Abstract
Rapid induction of transcription is known to be mediated by factors which bind DNA following post-translational modification. We report here that non-tyrosine phosphorylated (NTP)-Stat1 is involved in a cooperative interaction with Spi-1/PU.1 and IRF8 to form a pre-associated, poised complex is for IL1B gene induction. A double point mutation at a putative STAT binding site, which overlaps this composite Spi-1•IRF8 site located in the LPS and IL-1 response element (LILRE), inhibited human IL1B LPS-dependent reporter activity to about 10 percent of the control wild type vector. Chromatin immunoprecipitation revealed stimulation-independent constitutive binding of IRF8, Spi-1 and NTP-Stat1 at the LILRE, while binding of C/EBPβ was induced to an adjacent C/EBPβ site after LPS stimulation. In contrast to Stat1, IRF8 was tyrosine phosphorylated following LPS treatment. Supporting the involvement of NTP-Stat1, LPS-induced IL1B reporter activity in monocytes was enhanced by ectopic expression of NTP-Stat1Y701F. In contrast, co-expression of a Y211F IRF8 mutein functioned as a dominant-negative inhibitor of LPS-induced IL1B reporter activity. In vitro DNA binding using extracts from LPS-treated monocytes confirmed that the LILRE enhancer constitutively binds a trimolecular complex containing IRF8, Spi-1 and NTP-Stat1. Binding studies using in vitro-expressed proteins revealed that NTP-Stat1 enhanced the binding of Spi-1 and IRF8 to the LILRE. Co-expression of TRAF6, an LPS surrogate, with Spi-1 and IRF8 enhanced IL1B reporter activity in HEK293R cells, which was dramatically reduced when Y211F IRF8 was co-expressed. These results suggest that the rapid transcriptional induction of an important inflammatory gene is dependent upon constitutive cooperative binding of a Spi-1•IRF8•NTP-Stat1 complex to the LILRE, which primes the gene for immediate induction following IRF8 phosphorylation. Phosphorylation of chromatin pre-associated factors like IRF8 may be an important strategy for the rapid transcriptional activation of genes involved in innate immunity.
Keywords: DNA-binding proteins, Transcription factors, Interferon, Macrophages/Immunology, LIL-Stat, ETS, IRF, TLR signal transduction, Interleukin 1
1. Introduction
Components of the gram-negative bacterial cell wall, like lipopolysaccharide (LPS), are involved in the activation of monocytes, an important line of defense in innate immunity. LPS binds to Toll-like receptor 4 (TLR4) through LPS-binding protein (LBP), and leads to activation of TRAF6 through IRAK1 and MyD88 (Martin and Wesche, 2002). Spi-1/PU.1 (Spi-1) has been shown to be one of the critical transcription factors that constitutively binds to the human prointerleukin 1 (IL1B) gene transcription start-site proximal promoter (Kominato et al., 1995) and supports LPS induction derived from the action of the upstream induction sequence (UIS) enhancer located approximately 3 kbp upstream from the transcription start site (Shirakawa et al., 1993; Tsukada et al., 1994; Yang et al., 2000).
The IFNγ-activated JAK-STAT signal transduction pathway is important for mediating the antiviral, antiproliferative and immunomodulatory functions necessary for innate immunity. Tyrosine-phosphorylated (P-Tyr)-Stat1 forms P-Tyr-Stat1 homodimers, translocates to the nucleus and binds gamma interferon activation sequence (GAS) elements. IFNα or β-mediated activation, on the other hand, leads to the phosphorylation of Stat1 and Stat2, forming a heterodimer, which subsequently binds an interferon-responsive factor (IRF9), forming what is known as the ISGF3 complex, which binds an IFN-stimulated response element (ISRE) (Levy and Darnell, 2002). In contrast to IFNγ, LPS-mediated activation has not been consistently associated with P-Tyr-STAT binding to DNA. Earlier work by us and others revealed that human IL1B gene induction by LPS is strongly dependent upon sequences found between −2926 and −2729 upstream of the transcription start site (Fig. 1) involving a mechanism in which the bZIP factor C/EBPβ (also called NF-IL6) binds to a sequence within Region I of the enhancer (Fig. 1) as a heterodimer with one of two distinct members of the ATF bZIP subfamily and makes a protein-protein interaction with Spi-1 bound to the IL1B gene promoter (Chandra et al., 1995; Tsukada et al., 1994; Yang et al., 2000). We recently reported the nature of a specific interaction between Spi-1 and the C/EBPβ bZIP domain (Listman et al., 2005).
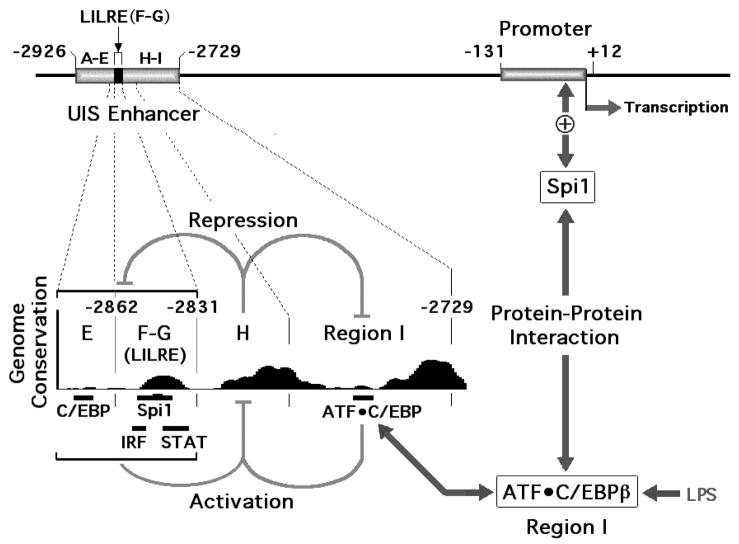
Fig. 1.
Human ILIB gene functional architecture. The LPS- and IL-1-responsive element (LILRE) is located between –2862 and –2831, within the upstream inducible sequence (UIS) enhancer. Utilizing the University of Santa Cruz genome browser (http://genome.ucsc.edu/), a high degree of homology can be detected among Human, Chimp, Dog, Mouse and Rat IL1B genes in the vicinity of the upstream induction sequence (UIS) enhancer of the human IL1B gene, as revealed by the Genome Conservation histogram plot. The plot reveals conservation at locations that correspond to potential and previously identified functional sites. Previously reported region designations (see text) are indicated by letters E through I. This figure also diagrammatically represents the cooperative behavior exhibited by sequences flanking the inhibitory Region H (gray inhibitor indicators) which in-turn support cooperativity between C/EBPβ bound to Region I of the UIS and Spi-1 bound to the basal monocyte-specific promoter (arrows and plus sign).
We have also previously reported (Tsukada et al., 1994) that various sub-regions of the enhancer (labeled as E, F–G, and I in Fig. 1) had independent activities that were inhibited in the presence of a potent silencer (contained within region H in Fig. 1). In the absence of region H, region I had almost 10-fold more activity than the other regions. However, it could not function in the presence of the inhibitory region H, unless the additional upstream regions were also present. Within the critical upstream region is a highly conserved 20 bp sequence (Fig. 1) that we have previously referred to as the LPS and IL-1 responsive element, or LILRE and reported that it contained both an ISRE and a specific GAS site which could bind a Stat1-like factor that we designated as LPS IL-1 Stat, or LIL-Stat (Tsukada et al., 1996). IRF8 is a transcription factor that has been reported to be critical for myeloid differentiation. Specifically, IRF8 functions as a heterodimeric complex with the myeloid-specific ETS factor Spi-1 critical for the expression of relevant genes (Escalante et al., 2002; Kanno et al., 2005; Laricchia-Robbio et al., 2005).
In studies presented here, we show that the LILRE binds Spi-1, IRF8 and NTP-Stat1 as a highly cooperative ternary protein complex which likely corresponds to the previously reported LIL-Stat. Examination of the nature of the proteins in this complex reveals rapid tyrosine phosphorylation of IRF8 following LPS treatment. Although tyrosine phosphorylation does not appear to be essential for cooperative binding to DNA, transfection experiments argue for a functional role in gene induction. We propose that the preassociation of this complex with chromatin primes IL1B for the rapid induction that has been consistently observed for this gene (Fenton et al., 1987; Fenton et al., 1988; Huang et al., 2001).
2. Materials and methods
2.1. Cell lines and cytokines
THP-1 cells were obtained from the American Type Culture Collection (ATCC). Cells were grown in endotoxin-free RPMI 1640 medium (ATCC TIB-202) supplemented with 2mM L-Glutamine, 10% heat-inactivated fetal bovine serum (Hyclone) and 0.05 mM 2-mercaptoethanol in humidified 5% CO2 and 95% air at 37°C. RAW 264.7 cells were also obtained from the American Type Culture Collection (ATCC). Cells were grown in DMEM (ATCC) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone). For stimulation with LPS, RAW 264.7 or THP-1 cells were treated with E. coli serotype 055:B5 LPS (Sigma Chemical Co., St. Louis, L2880) dissolved in endotoxin-free Dulbecco’s PBS at a maximum concentration of 10 μg/ml. Recombinant human IFNγ was purchased from Research Diagnostics, Inc, with less than 1Endotoxin Unit/μg of IFNγ. A final concentration of 16 ng/ml of IFNγ was used to stimulate THP-1 cells. HEK293R cells were derived from HEK293 (American Type Culture Collection CTRL-1573), as previously described (Wang et al., 2006).
2.2. Oligonucleotides and antibodies
All double-stranded oligonucleotides used for Electrophoretic mobility shift assay (EMSA) were custom ordered from Operon Biotechnologies (Valencia, CA). The LILRE (LPS, IL-1 responsive element) oligonucleotide was the same as previously reported (Tsukada et al., 1996), 5′-AGTCTTATAAGAGGTTTCACTTCCTGAGAGTCGA-3′. The GAS oligonucleotide was derived from the high-affinity Fc receptor for IgG (FcγRI) gene, 5′-CGATCGAGATGTATTTCCCAGAAAAGTCGA-3′; and the ISRE, 5′-TCGACGGCTTAGTTTCACTTTCCCTACTAT-3′. The LILRE point mutant oligonucleotide sequences mIRF8, mSpi-1/mSTAT(us), mSTAT(ds), and dmSTAT(ds) were identical to the wild type sequence, except that they contained the substitutions shown in Fig. 3A. The β1-A (ETS) oligo, 5′-TTAAAAAAGGGGAAGATTCC-3′ contains the β-globin B1-A site as reported (Galson and Housman, 1988). The Lambda probe from IgλL 3′ enhancer containing the Spi-1/IRF composite site was 5′-CTAGCGAGAAATAAAAGGAAGTGAAACCAAGA-3′ was used (Brass et al., 1996).
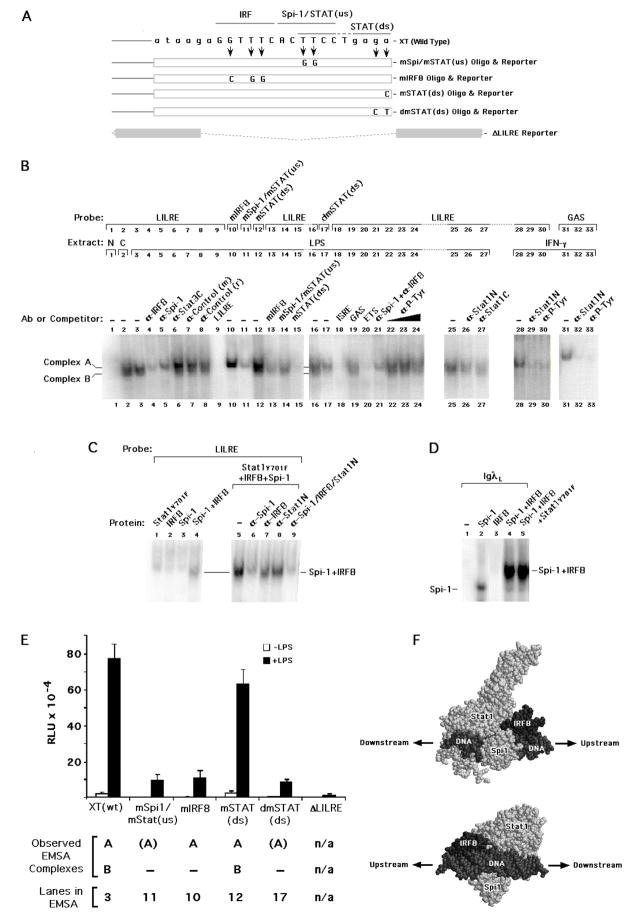
Fig. 3.
Functional and proposed structural nature of the LILRE. (A) Schematic showing the location of the individual mutations in binding sites within a long construct harboring a 3.76 kb region of the human IL1B gene, designated as XT. Similar mutations were made in the labeled probes and cold competitors used for EMSA. These sites include an IRF (mIRF) site, the Spi-1 site (which overlaps the upstream STAT/GAS half-site) designated as mSpi-1/mSTAT(us), and both single and double point mutations designated as mSTAT(ds) and dmSTAT(ds), respectively, in the downstream STAT half-site. An XT-derived construct in which the entire LILRE sequence was deleted is denoted as Δ LILRE. (B) Images of relevant EMSA data showing the binding of complexes from nuclear extracts derived from both control cells (labeled C) and cells exposed for 30 min to LPS or IFNγ. Binding studies used wild type and mutated (as indicated) LILRE or GAS α-32P-labeled probes at a final ionic strength of 50 mM Na+ plus K+. Null binding reactions (labeled N) did not contain extract. Antibodies were pre-incubated with nuclear extract on ice prior to the addition of labeled probe. Unlabeled competitor oligonucleotides were used at a 100-fold molar excess over the molar concentration of the probe. (C) EMSA showing the binding of in vitro expressed proteins to wild type LILRE α-32P-labeled probes at an ionic strength of 50 mM Na+, optimal for Spi-1 binding(Galson and Housman, 1988). Antibodies were added as indicated before addition of probe. (D) EMSA showing the binding of in vitro expressed proteins to IgλL α-32P-labeled probe. (E) Activity of wild type and mutant IL1B gene reporter in RAW 264.7 cells transfected with wild type XT, mIRF XT, mSpi-1/STAT(us) XT, mSTAT(ds) XT, dmSTAT(ds) XT, and ΔLILRE XT, each at 250 ng with total DNA adjusted to 300 ng with empty pGL3Basic vector. Cells were transfected with the indicated constructs and either left untreated or treated with 10 μg/ml LPS for 4 hrs. The values are derived from the average of three independent experiments. (F) Hypothetical arrangement of IRF8, Spi-1 and non-tyrosine phosphorylated (NTP) Stat1 bound to LILRE DNA. Proteins and DNA are labeled and represented as space filling models. The orientation of the proteins is indicated by arrows pointing upstream and downstream, either toward or away from, respectively, the transcription start site. The lower image is related to the upper via a 180° rotation about the Y axis plus a −70° rotation about the X axis.
Antibodies used in either EMSA or chromatin immunoprecipitation were derived from various commercial sources. Anti-Stat1 N-terminal antibodies were either a rabbit polyclonal directed against amino acids 1-194 of Stat1 (cat. no. 610120) from BD-Pharmingen Transduction Laboratories or a mouse monoclonal (clone SM2) against amino acids 8-23 of Stat1 (ab14743) from Abcam (Cambridge, MA). Rabbit polyclonal IgG antibodies purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) were: anti-Stat1α/β (M-22) against a common Stat1p84/p91 peptide (sc-592); anti-Stat1α (C-24) against the C-terminus of Stat1p91(sc-345X), anti-Spi-1 (T-21) against C-terminus (sc-352); anti-Spi-1 (H-135) against amino acids 1-135 (sc-22805); anti-IRF8/ICSBP (H-70) against amino acids 357-426 (sc-13043); anti-Stat3 (C-20) against C-terminus (sc-482); anti-Stat3 (K-15) against an internal region near the SH3 domain (sc-483); anti-Stat1α (M-23) against the unique C-terminus of Stat1p91(sc-591); anti-IRF9 (C-20) against the C-terminus (sc-496) and a pre-immune rabbit polyclonal IgG control (sc-2027). Polyclonal goat IgG antibodies from Santa Cruz included: anti-phosphotyrosine Stat1 (Tyr 701, sc-7988); and anti-IRF4 (M-17) against C-terminus (sc-6059). Mouse anti-phosphotyrosine monoclonal (PY99, sc-7020) and pre-immune mouse polyclonal antibody (sc-2025) were also from Santa Cruz. Mouse anti-phosphotyrosine monoclonal (4G10) from Upstate Biotechnology (05-321) was exclusively used for EMSA.
2.3. In vitro transcription/translation
Stat1, Stat1Y701F, Spi-1 and IRF-8 were translated in vitro using the TNT T7-based quick coupled transcription-translation kit (Promega) as per manufacturer’s instructions. The presence of transcript in the samples was ascertained by using fluorescent lysine incorporation (FluoroTect GreenLys) and SDS-PAGE electrophoresis, according to manufacturer’s instructions.
2.4. Preparation of nuclear extracts
Nuclear extracts were prepared as previously described (Shirakawa et al., 1993). Following incubation of cells with LPS (10μg/ml) for 30 min, 1×108 cells were harvested, washed with ice-cold Dulbecco’s PBS containing a protease inhibitor mixture (mixture P) consisting of: 1× protease inhibitor cocktail (Sigma Chemical Co., St. Louis, MO); 0.5 mM PMSF (diluted from freshly made stock); and 1 μg/ml each of chymotrypsin and antipain. Cells were then incubated in 5 ml of buffer A (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 300 mM sucrose; 0.1 mM EGTA; 0.5 mM DTT) containing mixture P and phosphatase inhibitor cocktails I and II (Sigma Chemical Co., St Louis, MO) for 5 min on ice, with occasional vortexing. After spinning, the cells were resuspended in 1 ml of ice-cold buffer A, with mixture P and the phosphatase inhibitors, and Dounce homogenized. The homogenate was spun for 30 sec at 3000 rpm, and nuclei were resuspended in 0.3 ml of buffer B (20 mM HEPES, pH 7.9; 1.5 mM MgCl2; 300 mM KCl; 0.1 mM EGTA; 20% glycerol; 0.5 mM DTT) with mixture P and phosphatase inhibitors per 1×108 cells, and gently rocked at 4°C for 30 min. The nuclei were spun at full speed in a microfuge at 4°C for 30 min, and the supernatant was transferred to a dialysis cassette with 7500 MW-cutoff (Pierce, Rockford, IL) and dialysed against 50 volumes of Buffer D (20 mM HEPES, pH 7.9; 100 mM KCl; 20% glycerol; 0.5 mM DTT), with mixture P and phosphatase inhibitors at 4°C for overnight. After dialysis, the samples were centrifuged at full speed (~14,000 rpm) at 4°C for 30 min. Protein concentration was measured using a Bio-Rad (Hercules, CA) assay kit and the supernatant aliquoted on dry ice and stored at −80°C.
2.4. Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described previously (Shirakawa et al., 1993). Briefly, for the binding reaction, equal volumes of nuclear extract containing equal cell numbers were pre-incubated in a total volume of 15–18 μl of binding buffer (10mM Tris-HCl [pH7.5], 1 mM EDTA, 1 mM β-mercaptoethanol, 4% glycerol, and 50 mM NaCl) and 0.2 μg poly (dI-dC). The cold competitors (100 fold molar excess of the wild type probe) were pre-incubated for 10 min at room temperature with the nuclear extract and buffer before the addition of the hot probe. Antibodies (1 μl or 3 μl for specific antibodies) were pre-incubated with nuclear extract and buffer for 30 min on ice before the addition of labeled probe. Finally, the α-32P-end-labeled probe (0.2 ng, about 20,000 cpm) was added and pre-incubated for 30 min at room temperature. Samples were run in a 4% polyacrylamide gel in 0.5× TBE buffer (45 mM Tris-borate and 1 mM EDTA) at 4°C. After drying, the image was captured using Typhoon Phosphoimager (Amersham Biosciences). EMSA probes were labeled by filling in 3′ recessed ends by DNA polymerase Klenow fragment using one α-32P-labeled deoxynucleoside triphosphate (dNTP) at 3000Ci/mmol (Amersham Biosciences), as described previously (Shirakawa et al., 1993). Unincorporated dNTP was removed with G-50 spin columns (Amersham Biosciences).
2.5. Chromatin immunoprecipitation (ChIP) assay
The ChIP assay kit (Upstate Biotech.) was used with some modifications. THP-1 cells were stimulated with either LPS or IFNγ for 30 min at a density of 0.5×106 cells/ml in 10 mls in 100 × 20 mm plates. After 30 min, stimulated and control cells were treated with 37% (w/w) formaldehyde solution (Fisher Scientific) to a final concentration of 1%, and incubated for 10 min at 37°C. Cells were scraped off on ice, and washed twice with ice-cold Dulbecco’s PBS, containing mixture P protease inhibitors and phosphatase inhibitor cocktails I and II (see preparation of nuclear extracts, above). Cells were lysed with 100 μl SDS Lysis buffer (from the Upstate kit), containing protease inhibitors and phosphatase inhibitors, and incubated for 30 min on ice. The samples were sonicated with a Fisher model 100 Sonic Dismembrator for 20 × 30 sec pulses to generate an average length of 400 bp chromatin fragments. Samples were diluted 10 fold in ChIP dilution buffer, with the necessary protease and phosphatase inhibitors added. Samples were pre-cleared to reduce non-specific background by adding protein A/G Plus agarose beads (Santa Cruz Biotech), pre-mixed with sheared salmon sperm DNA (2 μg/ml final) at 4°C for 30 min. Immunoprecipitation was carried out at 4°C overnight, with the respective antibodies at a final concentration of 8 μg/ml. The next day, protein A/G plus agarose beads and sheared salmon sperm DNA (2 μg/ml final) was added to collect the antibody-chromatin complexes for 1hr at 4°C with rotation. Beads were washed twice with each of the buffers provided in the kit. Chromatin was eluted from the beads with 200 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and DNA cross-links were reversed by incubating samples at 65°C overnight in a final concentration of 0.2M NaCl. The next day, samples were incubated with proteinase K, Tris-HCl and EDTA (see manufacturer’s instructions) at 45°C for 1 hr. DNA purification was carried out using a PCR purification MinElute kit (Qiagen) followed by PCR amplification using specific primers. The primers used were designed to amplify a 282 bp portion of the Upstream Induction Sequence (UIS) between −3058 and −2777 which contains the LILRE (Shirakawa et al., 1993). The upstream primer UIS282 (forward) was: 5′-CCATCTCTGCTTGTTCCA-3′, and the downstream primer UIS282 (reverse) was: 5′-AGTTCTGCCATGTCTCATCC-3′, The positive control primers were designed from the FcγRI gene, encompassing a GAS site (Paquette et al., 1995) which generated a 124 bp product. The FcγRI gene forward primer is: 5′-GATGGGCTAACAGGTATGA-3′ and FcγRI gene reverse primer is: 5′-GAGCCAAATTAGAAAAGAGG-3′. The double IP ChIP procedure was identical, except that after the first immunoprecipitation the eluent buffer was diluted 10 fold with ChIP dilution buffer (including the protease and phosphatase inhibitor cocktails) and the second immunoprecipitation was carried out. This was followed by elution and reversal of cross-links and PCR.
2.6. Real-time quantitative PCR and data analysis
Differences in DNA concentration obtained after immunoprecipitation of LPS treated and untreated samples was determined by real-time PCR using the ABI Prism 7700 sequence detector following the Applied Biosystems SYBR Green PCR Master Mix protocol. Real-time PCR was carried out in duplicate on equal quantities of stimulated and unstimulated DNA (ranging from 10 to100 pg of DNA). For each experiment, the threshold was set to cross a point at which amplification was linear and above the background. The fold difference of a given target sequence precipitated by a specific antibody, before and after stimulation by LPS was calculated by 2(Ct untreated IP)−(Ct LPS treated IP). The Ct values of equal quantitites of sonicated, and purified genomic input DNA derived from unstimulated and LPS stimulated THP-1 cells were almost the same (33.98 for unstimulated versus 33.73 for stimulated input DNA, each at 800pg respectively). Crosslinked, sonicated input DNA was prepared using a DNeasy kit (Qiagen), after Proteinase K and RNase A treatment.
2.7. DNA constructs
The XT-Luc IL1B vector was made from the BDC454 clone (Clark et al., 1986; Shirakawa et al., 1993) by doing a partial digestion with XbaI and TaqI to cut out the required length of human genomic IL1B DNA from −3757 downstream to +12. This 3.76 kbp sequence was inserted into the SmaI site of the pGL3Basic vector. The XT-Luc mutants, namely, mIRF8 LILRE XT, mSpi-1/STAT(us) LILRE XT, and mSTAT(ds) LILRE XT, as well as Δ LILRE XT, were made by using an XL QuikChange site-directed mutagenesis kit, using primers with the required mutations or deletions in them, and DpnI digestion of the original vector (Stratagene). The chimeric UIS enhancer CAT reporters were made by ligating the B-I sequence (−3132 to −2729) and the D–G sequence (−2925 to −2833) to the minimal (−59 to +105) murine c-fos promoter (Tsukada et al., 1994). The expression vector (in pcDNA 3) for wild type mouse IRF8, was a kind gift of Dr. Scott Plevy, University of Pittsburgh. The mouse IRF8Y211F and IRF8Y371F mutants were made from the wild type cDNA using specific primers containing the mutations, again using the XL QuikChange kit (Stratagene). Wild-type Stat3α expression vectors in pcDNA 3.1, as well as the mutants Stat3-Y705F and Stat3-Ser727Ala were made by Dr. Yasuhiro Yoshida (Yoshida et al., 2004). The wild type Stat1α expression vector was made by PCR cloning the ORF from the Stat1α (p91) cDNA in pBluescript I SK− vector using Pfu Turbo Polymerase (Stratagene). The PCR fragment was inserted into EcoRV-cut pcDNA3.1 (Invitrogen). All the Stat1α mutants, namely, Stat1α Y701F, S727A, R602L, K161A were made by using the QuikChange kit (Stratagene). Stat1β was also made by amplifying the ORF for Stat1β from the cDNA sequence of Stat1α in pBluescript I SK−. The amplified product was then inserted into EcoRV-digested pcDNA3.1. All constructs were verified by DNA sequencing.
2.8. Transfection and luciferase assays
Raw 264.7 cells were seeded at 3.4×104 cells/well in 24 well plates, 24 hours before the day of transfection. Cells were transfected with Calcium Phosphate reagent using the Mammalian Transfection kit (Stratagene). After 4 hours of incubation, cells were washed with serum-free DMEM and subjected to a 15% glycerol shock for 1.5 min, re-washed with serum-free DMEM once and incubated in DMEM containing 10% FBS for about 16 hours or overnight. The next day, cells were stimulated with the required doses of LPS for 4 hours. After washing with D-PBS, the cells were lysed at room temperature in Cell Culture Lysis buffer (Promega) for 15 min. The lysates were analysed with the luciferase reporter assay system (Promega), using Veritas Microplate Luminometer Software. THP-1 cells were transfected using DEAE-dextran plus heparin (Shirakawa et al., 1993). Transfection of HEK293R cells utilized Fugene 6 (Roche) as previously described (Wang et al., 2006).
2.9. Molecular modeling
The atomic coordinates for the IRF8•Spi-1 DNA complex (Escalante et al., 2002) were kindly provided by Dr. Aneel Aggarwal, Mount Sinai School of Medicine, New York, NY. Coordinates for the tyrosine phosphorylated Stat1 dimer DNA complex (Chen et al., 1998) were obtained from the Rutgers University Research Collaboratory for Structural Boinformatics (Berman et al., 2000), PDB code 1BF5. The two coordinate sets were loaded into RasMac (version 2.7.2.1) and independently rotated to align and overlap the DNA molecules for each complex in order to mimic the relative locations of the IRF8, Spi-1 and STAT GAS sites found within the LILRE. The Stat1 monomer bound to the upstream STAT(us) half-site was selected and rendered invisible in order to generate the hypothetical binding of monomeric Stat1 bound to the downstream STAT(ds) half-site.
3. Results
3.1. The LILRE is highly conserved and important for IL1B enhancer activity
The LILRE in region F–G is located in the Upstream Induction Sequence (UIS) enhancer of the human IL1B gene (Fig. 1). As depicted in Fig. 1, this response element cooperates with region E and region I (shown to bind an inducible heterodimeric ATF•C/EBPβ complex) to overcome inhibition by region H. The high degree of inter-species conservation for this sequence is consistent with a functional role, in IL1B gene regulation. Sequence analysis, in vitro binding, and transfection studies of the LILRE suggest the involvement of at least three different protein binding sites. These are: an ISRE (interferon stimulated response element) (Tsukada et al., 1996); a composite ETS•ISRE site that could bind a complex between the ETS family protein Spi-1 and either the IRF4 or IRF8 ISRE-binding proteins (Marecki et al., 1999; Marecki et al., 2001); and a GAS-like dyad site reported to bind a Stat1-like molecule (Tsukada et al., 1996; Tuyt et al., 1998) that could be a P-Tyr-Stat1 or Stat3 homo or hetero dimer (Lemmink et al., 2001).
The protein-DNA X-ray crystal structures for Spi-1•IRF4 binding to IgλL light chain gene 3′ enhancer (Escalante et al., 2002) and for both Stat1 and Stat3 binding to GAS site DNA (Becker et al., 1998; Chen et al., 1998) have been solved. The report on the Spi-1•IRF4 structure argued for high structural similarity between IRF4 and IRF8 and predicted a similar mode of interaction with Spi-1, which was subsequently demonstrated biochemically (Marecki et al., 2001). Using these structures, hypothetical DNA footprints can readily be predicted. Fig. 2A shows two-dimensional representations of the protein complexes derived from the three-dimensional data superimposed upon an oligonucleotide sequence containing the LILRE. Such shorthand representations are often used by structural biologists to permit a detailed display of the structural elements of an interaction without having to resort to the use of an interactive molecular modeling system. The LILRE contains an ETS•ISRE composite element (Fig. 2A, upper sequence) that is almost perfectly conserved in spacing and sequence with the Spi-1•IRF4 binding sequence derived from the IgλL light chain genes (denoted by caps). It also contains a GAS-like STAT dyad site to which a tyrosine-phosphorylated STAT dimer could bind (Fig. 2A, lower sequence), which is not found within the IgλL flanking sequence (differences indicated by arrows in upper sequence). However, it is immediately apparent from the two-dimensional footprints shown in Fig. 2A and from molecular modeling studies (not shown) that a classical tyrosine-phosphorylated STAT dimer cannot simultaneously bind along with Spi-1 to the LILRE, due to the almost complete overlap between the Spi-1 site and the upstream STAT (STAT(us)) half-site (compare sequences in Fig. 2A). Furthermore, while it may be possible, it is also unlikely that a STAT dimer and IRF8 can simultaneously bind, due to steric clashing between the IRF8 and the STAT monomer bound to the upstream STAT half-site (Fig. 2A, middle sequence). Consequently, at least two mutually exclusive binding configurations are predicted for these proteins complexed with the LILRE. One configuration is a hetero-complex of IRF8 and Spi-1 bound to DNA, as predicted from the related X-ray structure (Escalante et al., 2002), with either no STAT binding or only a STAT monomer (upper sequence). The other configuration would involve either IRF8 alone or a tyrosine-phosphorylated STAT dimer (lower sequence). It should be emphasized that IRF4 in the analogous IgλL and IgκL genes cannot bind well in the absence of Spi-1 (Brass et al., 1996).
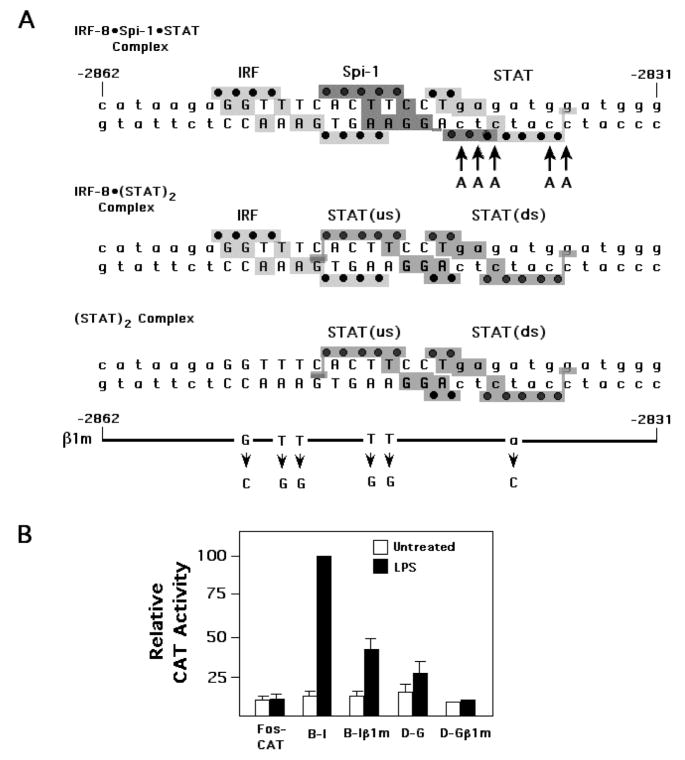
Fig. 2.
The LILRE is required for IL1B UIS function. (A) The LILRE sequence revealing predicted structural footprints for two alternative binding arrangements and nucleotide substitutions made in B–I and D–G β1m reporter constructs. The footprints were derived by examination of related structures reported for a Stat1-DNA complex (Chen et al., 1998) and a ternary complex of IRF4 and Spi-1 bound to IgλL DNA (Escalante et al., 2002). Gray regions reveal predicted structural contacts with bases (letters) and phosphates (black dots). Capital letters designate exact matches between the LILRE and the model IgλL sequence. The presence of five adenines (indicated with arrows) at the IgλL site distinguishes it from the LILRE. Two alternative footprints are presented, revealing that Spi-1 may not be able to bind in the presence of a Stat1 dimer because of extensive structural overlap (upper sequence), but might be able to bind as a complex with IRF along with a monomer of Stat1 bound to a single half-site (lower sequence). The 6-point β1m mutation in the UIS constructs is composed of the indicated base changes (horizontal bar with arrows) for the collective putative binding sites in the LILRE. (B) Reporter activity of wild type or mutant UIS enhancer constructs ligated to a heterologous murine c-fos Δ56 core promoter transfected into human THP-1 monocyte cells. The B–I construct contains sequence between −3132 and −2729 in the IL1B gene, while D–G contains sequence from −2925 to −2833. Cells were transfected with the indicated constructs and either left untreated or treated with 10 ng/ml LPS. The empty CAT vector containing the Δ56 fos promoter was used as a negative control. Data are derived from 3 independent experiments.
We originally identified the LILRE, and the overlapped binding of proteins to this sequence, using a collection of mutations during an early screening of the gene for regulatory regions (Shirakawa et al., 1993). At the time of that report, the nature of the proteins interacting with the sequence was unknown, and the biological relevance of this region was tested only in the context of a transiently transfected construct containing six base substitutions (β1m, as shown schematically at the bottom of Fig. 2A) within an IL1B gene fragment located between −3132 and −2729 ligated to a heterologous truncated murine c-fos promoter CAT reporter. The c-fos promoter was used because constructs containing both the IL1B enhancer and promoter do not function well in THP-1 cells in response to LPS (Gray et al., 1993; Shirakawa et al., 1993). Fig. 2B shows the activity of this original construct (labeled as B-Iβ1m) and the wild type construct (labeled B–I), transfected into both untreated and LPS treated THP-1 cells. This same mutation in the original c-fos promoter CAT reporter is now also shown to significantly affect the function of the shorter sequence comprising regions D through G (−2925 to −2831), one of two core activity regions of the IL1B UIS enhancer that we previously reported by deletion analysis (Tsukada et al., 1994). A portion of this region, located between −2862 and −2831, was subsequently also examined and demonstrated to be induced by IL-1, LPS, and IL-6, but not by either IL-4 or IFNγ (Tsukada et al., 1996).
3.2. LILRE constitutively binds Spi-1, IRF8 and Stat1 in vitro
In order to identify which proteins interact with the LILRE, DNA binding was examined by EMSA using THP-1 cell nuclear extracts and specific antibodies. A series of mutated LILRE oligonucleotides were generated (Fig. 3A) in order to dissect specific involvement of the putative IRF, Spi-1 and STAT binding sites by EMSA (Fig. 3B). In aggregate, these substitutions are the same as those previously used in the β1m reporters (Fig. 2), but were generated as three specific non-overlapping groups for both EMSA and the XT luciferase reporters. A double substitution (dmSTAT(ds)) was also introduced within the downstream STAT half-site in order to completely disrupt the nature of the STAT consensus sequence. Nuclear extracts from untreated cells and those treated with 10 μg/ml LPS for 30 min revealed two specific constitutive complexes (Complex A and B), as determined by competition with self and non-self unlabeled oligonucleotides (Fig. 3B, lanes 9 vs. 19). Complex B was completely abrogated by the addition of antibodies against either IRF8 (lane 4) or Spi-1 (lane 5), whereas, complex A was somewhat reduced by anti-IRF8, but not anti-Spi-1. Incubation with antibody specific for the C-terminus of Stat1α (lane 27) and against the common N-terminus of Stat1α and β (lane 26) reduced both complexes. This reactivity appeared less potent than that observed for either bona-fide IFNγ-activated Stat1 bound to a well characterized FcγRI gene GAS Stat1 binding site (lanes 31–33) or to the same IFNγ-activated extract incubated with the LILRE probe (lanes 28–30). Interestingly, an anti-phospho-tyrosine antibody had less effect on the complexes, as compared to the interferon γ control (compare lanes 22–24, with lanes 30 and 33, respectively). This antibody reactivity may be the result of IRF8 phosphorylation in LPS-stimulated extracts (see ChIP data, below). In contrast to Stat1, Stat3 did not appear to be present in either complex (lane 6).
Incubation of extract with α–32P-labeled mIRF8 LILRE probe, where the IRF-binding site was mutated (Fig. 3A, mIRF8), resulted only in the loss of complex B (Fig. 3B, lane 10). However, mutation of the Spi-1 site, which overlaps the upstream STAT half-site, in the LILRE (mSpi1/mSTAT(us)), resulted in complete loss of complex B, and a partial reduction in complex A (lane 11). This suggests that the Spi-1 site is central to the formation of both complexes, but is more critical for complex B. Competition of WT LILRE probe with either mIRF8 (lane 13) or mSpi-1/mSTAT(us) (lane 14) oligonucleotide was less effective for complex B than complex A, arguing that both sites are necessary for complex B formation and are less important individually for complex A. This suggests that a cooperative interaction between IRF8 and Spi-1 is required to form complex B. However, both the ISG54 ISRE binding sequence (Tsukada et al., 1996) and an oligonucleotide competitor containing an ETS factor binding site from the mouse β-globinM gene that avidly binds Spi-1 (Galson and Housman, 1988) compete effectively for both complexes (lane 18 and 20, respectively). This suggests the possibility that related, but distinct IRF and ETS family proteins may be involved in generating the two similar, but not identical complexes.
Addition of extract to labeled LILRE probe mutated at the downstream STAT half-site did not result in loss of binding for the single-point mutated (mSTAT(ds)) probe (Fig. 3B, lane 12), whereas the double mutation in this half-site (dmSTAT(ds)) resulted in the complete loss of complex B (lane 17). The mSTAT(ds) competitor completely abrogated both complexes on the WT LILRE probe (lane 15), further indicating that this single point mutation does not interfere with the formation of either complex. Interestingly, the FcγRI gene Stat1-binding GAS sequence does not compete (lane 19). Therefore, it appears that the LILRE does not bind a classical tyrosine-phosphorylated STAT dimer after LPS stimulation. Instead, the LILRE constitutively binds a STAT factor that makes a protein-protein interaction with either IRF8 or Spi-1 in complex B. We also tested the LPS-treated cell extracts used in Fig. 3B for binding to the FcγRI gene GAS probe. In contrast to the IFNγ-treated extracts, LPS did not induce any observable complex binding (not shown). However, as previously reported (Lemmink et al., 2001), the LILRE probe does appear to bind tyrosine-phosphorylated Stat1 when cells are treated with IFNγ (lanes 28–30). Consequently, we do not believe that LPS induces the formation of tyrosine-phosphorylated Stat1 dimers.
3.3. IRF8•Spi-1 binding to LILRE is enhanced by NTP-Stat1
In order to further characterize the nature of proteins bound to the LILRE, in vitro expressed proteins were used in an attempt to recapitulate the results obtained using nuclear extracts (Fig. 3C). In vitro expressed non-tyrosine phosphorylatable-Stat1 (Stat1 Y701F) alone did not bind to the LILRE probe (lane1). Similarly IRF8 (lane 2) as well as Spi-1 (lane 3) also did not yield relevant complexes when added individually. In contrast, Spi-1 and IRF8 together revealed some binding to the probe (lane 4). Therefore, Spi-1 and IRF8 cooperatively bind to LILRE to form a heterocomplex. This binding is similar to, but weaker than that reported for Spi-1 and IRF4 on the IgλL probe (Brass et al., 1996; Escalante et al., 2002). However, when Spi-1, IRF8 and Stat1 Y701F are added together, binding of this complex to LILRE is stronger (lane 5), and similar to complex B from nuclear extracts. The use of Spi-1 and IRF8 specific antibodies demonstrates that both proteins are involved in formation of this complex (lanes 6, 7, and 9). Importantly, the addition of anti-Stat1N antibody also diminished this in vitro-translated protein:DNA complex (lane 8), indicating that addition of Stat1Y701F protein facilitated the increased DNA binding by IRF8 and Spi-1. These data collectively argue for formation of a ternary complex involving Spi-1, IRF8 and NTP-Stat1 that cooperatively generates a DNA binding complex that both migrates and is antigenically similar to complex B. We also analyzed the binding of Spi-1 and IRF8 to a 32P-labeled IgλL 3′ enhancer (Brass et al., 1996) probe (Fig. 3D) under the same conditions as the LILRE (50 mM total Na+ and K+ ion). In contrast to the LILRE probe, and similar to that previously reported by others for IgλL (Brass et al., 1996), Spi-1 alone bound the IgλL probe (lane 2). Similar to IRF4, IRF8 alone did not bind (lane 3), but bound cooperatively in the presence of Spi-1 (lane 4). Addition of Stat1Y701F did not change the intensity of the Spi-1/IRF8 complex formed on IgλL (lane 5), indicating that Stat1 does not interact with the IgλL probe in association with Spi-1 and IRF-8. Therefore, the LILRE, which differs from IgλL by five nucleotides (indicated at the bottom of Fig. 2A), is a much weaker Spi-1•IRF8 composite site and is dependent upon NTP-Stat1 for efficient binding.
3.4. The LILRE is important for induction of an IL1B promoter reporter and complex B is the functional LILRE complex
The above results suggest that the LILRE region may be critical for IL1B activity. However, previously, activity was examined either in the context of relatively short constructs employing a heterologous promoter, as described above for Fig. 2, or with a longer construct containing the IL1B promoter in a system that generated constitutive, rather than inducible, gene transcription in the presence of ectopically expressed IRF and Spi-1 (Marecki et al., 2001). In order to determine the importance of the LILRE within the context of a long and inducible human IL1B gene sequence, a 3.76 kbp luciferase reporter construct (designated XT) containing both the UIS enhancer and the promoter was generated. Mutations within XT were made at the same specific sites introduced into the oligonucleotides used for EMSA (Fig. 3A). Since similar mutations were used to characterize biding by EMSA (Fig 3B and C) and function (Fig. 3E), a correlation could be made between the complexes formed and activity. The XT constructs contains both the IL1B enhancer and promoter, which do not function well together in THP-1 cells (Gray et al., 1993; Shirakawa et al., 1993), therefore, RAW264.7 cells were used for all XT reporter transfection studies. Cells transfected with wild type and the mutated reporters were stimulated with LPS and assayed for luciferase activity (Fig. 3E). Deletion of the entire LILRE within XT completely eliminated activity, reflecting the importance of this region in the context of the XT reporter (ΔLILRE XT). XT constructs with point mutations at either the Spi-1 (mSpi-1/STAT(us) XT), or at the IRF8 binding site (mIRF8 XT), or the double point mutation at the downstream STAT half-site (dmSTAT(ds)) showed much lower (<25%) total transcriptional activities than that of wild type XT although some induction was still observed. Of note, each of these mutations decreased formation of complex B, which includes IRF8, Spi-1, and a NTP-Stat1 monomer (Fig. 3B, lanes 10, 11, and 17). This result implies that when mutations cause any one of the 3 proteins to bind less well, cooperativity with the other two factors can still generate some binding, albeit not as much as WT. However, the total activity and the induction-fold of the XT reporter containing the single point mutation at the downstream STAT half-site (mSTAT(ds)) was close to wild type activity, consistent with the EMSA results (Fig. 3B and lane 12), demonstrating that this mutation does not affect transcription factor binding to the LILRE. These results, taken together with the predicted binding footprints shown in Fig. 2A and the EMSA results in Fig 3B and 3C, suggest that there may be multiple protein interactions at this site important for inducible activity and indicate that the most probable complex would include IRF8, Spi-1, and a NTP-Stat1 monomer (complex B).
3.5. Molecular modeling of the Spi-1/IRF-8/Stat1 molecular complex
The collective binding and activity data argue for the formation of an essential complex involving IRF8, Spi-1, and NTP-Stat1. In this complex, IRF8 and Spi-1 likely associate in a manner similar to that proposed for these proteins based upon the related structure for IRF4•Spi-1•DNA (Escalante et al., 2002). Using the reported X-ray crystal structures for IRF4•Spi-1 DNA binding domains simultaneously bound as a ternary complex with DNA (Escalante et al., 2002) and a DNA-bound Stat1 monomer derived from the structure of tyrosine-phosphorylated dimeric Stat1 bound to a DNA GAS site palindrome (Chen et al., 1998), we have generated a hypothetical structure of an IRF8•Spi-1•NTP-Stat1 complex with B-form DNA, using the spacing of the LILRE site (Fig. 3F). This hypothetical structure reveals that the LILRE has the potential to simultaneously accommodate all three DNA binding domains in a closely packed configuration with minimal atomic clashing. Furthermore, the model suggests the possibility that all three proteins could directly interact with LILRE DNA. If representative of the actual complex, the mutual proximity of all three proteins in this model suggests the possibility of a high degree of cooperativity, as reflected by the DNA binding data of Fig. 3C.
3.6. LILRE chromatin binds Spi-1, NTP-Stat1α and tyrosine–phosphorylated IRF8
Chromatin immunoprecipitation (ChIP) assays were carried out in order to examine changes in transcription factor binding at the LILRE within intact THP-1 monocyte chromatin. Both traditional semi-quantitative and quantitative real-time PCR were used to quantify the fold-change in binding for several factors following stimulation with either LPS or IFNγ. The analysis used upstream and downstream primers designed to amplify a 282 bp IL1B sequence between −3058 and −2777, which includes the LILRE. The fold increase (stimulated vs. unstimulated Ct values) in the binding for Spi-1, IRF-8, and Stat1 was approximately unity (Fig. 4) after 30 mins of LPS stimulation, suggesting that these factors are pre-associated on the LILRE at equal levels before and after LPS treatment. As a positive control, we also looked at the binding of the C/EBPβ (NFIL6) to an adjacent site to the LILRE (Fig. 4). Confirming our previous observation of inducible C/EBPβ binding after LPS stimulation of THP-1 cells (Tsukada et al., 1994), we saw an inducible increase in binding of C/EBPβ to this site at the IL1B enhancer. The level of target sequence pulled-down by Stat1 N-terminal antibody decreased after stimulation with LPS, possibly because the epitope for Stat1 N-terminus antibody is inaccessible due to complex formation with Spi-1 and IRF-8. Using a Stat3 internal antibody, it appeared that, although Stat3 binding to the LILRE was not observed by EMSA, it may bind to this region of IL1B in the cell. Therefore, like Stat1, it appears to be constitutively associated with chromatin in the vicinity of the LILRE. This could be due to the long average fragment sizes associated with the ChIP procedure (approximately 400 bp), as compared to the short length of the EMSA probes, and therefore, possibly due to Stat3 binding at an alternative site outside the LILRE.
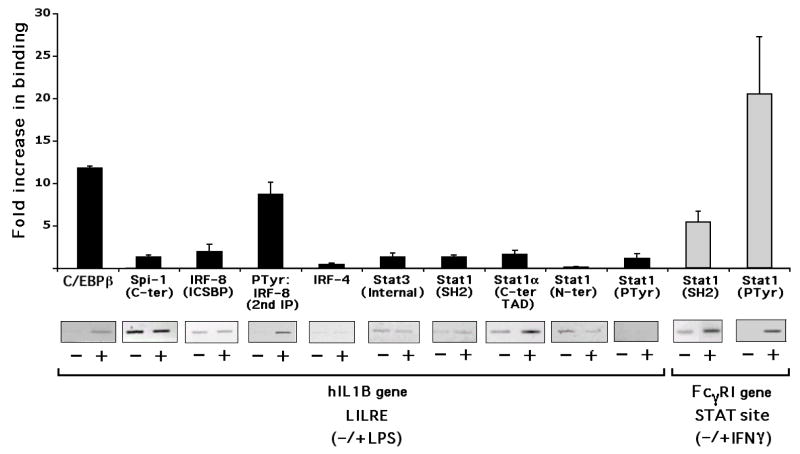
Fig. 4.
Differential binding of transcription factors at the IL1B upstream enhancer and FcγRI gene. ChIP reveals in vivo factor binding to LILRE chromatin. Proteins were cross-linked to DNA in unstimulated, LPS-treated (10 μg/ml for 30 min), and IFNγ-stimulated (16 ng/ml for 30 min) THP-1 cells. Immunoprecipitation was carried out with antibodies, as indicated along the X-axis, and analyzed by both quantitative PCR (histograms indicate the relative association in the presence vs. the absence of stimulation) as well as semi-quantitative PCR (gel images revealing appropriate-sized PCR amplicons in either the absence ‘−’ or presence ‘+’ of stimulant). LPS (black bars) and IFNγ (gray bars) stimulated samples are also indicated by brackets. Inducible C/EBPb binding to an adjacent C/EBPb binding-site, after LPS stimulation. Quantitative data were derived from two independent experiments.
Strikingly, chromatin-associated Stat1 is not tyrosine phosphorylated after LPS stimulation (Fig. 4, Stat1 PTyr). These constitutive binding results contrast with that observed for the FcγRI gene following IFNγ treatment of cells using specific primers for a region that contains a critical homodimeric Stat1 GAS site corresponding to the GAS probe sequence used for the in vitro binding studies shown in Fig. 3B (lane 31). In the case of FcγRI gene chromatin, Stat1 binding and tyrosine phosphorylation are both clearly induced by IFNγ (Fig. 4, Stat1 SH2 and Ptyr Stat1, on FcγRI gene). A supporting in vitro binding report by others (Lemmink et al., 2001) and our EMSA results (Fig. 3B, lanes 28–30), as well as our preliminary ChIP assay results (data not shown), indicate that the LILRE region in chromatin also binds Stat1 and P-Tyr-Stat1 in response to IFNγ. However, Stat1PTyr binding at the LILRE is not observed in response to LPS (Fig. 4). The in vitro binding of PTyr-Stat1 to the LILRE after IFNγ stimulation suggests the possible binding of a “classical” Stat1 homodimer to this site. However, stimulation of human monocytes with IFNγ alone did not activate human IL1B gene reporter activity (data not shown), indicating that binding of a “classical” homodimer to this site is ineffective for human IL1B gene induction.
IRF8, like Spi-1, appears to constitutively interact with LILRE chromatin (Fig. 4, IRF-8 antibody). In contrast, strong IRF4 binding could not be detected (Fig. 4, IRF-4). Previous work by others has shown that tyrosine phosphorylation of IRF8 inhibits direct DNA binding, while being required for heterodimer association with IRF1/2 (Sharf et al., 1997). Also, substitution of tyrosine 211 within the IRF association domain (IAD) leads to a defective molecule incapable of interaction with either IRF1/2 or Spi-1 (Levi et al., 2002). To determine whether chromatin-bound IRF8 might be tyrosine phosphorylated in vivo, two successive immunoprecipitations were executed, in which a general anti-phosphotyrosine antibody (P-Tyr) was used to pull down all tyrosine phosphorylated proteins followed by a second immunoprecipitation of the eluate with an anti-IRF8 antibody. PCR of the immunoprecipitated DNA suggested the possibility that IRF8 bound to the LILRE might be tyrosine phosphorylated, but only after LPS stimulation (Fig. 4, PTyr:IRF-8, 2nd IP).
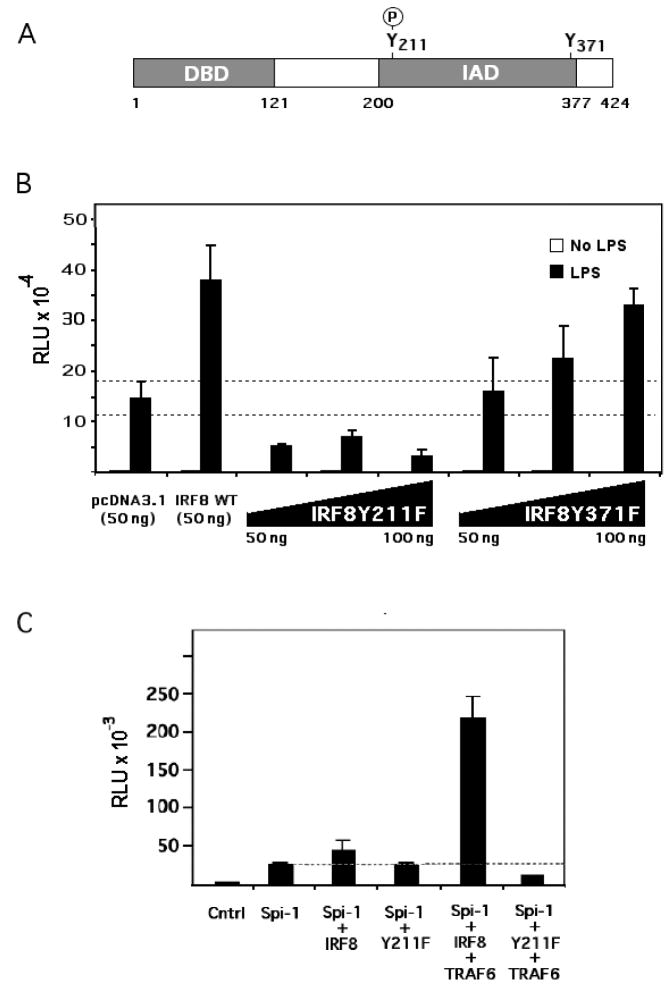
3.7. Tyrosine 211 of IRF8 is important for IL1B gene transcription
In order to determine which tyrosine residue(s) was phosphorylated and its relevance to IL1B induction, two tyrosine to phenylalanine IRF8 substituted expression vectors, Y211F and Y371F, were constructed (Fig. 5A). The tyrosine residue at 211 was shown to be conserved in many IRFs (Meraro et al., 1999), and the deletion of 38 amino acids including the tyrosine at 211 within the IRF association domain (IAD) of IRF8 resulted in inhibition of heterocomplex formation with Spi-1(Meraro et al., 1999). Tyrosine 371 is a conserved residue that has not been previously reported to be critical for activity. Mutant Y211F, Y371F and wild type IRF8 expression vectors were co-transfected along with the XT Human IL1B reporter into RAW 264.7 cells and stimulated with LPS. Fig. 5B shows that the XT reporter is activated by co-transfected IRF8 WT. However, co-transfection of the IRF8Y211F vector significantly decreased XT reporter activity, whereas co-transfection with IRF8Y371F did not significantly different from IRF8 WT. These data suggest that IRF8 tyrosine phosphorylation at Y211 may be an important component of LILRE function.
Fig. 5.
Tyrosine 211 of IRF8 is important for IL1B XT reporter activity. (A) Schematic diagram of IRF8 structure indicating the position of the tyrosine phosphorylated residue at 211. The DNA binding domain is abbreviated as DBD, while the interaction domain is abbreviated as IAD. (B) Activity of IL1B XT reporter after co-transfection of RAW 264.7 cells with either a wild type IRF8 (WT) or mutated Y211F and Y371F IRF8 expression vectors. Transfection used 50 ng of empty (pcDNA3.1) and wild type expression vectors, whereas mutated IRF8 cDNA expression vectors ranged in dose from 50 to 100 ng, as indicated. Cells were stimulated with 10 μg/ml of LPS for 4 hrs. (C) Activity of IL1B XT reporter transfected into HEK293R cells and co-transfected with the indicated expression vectors. Data indicate similar results from at least two independent experiments.
3.8. Phosphorylation of IRF8 is mediated via TRAF6 activation
The HEK293 cell line does not express either Spi-1 or IRF8, and has been previously reported to be capable of supporting IL1B gene induction by ectopic expression of these factors (Marecki et al., 2001). However, the study did not report any signal-dependent induction. In order to better define the involvement of specific factors in LILRE-dependent induction of IL1B, the XT reporter was co-transfected into HEK293 cells with expression vectors coding for Spi-1 and IRF8 in either the presence or absence of a TRAF6 expression vector acting as a surrogate for LPS signaling (Wang et al., 2006). As shown in Fig. 5C, Spi-1 and IRF8 could induce XT reporter activity which was greatly increased by TRAF6. However, co-expression of the dominant negative IRF8 mutant, IRF8Y211F, could not support XT reporter activation. This further supports a role for Tyr 211 of IRF8 in mediating LPS induction of IL1B.
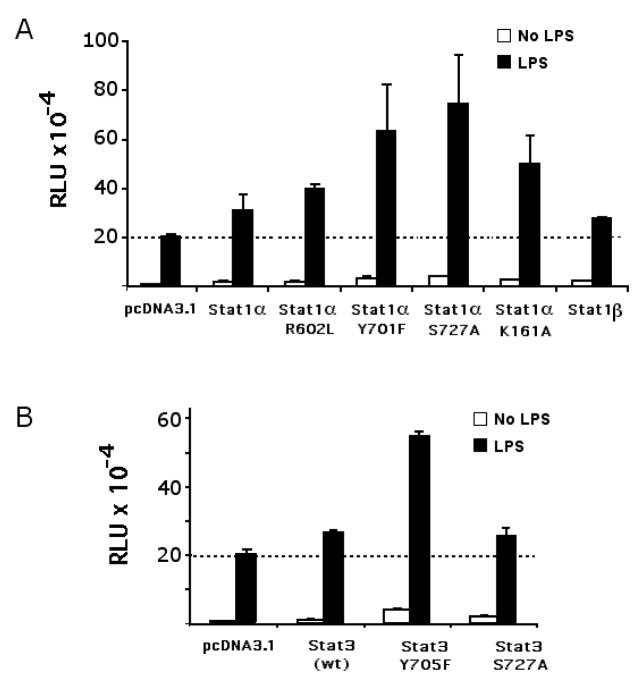
3.9. NTP-Stat1 increases IL1B induction
The ChIP and EMSA results both suggest that NTP-Stat1 may be recruited to the LILRE. Furthermore, mutation of the downstream STAT/GAS half-site in the LILRE significantly reduced the binding of the critical complex B (Fig. 3B, lane 17). Consequently, it appears as though NTP-Stat1 may be contributing to the binding of IRF8 and Spi-1 to the LILRE. Fig. 6A shows that co-expression of non-tyrosine phosphorylatable-Stat1Y701F enhance the activity of the XT reporter in an LPS-dependent manner, with respect to the co-expression of wild-type Stat1. These data are consistent with the involvement of NTP-Stat1 in LILRE function. Interestingly, serine 727, a phosphorylation site within the TAD important for maximal transactivation, and the presence of the TAD region, which is absent in Stat1β, may not be required for IL1B gene induction (Fig. 6A). It has been reported that the integrity of lysine 161 in Stat1 is required for interaction with IRF9 in the IFNα/β-activated ISGF3 complex (Horvath et al., 1996). However, co-transfection of XT reporter with an expression vector coding for Stat1αK161A resulted in increased activity, arguing that lysine 161 is not required for IL1B induction (Fig. 6A). The integrity of arginine 602 within the SH2 domain of Stat1, required for P-Tyr recognition (Shuai et al., 1993), like that of tyrosine 701, the P-Tyr phosphorylation site, is not required for activity (Fig. 6A). A similar approach using Stat3 expression vectors (Fig. 6B) shows that this STAT is also capable of activating the IL1B gene, and indicates that an NTP-Stat3 (as represented by Stat3 Y705F) is capable of playing a similar role to that of NTP-Stat1 for IL1B gene induction. Furthermore, the Stat3 S727A is as potent as wild-type Stat3, again indicating that phosphorylation of the STAT TAD is not necessary for IL1B gene induction.
Fig. 6.
Activity of wild type and mutated STAT expression vectors on IL1B XT. RAW264.7 cells were co-transfected with 250 ng IL1B XT reporter and empty (pcDNA 3.1) or Stat1α WT and mutant, or Stat3 WT and mutant cDNA expression vectors (50 ng), as indicated. Cells were stimulated with 10 ng/ml of LPS for 4 hrs. Data indicate similar results from at least two independent experiments.
4. Discussion
The data presented here suggest that Spi-1, IRF8 and an NTP-Stat1 bind cooperatively and constitutively to the LILRE site in the upstream induction sequence of the human IL1B gene to form the complex B observed by EMSA (Fig. 3). Although at least two complexes are observed to bind to this site in vitro, the ability of complex B to form correlates with the majority of LPS-dependent activity. This argues that a complex containing, in-part, a Spi-1•IRF association similar to that observed for the IgλL and 3′IgκL light-chain enhancers (Brass et al., 1996; Brass et al., 1999; Escalante et al., 2002; Perkel and Atchison, 1998) is important for activity. A Spi-1•IRF8 complex was previously reported to bind to the LILRE in unstimulated cells, but relevance to LPS gene induction was not addressed (Marecki et al., 2001). However, our observation of the presence of NTP-Stat1 binding to the LILRE is novel. In addition, the likely LPS-dependent tyrosine phosphorylation of IRF8, which we now report, suggests a possible activation mechanism important for the induction of transcription. It has been reported that such phosphorylation facilitates protein-protein interaction between Spi-1 and IRF8 (Levi et al., 2002).
Considerable effort has been focused on the mechanism for IFNγ-mediated JAK-STAT signaling, leading to Y701-phosphorylated Stat1 dimers binding directly to gamma-activated sequence (GAS) sites in specific genes (Levy and Darnell, 2002; Schindler et al., 1992). Both GAS half-sites are important for binding “classical” tyrosine phosphorylated-Stat1 dimers (Pearse et al., 1993). STAT homotypic dimerization takes place through reciprocal phosphotyrosine-Src-homology domain (SH2) interactions (Lau and Horvath, 2002; Shuai et al., 1994). However, unphosphorylated Stat1 and Stat3 are associated in unstimulated cells (Braunstein et al., 2003; Stancato et al., 1996), suggesting interaction by a distinct mechanism prior to tyrosine phosphorylation. Also, the nature of the ISGF3 complex, consisting of Stat1 (α or β isoforms), Stat2 and IRF9, induced after IFNα stimulation, has been known for some time (Stark et al., 1998). For ISGF3, Stat1 and 2 interact with IRF9 via the carboxy-terminal IRF9 interaction domain (IAD), which is partially conserved in IRF4, 8, and 9.
In the present study, the ability of NTP-Stat1 to form a cooperative in vitro complex with Spi-1 and IRF8 on the LILRE (Fig. 3C), argue for Spi-1 to be bound to the upstream STAT half-site and either an NTP-Stat1 monomer or an NTP-Stat1 homodimer to the downstream half-site with IRF8 bound next to the Spi-1. This cooperativity is similar to that reported for the low molecular mass polypeptide 2 (LMP2) gene, in which NTP-Stat1 binds to a STAT half-site only in cooperation with IRF1 binding to an adjacent upstream ISRE sequence (Chatterjee-Kishore et al., 2000b). Two interconverting structures have been reported for an NTP-Stat1 dimer not bound to DNA in which unlike P-Tyr-Stat1, the dimerization is through an N-terminal interaction (Mao et al., 2005; Zhong et al., 2005). However, it is not known if this structure can bind DNA or if NTP-STATs bind DNA as monomers with the help of other transcription factors such as Spi-1 and IRFs. We have attempted to model both the P-Tyr-Stat1 and NTP-Stat1 dimers on Spi-1-bound LILRE. However, in the case of P-Tyr-Stat1, steric clashing is excessive. Furthermore, the conformation of the reported NTP-Stat1 dimers appears to require some structural movement in order to accommodate the binding of DNA. Consequently, it is most likely that NTP-Stat1 cooperatively bound to the LILRE with Spi-1 and IRF8 is a monomer, as shown in Fig. 3F.
The first report of Stat1-induced transcription in the absence of tyrosine phosphorylation came from an analysis of constitutive expression of caspase genes like ICE, Cpp32 and Ich-1 (Kumar et al., 1997). This report demonstrated that apoptosis of Stat1-null U3A (transformed epithelial) cells was inhibited due to lack of Stat1α, and that a Y701F mutant of Stat1α restored apoptosis equally as well as wild type Stat1α. This suggests that NTP-Stat1α is required for the expression of these genes. It has also been reported that NTP-Stat1 binds to the LMP2 gene, to support constitutive transcription (Chatterjee-Kishore et al., 2000b). This report argued that NTP-Stat1 could bind as a dimer to LMP2 in vitro, but could not confirm this for a complex involving IRF1. In a parallel article by the same group (Chatterjee-Kishore et al., 2000a), NTP-Stat1 was shown to associate with IRF1 both in vivo and in vitro. Our report of the involvement of NTP-Stat3 as a cofactor in IL8 gene induction describes a mechanism by which activated NF-κBp65 homodimer can functionally utilize NTP-Stat3 (Yoshida et al., 2004). More recently, NTP-Stat3 has been reported to activate numerous genes when overexpressed in wild-type Stat3-null cells (Yang et al., 2005). Similarly, NTP-Stat1 can activate numerous genes, including IL1B (Chatterjee-Kishore et al., 2000b; Ramana et al., 2002). Our results now indicate that Stat1α bound constitutively to LILRE chromatin is not tyrosine phosphorylated either before or after LPS treatment. From our EMSA data showing the cooperative binding of all three in vitro translated proteins (Fig. 3C), we infer that NTP-Stat1 may make protein-protein interactions with either IRF8 and/or Spi-1 as in the predicted structure of complex B (Fig 3F) in which we posit that a monomeric NTP-Stat1 is involved.
Either IL-1 or LPS can induce IL1B in appropriate cell types (Ghezzi and Dinarello, 1988; Schindler et al., 1990; Shirakawa et al., 1993). However, in most circumstances neither IFNγ nor IL-6 alone is capable, but either one can synergize with (Cochran and Finch-Arietta, 1992; Held et al., 1999) or inhibit (Chujor et al., 1996) induction by LPS. The inhibition is particularly interesting in light of recent studies which suggest that IFNγ can induce IL1B in Stat1-null mice (Gil et al., 2001) but not in wild-type mice, suggesting that activated P-Tyr-Stat1 is involved in repression of IL1B induction (Ramana et al., 2002). This is consistent with the binding of a P-Tyr-Stat1 dimer to the LILRE after IFNγ stimulation (Fig. 3B, lanes 28–30), although IL1B gene reporter activity is negligible (data not shown). Furthermore, overexpression of NTP-Stat1 in Stat1-null mice results in constitutive induction of IL1B (Chatterjee-Kishore et al., 2000b). This suggests both a target site for P-Tyr-Stat dimer binding to the gene that inhibits expression and an induction role for NTP-Stat1. This concept is also confirmed by our results, where the LILRE binds NTP-Stat1 (under unstimulated conditions) and is important for active transcription of the human IL1B gene after LPS stimulation, although the same site also binds the “classical” homodimeric P-Tyr-Stat1 (anti-Stat1P-Tyr reactive, as in Fig. 3B) without gene activation when stimulated with IFNγ.
We had previously identified a novel Stat1-like ‘LIL-Stat’ protein binding to LILRE (Tsukada et al., 1996) that appeared to be tyrosine phosphorylated. In addition, our earlier report examined the effect of mutation of the downstream GAS-site using a reporter construct containing only the 23 bp LILRE with a heterologous murine c-fos promoter (Tsukada et al., 1996), rather than the long XT construct used in the present studies. Following our report, an independent group confirmed our observations, arguing for an identical complex constitutively binding the LILRE in extracts derived from acute myeloid leukemia patients (Tuyt et al., 1998). Subsequently, this group then reported the activation of classical Stat1 and Stat3 tyrosine-phosphorylated dimers binding to the LILRE following IFN-γ and IL-6 treatment, rather than LPS (Lemmink et al., 2001). Neither of these two studies attempted to examine IL1B reporter activity, although when used individually, neither IFNγ or IL-6 induce IL1B (not shown). We do not observe an increase in phospho-tyrosine Stat1 binding to LILRE chromatin after LPS stimulation (Fig. 4). This result is consistent with the finding that LPS is not a rapid and potent activator of Stat1 tyrosine phosphorylation (Rhee et al., 2003). However, NTP-Stat1 binds to LILRE chromatin prior to stimulation. In addition, LPS nuclear extracts did not reveal any binding to an FcγRI gene EMSA probe (not shown). Consequently, it is possible that the anti-P-Tyr antibody reactivity that we previously reported for in vitro complexes with the LILRE, and once again observe (Fig. 3B, lanes 22–24), may result from IRF8 phosphorylation following LPS treatment.
It has been reported that the integrity of lysine 161 in Stat1 is required for interaction with IRF9 in the IFNα/β-activated ISGF3 complex (Horvath et al., 1996). However, co-transfection of XT reporter with an expression vector coding for Stat1α K161A resulted in increased activity, as compared to wild type, suggesting that lysine 161 is not required for IL1B induction (Fig. 6A). Similarly, neither arginine 602, required for Stat1 SH2 domain P-Tyr recognition (Shuai et al., 1993), nor that of tyrosine 701, the P-Tyr phosphorylation site, is required (Fig. 6A). Finally, neither serine 727, a phosphorylation site within the TAD important for maximal transactivation, nor the TAD itself, which is absent in Stat1β, appears to be essential (Fig. 6A). A similar approach using Stat3 expression vectors did not reveal any inhibition, and also suggests that neither tyrosine nor serine phosphorylation is essential (Fig. 6B). This is further evidence that both Stat1 and Stat3 do not require tyrosine phosphorylation for LPS activation of the IL1B gene. The absence of any dominant-negative function for mutated forms of STAT suggests the possibility that these STAT molecules may play a role in supporting factor binding, rather than providing transactivation potential. This would be consistent with both the involvement of an NTP-STAT and the cooperative effect observed for the in vitro binding studies shown in Fig. 3C.
A previous report by others showed that monocyte extracts revealed binding of IRF8 and Spi-1 to the LILRE (Marecki et al., 2001), and that co-expression of these proteins cooperated to support IL1B reporter activity in the absence of LPS. Our ChIP and EMSA results reveal the cooperative binding of IRF8 and Spi-1 to the LILRE. Spi-1 alone does not bind LILRE probe, but is bound avidly by the IgλL probe under the same binding conditions. This is an important contrast between the two composite sites, arguing that the LILRE binds Spi-1 only in the presence of IRF-8. This difference may be due to the presence of five adenines in the IgλL (see Fig. 2A), located where the ETS domain wing interacts with the DNA backbone (Escalante et al., 2002; Galson et al., 1993; Kominato et al., 1995). In addition, we show that NTP-Stat1 binds to the LILRE and cooperatively supports IRF8 and Spi-1 binding. The binding of all these factors is constitutive. However, the putative tyrosine phosphorylation of IRF8 due to LPS stimulation, may be the factor that contributes to the potent inducibility of the human IL1B gene through TLR4. It has also been reported that the complex between Spi-1 and IRF8 that binds to IgλL gene enhancer-like critical composite sites in the myeloid CYBB and NCF2 genes requires IRF8 tyrosine phosphorylation for transcription (Kautz et al., 2001).
Another report demonstrated that tyrosine 211 within the IRF8 IAD domain is critical for interaction with Spi-1 (Levi et al., 2002). The interaction between IRF8 and Spi-1 appears to depend upon both the DBD and IAD (Brass et al., 1999; Escalante et al., 2002; Meraro et al., 1999). The most critical region of the IAD required for the interaction is the C-terminal third (Meraro et al., 1999), which contains tyrosine 211, conserved in both human and mouse IRF8. We show that Y211F substitution in IRF8 results in decreased XT reporter activity when cooexpressed, i.e. it is a dominant-negative. Tyrosine phosphorylation of IRF8 after LPS stimulation may cooperate with the LPS-induced binding of C/EBPβ (Shirakawa et al., 1993) to overcome the potent inhibition of the interposed Region H silencer located between the LILRE and the very potent C/EBPβ heterodimer binding site located within Region I, in the UIS (Tsukada et al., 1994). The absence of strong Stat3 binding to the LILRE in the LPS-stimulated nuclear extracts suggests that this factor may either not be present in complexes binding to the LILRE, or that the epitope may be blocked in some way. However, Stat3 was weakly detected in the ChIP, suggesting that there may either be another Stat3 binding site in the vicinity of the LILRE or that Stat3 occupies a small percentage of the LILRE sites within the cell population.
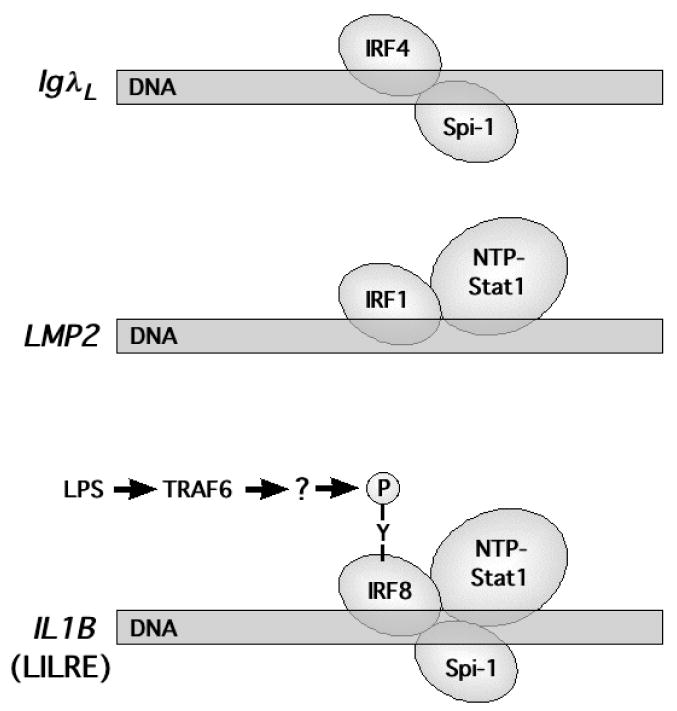
Fig. 7 schematically summarizes the hypothesized model for IRF8•Spi-1•NTP-Stat1 cooperative interactions with the IL1B LILRE and the proposed activation by LPS-dependent tyrosine phosphorylation of IRF8. Comparison is also made with the IRF binding sites found in the LMP2 and IgλL genes. We believe that several distinctions exist between the IRF8 complex in IL1B and those on the other two genes. First, the IL1B LILRE site is responsive to rapid signal-dependent induction, whereas the other two are constitutive. Also, the binding of Spi-1 to the LILRE is much weaker than to the IgλL site, resulting in dependence upon NTP-Stat1. Finally, it is unclear why the LMP2 gene, as reported (Chatterjee-Kishore et al., 2000b), appears to be constitutively expressed, whereas IL1B is inducible. Perhaps this is due to either the phosphorylation state not being appreciated or to the distinct sequence of the LMP2 site, which spaces the STAT binding sites one base-pair farther away from IRF and has a higher dependence upon the upstream, rather than the downstream, STAT half-site. It could also be due to the nature of the cell type in which IRF1 provides a distinct function in the absence of both IRF8 and Spi-1.
Fig. 7.
Comparison of composite IRF sites. Schematic representation of IRF composite sites referred to in the text. DNA (rectangles) are labeled according to the gene name (italics) and bound proteins are represented as ellipsoids. Also indicated is the LPS-dependent tyrosine phosphorylation of IRF8 for the IL1B gene.
Our observation that LPS induction of the human IL1B gene utilizes a chromatin pre-associated complex of Spi-1, IRF8, and NTP Stat1 that may rely upon tyrosine phosphorylation of one component (IRF8), may be a critical feature of a rapidly induced pro-inflammatory gene that must be “poised” or pre-programmed for the immediate transcription observed (Fenton et al., 1987; Fenton et al., 1988; Huang et al., 2001) in the face of a stress signal like bacterial infection. Very recently, such a poised architecture of the gene has been proposed by another group (Liang et al., 2006), which reported that the enhancer of the human IL1B gene is accessible to micrococcal nuclease even in the absence of transcription. This argues that the chromatin is in an open conformation. This result would be expected if transcription factors are pre-associated at the enhancer. Likewise, the pre-association of Stat1 with chromatin would also make this inflammatory gene more amenable for immediate transcription in the face of bacterial infection, as there would not be a requirement to phosphorylate or import Stat1 into the nucleus from the cytoplasm. It is possible that other pro-inflammatory and immediate-early genes are primed or poised in a similar way.
Acknowledgments
The authors wish to acknowledge Dr. Joseph C. Glorioso III in providing facilities and support and Dr. Scott E. Plevy for providing IRF8 cDNA. This work was supported by National Institutes of Health Grants CA06668544 and AI044122 and an award from the State of Pennsylvania Tobacco Settlement Fund to PEA and the University of Pittsburgh Start-up Fund to DLG.
Abbreviations
- IL1B
human gene coding for interleukin 1β
- C/EBP
CAAT enhancer binding protein
- ChIP
chromatin immunoprecipitation
- DBD
DNA binding domain
- EMSA
electrophoretic mobility assay
- ETS
erythroblastosis transformation specific
- GAS
gamma interferon activation sequence
- IAD
IRF association domain
- IFNGR
interferon gamma receptor
- IRF
interferon regulatory factor
- ISGF3
interferon stimulated gene factor 3
- ISRE
interferon stimulated response element
- LILRE
LPS and IL-1 response element
- LIL-Stat
LPS and IL-1 Stat
- LPS
lipopolysaccharide
- NF-IL6
nuclear factor for interleukin 6
- NTP-Stat1
non-tyrosine phosphorylated-Stat1
- P-Tyr
phosphotyrosine
- SH2
src homology 2
- STAT
signal transducer and activator of transcription (general designation for family of factors)
- Stat
a specific STAT family member as designated by the accompanying numeral
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nuc Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- Chandra G, Cogswell JP, Miller LR, Godlevski MM, Stinnett SW, Noel SL, Kadwell SH, Kost TA, Gray JG. Cyclic AMP signaling pathways are important in IL-1 beta transcriptional regulation. J Immunol. 1995;155:4535–4543. [PubMed] [Google Scholar]
- Chatterjee-Kishore M, van Den Akker F, Stark GR. Adenovirus E1A down-regulates LMP2 transcription by interfering with the binding of stat1 to IRF1. J Biol Chem. 2000a;275:20406–20411. doi: 10.1074/jbc.M001861200. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR, van Den Akker F. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000b;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JJ, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- Chujor CS, Klein L, Lam C. Selective inhibition of interleukin-1 beta gene expression in activated RAW 264.7 macrophages by interferon-gamma. Eur J Immunol. 1996;26:1253–1259. doi: 10.1002/eji.1830260611. [DOI] [PubMed] [Google Scholar]
- Clark BD, Collins KL, Gandy MS, Webb AC, Auron PE. Genomic sequence for human prointerleukin 1 beta: possible evolution from a reverse transcribed prointerleukin 1 alpha gene. Nuc Acids Res. 1986;14:7897–7914. doi: 10.1093/nar/14.20.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran FR, Finch-Arietta MB. Interleukin-6 can prime THP-1 macrophages for enhanced production of tumor necrosis factor-alpha in response to LPS. Immunopharmacology. 1992;23:97–103. doi: 10.1016/0162-3109(92)90033-9. [DOI] [PubMed] [Google Scholar]
- Escalante CR, Brass AL, Pongubala JM, Shatova E, Shen L, Singh H, Aggarwal AK. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol Cell. 2002;10:1097–1105. doi: 10.1016/s1097-2765(02)00703-7. [DOI] [PubMed] [Google Scholar]
- Fenton MJ, Clark BD, Collins KL, Webb AC, Rich A, Auron PE. Transcriptional regulation of the human prointerleukin 1β gene. J Immunol. 1987;138:3972–3979. [PubMed] [Google Scholar]
- Fenton MJ, Vermeulen MW, Clark BD, Webb AC, Auron PE. Human pro-IL-1β gene expression in monocytic cells is regulated by two distinct pathways. J Immunol. 1988;140:2267–2273. [PubMed] [Google Scholar]
- Galson DL, Hensold JO, Bishop TR, Schalling M, D’Andrea AD, Jones C, Auron PE, Housman DE. Mouse β-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol. 1993;13:2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galson DL, Housman DE. Detection of two tissue-specific DNA binding proteins with affinity for sites in the mouse β-globin intervening sequence 2. Mol Cell Biol. 1988;8:381–392. doi: 10.1128/mcb.8.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi P, Dinarello CA. IL-1 induces IL-1. III Specific inhibition of IL-1 production by IFN-gamma. J Immunol. 1988;140:4238–4244. [PubMed] [Google Scholar]
- Gil MP, Bohn E, O’Guin AK, Ramana CV, Levine B, Stark GR, Virgin HW, Schreiber RD. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JG, Chandra G, Clay WC, Stinnett SW, Haneline SA, Lorenz JJ, Patel IR, Wisely GB, Furdon PJ, Taylor JD, Kost TA. A CRE/ATF-like site in the upstream regulatory sequence of the human interleukin 1β gene is necessary for induction in U937 and THP-1 monocytic cell lines. Mol Cell Biol. 1993;13:6678–6689. doi: 10.1128/mcb.13.11.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held TK, Weihua X, Yuan L, Kalvakolanu DV, Cross AS. Gamma interferon augments macrophage activation by lipopolysaccharide by two distinct mechanisms, at the signal transduction level and via an autocrine mechanism involving tumor necrosis factor alpha and interleukin-1. Infect Immun. 1999;67:206–212. doi: 10.1128/iai.67.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CM, Stark GR, Kerr IM, Darnell JE., Jr Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, Lander ES, Hacohen N. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Levi BZ, Tamura T, Ozato K. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25:770–779. doi: 10.1089/jir.2005.25.770. [DOI] [PubMed] [Google Scholar]
- Kautz B, Kakar R, David E, Eklund EA. SHP1 protein-tyrosine phosphatase inhibits gp91PHOX and p67PHOX expression by inhibiting interaction of PU.1, IRF1, interferon consensus sequence-binding protein, and CREB-binding protein with homologous Cis elements in the CYBB and NCF2 genes. J Biol Chem. 2001;276:37868–37878. doi: 10.1074/jbc.M103381200. [DOI] [PubMed] [Google Scholar]
- Kominato Y, Galson DL, Waterman WR, Webb AC, Auron PE. Monocyte expression of the human prointerleukin 1 β gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1. Mol Cell Biol. 1995;15:58–68. [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Commane M, Flickinger TW, Horvath CM, Stark GR. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- Laricchia-Robbio L, Tamura T, Karpova T, Sprague BL, McNally JG, Ozato K. Partner-regulated interaction of IFN regulatory factor 8 with chromatin visualized in live macrophages. Proc Natl Acad Sci USA. 2005;102:14368–14373. doi: 10.1073/pnas.0504014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JF, Horvath CM. Mechanisms of Type I interferon cell signaling and STAT-mediated transcriptional responses. Mt Sinai J Med. 2002;69:156–168. [PubMed] [Google Scholar]
- Lemmink HH, Tuyt L, Knol G, Krikke E, Vellenga E. Identification of LIL-STAT in monocytic leukemia cells and monocytes after stimulation with interleukin-6 or interferon gamma. Blood. 2001;98:3849–3852. doi: 10.1182/blood.v98.13.3849. [DOI] [PubMed] [Google Scholar]
- Levi BZ, Hashmueli S, Gleit-Kielmanowicz M, Azriel A, Meraro D. ICSBP/IRF-8 transactivation: a tale of protein-protein interaction. J Interferon Cytokine Res. 2002;22:153–160. doi: 10.1089/107999002753452764. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Liang MD, Zhang Y, McDevit D, Marecki S, Nikolajczyk BS. The interleukin-1beta gene is transcribed from a poised promoter architecture in monocytes. J Biol Chem. 2006;281:9227–9237. doi: 10.1074/jbc.M510700200. [DOI] [PubMed] [Google Scholar]
- Listman JA, Wara-aswapati N, Race JE, Blystone LW, Walker-Kopp N, Yang Z, Auron PE. Conserved ETS domain arginines mediate DNA binding, nuclear localization, and a novel mode of bZIP interaction. J Biol Chem. 2005;280:41421–41428. doi: 10.1074/jbc.M509143200. [DOI] [PubMed] [Google Scholar]
- Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE, Jr, Chen X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Marecki S, Atchison ML, Fenton MJ. Differential expression and distinct functions of IFN regulatory factor 4 and IFN consensus sequence binding protein in macrophages. J Immunol. 1999;163:2713–2722. [PubMed] [Google Scholar]
- Marecki S, Riendeau CJ, Liang MD, Fenton MJ. PU.1 and multiple IFN regulatory factor proteins synergize to mediate transcriptional activation of the human IL-1 beta gene. J Immunol. 2001;166:6829–6838. doi: 10.4049/jimmunol.166.11.6829. [DOI] [PubMed] [Google Scholar]
- Martin MU, Wesche H. Summary and comparison of the signaling mechanisms of the Toll/interleukin-1 receptor family. Biochim Biophys Acta. 2002;1592:265–280. doi: 10.1016/s0167-4889(02)00320-8. [DOI] [PubMed] [Google Scholar]
- Meraro D, Hashmueli S, Koren B, Azriel A, Oumard A, Kirchhoff S, Hauser H, Nagulapalli S, Atchison ML, Levi BZ. Protein-protein and DNA-protein interactions affect the activity of lymphoid-specific IFN regulatory factors. J Immunol. 1999;163:6468–6478. [PubMed] [Google Scholar]
- Paquette RL, Minosa MR, Verma MC, Nimer SD, Koeffler HP. An interferon-gamma activation sequence mediates the transcriptional regulation of the IgG Fc receptor type IC gene by interferon-gamma. Mol Immunol. 1995;32:841–851. doi: 10.1016/0161-5890(95)00056-k. [DOI] [PubMed] [Google Scholar]
- Pearse RN, Feinman R, Shuai K, Darnell JE, Ravetch JV. Interferon γ-induced transcription of the high-affinity Fc receptor for IgG requires assembly of a complex that includes the 91-kDa subunit of transcription factor ISGF3. Proc Natl Acad Sci USA. 1993;90:4314–4318. doi: 10.1073/pnas.90.9.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel JM, Atchison ML. A two-step mechanism for recruitment of Pip by PU.1. J Immunol. 1998;160:241–252. [PubMed] [Google Scholar]
- Ramana CV, Gil MP, Schreiber RD, Stark GR. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Jones BW, Toshchakov V, Vogel SN, Fenton MJ. Toll-like receptors 2 and 4 activate STAT1 serine phosphorylation by distinct mechanisms in macrophages. J Biol Chem. 2003;278:22506–22512. doi: 10.1074/jbc.M208633200. [DOI] [PubMed] [Google Scholar]
- Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- Schindler R, Ghezzi P, Dinarello CA. IL-1 induces IL-1: IV. IFN-γ suppresses IL-1 but not lipopolysaccharide-induced transcription of IL-1. J Immunol. 1990;144:2216–2222. [PubMed] [Google Scholar]
- Sharf R, Meraro D, Azriel A, Thornton AM, Ozato K, Petricoin EF, Larner AC, Schaper F, Hauser H, Levi BZ. Phosphorylation events modulate the ability of interferon consensus sequence binding protein to interact with interferon regulatory factors and to bind DNA. J Biol Chem. 1997;272:9785–9792. doi: 10.1074/jbc.272.15.9785. [DOI] [PubMed] [Google Scholar]
- Shirakawa F, Saito K, Bonagura CA, Galson DL, Fenton MJ, Webb AC, Auron PE. The human prointerleukin 1β gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol Cell Biol. 1993;13:1332–1344. doi: 10.1128/mcb.13.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K, Horvath CM, Huang LHT, Qureshi SA, Cowburn D, Darnell JE. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- Shuai K, Ziemiecki A, Wilks AF, Harpur AG, Sadowski HB, Gilman MZ, Darnell JE. Polypeptide signalling to the nucleus through tyrosine phosphorylation of Jak and Stat proteins. Nature. 1993;366:580–583. doi: 10.1038/366580a0. [DOI] [PubMed] [Google Scholar]
- Stancato LF, David M, Carter-Su C, Larner AC, Pratt WB. Preassociation of STAT1 with STAT2 and STAT3 in separate signalling complexes prior to cytokine stimulation. J Biol Chem. 1996;271:4134–4137. doi: 10.1074/jbc.271.8.4134. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Tsukada J, Saito K, Waterman WR, Webb AC, Auron PE. Transcription factors NF-IL6 and CREB recognize a common essential site in the human prointerleukin 1β gene. Mol Cell Biol. 1994;14:7285–7297. doi: 10.1128/mcb.14.11.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada J, Waterman WR, Koyama Y, Webb AC, Auron PE. A novel STAT-like factor mediates lipopolysaccaride, interleukin 1 (IL-1), and IL-6 signaling and recognizes a gamma interferon activation site-like element in the IL1B gene. Mol Cell Biol. 1996;16:2183–2194. doi: 10.1128/mcb.16.5.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuyt LML, Bregman K, Lummen C, Dokter WHA, Vellenga E. Differential binding activity of the transcription factor LIL-Stat in immature and differentiated normal and leukemic myeloid cells. Blood. 1998;92:1364–1373. [PubMed] [Google Scholar]
- Wang KZ, Wara-Aswapati N, Boch JA, Yoshida Y, Hu CD, Galson DL, Auron PE. TRAF6 activation of PI 3-kinase-dependent cytoskeletal changes is cooperative with Ras and is mediated by an interaction with cytoplasmic Src. J Cell Sci. 2006;119:1579–1591. doi: 10.1242/jcs.02889. [DOI] [PubMed] [Google Scholar]
- Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- Yang Z, Wara-aswapati N, Chen C, Auron PE. Spi1/PU.1 and NF-IL6 (C/EBPβ) synergistically activate the IL-1β promoter via a protein-protein interaction. J Biol Chem. 2000;275:21272–21277. doi: 10.1074/jbc.M000145200. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Kumar A, Koyama Y, Peng H, Arman A, Boch JA, Auron PE. Interleukin 1 activates STAT3/nuclear factor-kappaB cross-talk via a unique TRAF6- and p65-dependent mechanism. J Biol Chem. 2004;279:1768–1776. doi: 10.1074/jbc.M311498200. [DOI] [PubMed] [Google Scholar]
- Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, Ren Z, Mao X, Chen X, Shuai K, Darnell JE., Jr Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci USA. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]