Disease reappears in the majority of leukemia patients who enter remission. For many years investigators have focused on detecting the "minimal residual disease" ultimately responsible for these relapses. Two primary methods, one based on polymerase chain reaction technology, the other on flow cytometry are in increasing use. This paper describes the application of such methodologies not only to chronic myeloid leukemia, childhood acute lymphoblastic leukemia, and acute promyelocytic leukemia but to other types of leukemia.
Keywords: minimal residual disease, leukemia, flow cytometry, standardization, monitoring
Abstract
Resistance to therapeutic agents is a major factor in the failure of cancer treatments. In leukemia, the resistant cells remaining in the bone marrow and/or peripheral blood constitute minimal residual disease and are detectable by highly sensitive assays when the patient appears to be in complete remission. Early detection of the expansion of residual cells permits clinical intervention with the aim of reversing the proliferation of resistant leukemic cells. Therefore, accurate and precise measurement of minimal residual disease can greatly enhance optimization of oncology patients' clinical management. This notion is supported by a large body of data among chronic myeloid leukemia patients, but minimal residual disease detection and monitoring is increasingly applied to other types of leukemia, and is starting to be a factor in decision-making for some therapeutic trials in childhood acute lymphoblastic leukemia. Here, from the solid ground of minimal residual disease detection in chronic myeloid leukemia, the current state of the art and development of molecular techniques in other leukemias and the growing field of multiparameter flow cytometry are reviewed in two separate parts reporting on the respective advances, advantages and pitfalls of these emerging methods.
Introduction
Rapid progress in understanding the etiology of hematologic malignancies and technological advances have in recent years increased the specificity and sensitivity of detection of malignant cells in patients who appeared to be cured or in remission by conventional techniques.1,2 Therefore patients’ therapeutic response can now be assessed by monitoring minimal residual disease (MRD) i.e. detection of malignant cells at ≥1×10−4 sensitivity, at sub-clinical levels.
However, MRD studies with sensitivities of 1×10−4 or higher have brought new challenges in differentiating malignant from normal cells and consequently, definition and clinical significance of remission and early relapse have become ambiguous. Thus, assigning prognostic value to MRD levels requires defined thresholds in selecting optimal therapeutic agent dose and the timing of hematopoietic stem cell transplantation (HSCT) or alternative drug; while conversely, treatment reduction could be considered in those predicted to have a favorable prognosis and thus minimize exposure to toxic agents.
Post induction therapy, one million or more leukemic cells may persist,2 even when the residual cells are undetectable, i.e. the patient appears to be in complete molecular remission (CMR). CMR is defined as failure to detect cancer cells by the most sensitive molecular methodology available, with acceptable control gene transcript numbers, e.g 10,000 ABL1 transcripts.3 Definition of CMR could be further refined by being valid only when leukemic cells are undetectable in three sequential samples one month apart, in addition to the prerequisite of an adequate number of control gene transcripts. In addition, an internationally recognized reference material enabling inter-laboratory comparison and an accurate assessment of the level of sensitivity achievable by a myriad of methodologies applied would strengthen definition of CMR. It is also generally accepted that informative MRD studies are best achieved when the peripheral blood leukocyte count is within normal limits4 with an adequate number of cells to achieve sensitivity of up to 1×10−4 to 1×10−5. For patients who had undergone HSCT, Mughal et al. defined molecular relapse (MR) as either three sequential samples, tested one month apart, with a BCR-ABL1/ABL ratio of 0.02% or showing clearly rising levels with the last two higher than 0.02%, or two results over a minimum period of four weeks higher than 0.05%.5 Thereby they were able to classify patients according to risk of progression. More generally, a confirmed one log increase in BCR-ABL1/ABL ratio or three consecutive increases is clinically significant. Timing and frequency of MRD is largely dependent upon clinical data and the aggressiveness of the leukemic clone, which is likely to vary between patients and diseases. A directive to perform regular close monitoring of peripheral and/or bone marrow at set time points may diminish the ambiguity and permit better inter-laboratory data comparison.
Tumor load, type of leukemia, whether disease specific marker is identifiable and technological limits will determine the optimum methodology for monitoring MRD. Whilst molecular monitoring targets disease-specific transcription of chimeric mRNA (e.g AML1-ETO) or utilizes somatic mutations, e.g. Nucleophosmin (NPM1) and/or B and T-cell clonal gene rearrangement, FCM detects the expression patterns of cell differentiation (CD) antigens thereby distinguishing leukemic cells from normal cells. Here we review the application of molecular and FCM methodologies, which are now also indispensible tools for diagnosis and monitoring chronic and acute leukemia.
Molecular studies
Polymerase chain reaction (PCR) has been eloquently exploited to detect and measure DNA sequences of interest. More recently, mRNA studies using reverse transcription PCR (RT-PCR) have become widespread. This approach brings junctional breakpoints separated by introns and exons into close proximity thereby enabling detection of different transcripts encoded by the same chromosomal translocation in a single multiplex PCR.6 Furthermore, RT-PCR detects RNA from viable cells and thus targets genes expressed that are likely to have functional role, directly or indirectly, in cellular proliferation.
Quantification of specific sequences of DNA has been greatly simplified by real time quantitative polymerase chain reaction (RQ-PCR),7 hereafter referred to as Q-PCR. In Q-PCR the rate of accumulation of amplicons is proportional to the number of target transcripts in the starting material during the exponential phase of the PCR. This technique also offers increased specificity with the inclusion of the third reporter labeled oligonucleotide probe using hydrolysis based technology, which anneals between forward and reverse primers.1 Hydrolysis is one of many methods now available for detection and quantification of target sequences.8
A sensitivity of 1×10−5 is achievable by Q-PCR but contamination is a major concern and hence strict working practices must be adhered to, e.g RNA extraction, cDNA synthesis and post PCR analysis must be geographically separated. Equally, false negatives due to a lack of mRNA or sub-optimum integrity of mRNA and/or cDNA must be controlled for. This is achieved by concomitantly measuring one of the ubiquitously expressed housekeeping genes, such as ABL1, BCR, β2-microglobulin, β-glucuronidase (GUSB) or glucose-6-phosphate dehydrogenase (G6PD).9–12
Gene rearrangement studies
The immunoglobulin (Ig) and T-cell receptor (TCR) gene rearrangements during normal B and T-lymphocyte development, respectively, generate unique fusions of variable, diversity and joining (VDJ) segments, interspersed by random nucleotide (N) insertion and/or deletion (Figure 1).13,14 These B and T-clonal recombinations generate patient-specific DNA length and sequences which represent ideal molecular markers for detection and quantification of leukemic cells among normal lymphocytes in remission samples. Whilst sensitive, the technology is susceptible to false negatives due to clonal evolution during natural history of the disease, thus some patients may relapse with a clone different to that observed at presentation. Furthermore, the sensitivity may be diminished through quenching by normal polyclonal B cells.15 The risk of false negatives can be diminished by targeting two Ig/TCR gene rearrangements when conducting MRD-PCR studies.
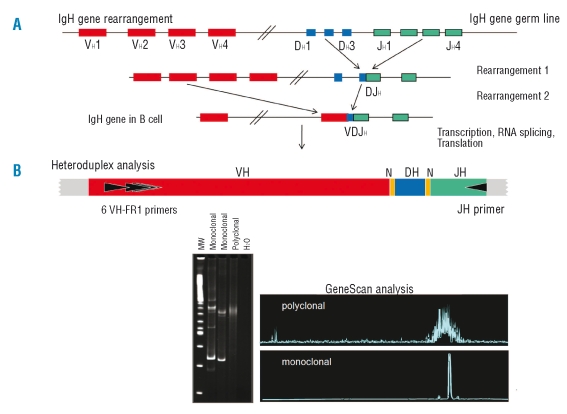
Figure 1.
IgH rearrangement and heteroduplex clonality studies by polymerase chain reaction (PCR). (A) A schematic representation of the IgH gene rearrangement is shown. Double strand DNA breaks are made to enable the V, D and J heavy immunoglobulin genes rearrangement in cells that are destined to become B lymphocytes and define these cells from others. Clonal population is detected when a group of cells with a unique VDJ spliced sequence expands such that it is detectable above the normal background of B-cell populations that have undergone rearrangement. (B) Heteroduplex analysis. (i) Schematic representation of the IgH rearranged gene (VH-DH-JH) is shown along with position of VH–family specific and JH consensus primers. The yellow lines represent the position of insertion and/or deletion of nucleotides at junctional regions of IgH. (ii) Ethdium stained acrylamide gel with results of a heteroduplex analysis is illustrated. In brief, the junctional region heterogeneity of PCR products of rearranged IgH or TCR genes is exploited to distinguish between monoclonal and polyclonal lymphoid B or T expansion. In heteroduplex studies PCR amplicons are heat denatured and rapidly cooled to induce homo- or heteroduplex. Samples with clonal lymphoid cells the PCR products of rearranged IgH or TCR genes yield homoduplex. In contrast samples with polyclonal lymphoid expansion the single strand PCR fragments leads to formation of heteroduplexes upon re-annealing. In samples with polyclonal and monoclonal expansion, both homoduplex and heteroduplex arise. Thus, because of the conformation differences the homo and heteroduplexes forms can be separated from each other by gel electrophoresis through non-denaturing acrylamide gels as shown. Homoduplexes migrate through the gel faster than the heteroduplexes with imperfect complimentary pairing. The latter form a background smear of slow migrating fragments. The homoduplex yields a relatively sharp discrete amplicon band. (iii) Automated clonal studies (florescent Genescan analysis). Polyclonal VH-DH-JH products form peaks reflecting a Gaussian distribution of average junctional region sizes in normal B lymphocytes. Monoclonal VH-DH-JH gene rearrangements form a discrete fluorescence peak, representing products of identical size. Adapted with permission from Van Dongen et al. Leukemia 2003;17:2257–17.
Consensus primer PCR and allele specific oligonucleotide PCR (ASO-PCR) are the two immunoglobulin (Ig) PCR strategies for MRD studies (Figure 1). The principle of the former is to amplify the third complementary-determining region (CDRIII) of the Ig gene, using a standard set of universal primers, a primer that recognizes a consensus sequence in the JH region and a primer for family specific framework regions (Figure 1). This qualitative method has sensitivity of 1×10−2 to 1×10−4. ASO-PCR utilizes primers designed to anneal to a unique patient specific Ig sequence and subsequently is used to monitor sequential samples in follow-up studies. This method overcomes the difficulty associated with the presence of normal polyclonal B cells and significantly improves the sensitivity of MRD studies. However, ASO-PCR is time consuming and expensive, despite the improvements following the introduction of Genescan which eliminates the need for polyacrylamide gels (Figure 1).15–18 Combination of ASO primers and consensus oligonucleotide probes make it accessible to Q-PCR, permitting precise quantification of MRD with a sensitivity of 1×10−4 to 1×10−5.
PCR-based studies have proved to be more sensitive than FCM, with around 10% of the cases having detectable clonal rearrangement in patients with disease below the FCM detection limit.19,20 Screening at intervals by PCR, with closer monitoring by ASO-PCR and FCM in parallel would reduce the risk of false negative data. Despite the complexity with improved inter-laboratory standardization the Ig-TCR studies15–18,21 are increasingly being seen as the gold standard for ALL MRD studies.
Standardization
Biggs and Denson in 1967, concerned with coumarin therapy, observed that a single scale properly applied at different centers would ensure safety and uniform dosage for a patient moving from one place to another and would greatly improve the standard of clinical trials carried out at more than one center.22 As a consequence, the International Normalised Ratio (INR) evolved. Similarly, in an era of multi-center clinical trials and wide accessibility of MRD studies by Q-PCR the need for standardization of MRD studies is essential. However, standardization of RNA based studies has proved to be complex because mRNA is labile and cDNA synthesis adds to the logistical difficulties in establishing inter-laboratory quality assurance. Such difficulties are negligible with the highly stable genomic DNA, as the latter can be made available easily and the quantity included in MRD assays can be measured with much greater accuracy than RNA. Hence, considerable progress has been made in standardization of Ig and TCR rearrangement studies.15–19,21 Furthermore, since the amount of DNA per cell can be calculated, reporting in terms of number of malignant cells is feasible.
Because of the difficulties associated with standardization of RNA studies, it is described here in greater detail, illustrated by BCR-ABL1. A meeting convened in 2002 in Bethesda, Washington, made some key recommendations when monitoring CML patients in the era of tyrosine kinase inhibitors9 and proposed an International Scale (IS) for BCR-ABL1 measurement based on the use of ABL1, BCR or GUSB control genes.9,23 This meeting, the Europe Against Cancer (EAC) consortium, as well as other investigators, highlighted the need for protocol standardization given inter-laboratory diversity of methodologies from collection of sample to final report.9,24–25 Despite achieving a consensus with key factors, e.g. use of random hexamers, processing of samples within 24 h, the lack of World Health Organisation (WHO) recognized primary reference material make inter-laboratory comparisons and accurate assessment of achievable sensitivity difficult. Thus, an urgent need for a WHO recognized External Quality Assurance (EQA) standard for the MRD targets soon became apparent. The establishment of EQA requires a suitable material that would enable all steps involved in MRD analysis to be monitored and one that is stable over a long period.26,27 Furthermore, it must be non-toxic, non-infectious and readily available. A number of investigators have suggested biological materials that might be suitable for an EQA scheme such as lyophilized cell lines that express the target genes or synthesis of protected RNA that can be reverse transcribed once it is heated to the appropriate temperature.19,20 Whilst these products mimic the starting material and therefore test the major aspects of MRD analysis, their long-term stability and suitability remain to be determined. The use of armored RNA as a reference material has enormous potential. In the absence of formally recognized WHO standard, consensus is emerging to use formulae applied in the landmark CML clinical phase II multi-center trial International Randomised study of Interferon and STI571 (IRIS) molecular report.23 Briefly, the international BCR-ABL1 scale (corresponding to the IRIS baseline) baseline is set at 100%, and major molecular response (MMR), is defined as 3-log reduction, i.e. 0.1%. By exchanging samples with a reference laboratory conversion factor (CF) the International Standardised Ratio (ISR) is calculated per formulae shown below.28 The baseline value must be defined by individual laboratories and this can vary between the centers. The ISR would probably benefit from the adoption of a recommended reference method and reagents, thereby further enhancing inter-laboratory comparisons.
Formulae for calculating the IS BCR-ABL1 ratio.28
BCR-ABL1:100
BCR-ABL1 MMR = 0.1
CF=MMR-IS ÷ MMR-LL
Then, LR = CF X Y
Where:
MMR:Major molecular response
IS:International standard
LL:Local lab BCR-ABL1 MMR
CF:Conversion factor
LR:Local international standardised BCR-ABL1 % ratio
Y:locally BCR-ABL1 % ratio for a given patient
In the following section the reported MRD findings for different types of leukemia are summarized.
Chronic myeloid leukemia
The clinical utility of MRD studies was proven in CML patients who had undergone HSCT3,29,30 and has been shown to be equally useful for tyrosine kinase inhibitors (TKI) treated patients.23,31–33 Indeed Q-PCR has largely replaced cytogenetic or FISH for closely monitoring CML patients’ response, particularly as a vast majority of patients achieve CCyR with TKI. Q-PCR also helps detect emerging TKI resistant clone much earlier than other methodologies.
Patients who do not achieve 1-log reduction at three months are said to have a reduced probability of achieving CCyR and/or MMR.31–35 More precisely, those who achieve less than 1-log reduction three months post therapy have a 13% chance of ever achieving MMR compared with >70% patients who have a deeper depletion at three months.31–36 Although long-term longitudinal studies are needed for confirmation, the published reports support the notion that early and close monitoring of patients assists in classifying patients likely to achieve MMR and permits the early detection of a resistant clone. A 2-fold increase in tumor load is reported to be suggestive of BCR-ABL1 kinase (KD) mutation, diminishing TKI efficacy.37 In addition, findings suggest that detection of KD mutation in patients who are in CCyR is associated with a significantly increased risk of cytogenetic relapse.38 By inference, together these observations suggest patients who experience a 2-fold increase in BCR-ABL1 transcripts have significantly increased risk of cytogenetic relapse. Furthermore, recent analysis implies patients in whom BCR-ABL1/ABL1 ratio is ≥0.05% have a statistically significant risk of loss of CCyR and progression free survival.39
Acute myeloid leukemia
Molecular monitoring by Q-PCR in AML is largely limited to fusion genes resulting from chromosomal aberrations, and exemplified by t(15;17),40 t(8;21)41 and inv(16),42 quantification of somatic mutations using mutation specific e.g NPM143 and aberrantly expressed genes e.g. ecotropic virus integration-1 (EVI1).44 The accumulating AML MRD data support the notion that such studies are an essential tool for relapse risk stratification of patients during treatment.45–47 A study among 70 APL patients showed that MRD levels after first consolidation therapy was a powerful predictor of relapse.48 Patients with residual disease ≥1×10−3 had a 10-fold increase of relapse at five years compared with those who had <1×10−3 (p=0.001).48
Rearrangements involving the core-binding factor AML1 and CBFβ resulting from t(8;21) and inv(16) are considered to be associated with good prognosis, and account for 15–20% of adult and pediatric AML cases. However, monitoring MRD in AML1-ETO and CBFβ-MYH1 patients is less than straightforward, as qualitative RT-PCR is often positive even when all other indicators are consistent with the patient being in long-term remission. This might be due to expression of AML1-ETO in non-leukemic stem cells, monocytes, and B cells in remission marrow, and/or in a fraction of B cells in leukemic marrow.49 These authors conclude that chimeric fusion gene is acquired in the hematopoietic stem cell and it is the acquisition of additional genetic lesions that lead to transformation of the affected stem cell.49 This implies the additional lesions arise downstream during differentiation or that AML1-ETO is only functional in more mature cells. Alternatively the persistence of AML1-ETO in long-term remission patients may reflect level of sensitivity and/or the number of cells that can be detected by RT-PCR. However, in either case, these observations highlight the need for quantification to assess the kinetics of the leukemic clone.50,51 More generally, reports suggest that a <1.0% (<1×10−3) MRD post-induction therapy correlates with good outcome.52–56
In the absence of disease-specific target molecular markers, the tumor suppressor gene expression levels, e.g WT-1, have been reported to be useful. WT-1 expression, normally highly regulated, is reported to be over-expressed in approximately 80% of AML patients and is therefore considered to be a specific feature of AML.57,58 There is evidence that all patients with higher levels of WT-1 in peripheral blood post induction therapy subsequently relapsed, with a median of 12 months after diagnosis. But, the notion that expression level at diagnosis is prognostic could not be confirmed.59 Furthermore, the significance of normalized WT-1 expression levels post induction therapy was less clear, as 21 of 48 these patients relapsed. These findings are supported by Ommen et al.60 The available studies indicate that WT-1 levels above normal levels, which may be seen in normal regenerating marrow, are associated with subsequent relapse. These data are supported by finding them to correlate with disease-specific fusion gene transcript numbers.57,58 Similarly, the overexpression of EVI1, mapping to 3q26, has been described in 8.0–20.0% of AML cases. The increased EVI1 expression is reported to correlate with worse prognosis and is therefore a useful marker for evaluation at diagnosis and follow-up studies.61–63 Furthermore, some AML patients express ME (ME+) resulting from intragenic fusion between MDS1 and EVI1, the former is 140 kb upstream of EVI1. Interestingly, patients who express EVI1 but are ME negative are reported to have poorer treatment response.44 Similarly, the preferentially expressed antigen of melanoma (PRAME) is over-expressed in 30–40% of cases and has been suggested as a possible marker too.63 PCR has also been applied in AML patients to detect mutations reported to have prognostic value, e.g. NPM143 and FLT3.64 An internal tandem duplication (ITD), that adds 5–100 base pairs to the juxtamembrane domain, is the most frequently observed FLT3 mutation.64–66 The presence of FLT3-ITD at diagnosis in AML is reported to be associated with a 8.5-fold higher frequency of MRD cells after the first course of chemotherapy compared to those with wild-type FLT3.67 This correlates with overall survival (OS), relapse free survival (RFS) and disease free survival (DFS), and if confirmed early evaluation of FLT3 inhibitor efficacy would be feasible. However, although these mutations are relatively common in normal karyoptype AML, their potential as MRD markers is unclear due to the need to design patient specific assays and mutant alleles instability.68–70
Exon 12 NPM1 mutations that displace the protein to the cytoplasm represent the most common genetic lesions observed in AML patients with normal karyotype. Three of the mutations account for 90% of all mutated cases.71 Reported data imply that unlike FLT3, NPM1 mutant alleles are stable and therefore a reliable MRD marker with prognostic value.72–75 Moreover, sensitivities of ≥10−5 have been reported to be achievable by targeting NPM1 mutations.71–74 The CCAAT/enhancer binding protein α (CEBPA) mutations, observed in approximately 10% of AML patients associated with good prognosis, have also been proposed as markers to monitor AML.75 In a study which included 149 patients’ samples analyzed at diagnosis and relapse, the CEBPA mutations were found to be stable, raising the possibility of patient specific Q-PCR MRD studies.75 Recently authors described the utility of CEBPA, NPMI and FLT3 mutations at diagnosis in cytogentically normal AML patients under the age of 60 to define those who might benefit from HSCT.76 Furthermore, FLT-3 mutation as predictor of relapse may be modified by NPM1.
Acute lymphoblastic leukemia
Philadelphia chromosome positive ALL patients’ leukemic clone kinetics can be monitored accurately and precisely by Q-PCR using BCR-ABL1 as the target.77,78 By these studies 42 patients could be classified into two groups as follows: good molecular responders with >2-log and >3-log reduction after induction and consolidation therapy, respectively, and poor molecular responders, with higher MRD levels at both time points. The two year OS was determined to be 48% and 0% for good and bad molecular responders (p=0.0026), respectively.78 The studies among Ph (+) ALL patients suggest that those expressing e1a2 BCR-ABL1 transcripts, representing approximately 70% of Ph(+) ALL, have a higher risk of relapse.79–81 The risk is estimated to be 8.7 compared to 2.2 for those expressing e13a2 and/or e14a2.81 The relative risk of relapse is estimated to be 4.4 among those patients who have detectable MRD compared with those in whom the BCR-ABL1 transcripts are undetectable at 4–6 months post transplant.81 Available data imply that MRD levels are higher in the bone marrow than peripheral blood; therefore BM samples should be tested at regular intervals.82 However, as PB based MRD studies are relatively non-invasive, patients are much more likely to acquiesce to them and therefore permit closer monitoring. This may offset the 1-log greater sensitivity achieved with BM samples. Thus, PB based analysis could be used as an indicator for timing of BM samples and confirmation of any minor changes. Furthermore, lymphocyte enrichment is essential to maximize sensitivity when analyzing PB samples from ALL patients.
Early clearance of leukemic cells is a favorable prognostic indicator in childhood ALL, whereas high levels of MRD at the end of induction therapy appear to be associated with a high risk of relapse.83–86 Available data imply that low molecular MRD after induction is a good prognostic factor in pediatric ALL, independent of other clinically relevant risk factors, such as age, blast count, immunophenotype, chromosomal aberrations at diagnosis and response to prednisone. Furthermore, investigators report that patients who relapse after remission and are again subjected to re-induction therapy have event free survival rates of 86% and 0% among those determined to have MRD levels of <1×10−3 and ≥1×10−3 by RQ-PCR (p<0.0001), respectively.87 The adult MRD ALL data is relatively limited but as in children, early depletion of tumor load after induction is prognostic of response to chemotherapy. A rapid decline in MRD levels, down to undetectable or <1×10−4 on days 11 and 24, during and after induction therapy, respectively, has been reported to be associated with low risk with three year DFS and OS of 100%. Conversely, patients with a decline of 1×10−4 reached at week 16 were at high risk, with a 3-year DFS and 3-year OS at 45.1%.88 In contrast Mortuza et al., reported that MRD positivity was associated with increased risk of relapse which is more pronounced at three and five months post induction.89 Significantly, the deletion of the IKZF1 gene, that encodes IKAROS Krupple family zinc finger transcription factor in ALL, is associated with poor prognosis independent of age, sex, cytogenetic findings, leukocyte count at diagnosis and MRD data.90
Chronic lymphocytic leukemia
Although patient management decisions are largely based upon clinical data, Rai91 and Binet92 staging has in recent years been superseded in stratification of CLL patients at diagnosis into good and bad prognosis by a myriad of markers.93 The most widely used molecular marker, the immunoglobulin variable region heavy-chain gene mutational status segregates CLL according to aggressiveness of the disease.94,95 The complexity of this assay led to the description of alternative surrogate markers. The most commonly assessed include ζ-chain associated protein kinase (ZAP-70),96 CD3897 and the presence of chromosomal aberrations (e.g. 11q and 17p and deletions)88,98 dysfunctional and/or mutated p53.99
Wierda et al. reported achieving 1×10−5 sensitivity when evaluating a chemotherapy regimen with fludarabine, cyclophosamide and rituximab (FCR) for CLL using patient specific PCR based assay.100 These investigators showed 21% of the patients assessed had undetectable MRD, with median time of progression of 44 months compared with 27 months with detectable CLL cells.100
The European Research Initiative in CLL (ERIC) concluded that a sensitivity of 1×10−4 was achievable either by ASO-PCR or FCM101 and defined MRD negativity as less than a single CLL cell in 10,000 leukocytes.102 However, it is unclear if MRD negativity is to be based on a single sample analysis or sequential samples. Given that ASO-PCR is theoretically 1-log more sensitive, it might be preferable to report MRD negativity as defined by ERIC, as undetectable as there may be up to a million or more CLL cells in samples which are reported to be negative by ASO-PCR. Significantly, it is suggested that PB samples are acceptable for monitoring most therapeutic agents, except perhaps alemtuzumab or rituximab, where bone marrow aspirate samples could be preferred, as antibody clears PB faster than BM.102
MRD data support the notion that CLL patients with undetectable disease post HSCT or immunotherapy have a greatly improved overall survival. These observations are consistent with early identification of patients at risk of relapse by monitoring MRD. In CLL patients treated with anti-CD52 antibody based therapy (CAM-PATH, alemtuzumab) or autologous transplant, positive MRD studies, FCM- or PCR-based, are highly indicative of subsequent relapse. Rawstron et al., reported that for 19 of 25 cases who achieved CR when treated with CAMPATH-1H antibody or autologous transplant with undetectable MRD, the EFS was >90%,103 whilst those with detectable MRD at time of CR subsequently relapsed.103 Similarly, Esteve et al.,104 reported that 4 of 5 (80%) patients with detectable MRD, while in CR following autologous transplant, eventually relapsed compared with 2 of 9 (22%) with undetectable MRD.104 Whilst these studies support the emerging consensus of the critical need for MRD studies in CLL, they require confirmation given the small sample numbers.
Flow cytometry
The notion that minimal residual disease (MRD) could be monitored by using the increasingly versatile and specific capacities of flow cytometry (FCM) emerged as early as the late 80s, following studies of normal mouse bone marrow.105
FCM has evolved gradually over the last 20 years, with advances in technology and software making it increasingly accessible for MRD detection, such that it is now becoming an essential tool for monitoring malignant clones.106 By contrast with molecular studies, it is of interest that FCM will explore viable cells. It is likely that this approach will develop increasingly in therapeutic trials, first for appraisal of the feasibility and informativity of the method, later probably as a therapeutic decision-indicator, as is already the case for molecular MRD in childhood ALL.107,108
This part of the review will first depict general considerations about the use of FCM in MRD detection, while recalling the major steps of the development of this technique, which sustain the rationale for its broader application.
Immunophenotypic patterns
From a strictly technological point of view, the construction of monoclonal antibody (Moab) combination panels applicable for the detection of MRD should consider four critical issues:
- lineage and maturation related molecular associations, in order to identify the coexpression of antigens normally mutually exclusive;
- cross-lineage expression, which may occur as part of the abnormal characteristics of the leukemic clone;
- differentiation antigens expression intensity, which has to be known from normal expression, in order to best choose the appropriate fluorochromes and avoid possible steric hindrance and quenching for antigens in close vicinity on the cell membrane.
These panels, may also behave differently when applied to normal versus leukemic cells and should be versatile enough to take these features into account.
Indeed, the challenge in FMC detection of MRD is to separate residual leukemic cells from non-malignant cells, including at stages when regenerating BM may contain more early maturation forms than normal samples at a steady stage of hematopoiesis. This presupposes both excellent immunophenotypic knowledge of the malignant clone and of normal bone marrow.
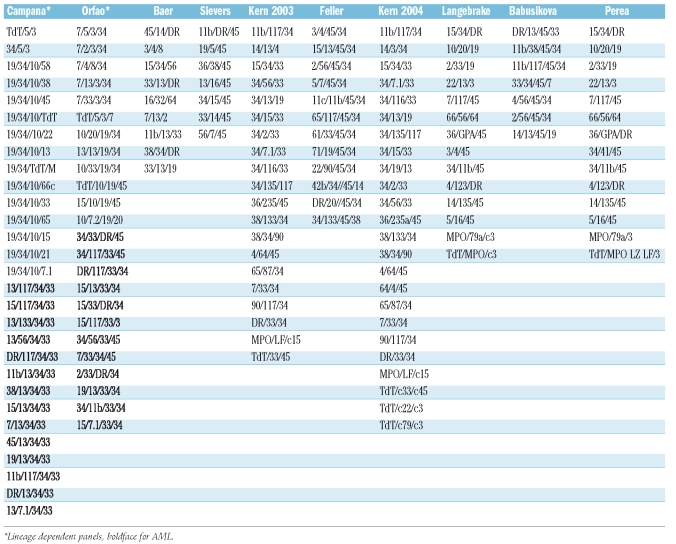
Leukemic cells can be similar to normal cells blocked in their differentiation and are recognized as such in FCM diagnosis by increased numbers compared to the scarcity of normal cells expressing the same immunophenotype. But, on closer examination, leukemic cells also frequently express abnormal maturation patterns. These immunophenotypic alterations have been referred to as leukemia associated immunophenotypic patterns (LAIP).109 They can be classified in four types: i) cross lineage or aberrant expression, ii) asynchronous expression, iii) overexpression and iv) lack of expression.109 The immunophenotypic diagnosis of leukemias must therefore be meticulous, in order to identify such patterns, which may subsequently be used to track the residual leukemic cells. The earliest contributions to tracking abnormal patterns in flow cytometry can be traced back to the early 1990s.110 Earlier attempts111 had used cytospin smears to identify the combined expression of cytoplasmic CD3 and terminal deoxynucleotidic transferase (TdT) in ALL. In 1990, Campana et al.112 reported on dilution experiments of AML cells coexpressing CD33 and CD7 in normal bone marrow, demonstrating the absence of such cells in normal BM and their retrieval in two-color FCM down to 1×10−3. Gross et al. in 1993,113 using dilutions of the cell line REH with 250 million of normal PBMC showed that such mixtures allowed retrieval of abnormal cells with a sensitivity of 1×10−6. This work also usefully defined the notion of empty spaces not occupied by normally maturing cells. This notion of empty spaces, however, is only significant following detailed studies of normal bone marrow with the same antibody combinations as designed for MRD detection, and the latter are clearly scarce and are not standardized in the literature.114–116 Table 1 provides an example of the large variety of proposals made in the literature to detect MRD in FCM.
Table 1.
Targeting blast cells
In 1997, Jennings and Foon,127 reporting on the diagnosis and follow-up of leukemia, stressed the need to use CD45 to discriminate cell subsets and most notably the immature/blastic cells with low side scatter (SSC) and low CD45 expression.128,129 Using this approach in a first gating for the selection of MRD within maturing cells in the BM is becoming increasingly common (Figure 2A). In the same paper,127 these authors also reviewed the description of aberrant immunophenotypes reported by a number of investigators, i.e. CD2 in t(15;17) AML or CD19 in CBF t(8;21). In 1999, Campana and Coustan Smith117 estimated that the top four combinations for acute leukemia MRD detection in flow were (i) CD19/CD34/CD10; (ii) CD13/CD33/CD34, iii) CD13/CD33/CD117 and iv) CD13/CD34/CD117. It infers that these immunophenotypic features should be assessed at diagnosis, in order to retain information as to the specific characteristics of a given leukemic clone in terms of coexpression and fluorescence intensity of differentiation antigens. It should, however, be noted that in some cases, these features may be lost over time.130–132 The subset of leukemic cells which persists will nevertheless retain a clonal quality, i.e. continue to present homogeneous features. These residual cells appear as a tight cluster of cells still with a frozen immunophenotype among maturing cells in FCM studies. Although no real consensus exists, it is commonly admitted that a cluster of between 10 and 100 MRD cells should be identified in a given sample to ensure that MRD cells have been seen.132 Thus, to achieve a sensitivity of between 1×10−4 to 1×10−5, approximately 105 to 106 leukocytes must be screened, stressing again the value of assessing MRD in samples with normalized cell counts.4
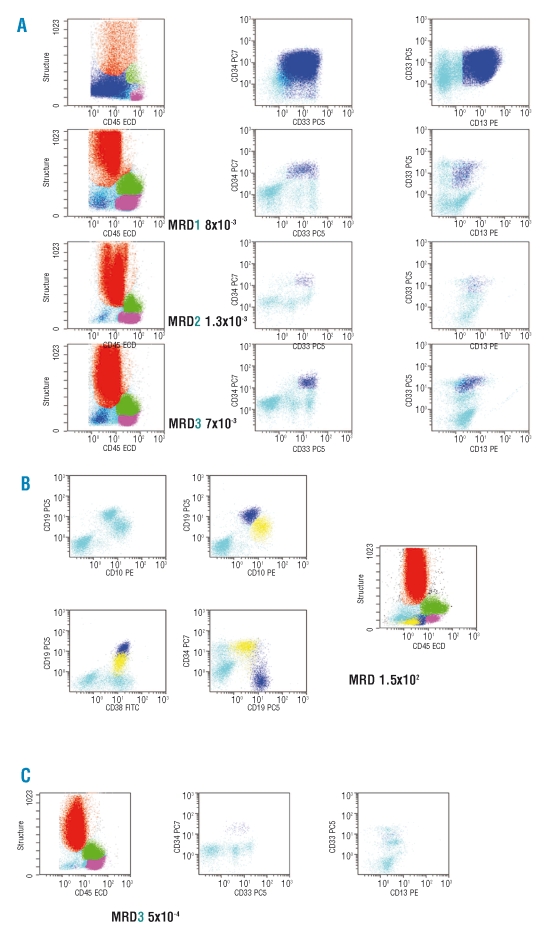
Figure 2.
Monitoring minimal residual disease (MRD) in FCM. (A) Backgating of leukemic cells on a CD45/SSC flow cytometry scatter-gram. The procedure of backgating was used to superimpose the population of cells identified at diagnosis and as MRD by multiparameter flow cytometry in bone marrow samples of a patient with AML. Color legend:granulocytes in red, monocytes in green, lymphocytes in purple, immature cells in cyan, MRD in dark blue. (B) Hematogones and B-ALL leukemic cells. Hematogones are shown in dark blue while the remaining blasts are displayed in yellow. Other colour code as above. (C) MRD in peripheral blood of an AML patient. Same patient as in panel A.
Data expression
Although not necessary, Ficoll enrichment in earlier studies was established as a standard procedure for samples from ALL patients. ALL MRD is thus often expressed as a proportion of mononuclear cells132 rather than as a proportion of leukocytes, as recommended for AML. More significantly, density gradient centrifugation may lead to loss of the MRD cells in AML studies. Therefore, in AML MRD studies, total cellular analysis following red cell lysis, especially in no-wash procedures, will provide the most clinically relevant picture. For these reasons this strategy is increasingly being adopted for ALL patients.
Bone marrow or peripheral blood?
For acute leukemia, there is usually 1-log difference between BM and PB analysis, with the highest MRD in BM.118,133,134 Thus, as with molecular based studies, PB could be exploited to establish MRD monitoring schedules with more frequent FCM investigations, with BM sampling being restricted to cases where MRD is repeatedly undetectable in PB samples.
Acute lymphoblastic leukemia
MRD detection in ALL, and especially childhood ALL, has been extensively explored with molecular tools derived from the specific rearrangements of the IgH or TCR, even leading to a new definition of remission.135 Concomitantly, FCM studies also developed, albeit with very heterogeneous panels, illustrating the broad potential of FCM in identifying ALL residual cells. The absence of consensus, even with multiparametric FCM, has probably delayed full recognition of this method as a valuable decision-making tool, while molecular detection of MRD was more readily applied.
The first series using flow cytometry appeared in the early 90s.110–112 Drach in 1991136 developed a two-color indirect fluorescence FCM assay combining surface staining and intranuclear labeling of TdT, reporting a sensitivity of 2×10−3. The same year, Imamura and Kuramoto137 reported on CD10/TdT staining in FCM in a small series of 6 ALL patients. In one of these patients, while considered to be in morphological remission, FCM detected about 2% of aberrant cells predicting later relapse. A new two-color FCM study from Drach in 1992138 included 24 ALL patients. Several authors then reported on two- then three-color approaches, using aberrant antigen expression on ALL blast cells.130,139
In 1998, a large study from Dario Campana's group140 described data that correlated well with other reported predictive features associated with either good or poor prognosis.141 The techniques used at this time were quite cumbersome yet began to consistently reach a sensitivity of 1×10−4.
The increasing application of FCM was also soon reflected in T-ALL studies.112,142
A number of investigators19,20,110,132,143–145 studied the correlation between FCM data and results obtained with PCR analyses, showing good correlation and emphasising the fact that FCM could be used for more patients.
Later on, the increasing sophistication of FCM led to more and more pertinent studies. Interestingly, Coustan-Smith et al.145 reported in 2006 on a simplified assay based on a three-color combination (CD19/CD10/CD34), which when applied at the post induction timepoint was highly significant for predicting relapse in childhood B-lineage ALL. In ALL studies, three FCM approaches have been mostly used.
Mike Loken group’s suggestion to identify patterns at variance with those expressed by normal bone marrow cells, was applied as early as 1998146 in a three color approach with CD45, systematically present in all combinations of monoclonal antibodies.
Another approach uses a minimal standardized panel for B-lineage ALL, also defined for normal cells such that it becomes possible to pinpoint abnormal events,144,147,148 More recently, a large study from the Children's Oncology Group149 confirmed the efficacy of a two-tube strategy in a series of over 2,000 children with a threshold of 1×10−4.
A third approach, based on the detection of LAIP, was mostly developed by Dario Campana and Elaine Coustan-Smith. The strategy of this group, applicable to common B-lineage ALL, is based on the detection of aberrant expression of a number of antigens on cells homogeneously defined by the combination of CD19, CD34 and CD10. In a large study in 2000, Coustan-Smith et al.150 reported that 204 of the 350 children who benefited from immunophenotyping at diagnosis exhibited an LAIP and 195 were included in MRD detection by FCM. The major difficulty in detecting MRD for patients with common B-II ALL (CD10+) is to differentiate blast cells from hematogones, which can be quite abundant in regenerating bone marrow. The combination CD34/CD19/CD10/CD38 is quite pertinent to differentiate these cell types, based on the difference in fluorescence intensity displayed by hematogones and blasts (Figure 2B).
The value of CD58 detection for FCM MRD analysis was confirmed in two series,151–152 supporting an earlier study by de Waele et al.153
The 2002 study by Coustan-Smith et al.118 emphasized the detection of MRD in peripheral blood by FCM.
The NOPHO Scandinavian study published in 2003 used tailored panels issued from the Biomed I concerted action. In this series of 70 children, a threshold of 1×10−4 was established as of prognostic value.154 Finally, a recent paper155 reported on the reproducibility of MRD detection in FCM in a multi-center study.
Acute myeloid leukemia
In 1997, San Miguel et al.156 published a landmark study carried out among 84 AML patients, demonstrating the prognostic value of thresholds of 5×10−3 MRD cells in the first remission BM and of 2×10−3 MRD cells at the end of intensification.
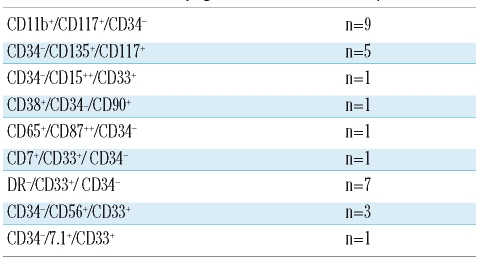
In a large study involving MRD assessments at a range of timepoints throughout treatment in 56 AML patients, Venditti et al.157 tested 437 BM samples and reported that the most frequent aberrations observed were CD33+/CD7+ (±CD34+) and CD34+/CD11b+ (±CD117+). Baer et al.120 and Langebrake et al.124 showed, using three-and four-color approaches, respectively, that LAIP shifts do not affect MRD monitoring in AML. Kern et al.122 endeavoured to define at least one LAIP per patient by comparison to 26 normal BM samples. LAIP were observed 140 times for 68 patients in 50 different patterns. Only one LAIP was seen in 28 cases. Among the 50 patterns reported, 10 included a lack of CD34 expression (Table 2). Feller et al.123 studied samples from 72 patients, which allowed for the identification of 122 LAIP (Table 3). Two further studies by the Munich group109,158 explored the possibility of shifting from an LAIP approach to a more comprehensive general strategy irrespective of the individual patient's features. This work confirmed the presence of LAIP patterns in normal bone marrow with a frequency of 7 to 3×10−4 cells. Feller et al.159 in 2005, used the same 12 tube panel as reported by Kern et al.122 in BM and mobilized PB samples from 54 patients, showing the presence of blasts in mobilized PB in AML. This was also reported in non-mobilized samples, by Maurillo et al.160
Table 2.
Combinations identifying LAIP with CD34 lack of expression.122
Table 3.
Cumulative incidence of LAIP in acute myeloid leukemia.123
It is generally accepted that in AML the presence of myeloid cells of clearly immature immunophenotype in PB is aberrant, especially if they obey the criteria of low SSC/low CD45 (Figure 2C). These observations make the development of MRD detection in AML very attractive in easy-to-obtain PB samples, thus, making a rapid turnaround of data possible. Perea et al.126 tested MRD in CBF AML and reported clinically discriminative thresholds of 1×10−3 for FCM and 10 copies for molecular studies. A less sensitive approach was reported after induction by Sievers et al.121
The impact of MRD assessment in AML is likely to evolve in the coming years. It might be of particular relevance in younger patients for whom allo-SCT is discussed, as suggested by the study from Laane et al.161
Chronic lymphocytic leukemia
In CLL, the recent development of aggressive but apparently successful therapies aroused a renewed interest in MRD detection. MRD studies in CLL patients are non-invasive as these can be performed using peripheral blood. However, normal B cells from the innate immune system, with immunophenotypic features akin to those of B-CLL exist physiologically. This theoretically decreases the sensitivity of MRD detection because the specificity threshold could be higher. However, little is known of the fate of these normal cells after CLL-directed chemotherapy. Among the first studies dealing with MRD in CLL, Rawstron et al.103 studied PB and BM with the aim of distinguishing CLL cells from the early B-cell population of hematogones in BM. This phenotype was expressed by less than 2% of normal B cells, or 1x10−4 leukocytes. Caballero et al.162 proposed a different four-color combination of CD23/CD79/CD19/CD5 which yielded a 1×10−3/1×10−4 sensitivity in BM. In 2006, Moreno et al.101 retained for a 10−4 threshold in both PB and BM the two combinations CD20/CD79b/CD19/CD5 and CD22/CD23/CD19/CD5.
Relying on clonality by assessing immunoglobulin light chains did not yield good results in their study. Maloum et al.163,164 in two different studies also retained the CD19/CD5/CD20/CD79b combination. Interestingly, Kay et al.165 published a clinically relevant study in which remaining CLL cells were solely identified by the two color combination CD5/CD19 on PBMC and applied a 1% threshold. More recently, however, Rawstron et al.166 reported on an international standardization approach recommending three combinations, respectively associating CD5/CD19 with CD20/CD38, CD81/CD22 and CD79b/CD43.
Chronic myeloid leukemia
Very few studies have considered flow cytometry for the detection of MRD in CML. Mention can be made of the work of Lanza et al.167 suggesting the interest of tracking CD56+ CD34+ cells.
Summary
The notion that monitoring MRD is highly useful in the stratification of patients according to risk of relapse by molecular and/or FCM is now widely accepted and increasingly incorporated in the follow-up design of multi-center trials, assessing novel therapeutic agents for rare disorders. Apart from this clinical application, MRD studies will undoubtedly also help in better understanding the kinetics of the leukemic clone and thereby the biology of the malignant cell. The value of reported prognostic markers, to be associated with aggressiveness of the malignant clone at diagnosis are open to conjecture. However, all indications are that they will prove to be useful. The identification of prognostic markers could be used to aid the timing and the frequency of MRD studies, although patients’ age, type of sample and leukemia will also influence the frequency of MRD studies. RQ-PCR and improvements in FCM have made these techniques highly useful, there is now a need for defined reference methods and robust EQA schemes which will help to further strengthen the utility of MRD studies.
In conclusion, MRD studies, molecular and FCM, through early detection of relapse at sub-clinical levels permit early clinical intervention, perhaps before early progenitor cells, including CD34+ cells, acquire genetic lesions that increase the aggressiveness of the clone. These methods are, therefore, highly useful in improving the prognosis of hematologyonclogy patients. Central to optimization of clinical management is preemptive modulation of therapy, for example, the need for more intensive therapy to overt the potential risk associated with MRD positivity post consolidation. Finally, genetic alterations at diagnosis or as the disease evolves or as a consequence of treatment, e.g BCR-ABL1 kinase domain mutation, affect the function and signaling molecules, transcription factors, growth-factor receptors and influence the response to treatment. As these genetic lesions are identified it will become increasingly time consuming and inefficient to detect and quantify the numerous markers in separate assays. Therefore, micro-array may be needed to be developed. This has the potential to provide a patient specific disease footprint that could be used to tailor patient specific therapy and optimize clinical management in conjunction with MRD studies.
Acknowledgments
we are grateful to members of the WP12 and WP10 of the European LeukemiaNet initiative for their support and critical review of the manuscript, Drs. G. Basso (Padua), P. Bettelheim (Vienna), D. Grimwade (London), W. Kern (Munich), F. Lanza (Ferrara), A. Porwit (Stockholm), A. Rawstron (Leeds), S. Richards (Leeds), G. Saglio (Turin), G. Schuurhuis (Rotterdam), T. Te Kronnie (Padua), and V. Van der Velden (Rotterdam).
Footnotes
Authorship and Disclosures
Both authors contributed equally to this paper.
The authors reported no potential conflicts of interest.
The authors would also like to thank Professors J Goldman (London) and N Cross (Salisbury) for their views and comments.
References
- 1.Kaeda J, Chase A, Goldman JM. Cytogenetic and Molecular monitoring of residual disease in chronic myeloid leukaemia. Acta Haematol. 2002;107:64–75. doi: 10.1159/000046635. [DOI] [PubMed] [Google Scholar]
- 2.Cross NCP. Assessing residual leukaemia. Baillière’s Clin Haematol. 1997;10:389–402. doi: 10.1016/s0950-3536(97)80014-5. [DOI] [PubMed] [Google Scholar]
- 3.Kaeda J, Derville O, Szydlo RM, Olavarria E, Dazzi F, Marin D, et al. Serial measurement of BCR-ABL transcripts in the peripheral blood after allogeneic stem cell transplant for chronic myeloid leukaemia. An attempt to define patients who may not require further therapy. Blood. 2006;107:4171. doi: 10.1182/blood-2005-08-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haferlach T, Kern W, Schnittger S, Schoch C. Modern diagnostics in acute leukaemias. Crit Rev Oncol Hematol. 2005;56:223–34. doi: 10.1016/j.critrevonc.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Mughal TI, Yong A, Szydlo RM, Dazzi F, Olavarria E, van Rhee F, et al. Molecular studies in patients with chronic myeloid leukaemia in remission 5 years after allogeneic stem cell transplant define the risk of subsequent relapse. Br J Haematol. 2001;115:569–74. doi: 10.1046/j.1365-2141.2001.03155.x. [DOI] [PubMed] [Google Scholar]
- 6.Cross NCP, Melo JV, Feng L, Goldman JM. An optimized multiplex polymerase chain reaction (PCR) for detection of BCR-ABL fusion mRNAs in haematological disorders. Leukemia. 1994;8:186–9. [PubMed] [Google Scholar]
- 7.Menisk E, van de Locht A, Scattenburg A. Quantitation of minimal residual disease in Philadelphia chromosome positive chronic myeloid leukaemia patients using real time quantitative RT-PCR. Br J Haematol. 1998;102:768–74. doi: 10.1046/j.1365-2141.1998.00823.x. [DOI] [PubMed] [Google Scholar]
- 8.Shipley GL. Introduction to real-time PCR. In: Dorak MT, editor. Advanced Methods. 2006. pp. 1–31. [Google Scholar]
- 9.Hughes TP, Deininger MW, Hochhaus A, Branford S, Radich JP, Kaeda J, et al. Monitoring CM patients responding to treatment with tyrosine kinase inhibitors - Review and recommendations for ‘harmonizing’ current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochhaus A, Lin F, Reiter A, Skladny H, Mason PJ, van Rhee F, et al. Quantification of residual disease in chronic myelogenous leukaemia patients on interferon-α therapy by competitive polymerase chain reaction. Blood. 1996;87:1549–55. [PubMed] [Google Scholar]
- 11.Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukaemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction. Leukemia. 2003;17:2474–86. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 12.Baccarani M, Saglio G, Goldman J, Hochhaus A, Simonsson B, Appelbaum F, et al. Evolving concepts in the management of chronic myeloid leukaemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2006;108:1809–20. doi: 10.1182/blood-2006-02-005686. [DOI] [PubMed] [Google Scholar]
- 13.Hozumi N, Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. J Immunol. 2004;173:4260–4. [PubMed] [Google Scholar]
- 14.Leder P, Max EE, Seidman JG. In: Immunology. Fougereau M, Dausset J, editors. Vol. 80. Academic; London: 1980. pp. 34–50. [Google Scholar]
- 15.van der Velden VH, Wijkhuijs JM, van Dongen Non-specific amplification of patient-specific Ig/TCR gene rearrangemenst depends on the time point during therapy: implications for minimal residual disease monitoring. Leukemia. 2008;22:641–4. doi: 10.1038/sj.leu.2404925. [DOI] [PubMed] [Google Scholar]
- 16.Langerak AW, Molina TJ, Lavender FL, Pearson D, Flohr T, Sambade C, et al. Polymerase chain reaction-based clonality testing in tissue samples with reactive lymphoproliferations: usefulness and pitfalls. A report of the BIOMED-2 Concerted Action MH4-CT98–3936. Leukemia. 2007;21:222–9. doi: 10.1038/sj.leu.2404482. [DOI] [PubMed] [Google Scholar]
- 17.van Dongen JJM, Langerak AW, Bruggemann M, Evans PAS, Hummel M, Lavender FL. Design and standardisation of PCR primers and protocols for detection of clonal immunoglobulin and T cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98–3936. Leukemia. 2003;12:2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 18.van der Velden VH, Cazzaniga G, Schrauder A, Hancock J, Bader P, Panzer-Grumayer ER, et al. European Study Group on MRD detection in ALL (ESG-MRD-ALL). Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21:604–11. doi: 10.1038/sj.leu.2404586. [DOI] [PubMed] [Google Scholar]
- 19.Kerst G, Kreyenberg H, Roth C, Well C, Dietz K, Coustan-Smith E, et al. Concurrent detection of minimal residual disease (MRD) in childhood acute lymphoblastic leukaemia by flow cytometry and real time PCR. Br J Haematol. 2005;128:774–82. doi: 10.1111/j.1365-2141.2005.05401.x. [DOI] [PubMed] [Google Scholar]
- 20.Neale GA, Coustan-Smith E, Stow P, Pan Q, Chen X, Pui CH, et al. Comparative analysis of flow cytometry and polymerase chain reaction for the detection of minimal residual disease in childhood acute lymphoblastic leukaemia. Leukemia. 2004;18:934–8. doi: 10.1038/sj.leu.2403348. [DOI] [PubMed] [Google Scholar]
- 21.Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stilgenbauer F, Davi F, et al. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukaemia. Leukemia. 2007;21:1–3. doi: 10.1038/sj.leu.2404457. [DOI] [PubMed] [Google Scholar]
- 22.Biggs R, Denson RWE. Standardisation of the one-stage prothrombin time for the control of anticoagulant therapy. Br Med J. 1967;1:84–8. doi: 10.1136/bmj.1.5532.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes TP, Kaeda J, Branford S, Rudzki Z, Hochhaus A, Hensley ML, et al. International Randomised Study of Interferon versus STI571 (IRIS) Study Group. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukaemia. N Engl J Med. 2003;349:1423–32. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]
- 24.Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N, et al. Standardisation and quality control studies of ‘real time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukaemia. Leukemia. 2003;17:2318–57. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 25.Branford S, Cross NCP, Hochhaus A, Radich J, Saglio G, Kaeda J, et al. Rationale for the recommendations for harmonizing current methodology for detecting BCR-ABL transcripts in patients with chronic myeloid leukaemia. Leukemia. 2006;20:1925–30. doi: 10.1038/sj.leu.2404388. [DOI] [PubMed] [Google Scholar]
- 26.Saldanha J, Silvy M, Beaufils N, Arlinghaus R, Barbany G, Branford S, et al. Characterization of a reference material for BCR-ABL (M-BCR) mRNA quantitation by real-time amplification assays: towards new standards for gene expression measurements. Leukemia. 2007;21:1481–7. doi: 10.1038/sj.leu.2404716. [DOI] [PubMed] [Google Scholar]
- 27.Walker-Peach CR, Winkler M, DuBois DB, Pasloske L. Ribonuclease-resistant RNA controls (Armored RNA) for reverse transcription-PCR, branched DNA, and genotyping assays for hepatitis C virus. Clin Chem. 1999;45:2079–85. [PubMed] [Google Scholar]
- 28.Branford S, Fletcher L, Cross NCP, Muller MC, Hochhaus A, Kim DW, et al. Desirable performance characteristics for BCR-ABL measurement on an international reporting scale to allow consistent interpretation of individual patient response and comparison of response rates between clinical trials. Blood. 2008;112:3330–8. doi: 10.1182/blood-2008-04-150680. [DOI] [PubMed] [Google Scholar]
- 29.Cross NC, Feng L, Chase A, Bungey J, Hughes TP, Goldman JM. Competative polymerase chain reaction to estimate the number of BCR-ABL transcripts in chronic myeloid leukaemia patients after bone marrow transplant. Blood. 1993;82:1929–36. [PubMed] [Google Scholar]
- 30.Cross NC, Hughes TP, Feng L, O’Shea P, Bungey J, Marks DI, et al. Minimal residual disease after allogeneic bone marrow transplantation for chronic myeloid leukaemia in first chronic phase: correlations with acute graft-versus-host disease and relapse. Br J Haematol. 1993;84:67–74. doi: 10.1111/j.1365-2141.1993.tb03026.x. [DOI] [PubMed] [Google Scholar]
- 31.Marin D, Kaeda J, Szydlo R, Saunders S, Fleming A, Howard J, et al. Monitoring patients in complete cytogenetic remission after treatment of CML in chronic phase with imatinib: patterns of residual leukaemia and prognostic factors for cytogenetic relapse. Leukemia. 2005;19:507–12. doi: 10.1038/sj.leu.2403664. [DOI] [PubMed] [Google Scholar]
- 32.Cortes J, Talpaz M, O’Brien S, Jones D, Luthra R, Shan J, et al. Molecular responses in patients with chronic myelogenous leukaemia in chronic phase treated with imatinib mesylate. Clin Cancer Res. 2005;11:3425–32. doi: 10.1158/1078-0432.CCR-04-2139. [DOI] [PubMed] [Google Scholar]
- 33.Kantarjian H, Schiffer C, Jones D, Cortes J. Monitoring the response and course of chronic myeloid leukaemia in the modern era of BCR-ABL tyrosine kinase inhibitors: Practical advice on the use and interpretation of monitoring methods. Blood. 2008;111:1774–80. doi: 10.1182/blood-2007-09-110189. [DOI] [PubMed] [Google Scholar]
- 34.Branford S, Rudzki Z, Harper A, Grigg A, Taylor K, Durrant S, et al. Imatinib produces significantly superior molecular responses compared to interferon α plus cytarabine in patients with newly diagnosed chronic myeloid leukaemia in chronic phase. Leukemia. 2003;17:2401–9. doi: 10.1038/sj.leu.2403158. [DOI] [PubMed] [Google Scholar]
- 35.Merx K, Müller MC, Kreil S, Lahaye T, Paschka P, Schoch C, et al. Early reduction of BCR-ABL mRNA transcript levels predicts cytogenetic response in chronic phase CML patients treated with imatinib after failure of interferon α. Leukemia. 2002;16:1579–83. doi: 10.1038/sj.leu.2402680. [DOI] [PubMed] [Google Scholar]
- 36.Wang L, Pearson K, Ferguson JE, Clarke RE. The early molecular response to imatinib predicts cytogenetic and clinical outcome in chronic myeloid leukaemia. Br J Haematol. 2003;120:990–9. doi: 10.1046/j.1365-2141.2003.04200.x. [DOI] [PubMed] [Google Scholar]
- 37.Branford S, Rudzki Z, Parkinson I, Grigg A, Taylor K, Seymour JF, et al. Real time quantitative PCR analysis can be used as a primary screen to identify patients with CML treated with imatinib who have BCR-ABL kinase domain mutations. Blood. 2004;104:2926–32. doi: 10.1182/blood-2004-03-1134. [DOI] [PubMed] [Google Scholar]
- 38.Khorashad JS, de Lavallade H, Apperley JF, Milojkovic D, Reid AG, Bua M, et al. Finding of kinase domain mutations in patients with chronic phase chronic myeloid leukaemia responding to imatinib may identify those at high risk of disease progression. J Clin Oncol. 2008;26:4806–13. doi: 10.1200/JCO.2008.16.9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marin David, Khorashad Jamshid S, Foroni Letizia, Milojkovic Dragana, Szydlo Richard, Reid Alistair G, Rezvani Katayoun, Bua Marco, Goldman John M, Apperley Jane F. Does a rise in the BCR-ABL1 transcript level identify chronic phase CML patients responding to imatinib who have a high risk of cytogenetic relapse? Br J Haem. 2009;145:373–5. doi: 10.1111/j.1365-2141.2009.07646.x. [DOI] [PubMed] [Google Scholar]
- 40.Warrell RP, Jr, de Thé H, Wang Z-Y, Degos L. Acute promyelocytic leukaemia. N Engl J Med. 1993;329:177–89. doi: 10.1056/NEJM199307153290307. [DOI] [PubMed] [Google Scholar]
- 41.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) Breakpoints on chromosome 21 in acute myeloid leukaemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–4. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shurtleff SA, Meyers S, Hiebert SW, Raimondi SC, Head DR, Willman CL, et al. Heterogeneity in CBFβ/MYH11 fusion messages encoded by the inv(16)(p13q22) and the t(16; 16)(p13;q22) in acute myelogenous leukaemia. Blood. 1995;85:3695–704. [PubMed] [Google Scholar]
- 43.Schnittger S, Schoch C, Kern W, Mecucci C, Tschulik C, Martelli MF, et al. Nucleophosmin gene mutations are predictors of favourable prognosis in acute meylogenous leukaemia with normal karyotype. Blood. 2005;106:3733–9. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 44.Lugthart S, van Drunen E, van Norden Y, van Hoven A, Erpelinck CA, Valk PJ, et al. High EVI1 levels predict adverse outcome in acute myeloid leukaemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111:4329–37. doi: 10.1182/blood-2007-10-119230. [DOI] [PubMed] [Google Scholar]
- 45.Diverio D, Rossi V, Avvisati G, De Santis S, Pistilli A, Pane F, et al. Early detection of relapse by prospective reverse transcriptase-polymerase chain reaction analysis of the PML/RARα fusion gene in patients with acute prolymphocytic leukaemia enrolled in the GIMEMA-AIEP multicentre ‘AIDA’ trial. Blood. 1998;92:784–9. [PubMed] [Google Scholar]
- 46.Lee S, Kim YJ, Eom KS, Min CK, Kim HJ, Cho SG, et al. The significance of minimal residual disease kinetics in adults with newly diagnosed PML-RARα-positive acute prolymphocytic leukaemia: results of a prospective trial. Haematologica. 2006;91:671–4. [PubMed] [Google Scholar]
- 47.Santamaria C, Chillón MC, Fernández C, Martín-Jiménez P, Balanzategui A, García Sanz R, et al. Using quantification of the PML-RARα transcript to stratify the risk of relapse in patients with acute promyelocytic leukaemia. Haematologica. 2007;92:315–22. doi: 10.3324/haematol.10734. [DOI] [PubMed] [Google Scholar]
- 48.Gallagher RE, Yeap BY, Bi W, Livak KJ, Beaubier N, Rao S, et al. Quantitative real-time RT-PCR analysis of PML-RAR α mRNA in acute prolymphocytic leukaemia: assessment of prognostic significance in adult patients from intergroup protocol 0129. Blood. 2003;101:2521–8. doi: 10.1182/blood-2002-05-1357. [DOI] [PubMed] [Google Scholar]
- 49.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing non-leukemic stem cells in acute myelogenous leukaemia with 8;21 chromosomal translocation. Proc Natl Acad Sci USA. 2000;97:7521–6. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugimoto T, Das H, Imoto S, Murayama T, Kajimoto K, Sugimoto T, et al. Quantitation of minimal residual disease in t(8;21)-positive acute myelogenous leukaemia patients using real-time quantitative RT-PCR. Am J Hematol. 2000;64:101–6. doi: 10.1002/(sici)1096-8652(200006)64:2<101::aid-ajh5>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 51.Buonamici S, Ottaviani E, Testoni N, Montefusco V, Visani G, Bonifazi F, et al. Real-time quantitation of minimal residual disease in inv(16)-positive acute myeloid leukaemia may indicate risk for clinical relapse and may identify patients in a curable state. Blood. 2002;99:443–9. doi: 10.1182/blood.v99.2.443. [DOI] [PubMed] [Google Scholar]
- 52.Krauter J, Gorlich K, Ottmann O, Lubbert M, Dohner H, Heit W, et al. Prognostic value of minimal residual disease quantification by real-time reverse transcriptase poly-merase chain reaction in patients with core binding factor leukaemias. J Clin Oncol. 2003;21:4413–22. doi: 10.1200/JCO.2003.03.166. [DOI] [PubMed] [Google Scholar]
- 53.Marcucci G, Caligiuri MA, Dohner H, Archer KJ, Schlenk RF, Döhner K, et al. Quantification of CBFβ/MYH11 fusion transcript by real time RT-PCR in patients with INV(16) acute myeloid leukaemia. Leukemia. 2001;15:1072–80. doi: 10.1038/sj.leu.2402159. [DOI] [PubMed] [Google Scholar]
- 54.Stentoft J, Hokland P, Ostergaard M, Hasle H, Nyvold CG. Minimal residual core binding factor AMLs by real time quantitative PCR–initial response to chemotherapy predicts event free survival and close monitoring of peripheral blood unravels the kinetics of relapse. Leuk Res. 2006;30:389–95. doi: 10.1016/j.leukres.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 55.Tobal K, Newton J, Macheta M, Chang J, Morgenstern G, Evans PA, et al. Molecular quantitation of minimal residual disease in acute myeloid leukaemia with t(8;21) can identify patients in durable remission and predict clinical relapse. Blood. 2000;95:815–9. [PubMed] [Google Scholar]
- 56.Lapillonne H, Renneville A, Auvrignon A, Flamant C, Blaise A, Perot C, et al. High WT1 expression after induction therapy predicts high risk of relapse and death in pediatric acute myeloid leukaemia. J Clin Oncol. 2006;24:1507–15. doi: 10.1200/JCO.2005.03.5303. [DOI] [PubMed] [Google Scholar]
- 57.Ostergaard M, Olesen LH, Hasle H, Kjeldsen E, Hokland P. WT1 gene expression: an excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukaemia patients – results from a single-centre study. Br J Haematol. 2004;125:590–600. doi: 10.1111/j.1365-2141.2004.04952.x. [DOI] [PubMed] [Google Scholar]
- 58.Trka J, Kalinova M, Hrusäk O, Zuna J, Krejci O, Madzo J, et al. Real-time quantitative PCR detection of WT1 gene expression in children with AML: prognostic significance, correlation with disease status and residual disease detection by flow cytometry. Leukemia. 2002;16:1381–9. doi: 10.1038/sj.leu.2402512. [DOI] [PubMed] [Google Scholar]
- 59.Cilloni D, Messa F, Arruga F, Defilippi I, Gottardi E, Fava M, et al. Early prediction of treatment outcome in acute myeloid leukaemia by measurement of WT1 transcript levels in peripheral blood samples collected after chemotherapy. Haematologica. 2008;93:921–4. doi: 10.3324/haematol.12165. [DOI] [PubMed] [Google Scholar]
- 60.Ommen HB, Nyvold CG, Braend-strup K, Andersen BL, Ommen IB, Hasle H, et al. Relapse prediction in acute myeloid leukaemia patients in complete remission using WT1 as a molecular marker: development of a mathematical model to predict time from molecular to clinical relapse and define optimal sampling intervals. Br J Haematol. 2008;141:782–91. doi: 10.1111/j.1365-2141.2008.07132.x. [DOI] [PubMed] [Google Scholar]
- 61.Nucifora G. The EVI1 gene in myeloid leukaemia. Leukemia. 1997;11:2022–31. doi: 10.1038/sj.leu.2400880. [DOI] [PubMed] [Google Scholar]
- 62.Weisser M, Kern W, Schoch C, Tschulik C, Hiddemann W, Haferlach T, et al. Reverse transcriptase polymerase chain reaction based quantification of the combined MDS-EVI1/EVI1 gene in acute myeloid leukaemia. Leuk Lymphoma. 2006;47:2645–7. doi: 10.1080/10428190600942561. [DOI] [PubMed] [Google Scholar]
- 63.Paydas S, Tanriverdi K, Yavus S, Disel U, Baslamisli F, Burgut R. PRAME mRNA levels in cases with acute leukaemia: clinical importance and future prospects. Am J Hematol. 2005;79:257–61. doi: 10.1002/ajh.20425. [DOI] [PubMed] [Google Scholar]
- 64.Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukaemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]
- 65.Beretta C, Gaipa G, Rossi V, Bernasconi S, Spinelli O, Dell’Oro MG, et al. Development of a quantitative-PCR method for specific FLT3/ITD monitoring in acute myeloid leukaemia. Leukemia. 2004;18:1441–4. doi: 10.1038/sj.leu.2403409. [DOI] [PubMed] [Google Scholar]
- 66.Stirewalt DL, Willman CL, Radich JP. Quantitative, real-time polymerase chain reactions for FLT3 internal tandem duplications are highly sensitive and specific. Leuk Res. 2001;25:1085–8. doi: 10.1016/s0145-2126(01)00087-x. [DOI] [PubMed] [Google Scholar]
- 67.Hess CJ, Feller N, Denkers F, Kelder A, Merle PA, Heinrich MC, et al. Correlation of minimal residual disease cell frequency with molecular genotype in patients with acute myeloid leukaemia. Haematologica. 2009;94:46–53. doi: 10.3324/haematol.13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kottaridis PD, Gale RE, Langabeer SE, Frew ME, Bowen DT, Linch DC. Studies of FLT3 mutations in paired presentation and relapse samples from patients with acute myeloid leukaemia: implications for the role of FLT3 mutations in leukemogenesis, minimal residual disease detection, and possible therapy with FLT3 inhibitors. Blood. 2002;100:2393–8. doi: 10.1182/blood-2002-02-0420. [DOI] [PubMed] [Google Scholar]
- 69.Shih LY, Huang CF, Wu JH, Lin TL, Dunn P, Wang PN, et al. Internal tandem duplication of FLT3 in relapsed acute myeloid leukaemia: a comparative analysis of bone marrow samples from 108 adult patients at diagnosis and relapse. Blood. 2002;100:2387–92. doi: 10.1182/blood-2002-01-0195. [DOI] [PubMed] [Google Scholar]
- 70.Cloos J, Goemans BF, Hess CJ, van Oostveen JW, Waisfisz Q, Corthals S, et al. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia. 2006;20:1217–20. doi: 10.1038/sj.leu.2404246. [DOI] [PubMed] [Google Scholar]
- 71.Gorello P, Cazzaniga G, Alberti F, Dell’Oro MG, Gottardi E, Specchia G, et al. Quantitative assessment of minimal residual disease in acute myeloid leukaemia carrying nucleoplasmin (NPM1) gene mutations. Leukemia. 2006;20:1103–8. doi: 10.1038/sj.leu.2404149. [DOI] [PubMed] [Google Scholar]
- 72.Chou WC, Tang JL, Wu SJ, Tsay W, Chou WC, Yao M, et al. Clinical implications of minimal residual disease monitoring by quantitative polymerase chain reaction in acute myeloid leukaemia patients bearing nucleophosmin (NPM1) mutations. Leukemia. 2007;21:998–1004. doi: 10.1038/sj.leu.2404637. [DOI] [PubMed] [Google Scholar]
- 73.Schnittger S, Kern W, Haferlach C. PCR-based MRD detection in NPM1 mutated AML: a prospective follow-up study in 97 patients [abstract] Haematologica. 2007;92 (Suppl 1):147. [Google Scholar]
- 74.Palmisano M, Grafone T, Ottaviani E, Testoni N, Baccarani M, Martinelli G. NPM1 mutations are more stable than FLT3 mutations during the course of disease in patients with acute myeloid leukaemia. Haematologica. 2007;92:1268–9. doi: 10.3324/haematol.11202. [DOI] [PubMed] [Google Scholar]
- 75.Shiha LY, Liang DC, Huang CF, Chang YT, Lai CL, Lin TH, et al. AML patients with CEBPα mutations mostly retain identical mutant patterns but frequently change in allelic distribution at relapse; a comparative analysis on paired diagnosis and relapse samples. Leukemia. 2006;20:604–9. doi: 10.1038/sj.leu.2404124. [DOI] [PubMed] [Google Scholar]
- 76.Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukaemia. N Engl J Med. 2008;358:1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 77.Yokota H, Tsuno NH, Tanaka Y, Fukui T, Kitamura K, Hirai H, et al. Quantification of minimal residual disease in patients with e1a2 BCR-ABL-positive acute lymphoblastic leukaemia using a real-time RT-PCR assay. Leukemia. 2002;16:1167–75. doi: 10.1038/sj.leu.2402483. [DOI] [PubMed] [Google Scholar]
- 78.Pane F, Cimino G, Izzo B, Camera A, Vitale A, Quintarelli C, et al. Significant reduction of the hybrid BCR/ABL transcripts after induction and consolidation therapy is a powerful predictor of treatment response in adult Philadelphia-positive acute lymphoblastic leukaemia. Leukemia. 2005;19:628–35. doi: 10.1038/sj.leu.2403683. [DOI] [PubMed] [Google Scholar]
- 79.Miyamura K, Tanimoto M, Morishima Y, Horibe K, Yamamoto K, Akatsuka M, et al. Detection of Philadelphia chromosome-positive acute lymphoblastic leukaemia by polymerase chain reaction: possible eradication of minimal residual disease by marrow transplantation. Blood. 1992;79:1366–70. [PubMed] [Google Scholar]
- 80.Radich J, Gehly G, Lee A, Avery R, Bryant E, Edmands S, et al. Detection of bcr-abl transcripts in Philadelphia chromosome-positive acute lymphoblastic leukaemia after marrow transplantation. Blood. 1997;89:2602–9. [PubMed] [Google Scholar]
- 81.Stirewalt DL, Guthrie KA, Beppu L, Bryant EM, Doney K, Gooley T, et al. Predictors of relapse and overall survival in Philadelphia chromosome-positive acute lymphoblastic leukaemia after transplantation. Biol Blood Marrow Transplant. 2003;9:206–12. doi: 10.1053/bbmt.2003.50025. [DOI] [PubMed] [Google Scholar]
- 82.van der Velden VH, Jacobs DC, Wijkhuijs AJ, Comans-Bitter WM, Willemse MJ, Hählen K, et al. Minimal residual disease levels in bone marrow and peripheral blood are comparable in children with T cell acute lymphoblastic leukaemia (ALL), but not in precursor-B-ALL. Leukemia. 2002;16:1432–6. doi: 10.1038/sj.leu.2402636. [DOI] [PubMed] [Google Scholar]
- 83.Gameiro P, Mortuza FY, Hoffbrand AV, Foroni L. Minimal residual disease monitoring in adult T-cell acute lymphoblastic leukaemia: a molecular based approach using T-cell receptor G and D gene rearrangements. Haematologica. 2002;87:1126–34. [PubMed] [Google Scholar]
- 84.Specchia G, Liso A, Pannunzio A, Albano F, Mestice A, Pastore D, et al. Molecular detection of minimal residual disease is associated with early relapse in adult acute lymphoblastic leukaemia. Haematologica. 2004;89:1271–3. [PubMed] [Google Scholar]
- 85.Toubai T, Tanaka J, Ota S, Mori A, Ibata M, Shono Y, et al. Minimal residual disease (MRD) monitoring using rearrangement of T-cell receptor and immunoglobulin H gene in the treatment of adult acute lymphoblastic leukaemia patients. Am J Hematol. 2005;80:181–7. doi: 10.1002/ajh.20461. [DOI] [PubMed] [Google Scholar]
- 86.Eckert C, Biondi A, Seeger K, Cazzaniga G, Hartman R, Beyermann B, et al. Prognostic value of minimal residual disease in relapsed childhood acute lymphoblastic leukaemia. Lancet. 2001;358:1239–41. doi: 10.1016/S0140-6736(01)06355-3. [DOI] [PubMed] [Google Scholar]
- 87.Bruggemann M, Raff T, Flohr T, Gökbuget N, Nakao M, Droese J, et al. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukaemia. Blood. 2006;107:1116–23. doi: 10.1182/blood-2005-07-2708. [DOI] [PubMed] [Google Scholar]
- 88.Dewald GW, Brockman SR, Paternoster SF, Bone ND, O’Fallon JR, Allmer C, et al. Chromosome anomalies detected by interphase fluorescence in situ hybridisation: correlation with significant biological features of B-cell chronic lymphocytic leukaemia. Br J Haematol. 2003;121:287–95. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- 89.Mortuza FY, Papaioannou M, Moreira IM, Coyle LA, Gameiro D, Prentice HG, et al. Minimal residual disease tests provide an independent predictor of clinical outcome in adult acute lymphoblastic leukaemia. J Clin Oncol. 2002;20:1094–104. doi: 10.1200/JCO.2002.20.4.1094. [DOI] [PubMed] [Google Scholar]
- 90.Mullighan C, Xiaoping S, Zang J, Radke I, Phillips LAA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukaemia. N Eng J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocyte leukaemia. Blood. 1975;46:219–34. doi: 10.1182/blood-2016-08-737650. [DOI] [PubMed] [Google Scholar]
- 92.Binet JL, Leporrier M, Dighiero G, Charron D, d’Athis P, Vaugier G, et al. Clinical staging system for chronic lymphocytic leukaemia: prognostic significance. Cancer. 1977;40:855–64. doi: 10.1002/1097-0142(197708)40:2<855::aid-cncr2820400239>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 93.Krober A, Seiler T, Benner A, Bullinger A, Bruckle E, Lichter P, et al. V(H) mutation status, CD38 expression levels, genomic aberrations and survival in chronic lymphocytic leukaemia. Blood. 2002;100:1410–6. [PubMed] [Google Scholar]
- 94.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukaemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 95.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukaemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 96.Rassenti LZ, Huynh L, Toy TL, Chen L, Keating MJ, Gribben JG, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukaemia. N Eng J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 97.Del Poeta G, Maurillo L, Venditti A, Buccissano F, Epiceno AM, Capelli G, et al. Clinical significance of CD38 expression in chronic lymphocytic leukaemia. Blood. 2001;98:2633–9. doi: 10.1182/blood.v98.9.2633. [DOI] [PubMed] [Google Scholar]
- 98.Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, et al. Prognostic subgroups in B-cell chronic lymphocytic leukaemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323:720–4. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 99.Oscier DG, Gardiner AC, Mould SJ, Glide S, Davis ZA, Ibbotson RE, et al. Multivariate analysis of prognostic factors in CLL: Clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–84. [PubMed] [Google Scholar]
- 100.Wierda W, O’Brien S, Wen S, Faderl S, Garcia-Manero G, Thomas D, et al. Chemoimmunotherpy with fludarabine, cylcophosphamide, and rituximab for relapsed and refractory chronic lymphocyte leukaemia. J Clin Oncol. 2005;23:4070–8. doi: 10.1200/JCO.2005.12.516. [DOI] [PubMed] [Google Scholar]
- 101.Moreno C, Villamor N, Colomer D, Esteve J, Gine E, Muntanola A, et al. Clinical significance of minimal residual disease, as assessed by different techniques, after stem-cell transplantation for chronic lymphocyte leukaemia. Blood. 2006;107:4563–9. doi: 10.1182/blood-2005-09-3634. [DOI] [PubMed] [Google Scholar]
- 102.Sayala H, Rawstron A, Hillmen Minimal residual disease assessment in chronic lymphocytic leukaemia. Best Prac Res Clin Haematol. 2007;20:499–512. doi: 10.1016/j.beha.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 103.Rawstron AC, Kennedy B, Evans PA, Davies FE, Richards SJ, Haynes AP, et al. Quantitation of minimal residual disease levels in chronic lymphocyte leukaemia using a sensitive flow cytometric assay improves the prediction of outcome and can be used to optimize therapy. Blood. 2001;98:29–35. doi: 10.1182/blood.v98.1.29. [DOI] [PubMed] [Google Scholar]
- 104.Esteve J, Villamor N, Colomer D, Montserrat E. Different clinical value of minimal residual disease after autologous and allogenic stem cell transplantation for chronic lymphocytic leukaemia. Blood. 2002;99:1873–4. doi: 10.1182/blood.v99.5.1873. [DOI] [PubMed] [Google Scholar]
- 105.Visser JW, Martens AC, Hagenbeek A. Detection of minimal residual disease in acute leukaemia by flow cytometry. Ann NY Acad Sci. 1986;468:268–75. doi: 10.1111/j.1749-6632.1986.tb42045.x. [DOI] [PubMed] [Google Scholar]
- 106.Vidriales MB, San-Miguel JF, Orfao A, Coustan-Smith E, Campana D. Minimal residual disease monitoring by flow cytometry. Best Pract Res Clin Haematol. 2003;16:599–612. doi: 10.1016/s1521-6926(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 107.Schrappe M, Beier R, Bürger B. New treatment strategies in childhood acute lymphoblastic leukaemia. Best Pract Res Clin Haematol. 2002;15:729–40. doi: 10.1053/beha.2002.0226. [DOI] [PubMed] [Google Scholar]
- 108.Cazzaniga G, Biondi A. Molceular monitoring of childhood acute lymphoblastic leukaemia using antigen receptor gene rearrangements and quantitative polymerase chain reaction technology. Haematologica. 2005;90:382–90. [PubMed] [Google Scholar]
- 109.Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T. Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukaemia. Blood. 2004;104:3078–85. doi: 10.1182/blood-2004-03-1036. [DOI] [PubMed] [Google Scholar]
- 110.Campana D, Yokota S, Coustan-Smith E, Hansen-Hagge TE, Janossy G, Bartram CR. The detection of residual acute lymphoblastic leukaemia cells with immunologic methods and polymerase chain reaction: a comparative study. Leukemia. 1990;4:609–14. [PubMed] [Google Scholar]
- 111.Bradstock KF, Janossy G, Tidman N, Papageorgiou ES, Prentice HG, Willoughby M, et al. Immunological monitoring of residual disease in treated thymic acute lymphoblastic leukaemia. Leuk Res. 1981;5:301–9. doi: 10.1016/0145-2126(81)90002-3. [DOI] [PubMed] [Google Scholar]
- 112.Campana D, Coustan-Smith E, Janossy G. The immunologic detection of minimal residual disease in acute leukaemia. Blood. 1990;76:163–71. [PubMed] [Google Scholar]
- 113.Gross HJ, Verwer B, Houck D, Recktenwald D. Detection of rare cells at a frequency of one per million by flow cytometry. Cytometry. 1993;14:519–26. doi: 10.1002/cyto.990140511. [DOI] [PubMed] [Google Scholar]
- 114.Lúcio P, Parreira A, van den Beemd MW, van Lochem EG, van Wering ER, Baars E, et al. Flow cytometric analysis of normal B cell differentiation: a frame of reference for the detection of minimal residual disease in precursor-B-ALL. Leukemia. 1999;13:419–27. doi: 10.1038/sj.leu.2401279. [DOI] [PubMed] [Google Scholar]
- 115.van Lochem EG, van der Velden VH, Wind HK, te Marvelde JG, Westerdaal NA, van Dongen JJ. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: reference patterns for age-related changes and disease-induced shifts. Cytometry B Clin Cytom. 2004;60:1–13. doi: 10.1002/cyto.b.20008. [DOI] [PubMed] [Google Scholar]
- 116.Wood B. 9-Color and 10-color flow cytometry in the clinical laboratory. Arch Pathol Lab Med. 2006;130:680–90. doi: 10.5858/2006-130-680-CACFCI. [DOI] [PubMed] [Google Scholar]
- 117.Campana D, Coustan-Smith E. Detection of minimal residual disease in acute leukaemia by flow cytometry. Cytometry. 1999;38:139–52. doi: 10.1002/(sici)1097-0320(19990815)38:4<139::aid-cyto1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 118.Coustan-Smith E, Sancho J, Hancock ML, Razzouk BI, Ribeiro RC, Rivera GK, et al. Use of peripheral blood instead of bone marrow to monitor residual disease in children with acute lymphoblastic leukaemia. Blood. 2002;100:2399–402. doi: 10.1182/blood-2002-04-1130. [DOI] [PubMed] [Google Scholar]
- 119.San Miguel JF, Vidriales MB, Orfão A. Immunological evaluation of minimal residual disease (MRD) in acute myeloid leukaemia (AML) Best Pract Res Clin Haematol. 2002;15:105–18. doi: 10.1053/beha.2001.0193. [DOI] [PubMed] [Google Scholar]
- 120.Baer MR, Stewart CC, Dodge RK, Leget G, Sulé N, Mrózek K, et al. High frequency of immunophenotype changes in acute myeloid leukaemia at relapse: implications for residual disease detection (Cancer and Leukaemia Group B Study 8361) Blood. 2001;97:3574–80. doi: 10.1182/blood.v97.11.3574. [DOI] [PubMed] [Google Scholar]
- 121.Sievers EL, Lange BJ, Alonzo TA, Gerbing RB, Bernstein ID, Smith FO, et al. Immunophenotypic evidence of leukemia after induction therapy predicts relapse: results from a prospective Children’s Cancer Group study of 252 patients with acute myeloid leukemia. Blood. 2003;101:3398–406. doi: 10.1182/blood-2002-10-3064. [DOI] [PubMed] [Google Scholar]
- 122.Kern W, Danhauser-Riedl S, Ratei R, Schnittger S, Schoch C, Kolb HJ, et al. Detection of minimal residual disease in unselected patients with acute myeloid leukaemia using multiparameter flow cytometry for definition of leukaemia-associated immunophenotypes and determination of their frequencies in normal bone marrow. Haematologica. 2003;88:646–53. [PubMed] [Google Scholar]
- 123.Feller N, van der Pol MA, van Stijn A, Weijers GW, Westra AH, Evertse BW, et al. MRD parameters using immunophenotypic detection methods are highly reliable in predicting survival in acute myeloid leukaemia. Leukemia. 2004;18:1380–90. doi: 10.1038/sj.leu.2403405. [DOI] [PubMed] [Google Scholar]
- 124.Langebrake C, Brinkmann I, Teigler- Schlegel A, Creutzig U, Griesinger F, Puhlmann U, et al. Immunophenotypic differences between diagnosis and relapse in childhood AML: Implications for MRD monitoring. Cytometry B Clin Cytom. 2005;63:1–9. doi: 10.1002/cyto.b.20037. [DOI] [PubMed] [Google Scholar]
- 125.Zelezníková T, Babusíková O. The impact of cell heterogeneity and immunophenotypic changes on monitoring minimal residual disease in acute myeloid leukemia. Neoplasma. 2006;53:500–6. [PubMed] [Google Scholar]
- 126.Perea G, Lasa A, Aventín A, Domingo A, Villamor N, Queipo de Llano MP, et al. Grupo Cooperativo para el Estudio y Tratamiento de las Leucemias Agudas y Miel. Prognostic value of minimal residual disease (MRD) in acute myeloid leukaemia (AML) with favorable cytogenetics [t(8;21) and inv(16)] Leukemia. 2006;20:87–94. doi: 10.1038/sj.leu.2404015. [DOI] [PubMed] [Google Scholar]
- 127.Jennings CD, Foon KA. Recent advances in flow cytometry: application to the diagnosis of hematologic malignancy. Blood. 1997;90:2863–92. [PubMed] [Google Scholar]
- 128.Borowitz MJ, Guenther KL, Shults KE, Stelzer GT. Immunophenotyping of acute leukaemia by flow cytometric analysis. Use of CD45 and right-angle light scatter to gate on leukemic blasts in three-color analysis. Am J Clin Pathol. 1993;100:534–40. doi: 10.1093/ajcp/100.5.534. [DOI] [PubMed] [Google Scholar]
- 129.Lacombe F, Durrieu F, Briais A, Dumain P, Belloc F, Bascans E, et al. Flow cytometry CD45 gating for immunophenotyping of acute myeloid leukaemia. Leukemia. 1997;11:1878–86. doi: 10.1038/sj.leu.2400847. [DOI] [PubMed] [Google Scholar]
- 130.Oelschlägel U, Nowak R, Schaub A, Köppel C, Herbst R, Mohr B, et al. Shift of aberrant antigen expression at relapse or at treatment failure in acute leukaemia. Cytometry. 2000;42:247–53. doi: 10.1002/1097-0320(20000815)42:4<247::aid-cyto5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 131.Gaipa G, Basso G, Maglia O, Leoni V, Faini A, Cazzaniga G, et al. I-BFM-ALL-FCM-MRD Study Group. Drug-induced immunophenotypic modulation in childhood ALL: implications for minimal residual disease detection. Leukemia. 2005;19:49–56. doi: 10.1038/sj.leu.2403559. [DOI] [PubMed] [Google Scholar]
- 132.Robillard N, Cavé H, Méchinaud F, Guidal C, Garnache-Ottou F, Rohrlich PS, et al. Four-color flow cytometry bypasses limitations of IG/TCR polymerase chain reaction for minimal residual disease detection in certain subsets of children with acute lymphoblastic leukaemia. Haematologica. 2005;90:1516–23. [PubMed] [Google Scholar]
- 133.Maurillo L, Buccisano F, Spagnoli A, Del Poeta G, Panetta P, Neri B, et al. Monitoring of minimal residual disease in adult acute myeloid leukaemia using peripheral blood as an alternative source to bone marrow. Haematologica. 2007;92:605–11. doi: 10.3324/haematol.10432. [DOI] [PubMed] [Google Scholar]
- 134.van der Velden VH, Jacobs DC, Wijkhuijs AJ, Comans-Bitter WM, Willemse MJ, Hählen K, et al. Minimal residual disease levels in bone marrow and peripheral blood are comparable in children with T cell acute lymphoblastic leukaemia (ALL), but not in precursor-B-ALL. Leukemia. 2002;16:1432–6. doi: 10.1038/sj.leu.2402636. [DOI] [PubMed] [Google Scholar]
- 135.Pui CH, Campana D. New definition of remission in childhood acute lymphoblastic leukaemia. Leukemia. 2000;14:783–5. doi: 10.1038/sj.leu.2401780. [DOI] [PubMed] [Google Scholar]
- 136.Drach J, Gattringer C, Huber H. Combined flow cytometric assessment of cell surface antigens and nuclear TdT for the detection of minimal residual disease in acute leukaemia. Br J Haematol. 1991;77:37–42. doi: 10.1111/j.1365-2141.1991.tb07945.x. [DOI] [PubMed] [Google Scholar]
- 137.Imamura N, Kuramoto A. Detection of minimal residual disease in acute lymphoblastic leukaemia by flow cytometry with monoclonal antibodies. Am J Hematol. 1991;38:332–4. doi: 10.1002/ajh.2830380416. [DOI] [PubMed] [Google Scholar]
- 138.Drach J, Drach D, Glassl H, Gattringer C, Huber H. Flow cytometric determination of atypical antigen expression in acute leukaemia for the study of minimal residual disease. Cytometry. 1992;13:893–901. doi: 10.1002/cyto.990130813. [DOI] [PubMed] [Google Scholar]
- 139.Syrjala M, Anttila VJ, Ruutu T, Jansson SE. Flow cytometric detection of residual disease in acute leukaemia by assaying blasts co-expressing myeloid and lymphatic antigens. Leukemia. 1994;8:1564–70. [PubMed] [Google Scholar]
- 140.Coustan-Smith E, Behm FG, Sanchez J, Boyett JM, Hancock ML, Raimondi SC, et al. Immunological detection of minimal residual disease in children with acute lymphoblastic leukaemia. Lancet. 1998;351:550–4. doi: 10.1016/S0140-6736(97)10295-1. [DOI] [PubMed] [Google Scholar]
- 141.Pui C-H. Childhood leukaemias. N Engl J Med. 1995;332:1618–30. doi: 10.1056/NEJM199506153322407. [DOI] [PubMed] [Google Scholar]
- 142.van Dongen JJ, Breit TM, Adriaansen HJ, Beishuizen A, Hooijkaas H. Detection of minimal residual disease in acute leukaemia by immunological marker analysis and polymerase chain reaction. Leukemia. 1992;6:47–59. [PubMed] [Google Scholar]
- 143.Malec M, van der Velden VH, Björklund E, Wijkhuijs JM, Söderhäll S, Mazur J, et al. Analysis of minimal residual disease in childhood acute lymphoblastic leukaemia: comparison between RQ-PCR analysis Ig/TcR gene rearrangements and multicolor flow cytometric immunophenotyping. Leukemia. 2004;18:1630–6. doi: 10.1038/sj.leu.2403444. [DOI] [PubMed] [Google Scholar]
- 144.Dworzak MN, Stolz F, Fröschl G, Printz D, Henn T, Fischer S, et al. Detection of residual disease in pediatric B-cell precursor acute lymphoblastic leukaemia by comparative phenotype mapping: a study of five cases controlled by genetic methods. Exp Hematol. 1999;27:673–81. doi: 10.1016/s0301-472x(98)00071-x. [DOI] [PubMed] [Google Scholar]
- 145.Coustan-Smith E, Ribeiro RC, Stow P, Zhou Y, Pui CH, Rivera GK, et al. A simplified flow cytometric assay identifies children with acute lymphoblastic leukaemia who have a superior clinical outcome. Blood. 2006;108:97–102. doi: 10.1182/blood-2006-01-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wells DA, Sale GE, Shulman HM, Myerson D, Bryant EM, Gooley T, et al. Multidimensional flow cytometry of marrow can differentiate leukemic from normal lymphoblasts and myeloblasts after chemotherapy and bone marrow transplantation. Am J Clin Pathol. 1998;110:84–94. doi: 10.1093/ajcp/110.1.84. [DOI] [PubMed] [Google Scholar]
- 147.Weir EG, Cowan K, LeBeau P, Borowitz MJ. A limited antibody panel can distinguish B-precursor acute lymphoblastic leukaemia from normal B precursors with four color flow cytometry: implications for residual disease detection. Leukemia. 1999;13:558–67. doi: 10.1038/sj.leu.2401364. [DOI] [PubMed] [Google Scholar]
- 148.Borowitz MJ, Pullen DJ, Winick N, Martin PL, Bowman WP, Camitta B. Comparison of diagnostic and relapse flow cytometry phenotypes in childhood acute lymphoblastic leukaemia: implications for residual disease detection: a report from the children’s oncology group. Cytometry B Clin Cytom. 2005;68:18–24. doi: 10.1002/cyto.b.20071. [DOI] [PubMed] [Google Scholar]
- 149.Borowitz MJ, Devidas M, Hunger SP, Bowman WP, Carroll AJ, Carroll WL, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukaemia and its relationship to other prognostic factors: A Children's Oncology Group study. Blood. 2008;111:5477–85. doi: 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Coustan-Smith E, Sancho J, Hancock ML, Boyett JM, Behm FG, Raimondi SC, et al. Clinical importance of minimal residual disease in childhood acute lymphoblastic leukaemia. Blood. 2000;96:2691–6. [PubMed] [Google Scholar]
- 151.Chen JS, Coustan-Smith E, Suzuki T, Neale GA, Mihara K, Pui CH, et al. Identification of novel markers for monitoring minimal residual disease in acute lymphoblastic leukaemia. Blood. 2001;97:2115–20. doi: 10.1182/blood.v97.7.2115. [DOI] [PubMed] [Google Scholar]
- 152.Veltroni M, De Zen L, Sanzari MC, Maglia O, Dworzak MN, Ratei R, et al. I-BFM-ALL-FCM-MRD-Study Group. Expression of CD58 in normal, regenerating and leukemic bone marrow B cells: implications for the detection of minimal residual disease in acute lymphocytic leukaemia. Haematologica. 2003;88:1245–52. [PubMed] [Google Scholar]
- 153.De Waele M, Renmans W, Jochmans K, Schots R, Lacor P, Trullemans F, et al. Different expression of adhesion molecules on CD34+ cells in AML and B-lineage ALL and their normal bone marrow counterparts. Eur J Haematol. 1999;63:192–201. doi: 10.1111/j.1600-0609.1999.tb01767.x. [DOI] [PubMed] [Google Scholar]
- 154.Björklund E, Mazur J, Söderhäll S, Porwit-MacDonald A. Flow cytometric follow-up of minimal residual disease in bone marrow gives prognostic information in children with acute lymphoblastic leukaemia. Leukemia. 2003;17:138–48. doi: 10.1038/sj.leu.2402736. [DOI] [PubMed] [Google Scholar]
- 155.Dworzak MN, Gaipa G, Ratei R, Veltroni M, Schumich A, Maglia O, et al. Standardization of flow cytometric minimal residual disease evaluation in acute lymphoblastic leukaemia: multicentric assessment is feasible. Cytometry B Clin Cytom. 2008;74:331–40. doi: 10.1002/cyto.b.20430. [DOI] [PubMed] [Google Scholar]
- 156.San Miguel JF, Martínez A, Macedo A, Vidriales MB, López-Berges C, González M, et al. Immunophenotyping investigation of minimal residual disease is a useful approach for predicting relapse in acute myeloid leukaemia patients. Blood. 1997;90:2465–70. [PubMed] [Google Scholar]
- 157.Venditti A, Buccisano F, Del Poeta G, Maurillo L, Tamburini A, Cox C, et al. Level of minimal residual disease after consolidation therapy predicts outcome in acute myeloid leukaemia. Blood. 2000;96:3948–52. [PubMed] [Google Scholar]
- 158.Bacher U, Kern W, Schoch C, Schnittger S, Hiddemann W, Haferlach T. Evaluation of complete disease remission in acute myeloid leukaemia: a prospective study based on cytomorphology, inter-phase fluorescence in situ hybridization, and immunophenotyping during follow-up in patients with acute myeloid leukaemia. Cancer. 2006;106:839–47. doi: 10.1002/cncr.21665. [DOI] [PubMed] [Google Scholar]
- 159.Feller N, Jansen-van der Weide MC, van der Pol MA, Westra GA, Ossenkoppele GJ, Schuurhuis GJ. Purging of peripheral blood stem cell transplants in AML: a predictive model based on minimal residual disease burden. Exp Hematol. 2005;33:120–30. doi: 10.1016/j.exphem.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 160.Maurillo L, Buccisano F, Spagnoli A, Del Poeta G, Panetta P, Neri B, et al. Monitoring of minimal residual disease in adult acute myeloid leukaemia using peripheral blood as an alternative source to bone marrow. Haematologica. 2007;92:605–11. doi: 10.3324/haematol.10432. [DOI] [PubMed] [Google Scholar]
- 161.Laane E, Derolf AR, Björklund E, Mazur J, Everaus H, Söderhäll S, et al. The effect of allogeneic stem cell transplantation on outcome in younger acute myeloid leukaemia patients with minimal residual disease detected by flow cytometry at the end of post-remission chemotherapy. Haematologica. 2006;91:833–6. [PubMed] [Google Scholar]
- 162.Caballero D, García-Marco JA, Martino R, Mateos V, Ribera JM, Sarrá J, et al. Allogeneic transplant with reduced intensity conditioning regimens may overcome the poor prognosis of B-cell chronic lymphocytic leukaemia with unmutated immunoglobulin variable heavy-chain gene and chromosomal abnormalities (11q- and 17p-) Clin Cancer Res. 2005;11:7757–63. doi: 10.1158/1078-0432.CCR-05-0941. [DOI] [PubMed] [Google Scholar]
- 163.Maloum K, Sutton L, Baudet S, Laurent C, Bonnemye P, Magnac C, et al. Novel flow-cytometric analysis based on BCD5+ subpopulations for the evaluation of minimal residual disease in chronic lymphocytic leukaemia. Br J Haematol. 2002;119:970–5. doi: 10.1046/j.1365-2141.2002.03956.x. [DOI] [PubMed] [Google Scholar]
- 164.Maloum K, Charlotte F, Divine M, Cazin B, Lesty C, Merle-Béral H French Cooperative Group on CLL. A comparison of the sensitivity of flow cytometry and bone marrow biopsy in the detection of minimal residual disease in chronic lymphocytic leukaemia. Haematologica. 2006;91:860–1. [PubMed] [Google Scholar]
- 165.Kay NE, Rai KR, O'Brien S. Chronic lymphocytic leukaemia: current and emerging treatment approaches. Clin Adv Hematol Oncol. 2006;4:1–10. [PubMed] [Google Scholar]
- 166.Rawstron AC, Villamore N, Ritgen M, Böttcher S, Ghia P, Zehnder JL, et al. International standardised approach for flow cytometric residual disease monitoring in chronic lymphocyte leukaemia. Leukemia. 2007;21:956–64. doi: 10.1038/sj.leu.2404584. [DOI] [PubMed] [Google Scholar]
- 167.Lanza F, Bi S, Castoldi G, Goldman JM. Abnormal expression of N-CAM (CD56) adhesion molecule on myeloid and progenitor cells from chronic myeloid leukaemia. Leukemia. 1993;7:1570–5. [PubMed] [Google Scholar]