Abstract
In higher eukaryotes, introns are usually required for efficient pre-mRNA processing. However, some viruses have alternative approaches involving posttranscriptional regulatory elements (PREs) to enhance intronless heterologous gene expression through enabling stability and 3′ end formation, and to facilitate the nucleocytoplasmic export of unspliced mRNAs. In the current study, we compared the human cytomegalovirus (hCMV) immediate/early (IE) intronA, as well as virus-derived PREs—the PRE of Hepatitis B virus (HPRE) and Woodchuck Hepatitis virus (WPRE) on their ability to enhance antigen gene expression in vitro and immune responses induced by DNA vaccination in animal. Among all the constructs, the plasmids carrying the HPRE element showed the highest gene expression level in both in vivo and in vitro models. During immunization of mice with low doses (10 μg) of HIV-1 DNA vaccine, only −intronA/+HPRE and +intronA/+HPRE vaccine constructs induced anti-Gag antibodies, although the −intronA/+WPRE construct also elicited antigen-specific cellular immune responses. In addition, pInHGag (+intronA/+HPRE) at a 10 μg dose could induce higher anti-Gag antibody level than that induced by pGag (−intronA/−HPRE) or pInGag (+intronA/−HPRE) at 40 μg dose (p < 0.05). Our data are useful for the optimization of heterologous expression and immunogenicity of DNA vaccines.
Introduction
Vaccines have played a substantial role in controlling epidemic diseases since Jenner's smallpox vaccine in 1796. In 1990 Wolff and his colleagues found that injecting plasmid DNA into mouse muscle could directly induce protein expression within muscle cells (Wolff et al., 1990). Further, they reported gene expression lasting for a year after intramuscular injection of plasmid DNA (Wolff et al., 1992). Subsequently, the efficacies of a number of other plasmid DNA vaccines to induce humoral and cell-mediated immune responses against influenza A virus, human immunodeficiency virus (HIV), hepatitis B virus, and malignant tumor have been demonstrated. The discovery of DNA immunization in 1990s fundamentally changed our view of the nature of a vaccine: a genetic material that encodes for antigens, rather than the actual antigens themselves, can serve as an effective vaccine component (Lu et al., 2008).
Antigen-encoding naked DNA is relatively safe, stable, and easy to produce, and can mediate sustained antigen expression in vivo (Donnelly et al., 1997; Gurunathan et al., 2000). In addition, because DNA vaccines do not elicit antibodies against dsDNA itself in the host, they can be administered repeatedly. However, the development of DNA vaccine candidates has been limited due to their relatively modest immunogenicity. Therefore, great effort has been made to enhance the immune response through optimization of promoter/enhancer elements or the coding sequence according to the codon-usage bias of the host to achieve higher gene expression level (Xu et al., 2001; Garmory et al., 2003). Other approaches to improve the immunogenicity of DNA vaccine include the application of high-efficiency gene delivery systems such as gene gun or in vivo electroporation device (Fynan et al., 1993; Fuller et al., 2006; Luckay et al., 2007; Sharpe et al., 2007; Tsang et al., 2007), the use of cytokine genetic adjuvants such as GM-CSF, IL-2, and IL-12 (Lori et al., 2006), the employment of innate immunomodulatory signals (Li et al., 2007), and the modification of the properties of antigen-presenting cells (Tsen et al., 2007). Because the antigen expression level would influence activation, strength, and the duration of the specific immune response, it is generally believed that optimization of antigen expression is one of the most promising approaches proposed to enhance the immunogenicity of DNA vaccine candidates.
Posttranscriptional regulatory elements (PREs) of Hepatitis B virus (HPRE) and Woodchuck Hepatitis virus (WPRE) are both hepadnaviral cis-acting RNA elements that can increase the accumulation of cytoplasmic mRNA of an intronless gene by promoting mRNA exportation from the nucleus to the cytoplasm, enhancing 3′ end processing and stability. Usually, unspliced mRNAs are exported into the cytoplasm with low efficiency. However, PRE elements function independently of virally encoded proteins, and it is believed that some cellular factors may interact with the PRE element and mediate its posttranscriptional transport (Donello et al., 1998). HPRE can functionally substitute for the HIV type 1 (HIV-1) Rev-Rev-responsive element (RRE) complex in a transient transfection reporter assay (Huang and Liang, 1993; Huang and Yen, 1995). WPRE, as has been demonstrated by others, has been successfully used for efficient expression of intronless genes from plasmid (Garg et al., 2004; Callendret et al., 2007).
In the current study, we constructed a series of DNA expression vectors carrying either the HPRE or the WPRE, with and without human cytomegalovirus (hCMV) immediate/early (IE) intronA. Wild-type HIV-1 CN54 Gag coding sequence, a prevailing strain in many areas of China, was used as a model antigen to compare both the gene expression–enhancing and immunogenicity-improving effects of different PREs with or without hCMV IE intronA on the DNA vaccine constructs. We demonstrated that HPRE significantly improved Gag gene expression in 293T cell. The highest level of gene expression was observed when both hCMV intronA and HPRE were present in the same plasmid. Further, this vaccine construct elicited higher cellular and humoral responses with a ¼ dosage in comparison with the Gag DNA vaccine construct carrying neither of the PRE elements.
Materials and Methods
Plasmid construction
pVR1012 Is an optimized mammalian expression vector containing intronA sequence of CMV promoter and BGH polyA signal,which was kindly provided by Dr. Gary Nabel from the Vaccine Research Center, NIAID, NIH (Bethesda, MD). A plasmid vector pCMV containing CMV promoter and BGH polyA signal was constructed from pVR1012 through deleting the CMV IE intron. The HPRE and WPRE elements were synthesized by overlapping PCR according the nucleotides 2641–3214 of GenBank accession no. X02763 and nucleotides 1093–1684 of GenBank accession no. J04514, respectively. The HPRE and WPRE PCR fragments were digested with XbaI and BglII and ligated into the XbaI-BglII–digested pVR1012 backbone or pCMV backbone to generate the vectors pInH, pInW, pH, and pW. The Gag gene of HIV-1 CN54 (97CN001 GenBank accession no. AF286226), a prevailing strain in many areas of China, was subcloned into the SalI-EcoRV sites of the vectors pCMV, pH, pW, pVR1012, pInH, and pInW, producing pGag, pHGag, pWGag, pInGag, pInHGag, and pInWGag, respectively. DNA used for transfection and vaccination was prepared using the EndoFree Plasmid Giga Kit (Qiagen, Hilden, Germany). The final endotoxin level was less than 0.1 EU/μg DNA (0.1 endotoxin units/μg plasmid DNA). The supercoiled plasmid was no less than 80% as detected by agarose gel electrophoresis.
Cell culture and plasmid transfection
293T Cells (ATCC, CRL-11268) were cultured in DMEM containing 10% fetal bovine serum and plated in six-well tissue culture plates at a density of 5 × 105 cells/well the day before transfection. pCMV, pGag, pHGag, pWGag, pInGag, pInHGag, and pInWGag were transfected with lipofectamine 2000 (Invitrogen, Carlsbad, CA) at a ratio of 5 μg lipofectamine 2000 to 2 μg plasmid DNA. The cell was harvested 48 h after transfection. Gag mRNA and protein expression were measured by real-time RT-PCR and Western blot, respectively.
Real-time RT-PCR
Total RNA from transfected cells was extracted using RNeasy Kit (Qiagen), amplified in duplicates using One Step SYBR PrimeScript™ RT-PCR Kit (Takara, Dalian, China), and detected using Applied Biosystems 7500 real-time PCR System (ABI, Foster city, CA). Gag RNA was amplified by RT-PCR using the primers GagF (5′-AGACAAGATAGAGGAAGAACAAAAC-3′) and GagR (5′-ATGTCTCCTACTGGAACAGGTGGGT-3′). Serving as an internal standard, β-actin RNA was amplified with primers actinF (5′-CCAGCCATGTACGTTGCTATC-3′) and actinR (5′-CAGGTCCAGACGCAGGATGGC-3′) for each sample.
Western blot assay for Gag expression
Forty-eight hours after transfection, cell lysates were denatured and subjected to denaturing SDS-PAGE and then blotted onto PVDF membrane (Millipore, Bedford, MA). Blocking was done with 5% defatted milk powder/PBS containing 0.05% Tween (PBST) for 2 h. HIV human serum and rabbit anti-β-actin polyconal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) were used as the detecting antibodies at 1:500 dilution incubated for 1 h. Subsequently, the membranes were washed with PBST and then incubated with HRP-labeled anti-human IgG (1:2000) and HRP-labeled anti-rabbit IgG (1:2000), respectively. After final wash, chemiluminescence reagent was applied to the membranes. Then, the Anti-Gag Western blot membranes were scanned and quantified using the Gel/Chem doc program Quantityone (Bio-Rad, Milan, Italy). Protein expression levels were obtained from chem images using the Quantity One software (v.4.5.1; Bio-Rad).
DNA immunization
Six- to eight-week-old female Balb/C mice were purchased from the Institute of Laboratory Animal Science, the Chinese Academy of Medical Sciences & Peking Union Medical College. Animals were used in compliance with institutional animal health and care regulations, and all procedures used in the experiments with animals were approved by the local Institutional Animal Care and Use Committee. Groups of eight Balb/C mice each were injected intramuscularly with 100 μL plasmid DNA (50 μL in each tibialis anterior muscle) in PBS. Mice were injected with 40 μg or 10 μg pGag, pHGag, pWGag, pInGag, pInHGag, and pInWGag or 40 μg pCMV (as negative control) plasmid DNA at weeks 0, 3, and 6.
ELISPOT assay
The ELISPOT assay described by BDTM ELISPOT Mouse IFN-γ ELISPOT Set (BD, San Diego, CA) protocol was modified to detect HIV-1 Gag-specific T-cell responses. Three weeks after the last immunization, mouse splenocytes were harvested. Ninety-six-well plates were coated at 4°C overnight with 10 μg/mL of anti-mouse IFN-γ in sterile PBS. The plates were washed four times with 200 μL/well PBST and blocked with RPMI 1640 containing 10% FBS at room temperature for 2 h. Mouse splenocytes were isolated, and RBCs were lysed according to the protocol described previously (Li et al., 2007). Splenocytes were then seeded into wells with 50 μL/well (5 × 105 cells/well) in addition to 50 μL Gag overlapping peptides (acid sequence: AAMQILKDTINEEAA) and incubated for 30 h at 37°C in 5% CO2. After incubation, the ELISPOT plates were developed according to the kit instruction. Finally, plates were air-dried, and the spot-forming cells (SFC) were quantified by a Bioreader-4000 automated ELISPOT reader (BioSys, Karben, Germany) and normalized for 106 splenocytes.
Enzyme-linked immunosorbent assay
Ninety-six-well flat-bottom plates (Costar, Corning, NY) were coated with purified recombinant p55 protein (a recombinant protein of HIV-1 CN54 strain was expressed in E. coli and purified to 95% purity) at a concentration of 0.01 μg/mL in coating buffer (0.012 M Na2CO3 and 0.038 M NaHCO3, pH 9.6) at 4°C overnight. Plates were washed five times with PBST, and blocked with 3% BSA in PBST at 37°C for 1 h. Mouse sera were serially twofold diluted in block solution (starting at 1:100), and 100 μL was added to each well. After incubation at 37°C for 1 h, the plates were washed five times with PBST and then incubated with 1:5000 diluted HRP-labeled goat anti-mouse IgG antibody (Santa Cruz Biotechnology) at 37°C for 1 h. After the final wash, 100 μL of fresh TMB substrate (Sigma, St. Louis, MO) was added per well, and plates were incubated for 5 min. The reaction was stopped by adding 25 μL of 2M H2SO4, when the optical density of the plate measured at 450 nm by Multiscan enzyme-linked immunosorbent assay (ELISA) plate reader (Thermo Life Sciences, Hampshire, United Kingdom).
Statistical analysis
Values were expressed as means ± standard deviations (SD). Analysis of differences in means between groups was conducted by one-way analysis of variance (ANOVA); p < 0.05 was considered significant.
Results
Construction of HIV-1 Gag expression vectors and Gag expression in transfected cells
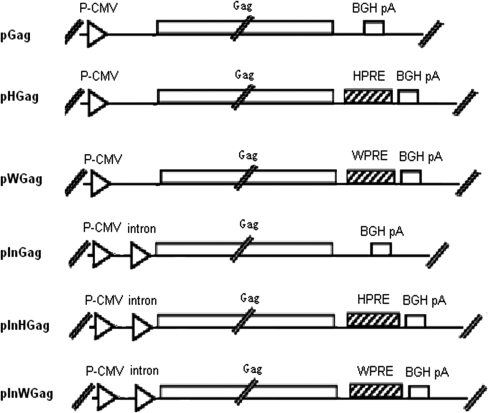
As depicted in Figure 1, a total of six Gag DNA vaccine plasmids were constructed with various combinations of CMV IE promoter/IntronA, PREs, and Gag gene of HIV-1 CN54. The RNA export elements of the HBV and Woodchuck Hepatitis virus, know as HPRE and WPRE (Donello et al., 1998; Heise et al., 2006), were inserted to upstream of the BGH poly A signal, respectively.
FIG. 1.
Schematic maps of Gag expression plasmids. P-CMV, human cytomegalovirus (hCMV) immediate/early (IE) promoter; intron, hCMV IE intronA; HPRE, hepatitis B virus posttranscriptional element; WPRE, Woodchuck hepatitis virus posttranscriptional element; BGH pA, bovine growth hormone polyadenylation signal.
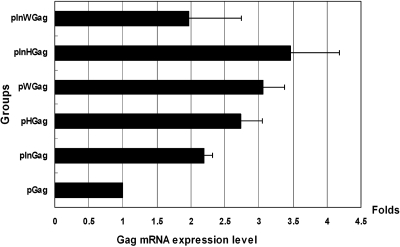
The Gag expression levels of these plasmids in transiently transfected 293T cells differed significantly as shown in Figure 2. Approximately 48 h after transfection, the cells were harvested, and total RNA was isolated. The amount of Gag transcripts in 293T cells transfected by the above DNA vaccine plasmids were determined by real-time RT-PCR. As internal standard, the amount of β-actin RNA was determined for each sample (Fig. 2).
FIG. 2.
Relative Gag RNA levels. Gag mRNA was analyzed by real-time RT-PCR using Gag-specific primers. Serving as internal standard, β-actin RNA level was determined by real-time RT-PCR using β-actin–specific primers. The Gag RNA level in pGag-transfected 293T cells was set to one, and the RNA levels in other plasmid-transfected cells are presented as relative values to that of 293T cells.
As shown by real-time RT-PCR (Fig. 2), the Gag RNA levels in 293T cells transfected by DNA plasmid with CMV-IE intronA and/or PREs were significantly higher than that observed in pGag-transfected cells. However, no significant differences were observed between these constructs carrying different PREs with or without intronA sequences. Therefore, our data confirmed previous findings that efficient expression of intronless genes from plasmid may require the presence of additional cis-acting regulatory modules within the expression cassettes, such as splice sites and hepadnaviral PREs (Callendret et al., 2007).
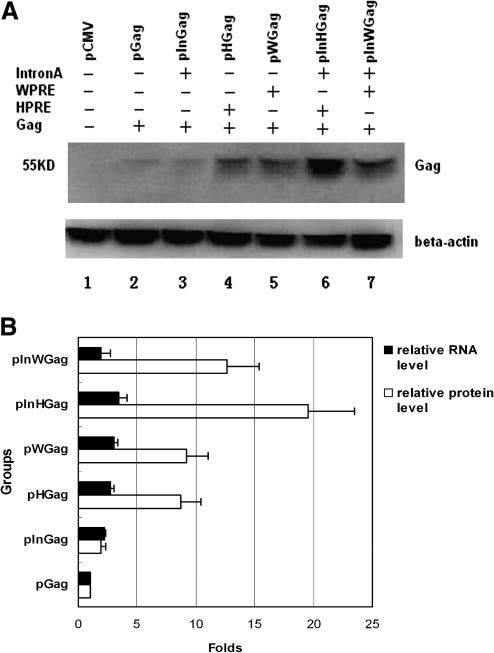
Subsequently, the cell extracts were subjected to SDS-PAGE and Western blot analysis. It is quite interesting that the Gag expression level of pInGag was very low regardless of the presence of CMV intronA sequence (Fig. 3A, lane 3). However, to a large extent, the Gag expression was significantly enhanced when the constructs included PREs (Fig. 3A, lanes 4 and 5). More excitingly, Gag expression was remarkably enhanced by +HPRE/+intronA DNA plasmid (Fig. 3A, lane 6). To analyze the effect of PREs on transgene expression in RNA processing steps, we compared the relative level of Gag RNA and protein (Fig. 3B). Note that more obvious expression enhancement effect of PRE could be observed in Gag protein accumulation rather than total RNA in the transfected cells. This result further confirmed that PREs are very efficient for intronless gene expression at the posttranscriptional level.
FIG. 3.
(A) Gag protein expression levels in plasmid-transfected 293T cells analyzed by Western blotting. β-Actin was used as an internal standard. The position of Gag protein is indicated on the side with molecular weight. (B) Relative quantitative presentation of Gag mRNA and protein expression levels in transiently transfected 293T cells. The expression levels of Gag mRNA or protein are normalized to the expression level of pGag, which is set as 1. The average of two experiments with SD is indicated.
Gag-specific humoral immune responses
pGag, pInGag, pHGag, pInHGag, pWGag, and pInWGag were intramuscularly immunized in Balb/C mice. Mice were injected three times with 3-week intervals. Each group was made up of eight mice, each received 10 μg or 40 μg DNA plasmid. Serving as negative control, a group of mice was injected with 40 μg of pCMV DNA.
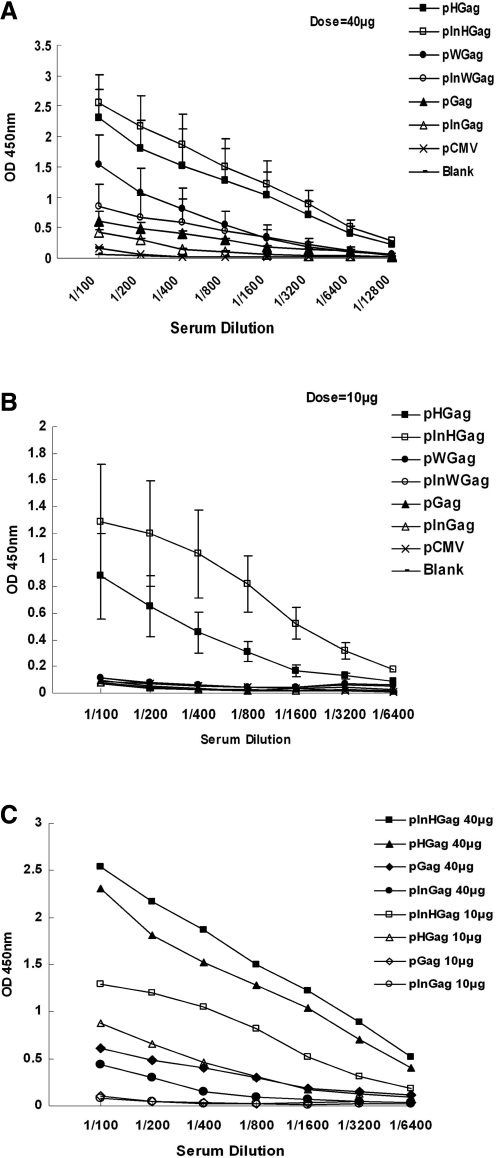
Three weeks after the final immunization, the sera were collected to detect HIV-1 Gag–specific IgG responses by ELISA assay.
At the 40 μg dose, each of the Gag expression plasmids induced anti-Gag IgG response. pInGag Raised weak anti-Gag response that was much lower than that induced by pWGag or pInWGag. pHGag And pInHGag elicited the highest antibody responses among all groups. At the dose of 10 μg of DNA, as demonstrated by Figure 4B, only pHGag and pInHGag induced satisfactory antibody responses. Parallel with in vitro gene expression data, both pGag and pInGag failed to induce any detectable anti-Gag antibodies. Somewhat to our surprise, the pWGag and pInWGag constructs could not induce anti-Gag IgG responses at the 10 μg dosage, although WPRE had enhanced the Gag gene expression in vitro.
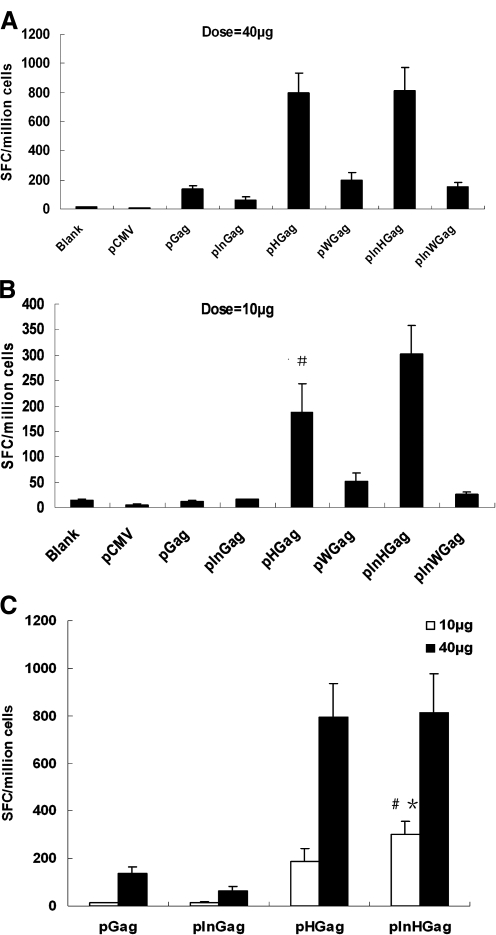
FIG. 4.
Anti-Gag antibody responses in mice immunized with Gag DNA vaccine plasmids with or without PRE or/and CMV IE IntronA elements. Balb/c mice were injected three times intramuscularly at 3-week intervals with 10 or 40 μg plasmid DNA as indicated, or with 40 μg of empty plasmid DNA pCMV. Serum samples from Balb/C mice (eight/group) were collected for ELISA 3 weeks after the final vaccination. (A) 40-μg dose. (B) 10-μg dose. The open symbols represent 10 μg dose, and the solid symbols represent 40 μg dose in Figure 4 (C). (We omit some error bars in Fig. 4 to avoid overlap.)
Our data also indicated that 10 μg dose of pInHGag induced much higher anti-Gag IgG response than 40 μg dose pInGag or pGag did (p < 0.05). Ten μg dose of pHGag elicited similar humoral response level compared with 40 μg dose of pGag (Fig. 4C).
Gag-specific cell-mediated immune responses
We next wanted to determine the effect that the DNA vaccine constructs, with PREs and/or CMV intronA, had on generation of Gag-specific T cell immune responses. Splenocytes from immunized mice were harvested 3 weeks after the third vaccination and stimulated with Gag peptide, and IFN-γ production was detected by ELISPOT analysis. As shown in Figure 5A, when mice were immunized with high-dose (40 μg) vaccines, all DNA vaccine constructs could elicit Gag-specific T cell responses. However, mice vaccinated with pHGag (+HPRE/−intronA) or pInHGag (+HPRE/+intronA) generated significantly higher Gag-specific T cell responses than mice vaccinated with other plasmid DNA without the HPRE element (p < 0.01).This result was consistent with that of the humoral immune responses.
FIG. 5.
Gag-specific IFN-γ T cells induced by DNA vaccination. Splenocytes were harvested and stimulated with Gag (No. 06042000010) peptide. The data are shown as means ± SD (n = 8 mice/group). (A) HPRE significantly improved Gag-specific T cell responses (p < 0.01). (B) #p < 0.05 compared with pWGag group. (C) #p < 0.05 compared with pGag group; *p < 0.01 compared with pInGag group.
From the data presented (Fig. 5B), it is clear that only pInHGag, pHGag, and pWGag could elicit Gag-specific cellular immune responses with 10 μg dose (p < 0.05). pInHGag DNA vaccine generated a significantly higher number of Gag-specific T cells (301 SFC/million splenocytes) than any other DNA vaccine without HPRE element did (p < 0.01). In addition, the number of Gag-specific T cells induced by DNA vaccines was 186 SFC/million splenocytes for pHGag and 51 SFC/million splenocytes for pWGag, indicating that pHGag elicited significantly higher (p < 0.05) cell-mediated immune responses than pWGag. It was clearly demonstrated that HPRE strongly enhances the immunogenicity of Gag DNA vaccine in mice, whereas WPRE has less effect.
At 10 μg low dosage, pInHGag induced a much higher Gag-specific cell immune response than 40 μg of pGag (p < 0.05) or pInGag (p < 0.01) did (Fig. 5C). Mice immunized with low-dose pHGag DNA plasmid generated similar levels of Gag-specific T cells compared with mice administrated with high-dose (40 μg/mice) pGag DNA plasmid (p > 0.05).
Discussion
Since the early 1990s, DNA vaccination has become an important approach against infectious diseases, malignant tumors, and allergy. Many efforts have been made to design and develop better therapeutic and prophylactic DNA vaccines. Most DNA vaccine optimization strategies have been focused on three areas: (1) to enhance in vivo delivery and transfection of DNA vaccine plasmids, (2) to develop both chemical and genetic adjuvants regulating the magnitude and balance of immune responses, and (3) to improve gene expression through optimizing the gene regulatory elements or coding sequences.
Among all these efforts, higher antigen expression strength is pursued mainly through optimization of the codon usage of the antigen gene and optimization of the gene expression regulatory elements at the transcription level. A few studies have demonstrated that some PREs such as the HPRE and the WPRE, when combined with appropriate promoter/enhancer elements, can also play an important role in the enhancement of transgene expression from retroviral vector through promoting the nuclear export of intronless transcripts (Zufferey et al., 1999; Gruh et al., 2008). Moreover, the WPRE has been shown to improve gene expression from nonviral vectors and enhance the immune efficacy induced by DNA vaccination (Pertl et al., 2003). As another better known gene expression enhancement approach, the involvement of RNA-splicing donor and receptor elements in the gene sequences has been considered to enhance gene expression both in vitro and in vivo through improving the efficiencies of several key steps in gene expression process, including the cytoplasmic transport of the spliced transcripts (Chapman et al., 1991; Norman et al., 1997; Le Hir et al., 2003).
However, some questions on the optimization of gene expression at the posttranscriptional level remain unanswered. First, the role of HPRE on gene expression and immunogenicity induced by DNA vaccine remains to be demonstrated. Second, the comparison of expression-enhancing effect of different PREs such as the WPRE, HPRE, and IntronA, especially in the context of a DNA vaccine, has not been reported. Third, whether PREs could substitute for CMV intronA or have a synergistic effect with it is unknown. Finally, previous studies have demonstrated that codon usage has become an important consideration in the design of gene-based HIV-1 vaccines (Wang et al., 2006). It is not clear, as compared with codon-optimized DNA vaccine, whether DNA vaccine carrying HIV gene with wild-type sequence could elicit comparable level of immune responses when the PRE and/or intron elements were introduced.
In the current study, we observed the enhancing effect of both the HPRE and WPRE, on antigen gene expression in transfected 293T cells, which is consistent with the results of some previous studies (Xu et al., 2003; Guang and Mertz, 2005; Klein et al., 2006; Mähönen et al., 2007). These PRE elements were able to functionally substitute for the HIV-1 Rev responsible element and mediate CRM1-dependent nuclear export pathway of intronless transcripts (Roth and Dobbelstein, 1997, Popa et al., 2002). However, the DNA vaccine constructs carrying HPRE showed significantly higher immunogenicity than those carrying WPRE. pInHGag Plasmid in general showed the strongest immunogenicity among all constructs. Immune responses induced by 10 μg pHGag were greater than those induced by 40 μg pGag (Figs. 4C and 5C). To our surprise, neither pWGag nor pInWGag plasmid elicited detectable level of anti-Gag antibody. Only weak cell-mediated immune responses could be detected in the pWGag group. This result is quite different from the previous reports (Garg et al., 2004; Callendret et al., 2007). The underlying mechanism that makes the difference remains to be revealed.
The hCMV intronA was demonstrated to be able to enhance expression of heterologous genes in vitro, especially when combined with the hCMV IE promoter/enhancer elements (Chapman et al., 1991). Other studies indicated that the effects of introns on expression were attributed primarily to their ability to alter the nucleocytoplasmic distribution of mRNAs by RNA splicing and enhancing the rate of RNA polyadenylation (Huang and Gorman, 1990; Lu and Cullen, 2003; Nott et al., 2003). In contrast, mRNAs of intronless genes could be efficiently processed if they contain cis-acting RNA elements such as the HPRE or WPRE. The current study showed that HPRE or WPRE had a greater effect on Gag gene expression in vitro than CMV intronA. Further, pInHGag carrying both the HPRE and IntronA elements induced the best immune responses among all groups, suggesting synergistic effect on gene expression.
It is known that the HIV type 1 (HIV-1) Gag and Env gene have inhibitory sequences that affect the accumulation and utilization of their mRNAs, but the inhibitory effect can be overcome by the RRE interaction (Hadzopoulou-Cladaras et al., 1989; Hammarskjöld et al., 1989; Schwartz et al., 1992). Codon-usage optimization of these genes can lead to Rev-independent expression and has been proved a very effective DNA vaccine optimization strategy. In our present study, we have found that HPRE, which has a similar regulatory pathway to HIV-1 RRE, enhanced the in vitro Gag expression and immunogenicity of Gag-based DNA vaccine in Balb/C mice. We also observed that 10 μg of pInHGag that carries both the intronA and HPRE induced robust anti-Gag IgG antibody response that was comparable to that induced by the same amount of codon-optimized Gag DNA vaccine (data not shown).
In summary, the current study has provided valuable information on the vector optimization strategies employing 5′intronA and 3′HPRE elements that could obviously improve HIV-1 gene expression and immunogenicity of a Gag-based DNA vaccine. Our data also demonstrated that DNA vaccine carrying an HIV gene with native codon usage could achieve impressive in vitro expression and in vivo immunogenicity in a Rev-independent manner if the elements were appropriately optimized at the posttranscriptional level. Further, our data highlight the application of this alternative gene optimization strategy in the high-throughput screening experiments involving native an HIV genes or the production of lentiviral vectors where the codon-optimized gene is not available.
Acknowledgments
The authors wish to thank Dr. Liliang Wang for his valuable suggestions on this study, and Yunfei Guo, Pei Chen, Hongyan Sun, and Ye Liu for their assistances in animal experiment. This work was supported by CIPRA Program (NIH Grant No. 1 U19 AI51915-02), Hi-tech Research and Development Program of China (“863 Program,” Grant No. 2003AA219100), and the National Key S&T Special Projects on Major Infectious Diseases (Grant No. 2008ZX10104).
Disclosure Statement
This article was authored by Jing Sun, Dingfeng Li, Yanling Hao, Yuwei Zhang, Wenling Fan, Jingjing Fu, Yunzhang Hu, Yong Liu, and Yiming Shao.
All authors have directly participated in the planning, execution, or analysis of this work. All authors have read and approved this version of the article.
The authors claim that no part of this article has been copyrighted or published previously and that none of the material in the article is under consideration for publication elsewhere. No conflict of interest exits in the submission of this article.
References
- Callendret B. Lorin V. Charneau P. Marianneau P. Contamin H. Betton J.M. van der Werf S. Escriou N. Heterologous viral RNA export elements improve expression of severe acute respiratory syndrome (SARS) coronavirus spike protein and protective efficacy of DNA vaccines against SARS. Virology. 2007;363:288–302. doi: 10.1016/j.virol.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman B.S. Thayer R.M. Vincent K.A. Haigwood N.L. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donello J.E. Loeb J.E. Hope T.J. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly J.J. Ulmer J.B. Shiver J.W. Liu M.A. DNA vaccines. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- Fuller D.H. Loudon P. Schmaljohn C. Preclinical and clinical progress of particle-mediated DNA vaccines for infectious diseases. Methods. 2006;40:86–97. doi: 10.1016/j.ymeth.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Fynan E.F. Webster R.G. Fuller D.H. Haynes J.R. Santoro J.C. Robinson H.L. DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc Natl Acad Sci USA. 1993;90:11478–11482. doi: 10.1073/pnas.90.24.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S. Oran A.E. Hon H. Jacob J. The hybrid cytomegalovirus enhancer/chicken beta-actin promoter along with woodchuck hepatitis virus posttranscriptional regulatory element enhances the protective efficacy of DNA vaccines. J Immunol. 2004;173:550–558. doi: 10.4049/jimmunol.173.1.550. [DOI] [PubMed] [Google Scholar]
- Garmory H.S. Brown K.A. Titball R.W. DNA vaccines: improving expression of antigens. Genet Vaccines Ther. 2003;1:1–5. doi: 10.1186/1479-0556-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruh I. Wunderlich S. Winkler M. Schwanke K. Heinke J. Blömer U. Ruhparwar A. Rohde B. Li R.K. Haverich A. Martin U. Human CMV immediate-early enhancer: a useful tool to enhance cell-type-specific expression from lentiviral vectors. J Gene Med. 2008;10:21–32. doi: 10.1002/jgm.1122. [DOI] [PubMed] [Google Scholar]
- Guang S. Mertz J.E. Pre-mRNA processing enhancer (PPE) elements from intronless genes play additional roles in mRNA biogenesis than do ones from intron-containing genes. Nucleic Acids Res. 2005;33:2215–2226. doi: 10.1093/nar/gki506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S. Klinman D.M. Seder R.A. DNA vaccines: immunology application, and optimization. Annu Rev Immunol. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- Hadzopoulou-Cladaras M. Felber B.K. Cladaras C. Athanassopoulos A. Tse A. Pavlakis G.N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the Env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjöld M.L. Heimer J. Hammarskjöld B. Sangwan I. Albert L. Rekosh D. Regulation of human immunodeficiency virus Env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise T. Sommer G. Reumann K. Meyer I. Will H. Schaal H. The hepatitis B virus PRE contains a splicing regulatory element. Nucleic Acids Res. 2006;34:353–363. doi: 10.1093/nar/gkj440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Liang T.J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.T. Gorman C.M. Intervening sequences increase efficiency of RNA 3' processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.M. Yen T.S. Role of the hepatitis B virus posttran-scriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. Ruttkowski B. Knapp E. Salmons B. Günzburg W.H. Hohenadl C. WPRE-mediated enhancement of gene expression is promoter and cell line specific. Gene. 2006;372:153–161. doi: 10.1016/j.gene.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Le Hir H. Nott A. Moore M.J. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Li D. Liu Y. Zhang Y. Xu J. Hong K. Sun M. Shao Y. Adjuvant effects of plasmid-generated hairpin RNA molecules on DNA vaccination. Vaccine. 2007;25:6992–7000. doi: 10.1016/j.vaccine.2007.06.046. [DOI] [PubMed] [Google Scholar]
- Lori F. Weiner D.B. Calarota S.A. Kelly L.M. Lisziewicz J. Cytokine-adjuvanted HIV-DNA vaccination strategies. Springer Semin Immunopathol. 2006;28:231–238. doi: 10.1007/s00281-006-0047-y. [DOI] [PubMed] [Google Scholar]
- Lu S. Cullen B.R. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. Wang S. Grimes-Serrano J.M. Current progress of DNA vaccine studies in humans. Expert Rev Vaccines. 2008;7:175–191. doi: 10.1586/14760584.7.2.175. [DOI] [PubMed] [Google Scholar]
- Luckay A. Sidhu M.K. Kjeken R. Megati S. Chong S.Y. Roopchand V. Garcia-Hand D. Abdullah R. Braun R. Montefiori D.C. Rosati M. Felber B.K. Pavlakis G.N. Mathiesen I. Israel Z.R. Eldridge J.H. Egan M.A. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in rhesus macaques. J Virol. 2007;81:5257–5269. doi: 10.1128/JVI.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen A.J. Airenne K.J. Purola S. Peltomaa E. Kaikkonen M.U. Riekkinen M.S. Heikura T. Kinnunen K. Roschier M.M. Wirth T. Ylä-Herttuala S. Posttranscriptional regulatory element boosts baculovirus-mediated gene expression in vertebrate cells. J Biotechnol. 2007;131:1–8. doi: 10.1016/j.jbiotec.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Norman J.A. Hobart P. Manthorpe M. Felgner P. Wheeler C. Development of improved vectors for DNA-based immunization and other gene therapy applications. Vaccine. 1997;15:801–803. doi: 10.1016/s0264-410x(96)00247-2. [DOI] [PubMed] [Google Scholar]
- Nott A. Meislin S.H. Moore M.J. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertl U. Wodrich H. Ruehlmann J.M. Gillies S.D. Lode H.N. Reisfeld R.A. Immunotherapy with a posttranscriptionally modified DNA vaccine induces complete protection against metastatic neuroblastoma. Blood. 2003;101:649–654. doi: 10.1182/blood-2002-02-0391. [DOI] [PubMed] [Google Scholar]
- Popa I. Harris M.E. Donello J.E. Hope T.J. CRM1-dependent function of a cis-acting RNA export element. Mol Cell Biol. 2002;22:2057–2067. doi: 10.1128/MCB.22.7.2057-2067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. Dobbelstein M. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IkappaB alpha. J Virol. 1997;71:8933–8939. doi: 10.1128/jvi.71.11.8933-8939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. Felber B.K. Pavlakis G.N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe M. Lynch D. Topham S. Major D. Wood J. Loudon P. Protection of mice from H5N1 influenza challenge by prophylactic DNA vaccination using particle mediated epidermal delivery. Vaccine. 2007;25:6392–6398. doi: 10.1016/j.vaccine.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Tsang C. Babiuk S. van Drunen Littel-van den Hurk S. Babiuk L.A. Griebel P. A single DNA immunization in combination with electroporation prolongs the primary immune response and maintains immune memory for six months. Vaccine. 2007;25:5485–5494. doi: 10.1016/j.vaccine.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Tsen S.W. Paik A.H. Hung C.F. Wu T.C. Enhancing DNA vaccine potency by modifying the properties of antigen-presenting cells. Expert Rev Vaccines. 2007;6:227–239. doi: 10.1586/14760584.6.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. Farfan-Arribas D.J. Shen S. Chou T.H. Hirsch A. He F. Lu S. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine. 2006;24:4531–4540. doi: 10.1016/j.vaccine.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Wolff J.A. Ludtke J.J. Acsadi G. Williams P. Jani A. Long-term persistence of plasmid DNA and foreign gene expression in mouse muscle. Hum Mol Genet. 1992;1:363–369. doi: 10.1093/hmg/1.6.363. [DOI] [PubMed] [Google Scholar]
- Wolff J.A. Malone R.W. Williams P. Chong W. Acsadi G. Jani A. Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- Xu Z.L. Mizuguchi H. Ishii-Watabe A. Uchida E. Mayumi T. Hayakawa T. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene. 2001;272:149–156. doi: 10.1016/s0378-1119(01)00550-9. [DOI] [PubMed] [Google Scholar]
- Xu Z.L. Mizuguchi H. Mayumi T. Hayakawa T. Woodchuck hepatitis virus posttranscriptional regulation element enhances transgene expression from adenovirus vectors. Biochim Biophys Acta. 2003;1621:266–271. doi: 10.1016/s0304-4165(03)00078-3. [DOI] [PubMed] [Google Scholar]
- Zufferey R. Donello J.E. Trono D. Hope T.J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]