Plants constantly face a plethora of abiotic and biotic stresses in their natural habitat. Adapting to such changes requires a great degree of phenotypic plasticity that is mainly determined by the plant's genome. We currently do not know how plants are able to integrate the multitude of partly synergistic/partly antagonistic environmental signals that enable them to respond properly under any given condition. What has become apparent, however, is that plants are capable of extensive reprogramming of their transcriptome in a highly dynamic and temporal manner. This regulation in response, leading to adaptive plasticity of plants in highly variable environments, is mainly achieved by enforcement of a network of various transcription factors (TFs). WRKY TFs are a large family of regulatory proteins forming such a network (Eulgem and Somssich, 2007). They are involved in various plant processes but most notably in coping with diverse biotic and abiotic stresses. In this update, we will restrict our attention to the role of WRKY TFs in plant immunity.
THE WRKY FACTORS
The WRKY TF superfamily consists of 74 and 109 members in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), respectively (Eulgem and Somssich, 2007; Ross et al., 2007). Members of this family contain at least one conserved DNA-binding region, designated the WRKY domain, comprising the highly conserved WRKYGQK peptide sequence and a zinc finger motif (CX4–7CX22–23HXH/C). This domain generally binds to the DNA element termed the W box (C/TTGACT/C), although alternative binding sites have been identified (Sun et al., 2003; Cai et al., 2008; Ciolkowski et al., 2008; van Verk et al., 2008). WRKY family members are divided into three groups based on the number of WRKY domains and certain features of the zinc finger-like motifs (Eulgem et al., 2000). The NMR solution structure revealed that the C-terminal WRKY domain of Arabidopsis WRKY4 consists of a four-stranded β-sheet, with a zinc-binding pocket formed by the conserved Cys/His residues located at one end of the β-sheet, and the WRKYGQK residues, corresponding to the most N-terminal β-strand (strand β-1), kinked in the middle of the sequence by the Gly residue (Yamasaki et al., 2005). The concave curvature of strand β-1 induced by this kink is predicted to enable this strand to deeply enter the DNA groove and make contact with bases of the W box element. The crystal structure of the extended WRKY domain of Arabidopsis WRKY1 (AtWRKY1-C) revealed that this domain is composed of a globular structure with five β-strands forming an antiparallel β-sheet with an additional novel zinc-binding site at one end (Duan et al., 2007). One should note, however, that no crystal structure information exists of a WRKY domain associated with its DNA-binding site or for a full-length WRKY protein.
WRKY factors were generally regarded as being plant specific, but their identification in the protist Giardia lamblia and the slime mold Dictyostelium discoideum imply an earlier origin (Ülker and Somssich, 2004; Pan et al., 2009). They may have evolutionary links with transposons such as Mutator-like elements and could have originated from a BED finger intermediate (an atypical zinc finger DNA-binding domain found both in cellular chromatin boundary element-binding proteins BEAF and DREF and in transposases from animals), although this is controversially debated (Babu et al., 2006; Yamasaki et al., 2008). Duplicated WRKY genes have been maintained in wild and cultivated plant species in the course of selection during domestication and polyploidization (Petitot et al., 2008). They have recently been associated with the viability of interploidy hybrids (Dilkes et al., 2008). Phylogenetic sequence analysis and comparative transcriptomics have revealed that they have retained their functions between monocots and dicots (Mangelsen et al., 2008). The majority of the analyzed WRKY genes respond to pathogen attack and to the endogenous signal molecule salicylic acid (SA; Eulgem and Somssich, 2007).
WRKY FACTORS IN DISEASE RESISTANCE NETWORKS
Plant innate immunity is composed of two interconnected branches: (1) PTI, or pathogen-associated molecular pattern (PAMP)-triggered immunity, which is initiated by the recognition of molecular signatures of many pathogens and often activates downstream mitogen-activated protein (MAP) kinase cascades and defense genes; and (2) ETI, or effector-triggered immunity, driven by plant disease resistance proteins (major R gene products) that recognize directly or indirectly specific pathogen-derived effectors (Chisholm et al., 2006). PTI and ETI activate local as well as systemic defense responses (called systemic acquired resistance [SAR]), which are modulated by phytohormones, especially jasmonic acid (JA) and SA (Durrant and Dong, 2004; Bostock, 2005). JA-dependent plant defenses are generally activated by necrotrophic pathogens and chewing insects, whereas SA-dependent defenses are often triggered by biotrophic pathogens. JA and SA signaling usually act antagonistically, but synergism between these two phytohormones has also been observed (Mur et al., 2006). These responses to pathogen attack require large-scale transcriptional reprogramming, including those of TF families such as WRKY genes (Eulgem, 2005; Ryu et al., 2006; Naoumkina et al., 2008).
WRKY TFs in the Arabidopsis World
Loss-of-function and gain-of-function studies in Arabidopsis have been pivotal in demonstrating that WRKY factors act in a complex defense response network as both positive and negative regulators (Eulgem and Somssich, 2007). AtWRKY52 (also designated RRS1) is a novel protein comprising structural features of nucleotide binding-Leu-rich repeat-type R gene products and a WRKY domain that confers wide-ranging resistance toward the bacterial wilt Ralstonia solanacearum (Deslandes et al., 2002). The discovery that AtWRKY52 physically interacts with its cognate bacterial effector PopP2 within the plant cell nucleus (Deslandes et al., 2003) helped to stimulate subsequent research clearly demonstrating the importance of nuclear trafficking for plant immunity (Caplan et al., 2008; Liu and Coaker, 2008). AtWRKY70 acts at a convergence point determining the balance between SA- and JA-dependent defense pathways as well as being required for R gene-mediated resistance (Li et al., 2006; Knoth et al., 2007). The indispensability of AtWRKY70 for JA and SA signaling, however, has recently been questioned (Ren et al., 2008).
Similarly, AtWRKY33 functions as a positive regulator of resistance toward the necrotrophic fungi Alternaria brassicicola and Botrytis cinerea (Zheng et al., 2006), and AtWRKY53 and AtWRKY70 both positively modulate SAR (Wang et al., 2006). Moreover, SA biosynthesis and expression of NONEXPRESSOR OF PR1 (NPR1), a key central regulator of SA-dependent defenses and SAR, also appear to be regulated by WRKY TFs (Yu et al., 2001). Two closely related WRKY TFs, AtWRKY3 and AtWRKY4, play a positive role in plant resistance toward necrotrophic pathogens, as Atwrky4, Atwrky3, and Atwrky3 wrky4 mutants showed increasing susceptibility toward the fungus B. cinerea, whereas overexpression of AtWRKY4 enhanced susceptibility toward the biotrophic bacterium Pseudomonas syringae (Lai et al., 2008).
Many WRKY TFs act as negative regulators of defense signaling, including AtWRKY7, -11, -17, -18, -23, -25, -27, -38, -40, -41, -48, -53, -58, -60, and -62. Showing functional redundancy, Atwrky7 along with Atwrky11 and Atwrky17 mutants were susceptible to virulent P. syringae (Journot-Catalino et al., 2006; Kim et al., 2006). Similarly, AtWRKY38 and AtWRKY62 also contribute negatively to basal resistance toward this bacterial pathogen (Kim et al., 2008). AtWRKY62 expression is induced by SA and JA in a NPR1-dependent manner. How AtWRKY62 alters JA/SA signaling remains unclear, since one study has shown that loss of AtWRKY62 function resulted in enhanced expression of JA-response genes, whereas AtWRKY62 overexpressor lines inhibited JA-response gene expression (Mao et al., 2007), while in a second study, elevated transcript levels of the SA-response gene PR1 were observed in the Atwrky62 mutant, whereas WRKY62 overexpression led to suppression of PR1 (Kim et al., 2008). AtWRKY48 also negatively influences basal resistance toward virulent P. syringae (Xing et al., 2008). Reduced bacterial growth in Atwrky48 mutants was associated with increased induction of PR1, whereas AtWRKY48 overexpressors showed the opposite phenotypes. AtWRKY58 acts downstream of NPR1, negatively regulating SAR (Wang et al., 2006). Recently, knockdown of AtWRKY23 expression was shown to decrease susceptibility toward the parasitic cyst nematode Heterodera schachtii (Grunewald et al., 2008). Mutation in AtWRKY27 resulted in delayed symptom development against R. solanacearum, possibly by affecting nitric oxide signaling, and vascular trafficking (Mukhtar et al., 2008).
The closely related WRKY TFs AtWRKY18, -40, and -60 have partly redundant functions in negatively regulating resistance to P. syringae (Xu et al., 2006). Interestingly, Atwrky18 wrky40 double mutants also displayed enhanced resistance to the powdery mildew pathogen Golovonomyces orontii (Shen et al., 2007). In contrast, Atwrky18 wrky40 and Atwrky18 wrky60 double mutants were more susceptible to B. cinerea (Xu et al., 2006), and AtWRKY18 alone appears also to have positive regulatory functions in SAR (Wang et al., 2006). Dual functionality in defense signaling was also observed for AtWRKY53. While Atwrky53 mutants showed delayed symptom development against R. solanacearum, such plants displayed increased susceptibility toward P. syringae (Murray et al., 2007; Hu et al., 2008). Dual functionality was also suggested for AtWRKY41. Arabidopsis plants overexpressing AtWRKY41 showed enhanced resistance toward virulent Pseudomonas but decreased resistance toward Erwinia carotovora (Higashi et al., 2008). However, Atwrky41 mutants did not display a differential phenotype. Intriguingly, expression of AtWRKY41 is specifically suppressed by a compatible strain of P. syringae in an effector-dependent manner. Finally, overexpression of AtWRKY25 resulted in increased disease symptoms to P. syringae infections, possibly by negatively regulating SA-mediated defense responses. However, Atwrky25 mutants supported normal growth of a virulent P. syringae strain (Zheng et al., 2007). Thus, as in the case for AtWRKY41, the in vivo relevance of such findings remains to be critically assessed.
Recent Developments in Rice
An increasing number of studies in other plants, particularly in rice, have strongly confirmed the importance of WRKY TFs in plant defense signaling. The rice genome contains more than 100 WRKY genes, often present in duplicated chromosomal regions, suggesting genome duplications as one of the mechanisms for the expansion of this family in this plant species (Ross et al., 2007; Ramamoorthy et al., 2008). The majority of these genes respond to (a)biotic stresses and various phytohormones (Ryu et al., 2006; Ramamoorthy et al., 2008). Individual WRKY members have been associated with pathogen defense, albeit with the caveat that the majority of such studies have employed strong ecotopic overexpressor lines. For example, overexpression of OsWRKY13 enhances resistance to the bacterial blight Xanthomonas oryzae pv oryzae (Xoo) and the fungal blast Magnaportha grisea. It exerts its function by activating SA-biosynthesis and SA-response genes while suppressing JA signaling (Qiu et al., 2007, 2008a). Similarly, OsWRKY53 overexpressor lines are more resistant to M. grisea and may act as a positive regulator of basal defense (Chujo et al., 2007). Expression of OsWRKY03 and OsWRKY71 is strongly induced by pathogen-mimicking stimuli, and these genes function upstream of OsNH1 (the rice ortholog of NPR1) in defense signaling (Liu et al., 2005, 2007). In the case of OsWRKY71, overexpressor lines display enhanced resistance to virulent Xoo (Liu et al., 2007). Ectopic expression of OsWRKY31 resulted in enhanced resistance to fungal blast, altered lateral root formation, and constitutive expression of two early auxin-response genes (Zhang et al., 2008a). Whether these two phenotypes are functionally linked remains to be determined. Moreover, corresponding OsWRKY31 RNA interference lines showed no altered disease phenotype. Enhanced resistance to M. grisea was observed with OsWRKY45 overexpressor lines but not with plants overexpressing OsWRKY19, -62, and -76 (Shimono et al., 2007). In this case, OsWRKY45 knockdown lines decreased resistance to this fungal blast. OsWRKY45 appears to act in SA signaling independent of NH1. Notably, ecotopic expression of OsWRKY45 in Arabidopsis resulted in plants with enhanced resistance to virulent P. syringae, increased PR1 expression, elevated tolerance to salt and drought stress, but decreased sensitivity toward abscisic acid signaling (Qiu and Yu, 2009). OsWRKY89 overexpression seems to positively contribute to resistance against fungal blast and the white-backed plant hopper Sogatella furcifera by regulating the wax content/deposition on the leaf surface. OsWRKY89 knockdown lines showed reduced wax content and increased susceptibility to M. grisea (Wang et al., 2007). Finally, OsWRKY62 was recently shown to be a negative regulator of both PTI and ETI. The rice gene Xa21 confers race-specific resistance to Xoo. Xa21 was shown to bind to OsWRKY62, and overexpression of one splice variant, OsWRKY62-1, compromised basal defense and Xa21-mediated resistance to Xoo and suppressed defense gene activation (Peng et al., 2008).
WRKY TFs in Other Plant Species
The number of WRKY genes identified in other recently sequenced plant genomes are 66 in papaya (Carica papaya), 104 in poplar (Populus spp.), 68 in sorghum (Sorghum bicolor), and 38 in the moss Physcomitrella patens. Currently, no data exist on the role of these factors in mediating plant immunity. Some isolated studies in other plant species, however, have been reported. Overexpression of grapevine (Vitis vinifera) VvWRKY1 in tobacco (Nicotiana tabacum) rendered plants susceptible toward a variety of fungi (Marchive et al., 2007), whereas ectopic expression of grapevine VvWRKY2 resulted in enhanced resistance to the necrotrophic fungi Alternaria tenuis, B. cinerea, and Pythium (Mzid et al., 2007). Similarly, CaWRKY1 from chili pepper (Capsicum annuum) appears to act as a negative regulator of defense, as virus-induced gene silencing of this gene decreased growth of Xanthomonas, whereas its overexpression resulted in enhanced hypersensitive cell death to P. syringae and Tobacco mosaic virus (Oh et al., 2008). In barley (Hordeum vulgare), MLA confers isolate-specific resistance to the powdery mildew Blumeria graminis. MLA was shown to physically interact in the nucleus with HvWRKY1 and -2, two repressors of PAMP-triggered basal defense, thereby interfering with WRKY repressor functions and leading to resistance against the powdery mildew fungus (Shen et al., 2007). In the native tobacco Nicotiana attenuata, two WRKY genes, NaWRKY3 and -6, were identified that coordinate JA-mediated defense responses to native herbivory. Silencing of NaWRKY3, NaWRKY6, or both rendered plants highly vulnerable to Manduca sexta attack (Skibbe et al., 2008). Finally, elicitor-triggered reprogramming of secondary metabolites in Medicago truncatula seems to involve several WRKY factors: overexpression of four WRKY genes in tobacco demonstrated their regulatory roles in lignin deposition, PR gene expression, and systemic defense responses against Tobacco mosaic virus (Naoumkina et al., 2008).
Overall, these findings highlight the importance of WRKY factors in transcriptionally reprogramming plant responses toward different invading pathogens (Supplemental Table S1). While some appear to positively influence the outcome of such plant-pathogen interactions, others actually appear to negatively influence it. This negative influence may be due to active targeting of the WRKY genes/factors, or products under their control, by certain pathogens. Manipulation of WRKY proteins by pathogen effectors may partly explain the existence of redundancy within the WRKY TF family as a reinforcement measure for essential regulatory functions. Coordinated modulation of positive- and negative-acting factors could also enable the proper amplitude and duration of the plant response during pathogen attack. Some key questions that need to be addressed in future WRKY research are as follows. (1) How are the WRKY genes themselves regulated? (2) With which cellular/nuclear components do they interact during defense signaling and during recruitment at specific target gene sites? (3) What are the exact targets of individual WRKY factors within the genome?
WHAT REGULATES THE WRKY NETWORK?
The last decade of research has clearly revealed that WRKY factors form a complex and highly interconnected regulatory network (Eulgem and Somssich, 2007). Such a network needs to be controlled at several levels.
Auto/Cross-Regulation by WRKY Genes
The majority of the Arabidopsis WRKY genes are themselves responsive to pathogenic stimuli and many contain numerous W box elements within their promoters (Eulgem and Somssich, 2007). This suggests that several WRKY genes are under direct positive or negative control by WRKY factors via specific feedback mechanisms (auto/cross-regulation). Studies in parsley (Petroselinum crispum) protoplast showed that a specific arrangement of W boxes within the promoter of PcWRKY1 determines its temporal expression upon PAMP treatment (Eulgem et al., 1999). Moreover, chromatin immunoprecipitation (ChIP) analysis confirmed PAMP-dependent in vivo binding of PcWRKY1 to its own promoter as well as to the defense-response gene PcPR10 (Turck et al., 2004). Additional cotransfection experiments have substantiated such a mode of regulation (Eulgem and Somssich, 2007; Lippok et al., 2007). Upon herbivore attack, NaWRKY6 transcript accumulation was shown to be dependent on NaWRKY3 expression (Skibbe et al., 2008). Moreover, physical interaction of related WRKY TFs may also be necessary for their efficient function, as evidenced by homodimer and heterodimer complex formation of Arabidopsis WRKY18, -40, and -60 in response to P. syringae (Xu et al., 2006).
Regulation via Other TFs and Proteins
Six distinct proteins, including OsWRKY13, were identified in a yeast one-hybrid screen that bind to functionally important cis-regulatory DNA elements within the rice OsWRKY13 promoter (Cai et al., 2008). Similar screens employing the AtWRKY53 promoter led to the identification of a MAP kinase kinase kinase (MEKK1). Interestingly, MEKK1 was also shown to interact with and to phosphorylate AtWRKY53 (Miao et al., 2007). The in vivo relevance of these interactions with respect to plant defense, however, remains to be tested. In Arabidopsis, expression of the key defense regulator NPR1 is controlled by unknown WRKY TFs (Yu et al., 2001). NPR1 does not bind DNA on its own but associates with TGA TFs to modulate SA-dependent genes and SAR (Durrant and Dong, 2004). Expression of at least nine WRKY genes, AtWRKY18, -38, -53, -54, -58, -59, -62, -66, and -70, is dependent on NPR1, suggesting that they may be under TGA factor control (Wang et al., 2006; Mao et al., 2007). In the case of AtWRKY51, ChIP and whole-genome arrays identified its promoter to be targeted by TGA2 in an SA-dependent manner (Thibaud-Nissen et al., 2006).
PTI involves tightly regulated MAP kinase signaling cascades. The D motif within several WRKY TFs contains consensus phosphorylation sites for MAP kinases, and several WRKY TFs have been shown to be phosphorylated in vitro (Kim and Zhang, 2004; Menke et al., 2005; Eulgem and Somssich, 2007; Popescu et al., 2009). Recently, the association of MAP Kinase4 (MPK4) with AtWRKY33 and a coupling factor, MKS1, within the plant cell nucleus was demonstrated (Qiu et al., 2008b). Upon virulent P. syringae infection, MPK4 is phosphorylated, thereby releasing MKS1 and WRKY33 and thus allowing recruitment of WRKY33 to target promoters.
Chromatin structure can locally and globally regulate gene expression. Interestingly, AtWRKY38 and -62 were found to interact with Histone Deacetylase19 (HDA19), a chromatin-remodeling factor that contributes to global transcriptional repression (Kim et al., 2008). Overexpression of HDA19 enhanced resistance to P. syringae, whereas the hda19 mutant was compromised in resistance. These are the opposite phenotypes obtained from similar studies with AtWRKY38 and -62, revealing yet another level of WRKY network regulation in fine-tuning the plant basal defense response (Kim et al., 2008).
The Small RNA-WRKY Interactome
Small RNAs (smRNAs) have emerged as a fundamental layer of regulation of gene expression. Plant smRNAs are broadly classified into micro RNAs (miRNAs) and small interfering RNAs (siRNAs). miRNAs are approximately 21 nucleotides and derived from the precursor-stem-loop structures encoded by distinguished miRNA genes (Voinnet, 2009); siRNAs are derived from double-stranded RNAs, in an RNA-directed RNA polymerase-dependent manner, and may be further classified as trans-acting siRNAs, repeat-associated siRNAs, and natural antisense transcript-derived siRNAs. High-throughput sequencing of the smRNA portion of the transcriptome revealed that a multitude of smRNAs accumulate in plants (Lu et al., 2005; Kasschau et al., 2007; Pandey et al., 2008). These 18- to 40-nucleotide-long smRNAs regulate gene expression posttranscriptionally in a process often called RNA interference, RNA silencing, or posttranscriptional gene silencing.
The importance of smRNAs in plant processes related to adaptation to (a)biotic stresses is increasingly becoming evident, and the endogenous plant-derived smRNAs probably have broad implications in posttranscriptionally regulating plant responses to pathogen attack (Navarro et al., 2006; Pandey and Baldwin, 2007; Voinnet, 2008). Phytohormone treatments induced the expression of several miRNAs in rice (Liu et al., 2009). Predicted targets for several miRNAs encode WRKY factors (Zhang et al., 2008b; S.P. Pandey and I.T. Baldwin, unpublished data), suggesting smRNA-mediated regulation of WRKY TFs. Conversely, several miRNA gene promoters are highly abundant in W box sequences, implicating WRKY TFs in their activation/repression (Zhou et al., 2008). Further evidence of a WRKY-smRNA interactome comes from our studies on AtWRKY18 and -40 in modulating responses to powdery mildew (S.P. Pandey, M. Roccaro, E. Logemann, and I.E. Somssich, unpublished data). Atwrky18 wrky40 double mutants are resistant to powdery mildew infection and strongly up-regulate the expression of SIMILAR TO RCD ONE5 (SRO5) upon infection, suggesting WRKY-dependent suppression of siRNA-generating loci. SRO5 along with PYRROLINE-5-CARBOXYLATE DEHYDROGENASE (an overlapping gene in the antisense orientation) generate 24- and 21-nucleotide siRNAs, which together are components of a regulatory loop controlling reactive oxygen species production and stress response (Borsani et al., 2005). Similar suppression of the host miRNA machinery by bacterially derived effector proteins has recently been demonstrated in Arabidopsis (Navarro et al., 2008).
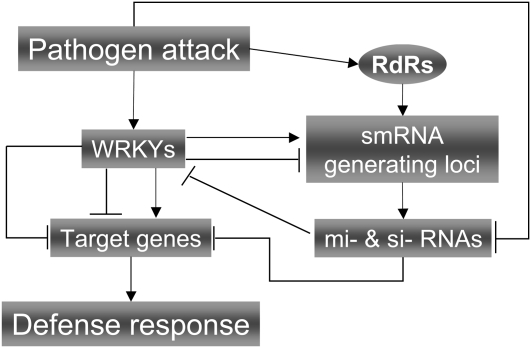
The current data point toward the existence of a WRKY-smRNA interactome, where on the one hand, pathogen attack triggers the expression of WRKY genes that regulate cellular smRNA populations, and on the other hand, several differentially regulated smRNAs modulate WRKY TF levels by targeting their transcripts (Fig. 1). This model certainly warrants further investigation.
Figure 1.
Modeling the WRKY-smRNA interactome during reprogramming of defense responses. During pathogen attack, smRNA-generating loci may be under the control of WRKY TFs; at the same time, WRKY abundance may be regulated by smRNAs. RdR, RNA-directed RNA polymerase.
PATHOGEN-DEPENDENT IN VIVO WRKY TF TARGETS IN THE POSTGENOMIC ERA
As with other large TF families, identification of all in vivo downstream targets of specific WRKY TFs is a highly challenging endeavor. Sequenced genomes reveal a widespread distribution of W box-like elements, but the biological relevance of these potential WRKY-binding sites remains unclear. Earlier target identification was limited to selected candidates on a gene-for-gene basis and rested mostly on ectopic expression of the respective WRKY gene in transient cotransfection assays. Development of the ChIP technology was a major step forward, allowing DNA-protein and protein-protein interactions to be studied under in vivo conditions (Massie and Mills, 2008). ChIP studies in parsley identified two PcWRKY1 target genes activated upon PAMP treatment (Turck et al., 2004). Similarly, PAD3, a gene encoding a key enzyme of camalexin biosynthesis, was detected as a direct target of AtWRKY33 following pathogen infection (Qiu et al., 2008b). Recently, using information derived from whole-genome microarrays followed by ChIP analyses, we identified two key regulators of plant defense as being direct targets of AtWRKY40 during powdery mildew infection (S.P. Pandey, M. Roccaro, E. Logemann, and I.E. Somssich, unpublished data).
A major limitation of previous studies was that the number of target genes that could be assayed was restricted. Recent developments expanding the use of ChIP-enriched DNA for hybridization to genomic microarrays (ChIP-chip) or for direct sequencing (ChIP-Seq) using second-generation high-throughput sequencing technology are opening the door to identify WRKY TF binding sites on a global level (Massie and Mills, 2008). Nevertheless, despite such progress, the task remains daunting both technically, starting with the quality of various specific antibodies and proper evaluation of the gigabits of sequencing information obtained, and because such in vivo interactions can be highly dynamic in both temporal and spatial terms.
CONCLUSION
WRKY TFs are indeed global regulators of host responses following challenge by phytopathogenic organisms. They participate in regulating defense gene expression at various levels, partly by directly modulating immediate downstream target genes, by activating or repressing other TF genes, and by regulating WRKY genes by means of feed-forward and feedback regulatory loops. Moreover, they also appear to interact with key chromatin-remodeling factors, thereby adding another layer of complexity to the WRKY network. WRKY factors can associate with MAP kinases in the nucleus, and MAP kinase cascades constitute key components of plant defense signaling. In yeast, the majority of terminal MAP kinases appear to be within the nucleus, associated with transcriptional complexes at target genes (Pokholok et al., 2006). Hence, one can expect that future studies will reveal additional nuclear functions of such WRKY-MAP kinase associations involving chromatin remodeling at target DNA sites. In addition, the involvement of WRKY TFs in modulating the expression of several miRNAs while at the same time their transcription is possibly partly under smRNA surveillance adds yet another dimension to the regulatory complexity that must be sorted out. Nevertheless, to fully understand regulation, we need to gain access to the full set of proteins associated with WRKY TFs at specific genomic loci. Indeed, promising technological advances combining DNA probes and mass spectrometry, such as proteomics of isolated chromatin segments and stable isotope labeling with amino acids, are starting to demonstrate that identification of TFs and associated proteins in vivo at given promoters may become feasible in the near future (Dèjardin and Kingston, 2009; Mittler et al., 2009).
The WRKY transcriptional network may provide the proper balance to respond quickly and efficiently to deter pathogens but at the same time to restrict defense responses that can be detrimental for plant growth and development. Elucidation of how WRKY TFs help to exert these functions will certainly be assisted in the near future by the ability to monitor specific WRKY TF interactions with DNA/chromatin on a global basis. This will allow us to construct testable hypotheses regarding how WRKY factors can influence diverse metabolic pathways and overall cellular physiology. At the same time, they will also provide us with valuable information on where and how coevolving pathogens impinge on this vast network to counteract host defenses and/or make use of it for their specific advantages.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Plant WRKY genes implicated in plant immunity.
Supplementary Material
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Imre E. Somssich (somssich@mpiz-koeln.mpg.de).
The online version of this article contains Web-only data.
References
- Babu MM, Iyer LM, Balaji S, Aravind L (2006) The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res 34 6505–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock RM (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43 545–580 [DOI] [PubMed] [Google Scholar]
- Cai M, Qiu D, Yuan T, Ding X, Li H, Duan L, Xu C, Li X, Wang S (2008) Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ 31 86–96 [DOI] [PubMed] [Google Scholar]
- Caplan J, Padmanabhan M, Dinesh-Kumar SP (2008) Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3 126–135 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Chujo T, Takai R, Akimoto-Tomiyama C, Ando S, Minami E, Nagamura Y, Kaku H, Shibuya N, Yasuda M, Nakashita H, et al (2007) Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim Biophys Acta 1769 497–505 [DOI] [PubMed] [Google Scholar]
- Ciolkowski I, Wanke D, Birkenbihl R, Somssich I (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol 68 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dèjardin J, Kingston RE (2009) Purification of proteins associated with specific genomic loci. Cell 136 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulières T, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA 99 2404–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilkes BP, Spielman M, Weizbauer R, Watson B, Burkart-Waco D, Scott RJ, Comai L (2008) The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol 6 e308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan MR, Nan J, Liang YH, Mao P, Lu L, Li L, Wei C, Lai L, Li Y, Su XD (2007) DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res 35 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10 71–78 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE (1999) Early nuclear events in plant defense: rapid gene activation by WRKY transcription factors. EMBO J 18 4689–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10 366–371 [DOI] [PubMed] [Google Scholar]
- Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, Inze D, Beeckman T, Gheysen G (2008) A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol 148 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi K, Ishiga Y, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y (2008) Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol Genet Genomics 279 303–312 [DOI] [PubMed] [Google Scholar]
- Hu J, Barlet X, Deslandes L, Hirsch J, Feng DX, Somssich I, Marco Y (2008) Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil-borne phytopathogenic bacterium, Ralstonia solanacearum. PLoS One 3 e2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5 e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Zhang S (2004) Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J 38 142–151 [DOI] [PubMed] [Google Scholar]
- Kim KC, Fan B, Chen Z (2006) Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol 142 1180–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth C, Ringler J, Dangl JL, Eulgem T (2007) Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant Microbe Interact 20 120–128 [DOI] [PubMed] [Google Scholar]
- Lai Z, Vinod KM, Zheng Z, Fan B, Chen Z (2008) Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol 8 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Tapio Palva E (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46 477–491 [DOI] [PubMed] [Google Scholar]
- Lippok B, Birkenbihl RP, Rivory G, Brümmer J, Schmelzer E, Logemann E, Somssich IE (2007) Expression of AtWRKY33 encoding a pathogen-/PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol Plant Microbe Interact 20 420–429 [DOI] [PubMed] [Google Scholar]
- Liu J, Coaker G (2008) Nuclear trafficking during plant innate immunity. Mol Plant 1 411–422 [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang YC, Wang CY, Luo YC, Huang QJ, Chen SY, Zhou H, Qu LH, Chen YQ (2009) Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett 583 723–728 [DOI] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164 969–979 [DOI] [PubMed] [Google Scholar]
- Liu XQ, Bai XQ, Qian Q, Wang XJ, Chen MS, Chu CC (2005) OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res 15 593–603 [DOI] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ (2005) Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569 [DOI] [PubMed] [Google Scholar]
- Mangelsen E, Kilian J, Berendzen K, Kolukisaoglu U, Harter K, Jansson C, Wanke D (2008) Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Duan M, Wei C, Li Y (2007) WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol 48 833–842 [DOI] [PubMed] [Google Scholar]
- Marchive C, Mzid R, Deluc L, Barrieu F, Pirrello J, Gauthier A, Corio-Costet MF, Regad F, Cailleteau B, Hamdi S, et al (2007) Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J Exp Bot 58 1999–2010 [DOI] [PubMed] [Google Scholar]
- Massie CE, Mills IG (2008) ChIPping away at gene regulation. EMBO Rep 9 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke FLH, Kang HG, Chen Z, Park JM, Kumar D, Klessig DF (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol Plant Microbe Interact 18 1027–1034 [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun T, Smykowski A, Zentgraf U (2007) Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol Biol 65 63–76 [DOI] [PubMed] [Google Scholar]
- Mittler G, Butter F, Mann M (2009) A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res 19 284–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar SM, Deslandes L, Auriac MC, Marco Y, Somssich IE (2008) The Arabidopsis transcription factor WRKY27 influences wilt disease symptom development caused by Ralstonia solanacearum. Plant J 56 935–947 [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C (2006) The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SL, Ingle RA, Petersen LN, Denby KJ (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 20 1431–1438 [DOI] [PubMed] [Google Scholar]
- Mzid R, Marchive C, Blancard D, Deluc L, Barrieu F, Corio-Costet MF, Drira N, Hamdi S, Lauvergeat V (2007) Overexpression of VvWRKY2 in tobacco enhances broad resistance to necrotrophic fungal pathogens. Physiol Plant 131 434–447 [DOI] [PubMed] [Google Scholar]
- Naoumkina M, He X, Dixon R (2008) Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol 8 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436–439 [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Baek KH, Park JM, Yi SY, Yu SH, Kamoun S, Choi D (2008) Capsicum annuum WRKY protein CaWRKY1 is a negative regulator of pathogen defense. New Phytol 177 977–989 [DOI] [PubMed] [Google Scholar]
- Pan Y-J, Cho C-C, Kao Y-Y, Sun C-H (2009) A novel WRKY-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. J Biol Chem 284 17975–17988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Baldwin IT (2007) RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuata to herbivore attack in nature. Plant J 50 40–53 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Shahi P, Gase K, Baldwin IT (2008) Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc Natl Acad Sci USA 105 4559–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC (2008) OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant 1 446–458 [DOI] [PubMed] [Google Scholar]
- Petitot AS, Lecouls AC, Fernandez D (2008) Sub-genomic origin and regulation patterns of a duplicated WRKY gene in the allotetraploid species Coffea arabica. Tree Genet Genomes 4 379–390 [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA (2006) Activated signal transduction kinases frequently occupy target genes. Science 313 533–536 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev 23 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20 492–499 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S (2008. a) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol Plant 1 538–551 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al (2008. b) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Yu D (2009) Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot 65 35–47 [Google Scholar]
- Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S (2008) A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol 49 865–879 [DOI] [PubMed] [Google Scholar]
- Ren CM, Zhu Q, Gao BD, Ke SY, Yu WC, Xie DX, Peng W (2008) Transcription factor WRKY70 displays important but no indispensable roles in jasmonate and salicylic acid signaling. J Integr Plant Biol 50 630–637 [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ (2007) The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol 49 827–842 [Google Scholar]
- Ryu HS, Han M, Lee SK, Cho JI, Ryoo N, Heu S, Lee YH, Bhoo S, Wang GL, Hahn TR, et al (2006) A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep 25 836–847 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H (2007) Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19 2064–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe M, Qu N, Galis I, Baldwin IT (2008) Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15 2076–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Wu H, Richmond T, Redman JC, Johnson C, Green R, Arias J, Town CD (2006) Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J 47 152–162 [DOI] [PubMed] [Google Scholar]
- Turck F, Zhou A, Somssich IE (2004) Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in parsley. Plant Cell 16 2573–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJM (2008) A novel WRKY transcription factor is required for induction of PR-1A gene expression by salicylic acid and bacterial elicitors. Plant Physiol 146 1983–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7 491–498 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2008) Post-transcriptional RNA silencing in plant-microbe interactions: a touch of robustness and versatility. Curr Opin Plant Biol 11 464–470 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136 669–687 [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol 65 799–815 [DOI] [PubMed] [Google Scholar]
- Xing DH, Lai ZB, Zheng ZY, Vinod KM, Fan BF, Chen ZX (2008) Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol Plant 1 459–470 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, et al (2005) Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17 944–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Watanabe S, Tateno M, Seki M, Shinozaki K, Yokoyama S (2008) Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol Biochem 46 394–401 [DOI] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peng Y, Guo Z (2008. a) Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res 18 508–521 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wei L, Zou X, Tao Y, Liu Z, Zheng Y (2008. b) Submergence-responsive microRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Ann Bot (Lond) 102 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Mosher S, Fan B, Klessig D, Chen Z (2007) Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol 7 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48 592–605 [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W (2008) Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim Biophys Acta 1779 780–788 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.