Abstract
Systemic acquired resistance (SAR) is a plant immune response induced by local necrotizing pathogen infections. Expression of SAR in Arabidopsis (Arabidopsis thaliana) plants correlates with accumulation of salicylic acid (SA) and up-regulation of Pathogenesis-Related (PR) genes. SA is an essential and sufficient signal for SAR. In a genetic screen to search for negative regulators of PR gene expression and SAR, we found a new mutant that is hypersensitive to SA and exhibits enhanced induction of PR genes and resistance against the virulent oomycete Hyaloperonospora arabidopsidis Noco2. The enhanced pathogen resistance in the mutant is Nonexpressor of PR genes1 independent. The mutant gene was identified by map-based cloning, and it encodes a protein with high homology to Replication Factor C Subunit3 (RFC3) of yeast and other eukaryotes; thus, the mutant was named rfc3-1. rfc3-1 mutant plants are smaller than wild-type plants and have narrower leaves and petals. On the epidermis of true leaves, there are fewer cells in rfc3-1 compared with the wild type. Cell production rate is reduced in rfc3-1 mutant roots, indicating that the mutated RFC3 slows down cell proliferation. As Replication Factor C is involved in replication-coupled chromatin assembly, our data suggest that chromatin assembly and remodeling may play important roles in the negative control of PR gene expression and SAR.
Plants have evolved inducible defense mechanisms to cope with infections by a wide range of microbial pathogens during plant-pathogen coevolutionary history (Jones and Dangl, 2006). In 1961, Ross found that tobacco (Nicotiana tabacum) plants challenged with Tobacco mosaic virus subsequently developed enhanced resistance to secondary infections in distal tissues (Ross, 1961). This spread of resistance throughout the plant was termed systemic acquired resistance (SAR). The response is long lasting, sometimes for the lifetime of the plant. SAR is also effective against a broad spectrum of pathogens, including viruses, bacteria, fungi, and oomycetes (Ryals et al., 1996; Durrant and Dong, 2004). During the onset of SAR, salicylic acid (SA) levels increase in both local and systemic tissues, accompanied by up-regulation of a set of Pathogenesis-Related (PR) genes. Because overexpression of a single PR gene is not sufficient to establish broad-spectrum resistance, it is believed that PR proteins enhance resistance by working in concert. In particular, PR-1 and β-1,3-glucanase (BGL2; also known as PR-2) have been widely used as molecular markers for SAR (Uknes et al., 1992; Bowling et al., 1994; Cao et al., 1994).
SA was found to be a necessary and sufficient signal for SAR. SAR can be brought on by exogenous application of SA or its analogs such as 2,6-dichloroisonicotinic acid (INA) and benzothiadiazole S-methyl ester. On the other hand, removal of SA by exogenous SA hydroxylase (NahG) or genetic mutations in the SA biosynthetic pathway, such as EDS5 and SID2, disable SAR (White, 1979; Métraux et al., 1991; Ward et al., 1991; Görlach et al., 1996; Lawton et al., 1996).
Nonexpressor of PR genes1 (NPR1) is a central positive regulator of SAR downstream of SA. When NPR1 is mutated, plants can no longer mount a SAR response even with induction of SA or INA (Cao et al., 1994). It functions through associations with TGA transcription factors to regulate PR gene expression and pathogen resistance (Durrant and Dong, 2004). A triple knockout mutant of TGA2, TGA5, and TGA6 displayed compromised SAR responses and increased basal PR gene expression, suggesting that TGA transcription factors have both positive and negative roles in regulating PR gene expression and SAR (Zhang et al., 2003).
As constitutive activation of defense is detrimental to plants, expression of PR genes is usually under tight control. In a previous study, an INA hypersensitive mutant, suppressor of npr1-1, inducible1 (sni1), was found to be a transcriptional repressor of PR gene expression that negatively regulates SAR (Li et al., 1999; Mosher et al., 2006). Mutant plants of sni1 exhibit a basal level of PR gene expression that is independent of NPR1, and this expression is further enhanced upon SAR induction. SNI1 encodes a protein with structural similarity to Armadillo repeat proteins potentially involved in scaffolding or protein-protein interactions. Chromatin immunoprecipitation experiments indicated that histone modification could be involved in SNI1 function (Mosher et al., 2006). When a genetic screen was conducted to search for genetic suppressors of sni1, it was found that a loss-of-function mutation in RAD51D suppresses sni1 phenotypes completely. Both SNI1 and RAD51D were found to play roles in PR gene transcription and DNA recombination (Durrant et al., 2007). To identify additional negative regulators of PR gene expression and SAR, we screened for mutants that are hypersensitive to SA induction and found one with similar phenotypes to sni1. This mutant has higher expression levels of PR genes and displays enhanced resistance against the virulent oomycete Hyaloperonospora arabidopsidis (H. a.) Noco2. The gene was identified by map-based cloning and found to encode a protein with high homology to Replication Factor C Subunit3 (RFC3) proteins.
RESULTS
Novel Genetic Screen to Search for Mutants That Are Hypersensitive to SA Induction
While exogenous application of SA induces high expression of PR genes and SAR, wild-type plants are almost nonresponsive to very low concentrations of SA. Since the previously reported sni1 mutant plants are hypersensitive to SA or INA induction, we reasoned that if similar negative regulators are mutated, we should be able to find mutants having similar phenotypes as sni1. We first built an individually harvested M2 population of ethylmethane sulfonate-mutagenized ecotype Columbia (Col) wild-type plants carrying the pBGL2-GUS reporter gene (Cao et al., 1994). Subsequently, we searched for mutants showing strong GUS staining when grown on Murashige and Skoog (MS) plates supplemented with 10 μm SA. This concentration of SA does not induce visible GUS staining on wild-type plants. Except for additional alleles of sni1 (S. Xia and X. Li, unpublished data), one mutant with enhanced inducible GUS staining was obtained, and it was later named rfc3-1 after we found its identity (Fig. 1A).
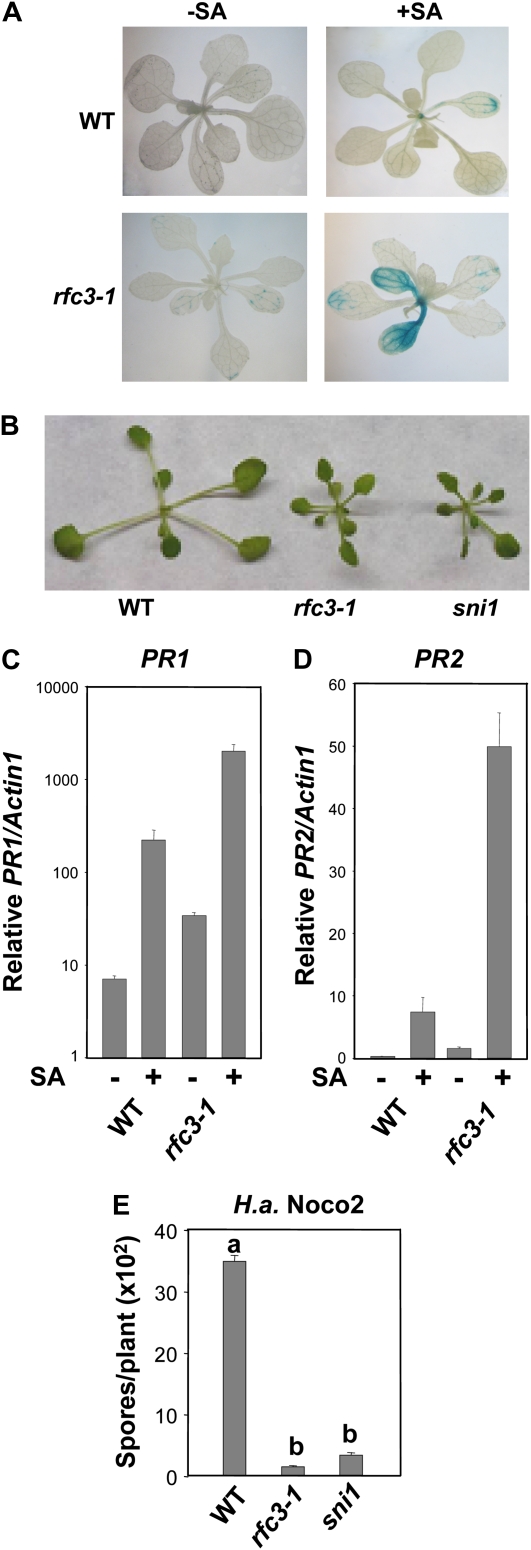
Figure 1.
Characterization of the rfc3-1 mutant. A, GUS staining of wild-type (WT) Col and rfc3-1 plants, both with the pBGL2-GUS reporter gene. Two-week-old seedlings grown on MS with or without 10 μm SA were stained for GUS activity as described previously (Bowling et al., 1994). B, Morphology of wild-type, rfc3-1, and sni1 plants. All plants were grown on soil and photographed when they were 4 weeks old. C and D, Relative PR1 (C) and PR2 (D) expression in wild-type and rfc3-1 plants. Two-week-old seedlings grown on MS with or without 50 μm SA were collected for RNA extraction and reverse transcribed to obtain total cDNA. The cDNA samples were normalized by real-time PCR with Actin1 (multiplied by 1,000 for clarity). Shown are means of three replicates ± sd. The PR1 expression of rfc3-1 is significantly higher than that of the Col wild type, with or without SA treatment (P < 0.01, t test). The expression level of PR2 in rfc3-1 with 50 μm SA is significantly higher than that of the wild type under the same conditions (P < 0.01), but there is no significant difference in PR2 expression between wild-type and rfc3-1 plants without SA induction (P > 0.05). The experiment was repeated once with similar results. E, Growth of H. a. Noco2 on wild-type, rfc3-1, and sni1 plants. Two-week-old seedlings were sprayed with H. a. Noco2 at a concentration of 5,000 spores mL−1 water. The infection was scored 7 d after inoculation. The values presented are averages of four replicates ± sd. Statistical differences among the samples are labeled with different letters (P < 0.01, ANOVA). The experiment was repeated three times with similar results.
Mutant rfc3-1 plants are smaller than wild-type plants and have similar morphology to sni1 (Fig. 1B). Real-time PCR analysis showed that the endogenous PR-1 (Fig. 1C) and PR-2 (Fig. 1D) expression levels in rfc3-1 were slightly higher than in the wild type without SA induction, and the expression levels of PR-1 and PR-2 are much higher under SA induction. In addition, the rfc3-1 mutant plants are more resistant to the virulent oomycete pathogen H. a. Noco2 (Fig. 1E). Thus, like SNI1, RFC3 plays a negative regulatory role in PR gene expression and SAR. When RFC3 is mutated, plants exhibit enhanced PR gene expression and hypersensitivity to SA induction.
When rfc3-1 (with pBGL2-GUS) was backcrossed with a wild-type pBGL2-GUS line, the F1 progeny exhibited wild-type morphology, indicating that rfc3-1 is a recessive mutation. Among 46 F2 progeny, 10 showed rfc3-1-like pBGL2-GUS staining, suggesting that the defect in rfc3-1 is caused by a single recessive mutation (3:1, χ2 = 0.26, P > 0.1).
The rfc3-1 npr1 Double Mutant Exhibits Similar Enhanced Resistance Phenotypes as rfc3-1
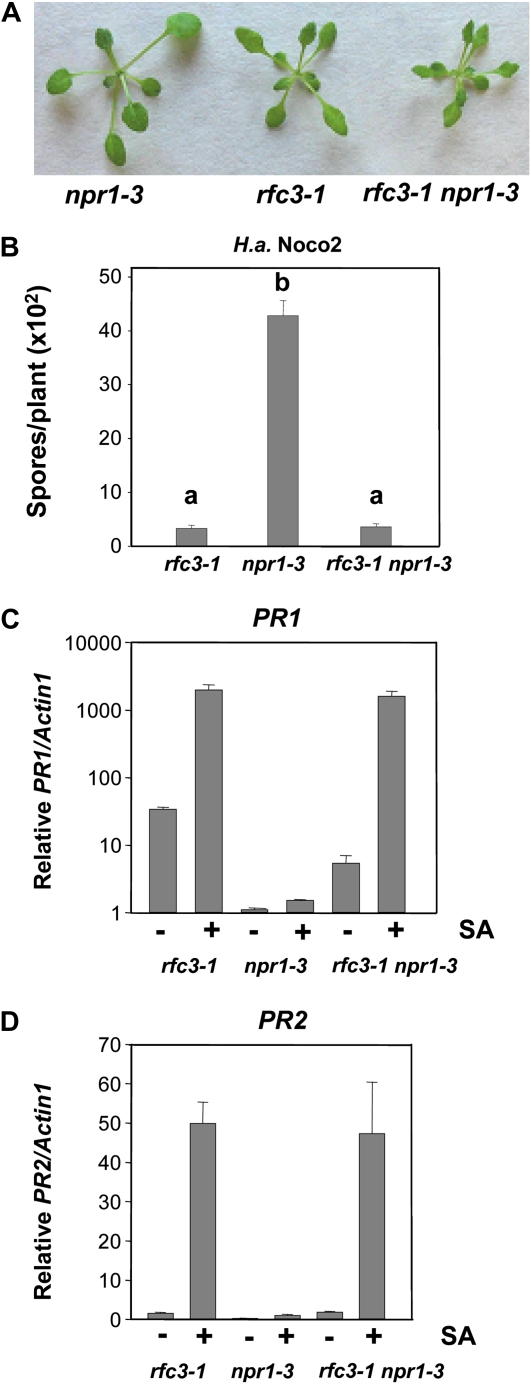
SNI1 is a repressor of PR gene expression, and the double mutant sni1 npr1 exhibits phenotypes similar to that of sni1 (Li et al., 1999). Since rfc3-1 has phenotypes very similar to sni1, we tested the relationship between RFC3 and NPR1 through generating a double rfc3-1 npr1-3 mutant. As shown in Figure 2A, the morphology of rfc3-1 npr1-3 is similar to that of the rfc3-1 single mutant. The rfc3-1 npr1-3 double mutant plants also exhibit enhanced resistance to H. a. Noco2 at the same level as rfc3-1 (Fig. 2B). Quantitative reverse transcription-PCR analysis showed that with or without SA induction, both the endogenous PR-1 (Fig. 2C) and PR-2 (Fig. 2D) expression levels in the rfc3-1 npr1-3 double mutant were more similar to those of the rfc3-1 single mutant. Thus, RFC3 probably also functions as a repressor to regulate PR gene expression. When RCF3 is mutated, the ability to express PR genes no longer requires the function of NPR1.
Figure 2.
Enhanced pathogen resistance and PR gene expression in the rfc3-1 npr1-3 double mutant. A, Morphology of npr1-3, rfc3-1, and rfc3-1 npr1-3 plants. All plants were grown on soil and photographed when they were 4 weeks old. B, Growth of H. a. Noco2 on npr1-3, rfc3-1, and rfc3-1 npr1-3 plants. The experiment was carried out as described in Figure 1E. Statistical differences among the samples are labeled with different letters (P < 0.01, ANOVA). The experiment was repeated three times with similar results. C and D, PR1 (C) and PR2 (D) expression in npr1-3, rfc3-1, and rfc3-1 npr1-3 plants with or without SA induction. The experiment was carried out as described in Figure 1C. PR1 and PR2 expression of rfc3-1 or rfc3-1 npr1-3 is significantly higher than that of npr1-3, with or without SA (P < 0.01, t test). There is no significant difference in PR1 or PR2 expression between rfc3-1 and rfc3-1 npr1-3 upon SA induction (P > 0.05). The experiments were repeated once with similar results. [See online article for color version of this figure.]
Map-Based Cloning of the rfc3-1 Mutant
To map the rfc3-1 mutation, rfc3-1 (in Col with pBGL2-GUS) was crossed with Landsberg erecta (Ler; with no pBGL2-GUS) to generate a segregating population. For crude mapping, 30 plants homozygous at the rfc3-1 locus were identified in the F2 progeny on the basis of rfc3-1 morphology. Linkage was found on the bottom of chromosome 1 between the markers T4O12 and T8K14. To avoid the possibility that the mutation causing the morphological phenotypes associated with rfc3-1 might be closely linked with the one causing the sni1-like defense phenotypes, lines that were homozygous for the pBGL2-GUS reporter gene and had wild-type morphology were genotyped with T4O12 and T8K14. Lines that are heterozygous at both markers should be heterozygous for rfc3-1, seeds of which were used for fine mapping. The presence of the homozygous pBGL2-GUS reporter gene enabled confident phenotyping of recombinants with GUS staining.
For fine mapping, 720 random F3 plants derived from F2 plants homozygous for the pBGL2-GUS reporter gene were genotyped with markers T4O12 and T8K14. A total of 53 recombinants between these two markers were identified. Further analysis of the 53 recombinants with additional markers in the region indicated that rfc3-1 was flanked by F2P24 and T5M16, with a 70-kb distance in between (Fig. 3A).
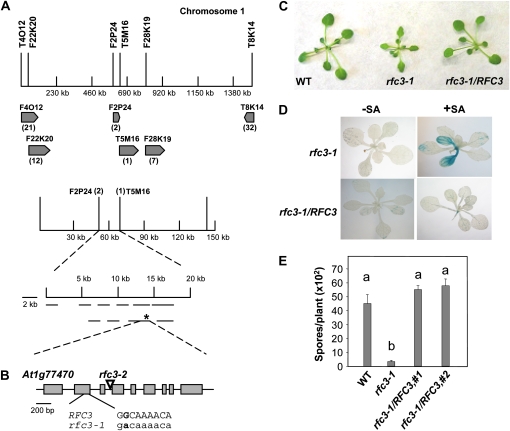
Figure 3.
Map-based cloning of rfc3-1. A, Map of the rfc3-1 locus on chromosome 1. Positions of the markers used for mapping are indicated. The two final flanking markers were F2P24 and T5M16. The rfc3-1 mutation is marked with an asterisk. B, Gene structure of RFC3. Exons are indicated with boxes, and introns are represented by lines. The region where the G-to-A mutation occurs in rfc3-1 is shown in detail. C, Morphology of the wild type (WT), rfc3-1, and rfc3-1 transformed with a genomic clone of RFC3 driven by its native promoter. Plants were grown on soil for 4 weeks before the photograph was taken. D, pBGL2-GUS reporter gene expression in rfc3-1 and rfc3-1 transformed with a genomic clone of RFC3 driven by its native promoter. Two-week-old seedlings grown on MS with or without 10 μm SA were stained for GUS activity as described previously (Bowling et al., 1994). E, Growth of H. a. Noco2 on the wild type, rfc3-1, and two independent transgenic rfc3-1 lines carrying genomic RFC3 driven by its native promoter. The experiment was carried out as described in Figure 1E. Statistical differences among the samples are labeled with different letters (P < 0.01, ANOVA). The experiment was repeated at least three times with similar results.
To identify the mutation in rfc3-1, primers were designed to sequence the coding regions of the genes between the final two flanking markers. A single G-to-A mutation was found in At1g77470. At1g77470 consists of nine exons. The mutation in rfc3-1 occurred in the second exon (Fig. 3B), changing the nonpolar aliphatic Gly-84 to a negatively charged Asp.
To confirm that the mutation found in rfc3-1 indeed causes the enhanced disease resistance phenotypes, we carried out complementation analysis using a genomic clone containing RFC3. As rfc3-1 homozygotes are partially sterile, the wild-type RFC3 clone was transformed into rfc3-1/RFC3 heterozygous plants. Transgenic lines homozygous for rfc3-1 were identified by PCR. As shown in Figure 3C, transgenic plants carrying RFC3 in the rfc3-1 background displayed wild-type morphology. Expression of the pBGL2-GUS reporter gene was also restored to wild-type levels (Fig. 3D). Furthermore, enhanced resistance against H. a. Noco2 in rfc3-1 was also lost in the transgenic plants (Fig. 3E). All of these data suggest that the wild-type RFC3 can complement the rfc3-1 mutation and that the G-to-A mutation found in rfc3-1 causes the SA hypersensitivity and enhanced resistance phenotypes in rfc3-1.
rfc3-1 Is a Partial Loss-of-Function Allele of RFC3
To determine whether the rfc3-1 mutation is a complete or partial loss-of-function mutation, we obtained an additional mutant allele of At1g77470 from the Arabidopsis Biological Resource Center. rfc3-2 (SAIL_401_E05) contains a T-DNA insertion in the third intron of At1g77470 (Fig. 3B). We could not identify any rfc3-2 homozygotes among 31 plants carrying the T-DNA insertion, indicating that the homozygotes of rfc3-2 are lethal. To test whether it is allelic to rfc3-1, a heterozygous rfc3-2/RFC3 plant was crossed with homozygous rfc3-1. The F1 plants with the rfc3-1/rfc3-2 genotype all had morphology similar to that of rfc3-1 (data not shown), indicating that rfc3-2 did not complement rfc3-1, and they carry mutations in the same gene. Since rfc3-2 is lethal whereas rfc3-1 is not, we deduced that the G-to-A mutation in rfc3-1 most likely is a partial loss-of-function mutation.
RFC3 Localizes to the Nucleus and Functions in Cell Replication and Proliferation
RFC3 encodes a protein of 369 amino acids with a molecular mass of 41.4 kD. It is highly similar to RFC3 in yeast and other eukaryotic species (Fig. 4). In eukaryotes, a RFC complex resides in the nucleus and contains one large subunit and four small subunits. RFC3 is one of the small subunits. There is only one copy of the RFC3 gene in the Arabidopsis genome. In most other eukaryotes, RFC3 also appears to be a single-copy gene. To determine the localization of RFC3, Arabidopsis mesophyll protoplasts were transfected with a construct expressing the RFC3-GFP fusion protein. Consistent with its localization in other organisms, the RFC3-GFP fusion protein localized to the nucleus (Fig. 5A), suggesting that RFC3 is a nuclear protein.
Figure 4.
Amino acid sequence alignment of AtRFC3 and its homologs in Oryza sativa (OsRFC3), Saccharomyces cerevisiae (ScRFC3), Drosophila melanogaster (DmRFC3), human (hRFC5), Mus musculus (MmRFC5), Xenopus laevis (XRFC5), and Danio rerio (DrRFC5). Identical amino acids are shaded in black, and similar amino acids are shaded in gray. Alignment was carried out using ebi ClustalW (http://www.ebi.ac.uk/clustalw/). Accession numbers are as follows: AtRFC3, NP_177871.1; OsRFC3, XM_468050.1; ScRFC3, NC_001146.3; DmRFC3, NM_135555.2; hRFC5, NM_007370.3; MmRFC5, Q9D0F6; XRFC5, BC072889.1; and DrRFC5, NM_001003862.1.
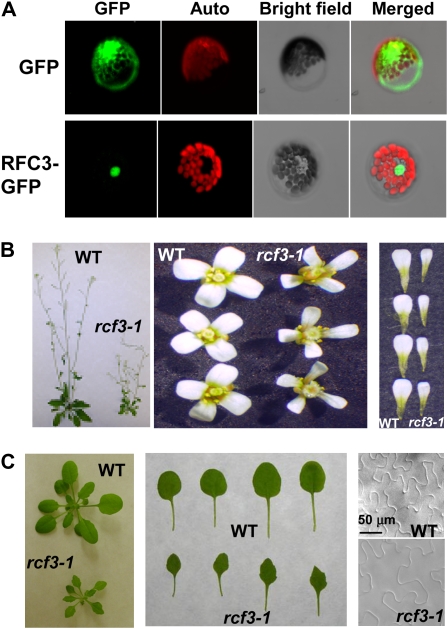
Figure 5.
Functional analysis of Arabidopsis RFC3. A, Localization of RFC3-GFP. Arabidopsis mesophyll protoplasts expressing the pG229-RFC3-GFP fusion proteins were examined by confocal microscopy, and the image shown was taken from a representative protoplast. The red fluorescence reflects chlorophyll autofluorescence. The nucleus is obvious from the bright-field image of the protoplast. pG229-GFP alone was used as a control. The experiment was repeated once with similar results. B, Morphology of mature plants, representative flowers, and petals of the wild type (WT) and rfc3-1. C, Morphology of young seedlings and leaves of the wild type and rfc3-1. The first to fourth true leaves of 20-d-old seedling were used to measure the epidermal cells with a microscope. The scale bar is the same for both images.
Since RFC3 encodes a putative replication factor, we tested whether the rfc3-1 mutant exhibits replication-related phenotypes. rfc3-1 plants are dwarfed and have smaller and narrower leaves compared with wild-type plants (Fig. 5B). The leaf blades of the mutant are almost half the size of wild-type blades (Fig. 5C). The flower petals are also smaller (Fig. 5B). To determine if the smaller leaves of the rfc3-1 mutant are caused by the smaller size of the cells, the epidermal cells of the true leaves were examined with a microscope. Surprisingly, the epidermal cells of the true leaves in rfc3-1 were much bigger than those of the corresponding wild-type true leaves (Fig. 5C). With larger cells and smaller leaves, the number of cells in rfc3-1 mutant leaves is likely lower than that of the wild type. Thus, smaller leaves of rfc3-1 are caused by reduced cell numbers rather than smaller sizes of the cells, indicating a crucial function of RFC3 in cell replication.
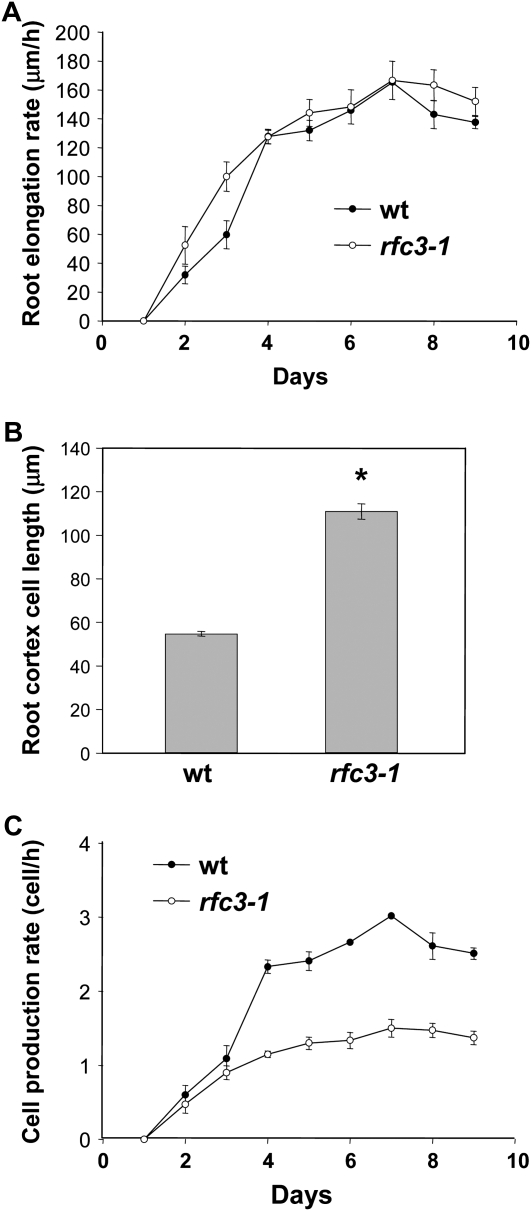
To determine whether the numbers of cells in other part of the plant are also reduced in the rfc3-1 mutant, root cortex cells of 10-d-old plants grown on MS plates were examined with the microscope. The root length of rfc3-1 mutant seedlings is slightly longer than that of the wild-type root (data not shown). The average root elongation rate of rfc3-1 is also higher than that of the wild-type root during the first 3 d, but from day 4, there is no significant difference between rfc3-1 and the wild-type plant (Fig. 6A). Further observation inside the root reveals that the root cortex cells in the root hair zone of rfc3-1 are significantly longer than those of the wild type, and quantitative data show that the length of cortex cells in rfc3-1 is twice the size of the wild-type cortex cells (Fig. 6B). As a result, the root cell production rate (number of cells produced per hour) of rfc3-1 is only half of that of the wild-type plants (Fig. 6C), indicating that mutation in RFC3 leads to defects in cell proliferation. Together with data from the leaf epidermal cells, our data suggest that the Arabidopsis RFC3 functions in the process of cell proliferation and replication and most likely is an ortholog of the yeast RFC3.
Figure 6.
Root growth and cell production in the wild type (wt) and the rfc3-1 mutant. A, Root elongation rate in wild-type and rfc3-1 seedlings. The values presented are averages of 30 replicates ± sd. Statistical analyses of root elongation rates of rfc3-1 compared with Col on each date revealed no statistically significant difference (P > 0.05, t test). B, Root cortex cell length in the root hair zone of wild-type and rfc3-1 plants. The values presented are averages of 50 replicates ± sd. The root cortex cell length of rfc3-1 is significantly longer than that of the Col wild type (P < 0.01, t test). C, Cell production rate (root elongation rate/root cortex cell length) of wild-type and rfc3-1 plants. Statistical analyses of cell production rates of rfc3-1 compared with Col on each date revealed statistically significant differences except for values at days 1, 2, and 3 (P < 0.01, t test).
DISCUSSION
Since constant defense is costly to plants without pathogen infection, tight negative control must be present to prevent the overactivation of the resistance mechanisms. SNI1 was identified earlier as a negative regulator of SAR (Li et al., 1999; Mosher et al., 2006). A mutation in SNI1 causes up-regulation of defense genes and hypersensitivity to the defense signal molecule SA. To identify more negative regulators of SAR like SNI1, a genetic screen was performed to search for mutants that are hypersensitive to SA induction. One recessive mutant, rfc3-1, was identified that displays similar morphology to sni1. rfc3-1 plants have higher PR gene expression than the wild type without SA induction, and under SA application they exhibit much higher sensitivity to the defense hormone. Mutation in SNI1 restored SAR in the npr1-1 background, and SNI1 was found to encode a Leu-rich nuclear transcriptional repressor protein (Li et al., 1999; Mosher et al., 2006). Similarly, rfc3-1 npr1 double mutant plants exhibit similar phenotypes as rfc3-1, suggesting that, like SNI1, RFC3 is a negative regulator of SAR and that this function of RFC3 does not require NPR1.
DNA replication is essential for all organisms with DNA genomes. RFC is a protein complex that can bind to a DNA template-primer junction and load the proliferating cell nuclear antigen clamp onto DNA with the assistance of ATP. This allows recruitment of DNA polymerase to the site of DNA synthesis. RFC plays essential roles in DNA replication and damage repair (for review, see Mossi and Hübscher, 1998). As a protein complex, RFC is conserved in all eukaryotes with one large subunit and four small subunits. AtRFC3 is highly homologous to the yeast RFC3 and the corresponding human RFC5 (Fig. 4). The Arabidopsis RFC3 was found to localize to the nucleus and is essential for plant survival, as a null mutant of RFC3 is lethal. Consistent with RFC3's function in replication, plants with the partial loss-of-function rfc3-1 allele exhibit smaller and narrower leaves due to the reduced number of cells, suggesting defects in replication. Moreover, the root cell production rate in rfc3-1 is only half that of wild-type plants (Fig. 6C), further indicating that rfc3-1 mutation slows down cell proliferation, most likely due to defects in replication.
One important question is how RFC3 regulates pathogen resistance in plants. Since the phenotypes of rfc3-1 are highly similar to those of snil, one possibility is that RFC3 may negatively regulate pathogen resistance through an interaction with SNI1. This interaction is probably not a direct protein-protein interaction, since we did not detect interactions between SNI1 and RFC3 in the yeast two-hybrid assays we performed (S. Xia and Y. Zhang, unpublished data). It has been suggested that SNI1 represses transcription through affecting chromatin modification (Mosher et al., 2006). Loss of SNI1 function leads to increased abundance of activating histone modifications such as acetylated histone H3 and methylated Lys-4 on histone H3 at the PR-1 promoter, which may make the chromatin at the promoter region adopt a more accessible conformation and lead to elevated gene expression (Mosher et al., 2006). During cell division, epigenetically defined chromatin structure is often propagated with high fidelity through replication-coupled chromatin assembly. Failure to transmit epigenetic modifications such as histone modifications and DNA methylations would lead to changes of gene expression patterns in the daughter cells. The rfc3-1 mutation probably causes defects in this process and leads to alterations of chromatin structure in the promoters of PR genes. In rfc3-1mutant plants, promoters of PR genes may adopt more accessible conformations, which result in elevated gene expression. We have performed rigorous chromatin immunoprecipitation experiments to test whether there is a detectable difference in TGA2 binding to the PR1 promoter between the wild type and rfc3-1. As expected, we were able to detect a clear difference between TGA binding to the PR1 promoter before and after INA induction. However, we did not observe a difference in binding between the wild type and rfc3-1 (data not shown). We believe that this could be due to the fact that the method we used is not sensitive enough to detect small differences in chromatin accessibility of the PR1 promoter between the wild type and rfc3-1 during defense.
The characterization and cloning of rfc3-1 suggest that RFC3 is required for DNA replication and cell proliferation in plants and probably contributes to chromatin assembly and remodeling that are required for negative regulation of PR gene expression and SAR. Further careful investigations into chromatin modifications in rfc3-1 may provide more detailed mechanistic insights in the future.
MATERIALS AND METHODS
Mutant Screen and Characterization
All Arabidopsis (Arabidopsis thaliana) plants were grown in a growth room under a 16-h-day (23°C)/8-h-night (21°C) regime. The pBGL2-GUS seeds (Bowling et al., 1994) were treated with ethylmethane sulfonate at a dose of 20 mm for 16 h. For the primary screen, individually harvested seeds from about 1,000 M1 plants were grown on MS medium supplemented with 10 μm INA. Half of the seedlings were tested for expression of the pBGL2-GUS reporter gene by GUS staining. Since the GUS staining procedure kills the seedlings, the remaining siblings of potential mutants were transplanted and their progeny were tested on MS with 10 μm SA for confirmation of SA hypersensitivity.
RNA used for gene expression analysis was extracted from 15-d-old plants grown on MS medium using the Totally RNA kit from Ambion. Reverse transcription was carried out using the Moloney murine leukemia virus reverse transcription kit (Takara). Real-time PCR was performed using the QuantiTect SYBR Green PCR kit from Qiagen. All data were normalized with Actin1. The primers used for amplification of Actin1, PR1, and PR2 were described previously (Zhang et al., 2003). Infection of wild-type and mutant plants with H. a. Noco2 was performed on 2-week-old seedlings as described previously (Li et al., 1999).
To obtain the rfc3-1 npr1-3 double mutant, an rfc3-1 homozygous plant was crossed with the npr1-3 mutant, and the specific PCR primers used to screen for the double mutant in the F2 are as follows: 5′-TATGGTCCTCCCGGTACTCA-3′ and 5′-TTGACGCCATATGCGTTGAC-3′ for rfc3-1; 5′-TATGGTCCTCCCGGTACTCG-3′ and 5′-TTGACGCCATATGCGTTGAC-3′ for wild-type RFC3; 5′-GACTCGGATGATATTGAGTTAG-3′ and 5′-TGTTCTCGTTTGTCTTCTGA-3′ for npr1-3; and 5′-GACTCGGATGATATTGAGTTAG-3′ and 5′-TGTTCTCGTTTGTCTTCTGG-3′ for wild-type NPR1.
Mapping of rfc3-1
To map the rfc3-1 mutation, rfc3-1 (in the Col background) was crossed with wild-type Ler. Crude mapping was performed on F2 plants homozygous for rfc3-1, and fine mapping was carried out on F3 plants derived from F2 plants that were heterozygous for rfc3-1 while carrying the homozygous pBGL2-GUS reporter gene. Both morphology and pBGL2-GUS reporter GUS staining of the progeny were used to confirm the phenotypes of the recombinants obtained.
The markers used for mapping were designed according to the Monsanto Arabidopsis polymorphism and Ler sequence collections (Jander et al., 2002). Marker T4O12 was amplified with primers 5′-CTGAAGAATCGAGCATTGCATC-3′ and 5′-CTGAAACAGACTGTTAGGCAAG-3′, and the Col fragment is 27 bp shorter than the Ler fragment. Marker F28K19 was amplified with primers 5′-CTTAATAAAGTTGGTTCAACCG-3′ and 5′-GTTGCCATTAGCAAGCTGTC-3′, and the Col fragment is 29 bp shorter than the Ler fragment. Marker F22K20 was amplified with primers 5′-ATGATCCGTGTGGAACTTAAC-3′ and 5′-CTTCACTCCAAGGACACAGC-3′, and the Col fragment is 17 bp shorter than the Ler fragment. Marker F2P24-2F2R was amplified with primers 5′-CAAGAAAGAAGTACATATTGGTC-3′ and 5′-AACTGGTCGCCAAACCTTGC3-′, and the Col fragment is 79 bp shorter than the Ler fragment. Marker T5M16-2 was amplified with primers 5′-CACCAAGTAATTACTTCCGAAG-3′ and 5′-TTAGTAGTGGCAATGCCACA-3′, and the Col fragment is 5 bp shorter than the Ler fragment. Marker T5M16-3 was amplified with primers 5′-GCTCGCTGACGTTGACCG-3′ and 5′-GTTATCTGTCAGACCCTACC-3′, and the Col fragment is 6 bp longer than the Ler fragment.
Transgene Complementation Analysis of rfc3-1
Since the RFC3 gene is relatively large, the full-length genomic sequence of RFC3 was divided into two fragments (fragments 1 and 2) for PCR amplification using Phusion high-fidelity PCR master mix (Finnzymes). The primer pairs used for amplifying fragments 1 and 2 were 5′-CGCGGATCCCGTCCTGCAAATGCTGATGA-3′ and 5′-CGGGAGCTCACCTATATGCTCACTGAAGG-3′ for fragment 1 and 5′-CGGGGTACCACATGGCTGGACCAGCAGAG-3′ with 5′-CGCGTCGACAGCTCACGCCCATCACAATG-3′ for fragment 2. The gel-purified PCR products were digested with BamHI and SacI (fragment 1) and KpnI and SalI (fragment 2). The digested fragments were ligated into pG229 (4.5 kp). The final constructs were confirmed by sequencing and transformed into Agrobacterium tumefaciens strain GV3101 together with the helper plasmid pSoup. Plants heterozygous with the rfc3/RFC3 genotype were transformed with Agrobacterium containing RFC3 using the floral dip method (Clough and Bent, 1998). The transformants were selected by the herbicide Basta. Transformants homozygous for rfc3-1 were identified by PCR.
Subcellular Localization of RFC3
To fuse RFC3 to the GFP gene, full-length RFC3 cDNA without the stop codon was amplified by PCR and cloned into the pG229-GFP vector. The resulting plasmid was sequenced to confirm that the fusion gene was in frame without PCR errors. For transient expression of the RFC3-GFP fusion proteins, pG229-RFC3-GFP was transfected into Arabidopsis mesophyll protoplasts according to a previously described protocol (Sheen, 2001). Green fluorescence was observed using confocal microscopy. pG229-GFP was used as a control.
Leaf Epidermal Cell and Root Cortex Cell Examination
Plant parts were photographed with a Leica dissecting microscope with a Canon Powershot s70 digital camera. The first to fourth true leaves of 21-d-old plants of the rfc3-1 mutant and the wild type were collected separately in Eppendorf tubes and treated with a chloral hydrate:glycerol:water solution (8:1:2) to clear the cells (Ohad et al., 1996). The epidermal cells on both the abaxial and adaxial leaf surfaces were photographed with a Zeiss Axiovert 200M microscope with an Axiocam HRm digital camera that permits visualization of optical sections within the leaf.
To examine cortex cells, seeds of rfc3-1 and the wild type were sown on the same MS plate according to Chen et al. (2003). After germination, the length of each root was measured and marked every 24 h. The roots of 10-d-old seedlings were harvested separately in Eppendorf tubes and treated with a chloral hydrate:glycerol:water solution (8:1:2), and the root cortex cells were examined with a Zeiss Axiovert 200M microscope.
Acknowledgments
We thank Jacquiline Monaghan for careful reading of the manuscript, Dongling Bi and Minghui Gao for assistance in confocal analysis of the transiently expressed RFC3-GFP in Arabidopsis mesophyll protoplasts, and Dr. Jane Parker for H. arabidopsidis strains.
This work was supported by the Natural Sciences Foundation of China (grant to Y.Z.) and the Natural Sciences and Engineering Research Council of Canada (grant to X.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xin Li (xinli@interchange.ubc.ca).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301 1728–1731 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong X (2007) Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci USA 104 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, et al (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10 71–82 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Clarke JD, Li Y, Dong X (1999) Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98 329–339 [DOI] [PubMed] [Google Scholar]
- Métraux JP, Ahl-Goy P, Staub T, Speich J, Steinemann A, Ryals J, Ward E (1991) Induced resistance in cucumber in response to 2,6-dichloroisonicotinic acid and pathogens. In H Hennecke, DPS Verma, eds, Advances in Molecular Genetics of Plant-Microbe Interactions, Vol 1. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 432–439
- Mosher RA, Durrant WE, Wang D, Song J, Dong X (2006) A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossi R, Hübscher U (1998) Clamping down on clamps and clamp loaders: the eukaryotic replication factor C. Eur J Biochem 254 209–216 [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu YC, Williams C, Repetti P, Fischer RL (1996) A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA 93 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF (1961) Systemic acquired resistance induced by localized virus infections in plants. Virology 14 340–358 [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RF (1979) Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99 410–412 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tessaro MJ, Lassner M, Li X (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]