Abstract
The bZIP transcription factor Nrf2 has emerged as a pivotal regulator of intracellular redox homeostasis by controlling the expression of many endogenous antioxidants and phase II detoxification enzymes. Upon oxidative stress, Nrf2 is induced at protein levels through redox-sensitive modifications on cysteine residues of Keap1, a component of the E3 ubiquitin ligase that targets Nrf2 for ubiquitin-dependent degradation. The mitogen activated protein kinases (MAPKs) have previously been proposed to regulate Nrf2 in response to oxidative stress. However, the exact role of MAPKs and the underlying molecular mechanism remain poorly defined. Here we report the first evidence that Nrf2 is phosphorylated in vivo by MAPKs. We have identified multiple serine/threonine residues as major targets of MAPK-mediated phosphorylation. Combined alanine substitution on those residues leads to a moderate decrease in the transcriptional activity of Nrf2, most likely due to a slight reduction in its nuclear accumulation. More importantly, Nrf2 protein stability, primarily controlled by Keap1, is not altered by Nrf2 phosphorylation in vivo. These data indicate that direct phosphorylation of Nrf2 by MAPKs has limited contribution in modulating Nrf2 activity. We suggest that MAPKs regulate the Nrf2 signaling pathway mainly through indirect mechanisms.
Introduction
Disruption of redox homeostasis is associated with the toxic effects of many environmental insults and pathogenesis of aging-related diseases such as cancer and neurodegenerative disorders [1], [2], [3], [4], [5]. Mammalian intracellular redox homeostasis is maintained mainly through transcriptional control of an array of antioxidative genes upon exposure to environmental insults that generate oxidative stress. As a key component of such a control system, the antioxidant response element (ARE) is a conservative cis-acting element found in the promoter regions of many genes encoding antioxidants and detoxification enzymes. The corresponding trans-acting factor for the ARE is the nuclear factor erythroid 2-related factor 2 (Nrf2), a protein belonging to the bZIP (basic-leucine zipper) transcription factor family [6], [7], [8].
The Nrf2-ARE system is responsible for both basal and inducible expression of many genes involved in antioxidant responses, such as NAD(P)H quinone oxidoreductase 1 (NQO1), heme oxygenase-1 (HO-1), glutathione S-transferase A1 (GSTA1, also known as GST-Ya in mouse), glutamate-cysteine ligase (also known as γ-glutamylcysteine synthetase, i.e. γGCS) modifier subunit (GCLM) and catalytic subunit (GCLC) [9], [10], [11], [12]. The importance of the Nrf2-ARE system is evidently demonstrated by findings that Nrf2 knockout mice display significantly increased sensitivity to chemical toxicants and carcinogens [13], [14].
Nrf2 is activated by a plethora of stress inducers, phyto-antioxidants, and pathological conditions. The current model of the molecular mechanism for Nrf2 activation is that oxidative stress modifies cysteine residues of Keap1, a component of the E3 ubiquitin ligase that targets Nrf2 for degradation, resulting in compromised E3 activity and enhanced Nrf2 protein stability. The subsequent elevation in Nrf2 protein levels leads to Nrf2 nuclear accumulation, increased ARE-Nrf2 binding, and transactivation of its downstream genes [15], [16], [17], [18], [19], [20], [21]. In addition to the primary mode of regulation on its protein stability controlled by Keap1, Nrf2 is also regulated at multiple levels such as nucleocytoplamic trafficking, acetylation, and phosphorylation [22], [23], [24], [25].
The mitogen-activated protein kinases (MAPKs) are a family of highly related kinases consisting of the extracellular signal-regulated protein kinases (ERKs), the c-jun N-terminal kinases (JNKs), the p38 kinases and other relatively less characterized kinases, all of which catalyze phosphorylation on either serine or threonine residues adjacent to a proline residue. MAPKs are activated in a cascade fashion, by MAP2Ks (also known as MKKs or MEKs) that are in turn activated by MAP3Ks (also known as MEKKs, including ASK1, TAK1, Rafs, etc.) [26]. The MAPK signaling system responds to diverse stimuli, including oxidative stress, and has been implicated in Nrf2 induction by many previous reports (Table 1, see Discussion) [24], [25]. However previous studies failed to address whether the observed effects of MAPKs on Nrf2 are through direct and specific phosphorylation events on Nrf2 or through indirect and less specific mechanisms. It is not even known whether or not MAPKs phosphorylate Nrf2 in vivo at all. Another question that remains unanswered is whether MAPKs affect Keap1-mediated control of Nrf2 protein stability in vivo. Therefore, the exact role of MAPKs in Nrf2 activation, as well as the underlying molecular mechanism, remains poorly understood.
Table 1. Summary of key findings by previous studies showing the involvement of MAPKs in Nrf2 activation.
| Manipulation of MAPKs | Readout of Nrf2 activity | Changes of Nrf2 activity | Nrf2 inducers | Ref |
| PD98059 or DN ERK2 | NQO1 enzyme, GSTA1 ARE-luc | 30–60% decrease | tBHQ, SF | [42] |
| PD98059 and SB202190 | GCS mRNA, ARE-luc, ARE binding | ∼100% decrease | PDTC | [43] |
| MEKK1, TAK1, ASK1, MKK4/6, JNK1 | GSTA1 ARE-luc | 3–18 fold increase | [44] | |
| p38 | GSTA1 ARE-luc | 30–40% decrease | tBHQ | [44] |
| SB203580, DN p38 or DN MKK3 | NQO1 enzyme, GSTA1 ARE-luc | 40–50% increase | BHA, SF, β-NF | [45] |
| SB202191 or DN p38α | HO-1 E1-luc | 70–80% decrease | CdCl2 | [46] |
| PD98059, DN JNK1/2 or DN ERK1/2 | HO-1 E1-luc | no change | CdCl2 | [46] |
| PD98059 and SB202191 | nuclear Nrf2 protein | 80–90% decrease | PDTC | [34] |
| JNK1 | ARE-luc | ∼6 fold increase | PEITC | [47] |
| DN JNK1 | ARE-luc | 20–70% decrease | PEITC | [47] |
| MEKK1, TAK1, ASK1, MEK1/5, ERK2/5 | ARE-luc | 1.5–5 fold increase | [35] | |
| U0126 or ERK1 knockout | ARE-luc, ARE binding, Nrf2 protein | block induction | hyperoxia | [48] |
| SP600125 or SB203580 | Nrf2 protein | block induction | gallic acid | [49] |
| Raf knockout | Nrf2 nulear entry | no change | curcumin | [50] |
| p38α, β, γ, δ | HO-1 promoter-luc | 20–80% decrease | SF | [33] |
| U0126, DN ERK2 or DN JNK1 | ARE-luc | 20–60% decrease | BHA | [51] |
| MKK4 and JNK1 | ARE-luc, HO-1 | 3 fold increase | PEITC | [52] |
| DN ERK2 or DN JNK1 | ARE-luc | 40–50% decrease | PEITC | [52] |
| PD98059 and SB203580 | nuclear Nrf2 protein | block induction | 4-HNE | [53] |
| U0126 or DN MEK | nuclear Nrf2 protein | block induction | PGG | [54] |
| PD169316 or p38 siRNA | HO-1 protein, nuclear Nrf2 protein | >50% decrease | PDT | [55] |
| PD98059 or SB203580 | Nrf2 nulear entry | block induction | quercectin | [56] |
| PD98059 or U0126 | nuclear Nrf2 protein, ARE-luc | ∼60% decrease | D3T | [57] |
| U0126 | Nrf2 nuclear entry, ARE-luc | block induction | Triphlorethol-A | [58] |
| PD98059 | nuclear Nrf2 protein | block induction | eupatilin | [59] |
| PD98059 | nuclear Nrf2, ARE-luc, ARE binding | block induction | MT-III | [60] |
| U0126, DN ERK1 or DN ERK2 | HO-1 protein, ARE-luc | 30–80% decrease | 15d-PGJ2 | [61] |
| U0126 | nuclear Nrf2 protein | block induction | EGCG | [62] |
Manipulation of MAPKs activities, by either using kinase inhibitors or genetic engineering of MAPKs by overexpressing wild-type or dominant-negative (DN) kinases, was used as the major approach to assess the role of MAPKs in Nrf2 activation. PD98059: MEK1 inhibitor; SB202190: p38 inhibitor; SB203580: p38 inhibitor; U0126 MEK1/2 inhibitor; SP6000125: JNK inihibitor; luc: luciferase reporter gene; PDTC: pyrrolidine dithiocarbamate; β-NF: beta-naphthoflavone; BHA: tert-butylhydroxyanisole; PEITC: phenethylisothiocyanates; PDT: photodynamic therapy; 4-HNE: 4-hydroxynonenal; D3T: 3H-1,2-dithiole-3-thione; PGG: 1,2,3,4,6-penta-O-galloyl-β-D-glucose; ISMC: ileal smooth muscle cell; MTIII: metallothionein-III; EGCG: Epigallocatechin gallate.
Here we report the first evidence indicating that Nrf2 is phosphorylated at several serine and threonine residues by MAPKs in vivo. Combined alanine substitution of those residues leads to only a very moderate decrease in the transcriptional activity of Nrf2, most likely due to a slight reduction in its nuclear accumulation. More importantly, Nrf2 protein stability, which is primarily controlled by Keap1, is not altered in vivo by Nrf2 phosphorylation. These data indicate that direct phosphorylation of Nrf2 by MAPKs has limited contribution in modulating the Nrf2 signaling pathway. We propose that MAPKs regulate Nrf2 mainly through indirect mechanisms.
Materials and Methods
Recombinant DNA molecules
Plasmids expressing JNK1/2, ERK2, p38, MEKK3 and MEKK4 were generous gifts from Dr. Richard R. Vaillancourt at University of Arizona. Plasmids for HA-Nrf2 and Keap1-CBD proteins have been previously described [27]. The Nrf2 mutants were generated by site-directed mutagenesis using the PCR and Dpn1 based method as described before [28]. The human NQO1-ARE TATA-Inr luciferase reporter plasmid and the mouse GSTA1-ARE TATA-Inr luciferase reporter plasmid were reported previously [28], [29]. Renilla luciferase plasmid pGL4.74[hRluc/TK] was purchased from Promega.
Cell culture, transfection and chemicals
HEK293T, MDA-MB-231 and NIH3T3 cells were purchased from ATCC. Cells were maintained in either Dulbecco's modified Eagle's medium (DMEM) or Eagle's minimal essential medium (MEM) in the presence of 10% fetal bovine serum (FBS). Transfections were performed with Lipofectamine Plus (Gibco BRL) according to the manufacturer's instructions.
Immunoblot, immunoprecipitation, and immunofluorescence analysis
Antibodies against the HA epitope (Santa Cruz), chitin-binding domain (CBD) (New England Biolab), α-tubulin (Santa Cruz), and phosphorylated serine/threonine followed by a proline (pS/TP) (Abcam) were purchased from commercial sources. For detection of protein expression in total cell lysates, cells were lysed in sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% Glycerol, 100 mM DTT, 0.1% bromophenol blue) 48 hours following transfection. For immunoprecipitation assays, cells were lysed in RIPA buffer (10 mM sodium phosphate pH [8.0], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Sigma). Cell lysates were pre-cleared with protein A beads and incubated with HA antibody-conjugated beads (Santa Cruz) or chitin beads (New England Biolab) overnight at 4°C. After washing three times with RIPA buffer, immunoprecipitated complexes were eluted in sample buffer by boiling for 5 minutes, electrophoresed through SDS-polyacrylamide gels, and subjected to immunoblot analysis. Indirect immunofluorescence analysis was performed as described before using anti-HA primary antibodies [28].
In vivo phosphorylation of Nrf2
To detect phosphorylated Nrf2 in vivo by immunoblot, transfected cells were lysed by boiling in a buffer containing 2% SDS, 150 mM NaCl, 10 mM Tris-HCl and 1 mM DTT. This rapid lysis procedure inactivated cellular phosphatases and therefore preserved phosphorylated-Nrf2 conjugates present in cells prior to lysis. These lysates were then diluted five-fold in buffer lacking SDS, followed by immunoprecipitation with HA antibody-conjugated beads.
Mass spectrometry
Mass spectrometry analysis was performed by the Harvard Taplin mass spectrometry core facility. Briefly, immunoprecipitated proteins were visualized by Coomassie staining and recoverd from the gel. Excised gel bands were cut into approximately 1 mm3 pieces. Gel pieces were then subjected to a modified in-gel trypsin digestion procedure [30], followed by washing and dehydration with acetonitrile for 10 min. After removal of acetonitrile, gel pieces were completely dried in a speed-vac, and rehydrated with 50 mM ammonium bicarbonate solution containing 12.5 ng/µl modified sequencing-grade trypsin (Promega, Madison, WI) at 4°C. After 45 min., the excess trypsin solution was removed and replaced with 50 mM ammonium bicarbonate solution to just cover the gel pieces. Samples were then incubated at 37°C overnight. Peptides were later extracted by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile and 5% acetic acid. The extracts were then dried in a speed-vac (∼1 hr). The samples were then reconstituted in 5–10 µl of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reverse-phase HPLC capillary column was created by packing 5 µm C18 spherical silica beads into a fused silica capillary (100 µm inner diameter x∼12 cm length) with a flame-drawn tip. After equilibrating the column each sample was loaded via a Famos auto sampler (LC Packings, San Francisco CA) onto the column. A gradient was formed and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). As each peptide was eluted they were subjected to electrospray ionization and then they entered into an LTQ Orbitrap mass spectrometer (ThermoFinnigan, San Jose, CA). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein or translated nucleotide databases with the acquired fragmentation pattern by the software program, Sequest (ThermoFinnigan, San Jose, CA).
Reporter gene assay
A plasmid encoding renilla luciferase was included in all samples to control for transfection efficiency. Reporter assays were performed using the Dual-Luciferase reporter gene assay system (Promega). Nrf2 inducers tert-butylhydroquinone (tBHQ), sulforaphane (SF), sodium arsenite (As), and cadmium chloride (Cd) were purchased from Sigma (St. Louis, MO). Hydrogen peroxide (H2O2) was purchased from Mallinckrodt (Phillipsburg, NJ). Oridonin (Orid, also known as rubescensin A) was purchased from LKT Laboratories (St. Paul, MN). 15-Deoxy-Δ-12, 14-prostaglandin J2 (PGJ2) was a kind gift from Dr. John W. Regan at University of Arizona.
qRT-PCR
The real-time RT-PCR analysis and primer/probe sequence information was previously described [28], [31], [32].
Statistical test
Student's t- test was used to determine the significant difference between two samples from three assays in reporter gene analysis and qPCR analysis.
Results
Nrf2 is phosphorylated in vivo at multiple sites by MAPKs
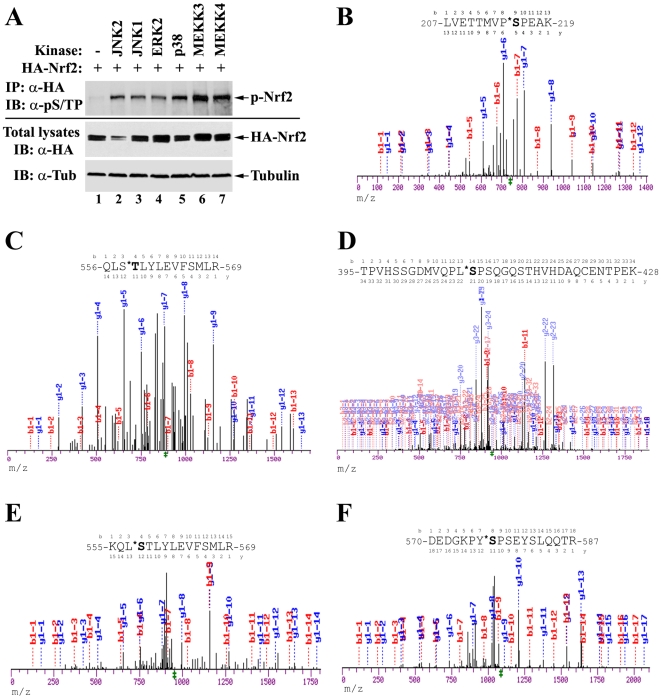
Purified recombinant GST-Nrf2 protein has been shown to be phosphorylated by immunoprecipitated p38 MAPKs in vitro [33]. However it is not clear whether Nrf2 is a bona fide substrate of MAPKs in vivo. To address this question, HEK293T cells were cotransfected with expression vectors for HA-tagged Nrf2 and the indicated MAPKs (Figure 1A). Cell lysates were immunoprecipitated with anti-HA antibodies. The immunoprecipitates were analyzed by immunoblot using antibodies that recognize phosphorylated serine or threonine residue adjacent to a proline (pS/TP), the consensus motif phosphorylated by MAPKs. The phosphorylation of Nrf2 is significantly enhanced in the presence of overexpressed MAPKs or their upstream kinases (Figure 1A, compare lane 1 with others). The phosphorylation antibody is specific to MAPK consensus sites, as demonstrated by its failure to recognize non-MAPK phosphorylation sites such as IKK sites (Figure S2).
Figure 1. Nrf2 is phosphorylated in vivo at multiple sites by MAPKs.
(A) HEK293T cells were cotransfected with expression vectors for HA-tagged Nrf2 and the indicated MAPKs. Cells were lysed in denaturing conditions. Cell lysates were diluted and immunoprecipitated with anti-HA antibodies. The immunoprecipitated protein was analyzed by immunoblot with antibodies specific for phosphorylated serine or threonine residues adjacent to a proline (pS/TP) (Abcam ab9344). p-Nrf2: phosphorylated Nrf2; Tub: tubulin. (B–F) Identification of distinct phosphorylation sites by tandem mass spectrometry analysis. Nrf2 proteins were purified through immunoprecipitation with anti-HA antibodies from HEK293T cells overexpressing HA-tagged Nrf2. The Nrf2 protein was size-separated by SDS-PAGE and visualized by Coomassie blue staining. The gel containing the Nrf2 protein was isolated and subjected to mass spectrometry analysis. m/z: mass to charge ratio.
To identify the phosphorylation sites on Nrf2 by endogenous kinases, HEK293T cells were transfected with an expression vector for HA-tagged human Nrf2. Nrf2 proteins were purified by immunoprecipitation with anti-HA antibodies, separated by SDS-PAGE, and visualized by coomassie blue staining. The gel containing the Nrf2 protein was isolated and subjected to mass spectrometry analysis using LC-MS/MS. Five phosphorylated serine or threonine residues (S215, S408, S558, T559 and S577) were identified (Figure 1B–F). Three of them (S215, S408, S577) fit consensus sites for MAPKs, while the other two (S558 and S559) do not. All five sites were subsequently characterized, individually and combined.
Abolishing Nrf2 phosphorylation by mutation causes a limited decrease in the transcriptional activity of Nrf2
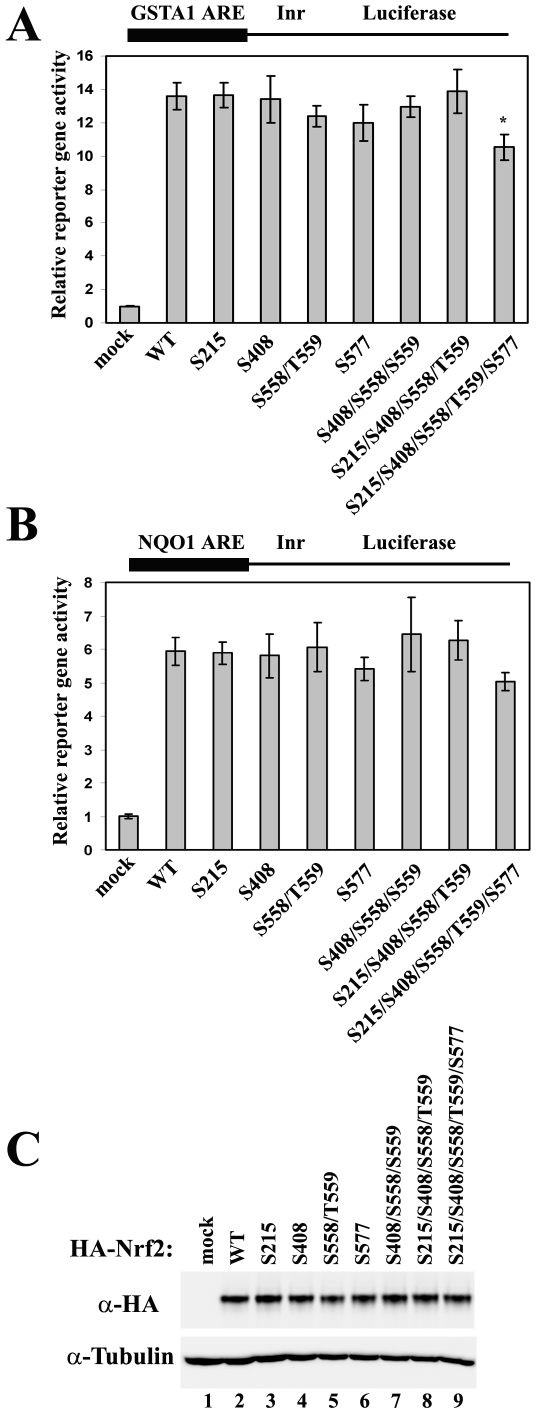
Genetic approaches were taken to explore the function of phosphorylation on Nrf2. Single or combined alanine substitutions on the phosphorylation sites were constructed, and the transcriptional activity of the resultant Nrf2 mutants were tested in the ARE-dependent reporter gene assays using the ARE sequence taken from either the NQO1 gene promoter (Figure 2A) or the GSTA1 gene promoter (Figure 2B). In both cases, only the combined mutation of all five phosphorylation sites caused a slight decrease in Nrf2 transcriptional activity (Figure 2A and 2B). More importantly, none of the mutations, whether single or combined, caused obvious changes in Nrf2 protein levels (Figure 2C).
Figure 2. Combined alanine substitution on all phosphorylation sites causes only a moderate decrease in the transcriptional activity of Nrf2.
(A) MDA-MB-231 cells were cotransfected with expression vectors for the GSTA1-ARE-dependent firefly luciferase, the TK-Renilla luciferase, and the indicated Nrf2 mutant. Luciferase reporter gene activities were analyzed using the Promega dual-luciferase reporter gene assay system. WT: wild-type; TK: thymidine kinase promoter; Inr: initiator. Relative luciferase activities and standard deviations were calculated from three independent assays. * p<0.05 when compared to WT. (B) Reporter gene assay was performed as described above using NQO1-ARE-dependent luciferase reporter gene construct in MDA-MB-231 cells. (C) Total cell lysates from one of the reporter gene assays were subjected to immunoblot analysis with anti-HA antibodies for detection of Nrf2.
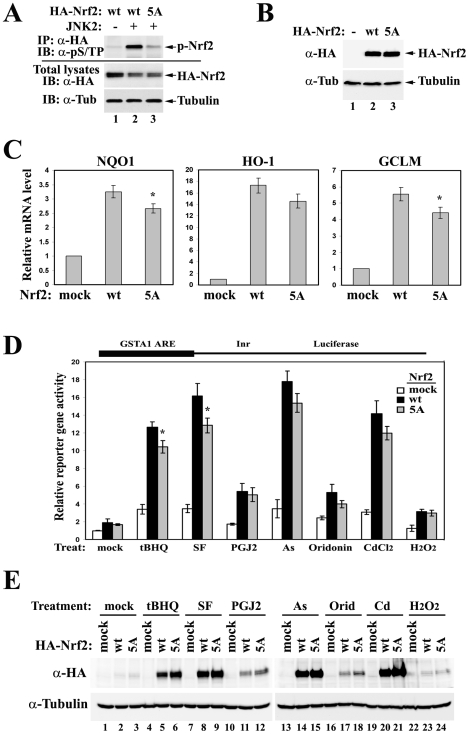
The mutant that has all five phosphorylation sites substituted by alanines was named “5A” and was subjected to subsequent analysis, since it seemed to have a slightly compromised ability in transactivating the ARE-dependent genes. Abolishment of Nrf2 phosphorylation in the Nrf2-5A mutant was confirmed by the in vivo phosphorylation assay (Figure 3A). All three MAPK consensus sites (S215, S408 and S577) seem to contribute to the overall phosphorylation level of Nrf2 in the presence of overexpressed MAPKs (Figure S1). If the antibody has no bias towards the context sequences around the consensus phosphorylation sites, S408 seems to be the most dominant site. None of the single mutations was able to abolish Nrf2 phosphorylation as well as the combined 5A mutation, suggesting a redundancy in phosphorylation sites (Figure S1). To further test the effects of phosphorylation on the transcriptional activity of Nrf2, mRNA expression of Nrf2 target genes was analyzed by qRT-PCR in HEK293T cells overexpressing either Nrf2-WT (wild-type) or Nrf2-5A mutant (Figure 3C). Loss of phosphorylation causes a minor or only moderate decrease in the expression of Nrf2 downstream gene NQO1, HO-1, and GCLM (Figure 3C), without affecting Nrf2 protein levels (Figure 3B and 3C).
Figure 3. Genetic disruption of Nrf2 phosphorylation slightly attenuates Nrf2 activities without affecting its protein levels.
(A) HEK293T cells were cotransfected with expression vectors for HA-tagged Nrf2 and JNK2. Cell lysates were immunoprecipitated with anti-HA antibodies. The immunoprecipitated protein was analyzed by immunoblot with antibodies specific for phosphorylated serine or threonine residue adjacent to a proline (pS/TP) (Abcam ab9344). wt: wild-type; 5A: the mutant with combined alanine substitution on all five phosphorylation sites. (B–C) HEK293T cells overexpressing either Nrf2-WT or Nrf2-5A were subjected to immunoblot (B) and qRT-PCR analysis (C). The error bars indicate the standard deviations calculated from triplicate samples. * p<0.05 when compared to wt. (D) MDA-MB-231 cells were cotransfected with expression vectors for the GSTA1-ARE-dependent luciferase reporter gene, Keap1, and the indicated Nrf2. Cells were treated with several Nrf2 inducers for 16 hrs prior to reporter gene analysis. tBHQ: tert-butylhydroquinone (50 µM); SF: sulforaphane (20 µM); PGJ2: 15-Deoxy-Δ-12,14-prostaglandin J2 (10 µM); As: sodium arsnite (20 µM); Orid: Oridonin (10 µM); Cd: cadmium chloride (20 µM); H2O2: hydrogen peroxide (500 µM). Error bars indicate standard deviations calculated from three independent assays. * p<0.05 when compared to wt in the same treatment group. (E) Total cell lysates from the above reporter gene assay were analyzed by immunoblot.
Phosphorylation of Nrf2 slightly increases the response of Nrf2 to several chemical inducers without affecting Keap1-mediated control of Nrf2 protein levels
To test whether phosphorylation of Nrf2 alters its behavior in response to different inducers, reporter gene analysis was performed on MDA-MB-231 cells overexpressing Keap1 and either Nrf2-WT or Nrf2-5A. Cells were treated with different Nrf2 inducers for 16 hrs prior to analysis. Altered response between Nrf2-WT and Nrf2-5A was only observed in the presence of relatively potent Nrf2 inducers (Figure 3D). The protein levels of Nrf2-WT and Nrf2-5A were increased to a similar extent in response to all inducers tested, suggesting that phosphorylation does not have any effect on Nrf2 protein stability that is primarily controlled by Keap1 (Figure 3E).
Phosphorylation of Nrf2 by MAPKs does not affect the interaction between Nrf2 and Keap1 in vivo
Keum et al. have reported that phosphorylation of Nrf2 by p38 MAPKs increased the interaction between Nrf2 and Keap1 in an in vitro GST-pulldown experiment [33]. To test whether similar effects can be observed in vivo, HEK293T cells were co-transfected with an expression vector for either HA-tagged Nrf2-WT or Nrf2-5A, along with expression vectors for CBD-tagged Keap1, and the indicated MAPK. Cell lysates were pulled-down with chitin beads and the precipitated protein complexes were analyzed by immunoblot with anti-HA and anti-CBD antibodies (Figure 4A). The interaction between Nrf2 and Keap1 was not affected by the phosphorylation status of Nrf2, consistent with the finding that phoshorylation of Nrf2 does not affect its protein stability (Figure 3E).
Figure 4. Phosphorylation of Nrf2 moderately enhances its nuclear accumulation without affecting the interaction between Nrf2 and Keap1.
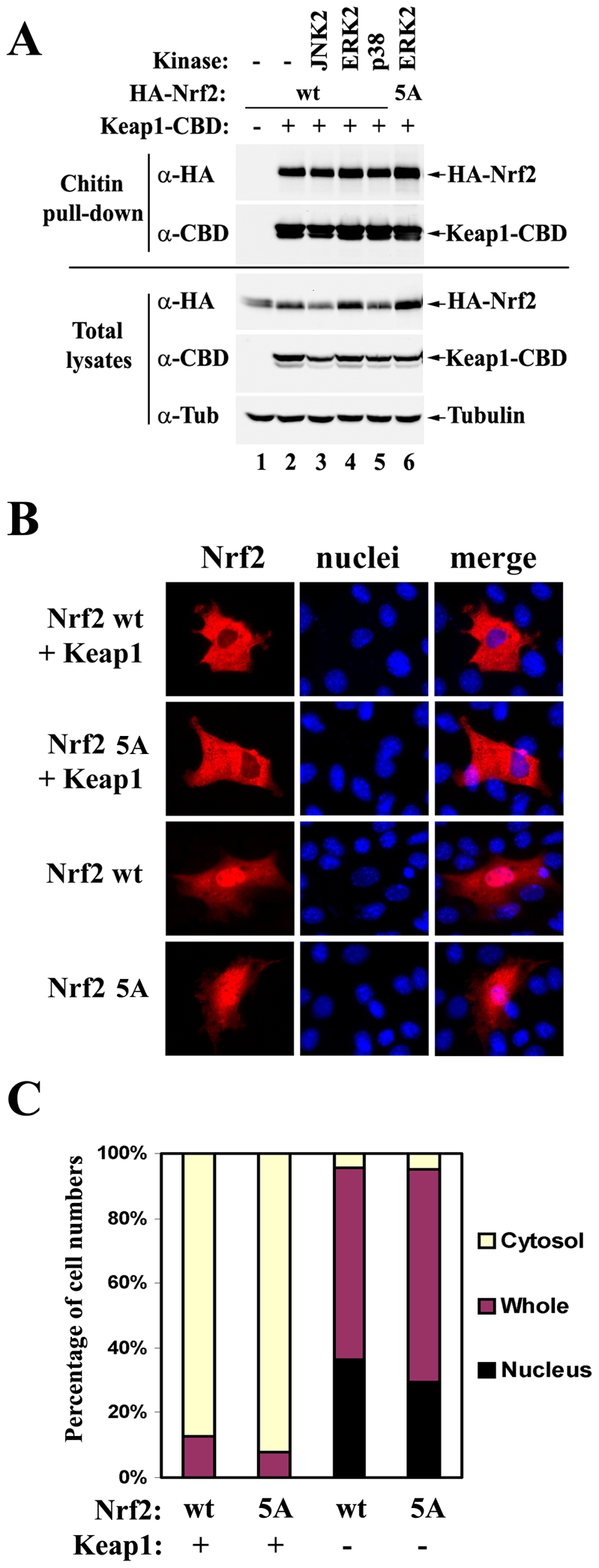
(A) HEK293T cells were cotransfected with expression vectors for the indicated MAPKs, HA-tagged Nrf2, and CBD-tagged Keap1. Cells were harvested and lysed in RIPA buffer. Cell lysates were pulled-down with chitin beads and analyzed by immunoblot with anti-HA and anti-CBD antibodies. CBD: chitin binding domain; wt: wild-type; 5A: mutant with combined alanine substitution on all five phosphorylation sites. (B) NIH3T3 cells were transfected with expression vectors for the indicated Nrf2 protein and Keap1. The subcellular localization of Nrf2 was determined by indirect immunofluorescence analysis with anti-HA antibodies. (C) Quantification analysis of the immunofluorescence assay. At least 100 positively stained cells were examined by fluorescence microscopy. Percentages of cells in which Nrf2 was localized predominantly in the cytosol, the whole cell, or the nucleus were presented as a bar graph.
Phosphorylation of Nrf2 moderately enhances its nuclear accumulation
Nuclear translocation of Nrf2 protein is essential for Nrf2 to transactivate its downstream genes. To test whether phosphorylation of Nrf2 alters its nuclear accumulation, NIH3T3 cells were transfected with the expression vector for Nrf2-WT or Nrf2-5A in the presence or absence of the expression vector for Keap1 to mimic basal or induced conditions, respectively. The subcellular localization of Nrf2 was determined by indirect immunofluorescence analysis with anti-HA antibodies. There was a slight decrease in nuclear accumulation of Nrf2-5A, compared to Nrf2-WT, in the absence of Keap1, as shown in representative pictures (Figure 4B) and quantification data (Figure 4C).
Discussion
Many previous studies have shown that MAPKs play a role in the induction of Nrf2. Table 1 summarizes key findings of all previous studies that support the involvement of MAPKs in Nrf2 activation. Manipulation of the catalytic activity of MAPKs, either through pharmaceutical intervention (kinase inhibitors) or genetic engineering (overexpression of wild-type or dominant-negative kinases, siRNA-mediated knockdown, or gene knockout), coupled with a readout of Nrf2 activity (ARE-dependent luciferase reporter gene, or Nrf2 nuclear accumulation), was used as the major approach to address the potential role of MAPKs on Nrf2 (Table 1). All three well-characterized categories of MAPKs (ERKs, JNKs, and p38) have been implicated in modulating Nrf2, with some contradictory results among different groups. Since MAPKs regulate bewilderingly diverse cellular programs, it is not clear whether the observed effects of MAPKs on Nrf2 are highly specific or just bystander effects. It remains elusive whether MAPKs regulate Nrf2 through direct phosphorylation of Nrf2 or through indirect mechanisms. Furthermore, it is not known whether or not MAPKs phosphorylate Nrf2 in vivo at all. Another key question that remains open is whether MAPKs affect Nrf2 protein stability that is primarily controlled by Keap1. Therefore, the exact role of MAPKs in Nrf2 activation, as well as the underlying molecular mechanism, remains poorly understood.
In this study, we report the first evidence that Nrf2 is phosphorylated by MAPKs in vivo. We have identified the major phosphorylation sites and used site-mutagenesis to decipher the function of direct phosphorylation of Nrf2 by MAPKs. As demonstrated by data obtained from ARE-dependent luciferase reporter gene assay and qRT-PCR analysis, a loss of Nrf2 phosphorylation caused only moderate changes in the transcriptional activity of Nrf2 (Figure 2 and 3). The Keap1-mediated control of Nrf2 protein levels was not affected by Nrf2 phosphorylation in vivo under both basal and induced conditions (Figure 3E), nor was the interaction between Nrf2 and Keap1 (Figure 4A). However, the nuclear accumulation of Nrf2 was slightly enhanced by its phosphorylation (Figure 4B and 4C). We concluded that direct phosphorylation of Nrf2 by MAPKs has a limited contribution in regulating the Nrf2-dependent antioxidant responses.
We are aware of the fact that our mass spectrometry analysis may not have identified all phosphorylated residues on Nrf2. There might be signal-induced Nrf2 phosphorylation on specific sites that were missed in the current identification procedure whereby overexpressed Nrf2 protein itself was purified from HEK293T cells under basal conditions followed by LC-MS/MS. However, the fact that the Nrf2-5A mutant has significantly decreased phosphorylation levels compared to wild-type Nrf2 in the presence of overexpressed JNK2 suggests that these five residues are the major targets of phosphorylation in vivo (Figure 3A). Neither inducible ARE-dependent transcription nor inducible Nrf2 protein levels was dramatically altered by combined mutations on the identified phosphorylation sites in the presence of several Nrf2 inducers such as tBHQ, sulforaphane, PGJ2, arsenite or cadmium, suggesting that phosphorylation at these sites has limited contribution in regulating inducible Nrf2 signaling (Figure 3D and 3F). Additionally, in an experiment without using mutations, overexpression of MAPKs failed to cause any observable effect on the interaction between wild-type Nrf2 and Keap1 (Figure 4A, lane 1–5), although Nrf2 phosphorylation was significantly enhanced under the same conditions (Figure 1A). Collectively, these data suggest that direct phosphorylation of Nrf2 plays a limited role in regulating Nrf2 activity.
Consistent with our data, Zipper and Mulcahy showed that mutational disruption of six MAPK consensus sites in Nrf2 (including S215, S408, S577 that were also characterized in this study) failed to significantly change the GCLM ARE-dependent reporter gene activities in the absence or presence of pyrrolidine dithiocarbamate (PDTC), an Nrf2 inducer [34]. Here we provide additional evidence that mutational disruption of these sites does not dramatically alter the endogenous mRNA levels of NQO1, HO-1 or GCLM (Figure 3C), excluding possibilities that phosphorylation might interfere with molecular events that are specific to native chromosomal architectures. In addition, we showed that neither Nrf2 protein levels nor ARE-dependent luciferase activities were dramatically altered by the combined mutation in cells either left untreated or treated with divergent Nrf2 inducers, such as tBHQ, sulforaphane, PGJ2, arsnite, oridonin, cadmium and H2O2 (Figure 3E), making it a much less attractive hypothesis that phosphorylation might be involved in the action of some particular Nrf2 inducers, if not all. Furthermore, we performed a co-immunoprecipitation assay and directly demonstrated that the physical interaction between Nrf2 and Keap1 was not altered in vivo in the presence of overexpressed MAPKs or with Nrf2 phosphorylation-deficient mutants (Figure 4A). Another previous report from Kong and colleagues also showed that site-directed mutagenesis of Nrf2 at several MAPK consensus sites did not affect the transactivation activity of Nrf2, although these sites do not overlap with the sites characterized in our present study [35]. Taken together, MAPK-mediated regulation of the Nrf2 signaling pathway is not likely to be through direct phosphorylation of Nrf2.
One possible indirect mechanism is translational control on Nrf2 protein synthesis by MAPKs. MAPKs have been shown to modulate mTOR signaling pathways in controlling eukaryotic initiation factor 4 (eIF4) complex assembly, a critical step in Cap-dependent translational initiation, through inhibiting tuberous sclerosis complex 2 (TSC2) and activating 90-kD ribosomal protein S6 kinases (RSKs) [36], [37]. MAPKs also promote phosphorylation of eIF4E, eIF4B-BP1, and translation elongation factor 2 kinase (eEF2 kinase) through MAPK-interacting kinase 1 and 2 (Mnk1/2), mitogen- and stress-activated protein kinase 1 (Msk1), and MAPK-activated kinase 2/3 (MK2/3), respectively, which is thought to enhance translation of some mRNAs [36], [37], [38], [39]. On the other hand, Nrf2 has been shown to be regulated at the translational level under several conditions [40], [41]. Thus, MAPKs-mediated indirect control of Nrf2 protein synthesis, along with the moderate role of direct phosphorylation on Nrf2, may represent the underlying mechanism by which MAPKs regulates the Nrf2 signaling pathway.
Supporting Information
HEK293T cells were co-transfected with expression vectors for HA-Nrf2 wild-type or mutants and different kinases. Cell lysates were collected in denaturing conditions, and phosphorylation analysis was performed as described in Fig. 1A and 3A. All three sites (S215, S408 and S577) contribute to the overall phosphorylation level of Nrf2 in the presence of all four kinases.
(0.81 MB TIF)
HEK293T cells were co-transfected with expression vectors for NF-κB p65, along upstream kinases IKKα and IKKβ in HEK293T cells. Phosphorylation of p65 at S536 by IKKs was confirmed by an antibody that recognizes this site-specific modification (Cell Signaling, Cat# 3033S). The antibody for phospho-S/TP did not pick up any signal above background, indicating the specificity of this antibody.
(0.83 MB TIF)
Acknowledgments
We thank Drs. R. R. Vaillancourt and J. W. Regan for their kind help and reagents. Thank N. F. Villeneuve and A. Lau for editing of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: NIEHS 1RO1ES015010, ACS RSG-07-154, SWEHSC ES006694. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yang CS, Landau JM, Huang MT, Newmark HL. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu Rev Nutr. 2001;21:381–406. doi: 10.1146/annurev.nutr.21.1.381. [DOI] [PubMed] [Google Scholar]
- 2.Kanwar JR. Anti-inflammatory immunotherapy for multiple sclerosis/experimental autoimmune encephalomyelitis (EAE) disease. Curr Med Chem. 2005;12:2947–2962. doi: 10.2174/092986705774462833. [DOI] [PubMed] [Google Scholar]
- 3.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 4.Ruef J, Peter K, Nordt TK, Runge MS, Kubler W, et al. Oxidative stress and atherosclerosis: its relationship to growth factors, thrombus formation and therapeutic approaches. Thromb Haemost. 1999;82(Suppl 1):32–37. [PubMed] [Google Scholar]
- 5.Simonian NA, Coyle JT. Oxidative stress in neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 1996;36:83–106. doi: 10.1146/annurev.pa.36.040196.000503. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 7.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, et al. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 10.Moinova HR, Mulcahy RT. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochem Biophys Res Commun. 1999;261:661–668. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- 11.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, et al. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 13.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in Nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, et al. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 22.Hayes JD, McMahon M. Nrf2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by p300/CBP Augments Promoter-specific DNA Binding of Nrf2 during Antioxidant Response. Mol Cell Biol. 2009 doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCubrey JA, Lahair MM, Franklin RA. Reactive oxygen species-induced activation of the MAP kinase signaling pathways. Antioxid Redox Signal. 2006;8:1775–1789. doi: 10.1089/ars.2006.8.1775. [DOI] [PubMed] [Google Scholar]
- 25.Owuor ED, Kong AN. Antioxidants and oxidants regulated signal transduction pathways. Biochem Pharmacol. 2002;64:765–770. doi: 10.1016/s0006-2952(02)01137-1. [DOI] [PubMed] [Google Scholar]
- 26.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 27.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang XJ, Sun Z, Chen W, Eblin KE, Gandolfi JA, et al. Nrf2 protects human bladder urothelial cells from arsenite and monomethylarsonous acid toxicity. Toxicol Appl Pharmacol. 2007;225:206–213. doi: 10.1016/j.taap.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 31.Wang XJ, Sun Z, Chen W, Li Y, Villeneuve NF, et al. Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction. Toxicol Appl Pharmacol. 2008;230:383–389. doi: 10.1016/j.taap.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keum YS, Yu S, Chang PP, Yuan X, Kim JH, et al. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006;66:8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 34.Zipper LM, Mulcahy RT. Erk activation is required for Nrf2 nuclear localization during pyrrolidine dithiocarbamate induction of glutamate cysteine ligase modulatory gene expression in HepG2 cells. Toxicol Sci. 2003;73:124–134. doi: 10.1093/toxsci/kfg083. [DOI] [PubMed] [Google Scholar]
- 35.Shen G, Hebbar V, Nair S, Xu C, Li W, et al. Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J Biol Chem. 2004;279:23052–23060. doi: 10.1074/jbc.M401368200. [DOI] [PubMed] [Google Scholar]
- 36.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raught B, Gingras A-C. Signaling to translation initiation. In: Mathews MB, NS, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 369–400. [Google Scholar]
- 39.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 40.Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. Translational control of Nrf2 protein in activation of antioxidant response by oxidants. Mol Pharmacol. 2007;72:1074–1081. doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 41.Wang X-J, Zhang DD. Ectodermal-neural cortex 1 down-regulates Nrf2 at the translational level. 2009 doi: 10.1371/journal.pone.0005492. (in revision for PLoS ONE) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu R, Lei W, Mandlekar S, Weber MJ, Der CJ, et al. Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. J Biol Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 43.Zipper LM, Mulcahy RT. Inhibition of ERK and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem Biophys Res Commun. 2000;278:484–492. doi: 10.1006/bbrc.2000.3830. [DOI] [PubMed] [Google Scholar]
- 44.Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, et al. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem. 2000;275:39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]
- 45.Yu R, Mandlekar S, Lei W, Fahl WE, Tan TH, et al. p38 mitogen-activated protein kinase negatively regulates the induction of phase II drug-metabolizing enzymes that detoxify carcinogens. J Biol Chem. 2000;275:2322–2327. doi: 10.1074/jbc.275.4.2322. [DOI] [PubMed] [Google Scholar]
- 46.Alam J, Wicks C, Stewart D, Gong P, Touchard C, et al. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J Biol Chem. 2000;275:27694–27702. doi: 10.1074/jbc.M004729200. [DOI] [PubMed] [Google Scholar]
- 47.Keum YS, Owuor ED, Kim BR, Hu R, Kong AN. Involvement of Nrf2 and JNK1 in the activation of antioxidant responsive element (ARE) by chemopreventive agent phenethyl isothiocyanate (PEITC). Pharm Res. 2003;20:1351–1356. doi: 10.1023/a:1025737622815. [DOI] [PubMed] [Google Scholar]
- 48.Papaiahgari S, Kleeberger SR, Cho HY, Kalvakolanu DV, Reddy SP. NADPH oxidase and ERK signaling regulates hyperoxia-induced Nrf2-ARE transcriptional response in pulmonary epithelial cells. J Biol Chem. 2004;279:42302–42312. doi: 10.1074/jbc.M408275200. [DOI] [PubMed] [Google Scholar]
- 49.Yeh CT, Yen GC. Involvement of p38 MAPK and Nrf2 in phenolic acid-induced P-form phenol sulfotransferase expression in human hepatoma HepG2 cells. Carcinogenesis. 2006;27:1008–1017. doi: 10.1093/carcin/bgi281. [DOI] [PubMed] [Google Scholar]
- 50.Andreadi CK, Howells LM, Atherfold PA, Manson MM. Involvement of Nrf2, p38, B-Raf, and nuclear factor-kappaB, but not phosphatidylinositol 3-kinase, in induction of hemeoxygenase-1 by dietary polyphenols. Mol Pharmacol. 2006;69:1033–1040. doi: 10.1124/mol.105.018374. [DOI] [PubMed] [Google Scholar]
- 51.Yuan X, Xu C, Pan Z, Keum YS, Kim JH, et al. Butylated hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol Carcinog. 2006;45:841–850. doi: 10.1002/mc.20234. [DOI] [PubMed] [Google Scholar]
- 52.Xu C, Yuan X, Pan Z, Shen G, Kim JH, et al. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther. 2006;5:1918–1926. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H, Liu H, Iles KE, Liu RM, Postlethwait EM, et al. 4-Hydroxynonenal Induces Rat Gamma-glutamyl Transpeptidase through MAPK Mediated EpRE/Nrf2 Signaling. Am J Respir Cell Mol Biol. 2005 doi: 10.1165/rcmb.2005-0280OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pae HO, Oh GS, Jeong SO, Jeong GS, Lee BS, et al. 1,2,3,4,6-penta-O-galloyl-beta-D-glucose up-regulates heme oxygenase-1 expression by stimulating Nrf2 nuclear translocation in an extracellular signal-regulated kinase-dependent manner in HepG2 cells. World J Gastroenterol. 2006;12:214–221. doi: 10.3748/wjg.v12.i2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kocanova S, Buytaert E, Matroule JY, Piette J, Golab J, et al. Induction of heme-oxygenase 1 requires the p38MAPK and PI3K pathways and suppresses apoptotic cell death following hypericin-mediated photodynamic therapy. Apoptosis. 2007;12:731–741. doi: 10.1007/s10495-006-0016-x. [DOI] [PubMed] [Google Scholar]
- 56.Yao P, Nussler A, Liu L, Hao L, Song F, et al. Quercetin protects human hepatocytes from ethanol-derived oxidative stress by inducing heme oxygenase-1 via the MAPK/Nrf2 pathways. J Hepatol. 2007;47:253–261. doi: 10.1016/j.jhep.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 57.Manandhar S, Cho JM, Kim JA, Kensler TW, Kwak MK. Induction of Nrf2-regulated genes by 3H-1, 2-dithiole-3-thione through the ERK signaling pathway in murine keratinocytes. Eur J Pharmacol. 2007;577:17–27. doi: 10.1016/j.ejphar.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 58.Kang KA, Lee KH, Park JW, Lee NH, Na HK, et al. Triphlorethol-A induces heme oxygenase-1 via activation of ERK and NF-E2 related factor 2 transcription factor. FEBS Lett. 2007;581:2000–2008. doi: 10.1016/j.febslet.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 59.Song HJ, Shin CY, Oh TY, Sohn UD. The protective effect of eupatilin on indomethacin-induced cell damage in cultured feline ileal smooth muscle cells: involvement of HO-1 and ERK. J Ethnopharmacol. 2008;118:94–101. doi: 10.1016/j.jep.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 60.Hwang YP, Kim HG, Han EH, Jeong HG. Metallothionein-III protects against 6-hydroxydopamine-induced oxidative stress by increasing expression of heme oxygenase-1 in a PI3K and ERK/Nrf2-dependent manner. Toxicol Appl Pharmacol. 2008;231:318–327. doi: 10.1016/j.taap.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 61.Kim JW, Li MH, Jang JH, Na HK, Song NY, et al. 15-Deoxy-Delta(12,14)-prostaglandin J(2) rescues PC12 cells from H2O2-induced apoptosis through Nrf2-mediated upregulation of heme oxygenase-1: potential roles of Akt and ERK1/2. Biochem Pharmacol. 2008;76:1577–1589. doi: 10.1016/j.bcp.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 62.Na HK, Kim EH, Jung JH, Lee HH, Hyun JW, et al. (−)-Epigallocatechin gallate induces Nrf2-mediated antioxidant enzyme expression via activation of PI3K and ERK in human mammary epithelial cells. Arch Biochem Biophys. 2008;476:171–177. doi: 10.1016/j.abb.2008.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HEK293T cells were co-transfected with expression vectors for HA-Nrf2 wild-type or mutants and different kinases. Cell lysates were collected in denaturing conditions, and phosphorylation analysis was performed as described in Fig. 1A and 3A. All three sites (S215, S408 and S577) contribute to the overall phosphorylation level of Nrf2 in the presence of all four kinases.
(0.81 MB TIF)
HEK293T cells were co-transfected with expression vectors for NF-κB p65, along upstream kinases IKKα and IKKβ in HEK293T cells. Phosphorylation of p65 at S536 by IKKs was confirmed by an antibody that recognizes this site-specific modification (Cell Signaling, Cat# 3033S). The antibody for phospho-S/TP did not pick up any signal above background, indicating the specificity of this antibody.
(0.83 MB TIF)