic soluble peptides [22]. Thus, substantial activity of the GPI-tethered t-PDF isoforms suggests that membrane tethering permits t-PDF to activate PDFR cell-autonomously by creating an effective concentration of non-amidated PDF moiety at its receptor.
The absence of activity of t-PDF-SCR (Figure 2A) when expressed on the cell-surface at levels identical to those of t-PDF-ML (Figure 2B) importantly indicates the dependence of activation of its cognate receptor on the particular amino acid sequence of the peptide moiety in the chimeric t-peptide. We also co-expressed t-peptides with closely related but non-cognate receptors. When co-expressed with either PDFR, DH31R, or DH44R (receptors for the fly peptides DH31 and DH44, respectively) [23, 24], t-PDF-ML only activates PDFR (Figure 2C). Conversely, t-DH31-ML only activates DH31R, and not PDFR or DH44R (Figure 2D). These results indicate that t-peptide ligands exhibit appropriate pharmacological specificity for their receptors.
GPI-tethered PDF is a pharmacologically specific, cell-autonomous activator of PDFR in vivo
We generated transgenic flies expressing various t-PDF isoforms or other t-peptides in all circadian clock neurons, by using the UAS-GAL4 binary expression system [25]. UAS-t-peptide transgenic flies are mated to tim(UAS)-GAL4 transgenic flies to produce progeny expressing t-peptide in all circadian clock neurons. t-PDF-ML expression by neurons in vivo can be confirmed via immunofluorescence detection of the Myc epitope tag constituting part of the linker domain (Supplemental Figure 2). The free-running circadian locomotor rhythm in constant darkness (DD) of each fly is categorized as either rhythmic, arrhythmic, or complex rhythmic (which occurs when an individual fly exhibits multiple rhythms of locomotor activity free-running simultaneously with different periods).
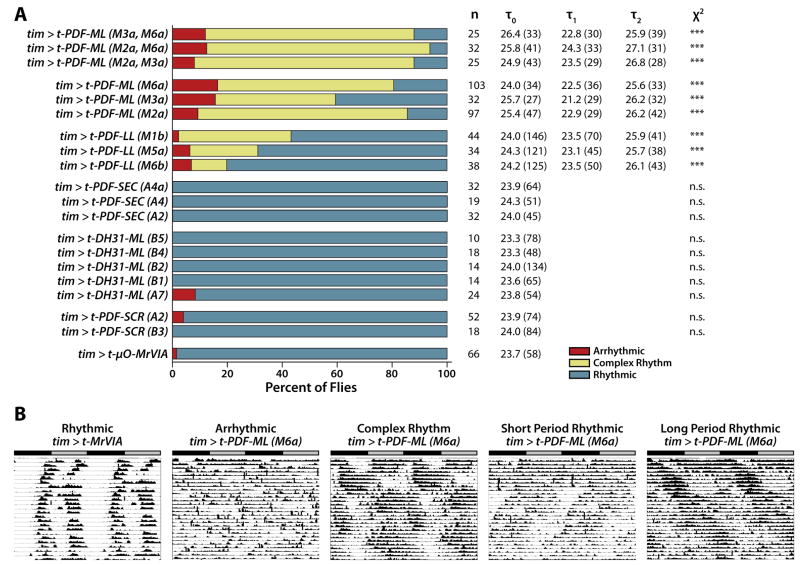
t-PDF-ML or t-PDF-LL each induce complex free-running locomotor rhythms when expressed in all clock neurons, in comparison to negative-control flies expressing t-μO-MrVIA, a GPI-tethered cone snail Na+ channel toxin with no activity in Drosophila [26]. The induction of complex free-running locomotor rhythms by t-PDF expression in all clock neurons is consistent with activation of PDFR in circadian clock neurons, as a variety of other experimental manipulations that lead to high levels of PDF signaling in the clock circuit also induce complex rhythms [14, 15]. Furthermore, the complex rhythm phenotype induced by t-PDF expression is unlike the phenotype induced by experimental manipulations that decrease PDF signaling in the clock circuit, which is a combination of arrhythmicity and weak short-period rhythms [6, 17–19, 26, 27].
Unlike t-PDF-ML and t-PDF-LL, constitutive expression in all clock neurons of either t-PDF-SCR or t-DH31-ML fails to induce free-running locomotor phenotypes (Figure 3). t-PDF-ML expression from any of three independent UAS-t-PDF-ML transgenes induces a higher proportion of complex rhythmic flies than expression of t-PDF-LL from any of three independent UAS-t-PDF-LL transgenes, and t-PDF-SEC has no effect (Figure 3). t-PDF-ML expression using a wide variety of other neuronal and glial GAL4 driver lines induces no free-running circadian phenotype (Supplemental Table). This includes the Mz1525 or Mz1366 GAL4 driver lines, which do induce strong complex rhythms when used to express the native amidated secreted form of PDF in neurosecretory cells that project to the region of dorsal clock neurons [15]. The rank ordering of t-PDF isoform bioactivity when expressed in circadian clock neurons, and the absence of bioactivity when expressed in a wide variety of different expression patterns in the CNS but not in circadian clock neurons, taken together indicate that t-PDF activation of PDFR is cell-autonomous in vivo, with t-PDF activating PDFR only in the cells in which t-PDF is expressed and not in neighboring cells.
Figure 3. t-PDF induces complex locomotor rhythms when expressed in vivo in all circadian clock neurons of transgenic flies.
Male flies bearing UAS-t-peptide transgenes are mated to female flies bearing a tim(UAS)-GAL4 transgene to produce progeny expressing t-peptide in all circadian clock neurons. Free-running locomotor rhythms of individual male progeny entrained in 12h:12h LD conditions and then released into DD are categorized as rhythmic, complex rhythmic, or arrhythmic, and free-running periods are assigned, using Lomb-Scargle periodogram analysis. (A) t-PDF-ML is more active in vivo than t-PDF-LL, while t-PDF-SEC and t-PDF-SCR have no activity, thus recapitulating the relative in vitro activities shown in Figure 2. t-DH31-ML, while active against DH31R in vitro, does not influence free-running locomotor rhythms when expressed in vivo in circadian clock neurons. Bar graph depicts proportions of rhythmic (blue), complex rhythmic (yellow), and arrhythmic (red) flies of the indicated genotypes, with the notations in parentheses referring to specific chromosomal insertions, or combinations of two chromosomal insertions, of the UAS-t-peptide transgenes. n indicates the number of individual flies assayed, τ0 indicates the average single free-running period of rhythmic flies, τ1 the average shorter free-running period of complex rhythmic flies, τ2 the average longer free-running period of complex rhythmic flies, and χ2 the significance of χ2 statistical comparison of the proportions for that genotype with that of tim > t-μO-MrVIA flies expressing a tethered conotoxin that has no activity in flies (***, p < 0.001). Average Lomb-Scargle periodogram powers are in parentheses following each free-running period component. (B) Representative free-running locomotor actograms of individual flies with the indicated phenotypes and genotypes. The gray bar indicates subjective day, and the black bar subjective night.
t-PDF expression in all clock neurons induces strong free-running rhythms in pdf01 null-mutant flies
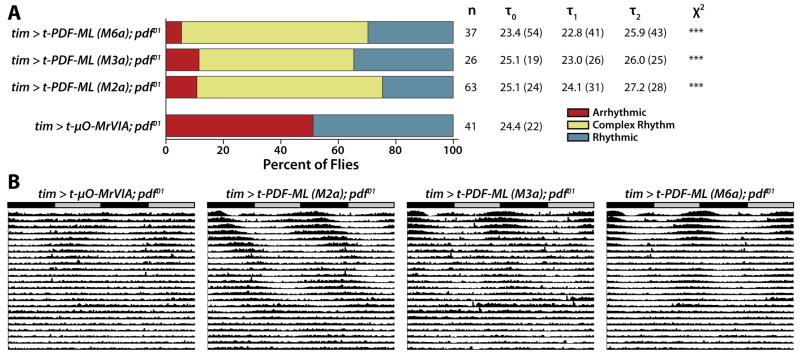
pdf01 null-mutant flies exhibit severely disrupted circadian rhythms, including a combination of arrhythmicity and weak rhythmicity while free-running in DD [6]. To address whether PDF functions solely to communicate circadian phase information from the PDF-secreting LNVs to PDFR-expressing clock neurons, or whether PDF signals also could gate the ability of PDFR-expressing clock neurons to drive locomotor rhythms, we expressed t-PDF-ML in all circadian clock neurons of pdf01 null-mutant flies. As expected, negative control pdf01 flies expressing inert t-μO-MrVIA exhibit severe deficits in free-running locomotor rhythms, with ~50% arrhythmicity in DD (Figure 4). In contrast, pdf01 flies expressing t-PDF-ML in all clock neurons from any of three independent chromosomal insertions exhibit very little arrhythmicity in DD, with only ~10% of flies arrhythmic, and instead most are complex rhythmic (Figure 4A). Comparing the averaged actograms of the negative-control and t-PDF-ML-expressing flies shows clear induction of strong free-running rhythmicity in DD, particularly apparent in the first week following transfer from entraining LD conditions to DD, before individual flies have had the opportunity to drift substantially out of phase with one another (Figure 4B).
Figure 4. t-PDF expression in all clock neurons suppresses free-running arrhythmicity induced by pdf01 null mutation.
Male pdf01 null-mutant flies bearing UAS-t-peptide transgenes are mated to female pdf01 null-mutant flies bearing tim(UAS)-GAL4 transgene to produce pdf01 null-mutant flies expressing t-peptide in all clock neurons. (A) Negative control pdf01 null-mutant flies expressing the inert t-μO-MrVIA conotoxin exhibit approximately 50% arrhythmicity, with the rhythmic flies exhibiting very weak rhythms, consistent with numerous published reports (see text). In contrast, very few pdf01 flies constitutively expressing t-PDF-ML in all clock neurons are arrhythmic, and instead predominately exhibit complex rhythms (***, p < 0.001, χ2 test comparing each t-PDF-ML-expressing genotype to the t-μO-MrVIA-expressing control). (B) Averaged free-running DD actograms of all flies tested of the indicated genotypes. The induction of strong rhythmicity in pdf01 null-mutant flies by t-PDF-ML expression in all clock neurons is particularly apparent over the first week in DD, before individual flies have had the opportunity to drift out of phase with one another, thereby dispersing the population activity pattern depicted in the averaged actogram.
While PDFR activation induces strong rhythms in pdf01 null-mutant flies, these rhythms are abnormal complex rhythms. This indicates that normal free-running rhythmicity requires not just PDFR activation per se, but temporally regulated PDFR activation driven by rhythmic PDF secretion by the LNVs. It also suggests that the circadian deficits of pdf01 flies are not solely due to the absence of PDF-mediated transfer of phase information per se from PDF-secreting LNVs to PDFR-expressing neurons, but that PDF signals also gate the ability of non-LNV PDFR-expressing clock neurons to drive locomotor rhythms themselves.
Cellular dissection of PDFR function via t-PDF expression in subsets of clock neurons
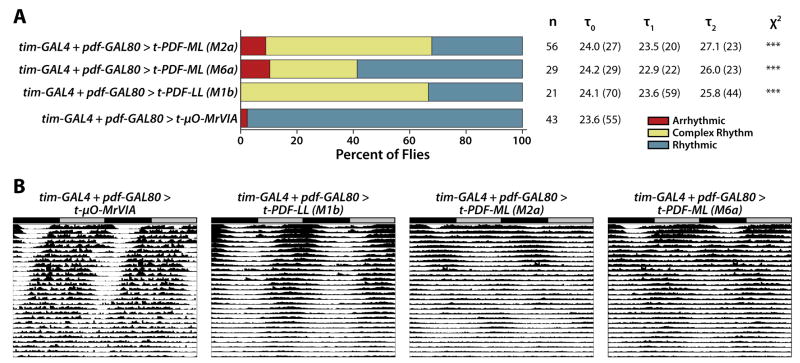
In order to dissect functional roles of PDFR activation in distinct subsets of clock neurons, we expressed t-PDF-ML using various GAL4 drivers and a pdf-GAL80 repressor transgene, which prevents GAL4 from activating transcription of UAS transgenes in the PDF-expressing LNVs [7]. When expressed solely in the PDF-expressing LNVs of pdfWT flies using a pdf-GAL4 driver [6], t-PDF-ML induces a modest, but statistically significant, degree of free-running arrhythmicity (Supplemental Table). When t-PDF-ML is expressed in the LNVs of pdf01 flies, free-running rhythms are unaffected (Supplemental Table). We then performed the converse experiment, expressing t-PDF-ML in all clock neurons other than the PDF-expressing LNVs, by generating flies simultaneously possessing tim(UAS)-GAL4, pdf-GAL80, and UAS-t-PDF transgenes (Figure 5). Flies expressing t-PDF solely in non-LNV clock neurons exhibit complex free-running locomotor rhythms similar to those induced by t-PDF expression in all clock neurons (Figure 3). These results suggest that PDFR in dorsal LND and DN clock neurons is more functionally important for circadian rhythms than in the PDF-secreting LNVs themselves.
Figure 5. t-PDF expression in all clock neurons except for the PDF-secreting LNV subset induces complex locomotor rhythms.
Male flies bearing UAS-t-peptide transgenes are mated to female flies bearing both tim(UAS)-GAL4 and pdf-GAL80 (which suppresses GAL4 activation of UAS transgene expression in the PDF-secreting LNVs) transgenes to produce progeny expressing t-peptide in all circadian clock neurons except the PDF-secreting LNVs. (A) Proportions of locomotor phenotypes are different between each t-PDF-expressing genotype and the t-μO-MrVIA-expressing control (***, p < 0.001, χ2 test). (B) Averaged actograms of flies of the indicated genotypes.
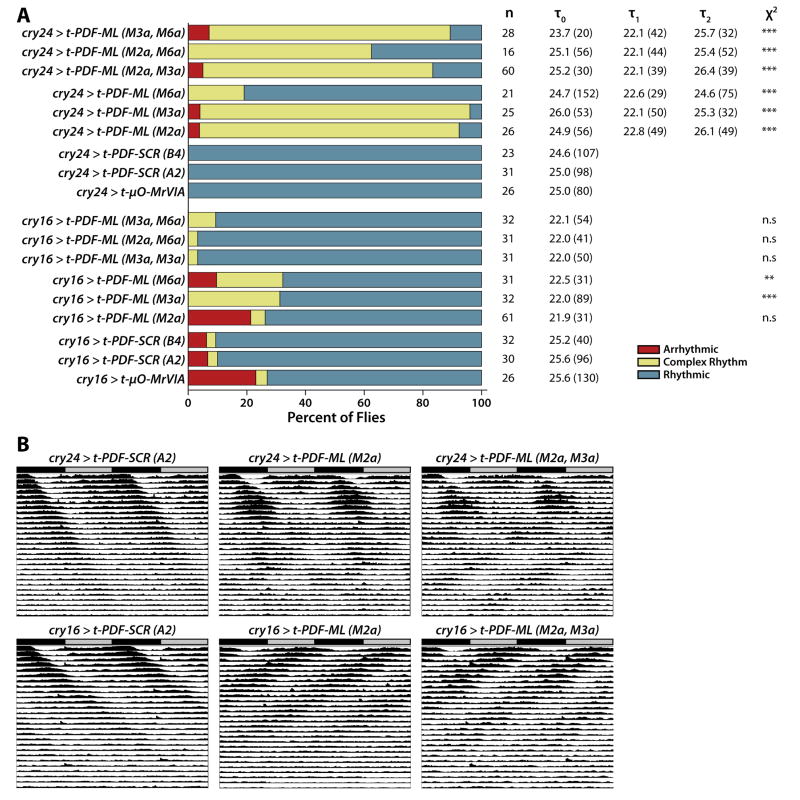
We also expressed t-PDF-ML in distinct partially overlapping subsets of clock neurons (and some non-clock neurons) using cry16-GAL4 and cry24-GAL4 drivers, which are two independent chromosomal insertions of the same transgene based on the Cryptochrome promoter [28]. Using nuclear GFP as a marker for driver expression and co-staining for various marker antigens, we have analyzed the expression patterns of the cry16-GAL4 and cry24-GAL4 drivers (Supplemental Figures 4–7). These drivers are both active in all PDF-expressing LNVs, all LNDs, two-three large DN3s, both DN2s, both anterior DN1s, and ring neurons of the central complex. The cry16-GAL4 driver is active in many glia, while cry24-GAL4 is not. The two drivers have different expression patterns in the posterior DN1s, with cry24-GAL4 almost always active in four or five cells of this group, while cry16-GAL4 is expressed more variably, in from two to six cells. When expressed using the cry24-GAL4 driver, t-PDF-ML induces complex free-running locomotor rhythms with a short period of 22–23 hours and a long period of 25–26 hours (Figure 6). This is very similar to the effect of t-PDF-ML expression in all clock neurons using the tim(UAS)-GAL4 promoter (Figure 3). In contrast, when expressed using the cry16-GAL4 driver, t-PDF-ML expression induces only a modest degree of complex rhythmicity—and even less when expressed at higher levels from two independent UAS transgenes simultaneously—and rather induces dramatic period shortening of 3–4 hours (Figure 6). Note that the longer than normal period of the negative control t-PDF-SCR- and t-μO-MrVIA-expressing flies is due for unknown reasons to the cry-GAL4 transgenes themselves, as has been previously reported [28]. This difference in the effect of t-PDF expression driven by cry16-GAL4 and cry24-GAL4 suggests that PDFR activation in some clock neurons accelerates circadian oscillation, while in other clock neurons it decelerates circadian oscillation.
Figure 6. t-PDF expression in distinct partially overlapping subsets of clock neurons induces either single short-period rhythmicity or complex rhythmicity.
Male flies bearing UAS-t-peptide transgenes are mated to cry24-GAL4 or cry16-GAL4 transgenes, which drive expression in distinct partially overlapping subsets of clock neurons. (A) t-PDF-ML expressed using cry24-GAL4 driver induces complex free-running locomotor rhythms similar to those induced using tim(UAS)-GAL4 driver (Figure 3). In contrast, t-PDF-ML expression using cry16-GAL4 driver induces only a modest degree of complex rhythmicity, and almost none when expressed at a higher dose simultaneously from two independent UAS-t-PDF-ML chromosomal insertions; rather, driving t-PDF-ML expression with cry16-GAL4 induces dramatic shortening of free-running period from ~25.5 hours in negative control flies expressing either t-PDF-SCR or t-μO-MrVIA (long-period phenotypes due to the cry16-GAL4 and cry24-GAL4 transgenes themselves have been previously reported; see text) to ~22 hours in flies expressing t-PDF-ML (overall ANOVA p < 0.001; p < 0.05 for paired comparisons to appropriate pooled controls using the Bonferroni Versus Control Test). ***, p < 0.001, **, p < 0.005, χ2 test comparing each t-PDF-ML-expressing genotype to the appropriate pooled controls. (B) Averaged actograms of flies of the indicated genotypes demonstrate clearly the induction of complex rhythmicity by t-PDF-ML expressed using cry24-GAL4 and short-period rhythmicity using cry16-GAL4.
Discussion
While there is substantial evidence that PDF signaling in the circadian control circuit is important for the intercellular communication of phase information, the specific functional role(s) of PDFR activation in particular subsets of PDFR-expressing clock neurons remains unknown. To address this important question, we activated PDFR in different subsets of circadian clock neurons in pdfWT and pdf01 null-mutant flies. When expressed solely in the LNVs themselves, t-PDF has only modest effects on rhythmic behavior (Supplemental Table). This suggests that—while the sLNV subset of LNVs expresses functional PDFR capable of inducing cAMP increases upon activation [20]—PDF signaling to the LNVs themselves does not strongly influence circadian rhythm generation. In contrast, t-PDF expression solely in the non-LNV dorsal LND and DN clock neurons induces complex rhythms (Figure 5) very similar to those induced by t-PDF expression in all clock neurons (Figure 3). This indicates an important role for PDFR activation in dorsal clock neurons for rhythm generation.
When t-PDF is expressed in all circadian clock neurons of pdf01 null-mutant flies, there is strong suppression of the substantial free-running arrhythmicity induced by the absence of LNV PDF secretion (Figure 4). After about one week in DD, the induced rhythms manifest themselves as complex rhythms (Figure 4B). This indicates that cell-autonomous PDFR activation in the circadian control circuit can substitute for native intercellular PDF signals in permitting strong free-running rhythmicity in DD, and suggests an important role for PDF signaling in addition to intercellular communication of clock phase per se in rhythm generation in the normal situation. There are a few other manipulations of the Drosophila circadian control circuit that have resulted in induction of rhythmicity in pdf01 null-mutant flies. In DD, electrical hyperexcitation of the LNVs themselves induces partial suppression of arrhythmicity in pdf01 flies [29]. In constant light (LL) conditions, where wild-type flies are arrhythmic, certain genetic manipulations allow dorsal clock neurons to drive locomotor rhythms even in pdf01 null-mutant flies, suggesting that dorsal neurons can function as PDF-independent pacemakers under some conditions [8, 9], although in another genetic context dorsal neurons appear to require LNV PDF secretion to drive rhythms [10].
These results have been interpreted as suggesting that darkness suppresses the ability of dorsal E clock neurons to drive locomotor rhythms while light unveils it, and vice versa for the LNV M cells, with light suppressing their ability to drive locomotor rhythms and darkness unveiling it [8–10]. This makes sense given earlier findings that LNVs appear to generate the morning anticipatory peak, while dorsal neurons do the same for the evening peak [5, 7, 26]. Our results thus suggest that PDFR activation permits dorsal neurons to drive strong rhythms in DD—in the absence of light—with or without LNV PDF secretion and supports the hypothesis that in normal flies in LD PDFR activation in the morning [when LNVs are most electrically excitable and PDF secretion is thus expected to be greatest, 30, 31] provides a gating signal that allows the dorsal neurons to drive rhythmicity and generate the evening peak. Thus, rhythmic PDF secretion by the LNV M cells not only determines the phase of morning anticipation [26] and likely provides a daily phase resetting signal to dorsal E cells [11], but also provides a timed gating signal to PDFR-expressing dorsal E cells allowing them to “take the reins” and generate the evening anticipatory locomotor peak. pdf01 null-mutant flies still generate a phase-advanced evening peak in LD [6], thus indicating that light and PDFR activation are parallel gating signals each capable of allowing dorsal clock neurons to drive rhythmicity. This makes sense in light of the observation that in short photoperiods, LNV M cells set the phase of the evening peak [10], that light can permit dorsal clock neurons to drive robust free-running rhythms either in the presence or absence of PDF [8–10, 32]. It also explains why the evening peak still occurs robustly in DD, in the absence of light [see, e.g., 11].
To further dissect the responses of particular subsets of dorsal clock neurons to PDFR activation, we expressed t-PDF in partially overlapping expression patterns using the cry16-GAL4 and cry24-GAL4 drivers. The effects of t-PDF expression using these two drivers are dramatically different. In the case of cry16-GAL4, t-PDF expression induces only a modest degree of complex rhythmicity—and almost none when expressed at higher levels using multiple UAS transgenes—and rather induces strong period shortening of 3–4 hours (Figure 6). In contrast, t-PDF expression using cry24-GAL4 leads to complex rhythms (Figure 6) very similar to those induced by t-PDF expression in all clock neurons or solely in dorsal clock neurons (Figures 3, 5). In combination with the absence of substantial effects on free-running period of t-PDF expression solely in LNVs (Supplemental Table), this difference in the effect of t-PDF expression driven by cry16-GAL4 and cry24-GAL4 suggests that PDFR activation in some dorsal neurons—LNDs and/or DNs—that are commonly expressed between cry16-GAL4 and cry24-GAL4 accelerates circadian oscillation, while in a subset of posterior DN1s that is presumably expressed only in cry24-GAL4 it decelerates circadian oscillation. This is consistent with the recent observation that mutations severely affecting the gross morphology of the Drosophila brain and thereby inducing increases in the density of PDF-containing neural processes in different parts of the brain differentially affect cellular free-running period, decelerating Cry-positive LNDs and one subset of DNs, and accelerating Cry-negative LNDs and a complementary subset of DNs [33, 34].
Conclusions
Our studies demonstrate the utility of the t-peptide technology for dissecting the cellular basis for neuropeptide signaling within a behavioral control circuit and raise the possibility that it will provide a generally applicable approach for cellular dissection of peptide signaling in a variety of neural circuits, non-neural tissues, and organisms. In relation to the latter, we have determined that t-peptide versions of various mammalian neuropeptides activate their GPCRs when co-expressed in mammalian tissue culture cells [35]. In the context of the Drosophila circadian control circuit, we have used the t-peptide system to provide support for the hypothesis that rhythmic PDF secretion by the LNVs not only determines the phase of morning anticipation and provides a daily resetting signal to dorsal E cells, but also provides a gating signal to PDFR-expressing dorsal clock neurons sufficient to allow them to “take the reins” and drive rhythmic locomotor activity. Our studies also implicate the question of the molecular mechanisms whereby PDFR activation and consequent cAMP increases can accelerate circadian timekeeping in some clock neurons while decelerating it in others. Future studies are required to determine (1) the cellular events induced by PDFR activation that allow dorsal neurons to drive locomotor rhythms, (2) the specific identities of the accelerated and decelerated dorsal neurons, and (3) the molecular mechanisms that underlie their differential responses to PDFR activation.
Materials and Methods
t-Peptide cDNAs
All t-peptide cDNAs were chemically synthesized using optimal Drosophila codon usage and with an optimal Drosophila Kozak translation initation site upstream of the start methionine (CAAA). Encoded t-peptides are as follows.
t-PDF-ML: MSALLILALVGAAVANSELINSLLSLPKNMNDAGNEQKLISEEDLGNGAGFATPV TLALVPALLATFWSLL*
t-PDF-LL: MSALLILALVGAAVANSELINSLLSLPKNMNDAGNGNGNGNGEQKLISEEDLGN GNGNGNGNGNGNGDGNGGALCGAGFATPVTLALVPALLATFWSLL*
t-PDF-SEC: MSALLILALVGAAVANSELINSLLSLPKNMNDAGNEQKLISEEDLGN*
t-PDF-SCR: MSALLILALVGAAVANLKNSISLEDLPLAMSNNGNEQKLISEEDLGNGAGFATPV TLALVPALLATFWSLL*
t-DH31-ML: MSALLILALVGAAVATVDFGLARGYSGTQEAKHRMGLAAANFAGGPGNEQKLI SEEDLGNGAGFATPVTLALVPALLATFWSLL*
For transfection into cultured HEK293 cells, these cDNAs were cloned into the pCDNA3.1(+) expression vector, and for generation of transgenic Drosophila they were cloned into pUAST [25].
Tissue Culture Experiments
Cell culture media, fetal bovine serum and the LipofectamineR transfection reagent were obtained from Invitrogen (Carlsbad, CA). Peroxidase-conjugated, rabbit polyclonal antibody directed against the c-Myc epitope was and BM-blue (3.3′-5,5′-tetramethylbenzidine), a peroxidase substrate, were purchased from Abcam (catalog No. ab19312) and Roche Applied Science (Indianapolis, IN), respectively. Drosophila PDF and DH31 receptor cDNAs are as previously reported [17].
Cell Culture
Human embryonic kidney (HEK) 293 cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, catalog No. 12100-038) supplemented with 10% fetal bovine serum, 100 U/ml penicillin G and 100 μg/ml streptomycin. The cells were maintained at 37°C in a humidified environment containing 5 % CO2.
Luciferase Reporter Gene Assay
Receptor-mediated signaling was assessed using a luciferase assay as previously described [36]. In brief, HEK293 cells were plated in 96-well white clear bottom plates (Costar, Corning, NY) at a density of 1500 cells per well and grown for 2 days (~ 80% confluency). Cells were then transiently transfected with (i) either pcDNA1 (empty vector) or a cDNA encoding the wild-type receptor (PDFR, DH31R, DH44R), (ii) increasing amounts of a cDNA encoding the tethered ligand and (iii) a reporter gene construct consisting of six tandem repeats of the cAMP-response element (CRE6x) ligated upstream from a reporter gene encoding firefly luciferase [36]. Forty-eight hours following transfection, cells were lysed with LucliteR reagent (PerkinElmer, Boston, MA) and luciferase activity was quantified using a TopCountR Microplate Luminescence Counter (PerkinElmer, Boston, MA).
Evaluation of Receptor Expression Using ELISA
The expression levels of the tethered PDF constructs were measured using a procedure previously described [37]. In brief, HEK293 cells grown in 96-well plates were transiently transfected with either pcDNA1 or a cDNA encoding the tethered PDF constructs. Forty-eight hours post-transfection, cells were washed once with phosphate buffered saline (PBS) (pH 7.4) and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After washing with 100 mM glycine in PBS, the cells were incubated for 30 min in blocking solution (PBS containing 20% bovine serum). A horseradish peroxidase (HRP)-conjugated polyclonal antibody directed against the c-Myc epitope (1:1500 dilution in blocking solution) was then added to the cells. After 1 hour the cells were washed five times with PBS and BM-blue solution (50 μl per well) was added and incubated for 30 min at room temperature. Conversion of this substrate by antibody-linked HRP was terminated by adding 2.0 M sulfuric acid (50 μl per well). Converted substrate (indicating the amount of bound antibody) was assessed by measuring light absorbance at 450 nm using a SpectraMaxR microplate reader (Molecular Devices, Sunnyvale, CA).
Fly Strains and Crosses
All crosses and behavioral experiments were performed at 25 °C. Multiple independent chromosomal insertions of UAS-t-peptide transgenes were obtained using standard embryo injection techniques, and some were recombined using classical genetic methods to generate chromosomes bearing two independent insertions. Driver or suppressor lines have all been previously reported: pdf-GAL4 [6], tim(UAS)-GAL4 [38], pdf-GAL80 [7], cry16-GAL4 and cry24-GAL4 [28], Mz1366-GAL4 and Mz1525-GAL4 [15], repo-GAL4 [39] (also see Supplemental Table).
Behavioral Assays
Free-running and entrained rhythms of locomotor activity of individual flies were assayed using an automated Trikinetics infrared beam-crossing monitor system, and data were analyzed with double-plotted actograms, Lomb-Scargle periodograms, and normalized averaged activity histograms, all as previously described [26]. Male flies are placed in locomotor activity monitor tubes 2–5 days after eclosion, maintained in entraining 12h:12h LD conditions for ~5 days, and then released into DD conditions for assay of free-running behavior.
Statistics
Proportions of rhythmic, arrhythmic, and complex rhythmic flies were compared between genotypes using χ2 test. Average free-running periods were compared between genotypes using ANOVA and the Bonferroni Versus Control Test for controlling experiment-wide p for multiple comparisons.
Supplementary Material
Acknowledgments
We thank P. Taghert for peptide receptor cDNA clones. We thank P. Taghert, J. Blau, M. Rosbash, R. Allada, C. Helfrich-Forster, and R. Jackson for fly stocks. We thank E. McCarthy, D. Clyne, M. Kunst, and J. Blau for valuable comments on the manuscript. Work in the laboratory of M.N.N. is supported in part by the Whitehall Foundation and NINDS (R01NS055035, R01NS056443, R21NS058330). C.C. is supported by NIGMS (T32GM007527). Work in the laboratory of A.S.K. is in part supported by NIDA (R01DA020415) and NIDDK (R01DK072497, R01DK074075). J.-P.F. is supported by a research fellowship from the Fond de la Recherche en Santé du Québec.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 2.Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- 3.Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- 4.Mertens I, Husson SJ, Janssen T, Lindemans M, Schoofs L. PACAP and PDF signaling in the regulation of mammalian and insect circadian rhythms. Peptides. 2007;28:1775–1783. doi: 10.1016/j.peptides.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 6.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 7.Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 8.Murad A, Emery-Le M, Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron. 2007;53:689–701. doi: 10.1016/j.neuron.2007.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 2007;5:e315. doi: 10.1371/journal.pbio.0050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoleru D, Nawathean P, Fernandez Mde L, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 11.Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 12.Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helfrich-Forster C, Tauber M, Park JH, Muhlig-Versen M, Schneuwly S, Hofbauer A. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J Neurosci. 2000;20:3339–3353. doi: 10.1523/JNEUROSCI.20-09-03339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Cao G, Pavlicek B, Luo X, Nitabach MN. Phase coupling of a circadian neuropeptide with rest/activity rhythms detected using a membrane-tethered spider toxin. PLoS Biol. 2008;6:e273. doi: 10.1371/journal.pbio.0060273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, Bae E, Kim J. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 20.Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibañez-Tallon I, Wen H, Miwa JM, Xing J, Tekinay AB, Ono F, Brehm P, Heintz N. Tethering naturally occurring Peptide toxins for cell-autonomous modulation of ion channels and receptors in vivo. Neuron. 2004;43:305–311. doi: 10.1016/j.neuron.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Riehm JP, Rao KR, Semmes OJ, Jorenby WH, Hintz MF, Zahnow CA. C-terminal deletion analogs of a crustacean pigment-dispersing hormone. Peptides. 1985;6:1051–1056. doi: 10.1016/0196-9781(85)90427-9. [DOI] [PubMed] [Google Scholar]
- 23.Johnson EC, Bohn LM, Taghert PH. Drosophila CG8422 encodes a functional diuretic hormone receptor. J Exp Biol. 2004;207:743–748. doi: 10.1242/jeb.00818. [DOI] [PubMed] [Google Scholar]
- 24.Johnson EC, Shafer OT, Trigg JS, Park J, Schooley DA, Dow JA, Taghert PH. A novel diuretic hormone receptor in Drosophila: evidence for conservation of CGRP signaling. J Exp Biol. 2005;208:1239–1246. doi: 10.1242/jeb.01529. [DOI] [PubMed] [Google Scholar]
- 25.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Cao G, Nitabach MN. Electrical silencing of PDF neurons advances the phase of non-PDF clock neurons in Drosophila. J Biol Rhythms. 2008;23:117–128. doi: 10.1177/0748730407312984. [DOI] [PubMed] [Google Scholar]
- 27.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113:755–766. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]
- 29.Sheeba V, Sharma VK, Gu H, Chou YT, O’Dowd DK, Holmes TC. Pigment dispersing factor-dependent and -independent circadian locomotor behavioral rhythms. J Neurosci. 2008;28:217–227. doi: 10.1523/JNEUROSCI.4087-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2008;99:976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rieger D, Shafer OT, Tomioka K, Helfrich-Forster C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J Neurosci. 2006;26:2531–2543. doi: 10.1523/JNEUROSCI.1234-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshii T, Wulbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Forster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wulbeck C, Grieshaber E, Helfrich-Forster C. Pigment-dispersing factor (PDF) has different effects on Drosophila’s circadian clocks in the accessory medulla and in the dorsal brain. J Biol Rhythms. 2008;23:409–424. doi: 10.1177/0748730408322699. [DOI] [PubMed] [Google Scholar]
- 35.Fortin JP, Zhu Y, Choi C, Beinborn M, Nitabach MN, Kopin AS. Membrane-tethered ligands are effective probes for exploring class B1 G protein-coupled receptor function. Proc Natl Acad Sci U S A. 2009;106:8049–8054. doi: 10.1073/pnas.0900149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al-Fulaij MA, Ren Y, Beinborn M, Kopin AS. Identification of amino acid determinants of dopamine 2 receptor synthetic agonist function. J Pharmacol Exp Ther. 2007;321:298–307. doi: 10.1124/jpet.106.116384. [DOI] [PubMed] [Google Scholar]
- 37.Shinyama H, Masuzaki H, Fang H, Flier JS. Regulation of melanocortin-4 receptor signaling: agonist-mediated desensitization and internalization. Endocrinology. 2003;144:1301–1314. doi: 10.1210/en.2002-220931. [DOI] [PubMed] [Google Scholar]
- 38.Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 39.Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.