Abstract
Plant hormones regulate growth and responses to environmental change. Hormone action ultimately modifies cellular physiological processes and gene activity. To facilitate transcriptome evaluation of novel mutants and environmental responses, there is a need to rapidly assess the possible contribution of hormone action to changes in the levels of gene transcripts. We developed a vector-based algorithm that rapidly compares lists of transcripts yielding correlation values. The application as described here, called HORMONOMETER, was used to analyze hormone-related activity in a transcriptome of Arabidopsis (Arabidopsis thaliana). The veracity of the resultant analysis was established by comparison with cognate and noncognate hormone transcriptomes as well as with mutants and selected plant-environment interactions. The HORMONOMETER accurately predicted correlations between hormone action and biosynthetic mutants for which transcriptome data are available. A high degree of correlation was detected between many hormones, particularly at early time points of hormone action. Unforeseen complexity was detected in the analysis of mutants and in plant-herbivore interactions. The HORMONOMETER provides a diagnostic tool for evaluating the physiological state of being of the plant from the point of view of transcripts regulated by hormones and yields biological insight into the multiple response components that enable plant adaptation to the environment. A Web-based interface has been developed to facilitate external interfacing with this platform.
Mutations that lead to aberrant phenotypes or to altered physiological responses are major tools in delineating gene function. Such mutations generally lead to varying degrees of primary and secondary transcriptome effects when compared with the transcriptomes of wild-type tissue and can be useful to establish pathways. For example, mutant lines in yeast were used to extract gene signatures of drug applications and thus identify primary and secondary drug target effects (Marton et al., 1998). In addition, environmental input can affect gene expression. Hence, monitoring the genome-wide expression of transcripts from these mutants or treated plants offers a useful portal to the initial understanding of gene functions. The concerted behavior of transcripts is commonly analyzed by hierarchical cluster analysis that extracts the degree of biological relatedness (Eisen et al., 1998). In Arabidopsis (Arabidopsis thaliana), an ever-growing data set (e.g. 3,110 microarray experiments compiled by Genevestigator version 3; https://www.genevestigator.ethz.ch/gv/index.jsp) can provide highly refined and elaborate hierarchical structures.
However, insightful understanding into the effect of a particular mutation can also be drawn by simplifying and limiting the choice of the databases used for comparison. As hormonal changes will initiate or reflect many plant responses, a choice database would be one that specializes in the transcriptome response to hormones and growth regulators. Thus, deriving a coherent hormone signature to a particular mutant or physiological state can be a useful starting point for further investigation. The large-scale transcriptome database developed by the AtGenExpress international consortium has provided detailed developmental (Schmid et al., 2005), abiotic stress (Kilian et al., 2007), and hormonal response (Goda et al., 2008) data sets. The hormonal data sets are based on the treatment of 7-d-old seedlings with hormones or small molecules. These include examples of the major growth regulator groups: auxin, indole-3-acetic acid (IAA); cytokinin, zeatin (ZEA); gibberellin (GA); brassinosteroids (BRs); abscisic acid (ABA); jasmonate (JA); ethylene, 1-aminocyclopropane-1-carboxylic acid (ACC); and salicylic acid (SA). The data generated was further validated by comparison with mutants and with hormone inhibitor applications (Goda et al., 2008).
To utilize such databases, a variety of bioinformatic techniques have been applied that specialize in comparing the concerted and usually major changes in gene expression. Each technique has its advantages and disadvantages. For example, Goda et al. (2008) have harnessed the hormonal data sets by hierarchical cluster analysis (Eisen et al., 1998) to help retrieve coexpressed transcripts and thus enhance gene function discovery. In addition, they show examples of Pearson correlation between select experiments based on a relatively small number of transcripts that survived stringent statistical constraints. In yet another method, preset gene thresholds were used to create select gene lists that were compared in order to describe functional overlap (Nemhauser et al., 2006). This was further refined in a method called functional association by response overlap (FARO; Nielsen et al., 2007). The strength of an association was determined by the size of the overlap between the experiment and a compendium of ranked gene expression responses. The qualitative nonparametric rank-based pattern-matching scheme in FARO is a simplification of the “connectivity map concept” found to be useful in human drug discovery (Lamb et al., 2006). There is a need for a tool whereby user-defined input can be compared by robust methodology to provide quantitative gene expression comparisons.
Transcriptome results of different experiments can be compared by vector-refined correlation. In this method, each experiment is represented as a vector in a space where every gene assayed represents a separate axis and the fold changes of the gene expression determine the geometric coordinates of the experiments. The analysis offers a rigorous transcript-by-transcript comparison by defining each transcript's “similarity” and setting up correlation tables. The final correlation value represents an unbiased factor of the number of transcripts scanned by the signature that essentially measures how two experiments (represented as two vectors in the space described above) are similar across the changing pattern of each individual gene expression measurement. Thus, even small changes, compiled over a large number of genes, can affect the correlation score. A variation of this method was first applied to the analysis of Saccharomyces cerevisiae and showed robust data management of yeast environmental responses (Kuruvilla et al., 2002). Here, we apply vector-refined correlation to interpret transcriptome response through a multihormonal lens composed of indexed transcript responses to different hormone treatments. The application is called HORMONOMETER. The resultant correlation values are projected onto a simple color-coded clustergram that helps identify the key hormone pathways that have been activated in a particular experiment. We first validate and calibrate HORMONOMETER by applying it to the hormone data sets themselves and to the transcriptome of a series of mutants. We then show examples of how HORMONOMETER can be applied to the analysis of complex environmental interactions.
RESULTS AND DISCUSSION
Building Indexes of Hormone Action and Calculating Correlations
The hormone signature was built in a two-step process. In the first step, a hormone expression index was compiled for each time point of the hormone treatments. In the subsequent step, the index was correlated by vector analysis to any transcriptome measurement to obtain a singular correlation value that is part of a hormone signature. The process was repeated for all of the hormone indexes. Vector analysis essentially measures how the changed gene expression pattern of a particular experiment is similar to the hormone index by computing the angle between two vectors generated by all of the participating transcripts. The numeral 1 indicates a complete correlation in terms of direction and intensity of the hormone index with the queried experiment, the numeral 0 indicates no correlation, and the numeral −1 indicates the highest possible anticorrelation for each transcript in the index. The angle between two vectors is essentially analogous to the Pearson correlation coefficient (Kuruvilla et al., 2002). For example, the Pearson correlation is used in the Sample Angler application, which identifies samples exhibiting similar expression profiles to a data set of transcriptomes and provides a correlation value (http://www.bar.utoronto.ca/). The difference between vector analysis and Pearson correlation is that the latter uses centered data points, meaning that it normalizes the values of the vectors to their arithmetical means. For example, in Pearson's correlation, a set of transcripts with expression values of 2-, 3-, and 4-fold is the same as the expression values 5-, 6-, and 7-fold. In our case, in which we extract a subset of transcripts from an experiment that matches the particular index, we chose not to center the data. A detailed description of the algorithm and other statistical considerations, such as the application of false discovery rate (FDR; Benjamini and Hochberg, 1995), are elaborated in “Materials and Methods.” In addition, a simplified example of how the vector correlation is calculated is provided in Supplemental Figure S1.
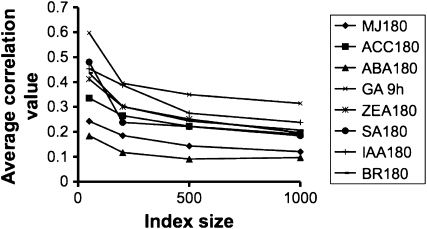
In order to arrive at an optimal index size for each hormone treatment, we analyzed the contribution of changed transcripts to the correlation value. In theory, an index can be composed for all of the transcripts that show significant change (e.g. P < 0.05); however, in practice, in any experiment the overwhelming majority of the transcripts remain nearly unchanged after normalization, and for economy of computation most information is gained from building an index of limited size. To arrive at an optimal hormone index size, we noted that when the average vector correlation values were computed using different size indexes, the values rapidly converged and hardly changed between index sizes of 500 and 1,000 (Fig. 1). This is due to the fact that the maximum number of detectably changed genes is generally less than 1,000 for most treatments (Goda et al., 2008). Furthermore, the screening of larger numbers of transcripts in the queried experiment with an index that is composed of transcripts that essentially do not change will compare transcripts that change randomly (up and down) and do not contribute to the correlation. Therefore, we adopted 1,000 as the index size for all hormones. The indexed transcripts for each treatment (hormones and time points) are tabulated in Supplemental Table S4.
Figure 1.
Average correlation values obtained for varying index sizes. Index sizes of 50, 200, 500, and 1,000 were extracted by arranging the values of statistically valid changes by decreasing absolute fold change values (i.e. the up- and down-regulated transcripts of ANOVA, P < 0.05). They were correlated to all noncognate hormone experiments, and the average correlation value is shown.
Vector-Refined Correlation Yields Robust Hormone Signatures
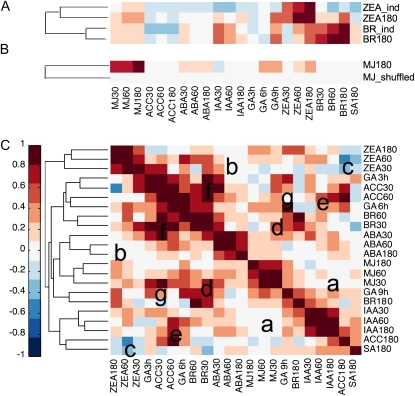
To illustrate the use of HORMONOMETER, we first calculated the correlation values of BR after 180 min (BR180) and ZEA after 180 min (ZEA180). These were chosen because independent applications that are not part of AtGenExpress are available. The calculated correlation results were then used as the input to the Matlab function “clustergram” from the bioinformatics toolbox (http://www.mathworks.com/products/matlab/). The clustergram arranges the experiments using hierarchical clustering with a Euclidean distance metric and average linkages to generate a hierarchical tree, as shown in Figure 2A. In order to facilitate visualization of the results, the range of correlations is color coded for positive (red) neutral (white), and negative (blue) correlation. The index used is shown on the x axis, while the experiments screened are on the y axis. Supplemental Table S1A contains the exact numerical values and number of transcripts scanned in each case. Examination of BR180 shows that it yields the expected value of 1 for its cognate index and lower values for the other, shorter BR time points. The values obtained with the other indexes range from negative to positive and represent hormonal cross talk that will be discussed further below. In an independent experiment, a higher concentration of BR (BR_ind; 1 μm as opposed to 10 nm in BR180) was applied to seedlings of identical age (7 d old) and examined after 180 min. The results (Fig. 2A; numerical values in Supplemental Table S1A) are very similar to those obtained with BR180. In contrast, in an independent experiment in which ZEA was applied at higher concentrations (ZEA_ind; 20 μm compared with 1 μm in ZEA180) and to older plants (21 d compared with 7 d), considerably less overlap was obtained. In this case, the ZEA signatures remain prominent, although results of other signatures show change, yet the hierarchical designation to ZEA180 is correct. The results indicate that the HORMONOMETER is sensitive to the seedling's age, likely reporting an age-dependent transcriptome reactivity to exogenous hormone applications.
Figure 2.
Hierarchical clustering matrix for correlation between gene expression profiles under different hormone treatments. The indexes are shown on the horizontal axis. The experiments screened by HORMONOMETER are shown on the vertical axis. A, Correlation values between independent hormone treatment experiments and the hormone expression index. Treatments are as follows: ZEA180, ZEA (cytokinin) at 1 μm, 180 min, 7-d-old seedlings; ZEA_ind, ZEA at 20 μm, 21-d-old seedlings; BR180, BRs at 10 nm, 180 min; BR_ind, BRs at 1 μm, 180 min. B, Correlation values between the hormone expression index and MJ180 hormone treatment and shuffled values of the MJ180 hormone treatment (MJ180_shuffled). C, Correlation values between the hormone expression index and the original hormone treatments used to build the indexes. The experiments listed on the vertical axis are clustered together according to the similarity of their profiles (the entire range of correlation values for each experiment). The hormone indexes on the horizontal axis are listed in the same order as the hierarchical clustering to emphasize correlations between cognate and noncognate hormones. The lowercase letters indicate areas as explained in the text.
Calibration of HORMONOMETER Values
A characteristic of statistical comparisons is that they always generate a number; the question then is, can biological meaning be attached to the levels of correlation values generated by the vector methodology? In order to approach this question, we analyzed the hormone data sets generated for AtGenExpress (Goda et al., 2008). As shown in Figure 2C, the expected correlations of value 1 appear along the diagonal as the index is used to analyze its own transcriptome data. The numerical values and number of transcripts screened are shown in Supplemental Table S1B. Generally, a three-by-three matrix (due to the three time points) surrounds the mid-time-point value and reflects the expected high cross-correlation between the different time points of the same hormone treatment. Note that the correlations are not necessarily symmetrical (above and below the diagonal). This is due to the fact that indexes presented on the horizontal axis comprise a select fraction of data in the experiments presented on the vertical axis; hence, the reciprocal correlations will differ slightly.
In some hormone treatments, the early, intermediate, and late treatments are highly correlated. For example, for IAA, they are all above the value of 0.65. In other cases, the corresponding values are lower (e.g. only above 0.4 for the GA time points). In those cases, discussed below, cognate hormone treatments do not cluster together, contributing, in part, to off-diagonal results. Inspection of the range of correlation values obtained in the different time points for the same hormone treatment can serve as a benchmark value used to “calibrate” the meaning of a “strong” vector-derived correlation value. In this case, values above 0.4, which represent the lowest correlation value among the different cognate hormone applications, signify the beginning of strong correlations. To estimate the lowest correlation values that need to be considered, one can examine the values obtained from comparing the indexes to randomized databases. To this end, the fold induction values and the P values of the MJ180 (for methyl jasmonate at 180 min) treatment were shuffled and screened by all of the hormone indexes. The MJ180 treatment was chosen because this treatment was one of the most effective treatments in terms of the number of transcripts that changed. The average of the resultant correlation scores was near 0 (Fig. 2B; average value = 0.05, σ = 0.04; Supplemental Table S1B). Thus, if correlation values of a particular experiment are distributed normally, values beyond 3.2906σ sds (i.e. 0.13) have only a 0.1% chance of being random. In summary, correlation numbers above 0.4 are seen between cognate hormone treatments and can be considered strong correlations, while numbers between 0 and 0.13 can be the result of chance and need not be considered. The significance of negative correlation values is discussed below.
Vector-Refined Correlation of Hormone Signatures Shows Overlap in Hormone Action
Alternative views have been reported using the hormone data generated by AtGenExpress (Goda et al., 2008); one conclusion states that hormones influence a low number of common target genes (Nemhauser et al., 2006), whereas another analysis showed that a significant amount of overlap exists between genes activated by different hormones (Goda et al., 2008). Low correlations are indicated in areas a and b (Fig. 2C). They indicate that IAA has no MJ signature and that ZEA has practically no ABA signature. However, other areas (Fig. 2C, areas e–g) depict regions of strong signature cross-correlations between noncognate hormone treatments. Thus, any high correlation values that are off the diagonal lend clear evidence to the conclusion that strong noncognate interactions exist. In addition, as exemplified in area c (Fig. 2B), negative correlations exist between ZEA and late treatments of ACC and SA. Furthermore, hierarchical distribution shows that time points for BR, ACC, and GA do not always cosegregate. The most promiscuous correlations are associated with early MJ and all of the BR treatments. The overlaps detected here are somewhat analogous to the analysis carried out by pairwise comparisons with Fisher's exact test by Goda et al. (2008). However, the vectorial analysis and subsequent clustergram presentation lend a global viewpoint and uniform quantification basis to the degree of hormone cross-correlations.
Cross-correlation of signatures for hormone action can arise due to many scenarios, for example: (1) the direct activation by one hormone of other hormone biosynthetic pathways; (2) increased sensitivity to existing basal hormone levels; and (3) independent activation of a subset of the signaling pathways by bifurcating signaling input. In many cases, the correlations observed are strongest for the earlier time points measured and would seem to argue against the scenario of direct activation of noncognate hormone biosynthesis. However, in the case of overlap detected at later times (e.g. measurements at 3 h), direct activation is a distinct possibility. Indeed, partial overlap between hormonal signatures is supported in many cases by experimental observations. For example, BR plays a role in apical hook formation, as does ethylene, which has been suggested to jointly control BR biosynthesis (De Grauwe et al., 2005). In addition, BR has been shown to directly down-regulate the level of ACC oxidase and to up-regulate JA biosynthetic enzymes (Mussig et al., 2000; Deng et al., 2007), consistent with the cross-activation detected here. The significant correlation of BR with early IAA and GA responses is also readily detected in the signature. This is expected, as BRs are able to rescue GA-deficient mutants, which normally fail to germinate, and BRs can act synergistically with IAA via common elements in the promoters of the transcripts (Nemhauser et al., 2004) and also additively with GAs (Mandava, 1988).
An antagonistic relationship between GA and cytokinins and ABA has been recorded (summarized in Weiss and Ori, 2007) and is observed here by low correlation values with ABA180 and ZEA signatures. In contrast, the up-regulation of GA biosynthetic genes by auxin is evident in the presence of GA signatures in the IAA applications. A clear positive BR signature is present in GA applications, as is also true for the reciprocal signature of GA in BR applications. Yet, for some specific genes, a reciprocal relationship between GA and BR was recorded (Bouquin et al., 2001). Application of BR can induce GA biosynthetic genes (Olszewski et al., 2002), and the overall effect as observed here is one of cross-activation.
SA application appears to have an IAA signature. Yet, SA has been shown to desensitize the plant to some auxin responses; indeed, some pathogens actively secrete auxin to achieve SA repression, enhancing their infectivity (Wang et al., 2007). However, the continuous and unambiguous coverage offered by the vector-based analysis shows that, on a global level, not only is the expected negative correlation for IAA absent but a positive auxin signature appears. Thus, the vectorial approach reveals unappreciated selectivity in the repression that auxin supplies to SA-derived processes. In addition, a significant ethylene response (ACC180 signature) in SA application is evident that was previously overlooked. The contribution of enhanced ethylene responses brought about by SA may be as significant as the auxin response and could contribute to the selectivity wrought by SA application.
A clear example of the effect of sequential hormone activation is found in the ethylene mediation of increased auxin levels by the induction of auxin biosynthetic genes (Stepanova et al., 2007; Swarup et al., 2007). Inspection of Figure 2C reveals that ACC180 application shows robust association with auxin late gene profiles. Indeed, ethylene was recently shown to directly influence local auxin biosynthesis through activation of the WEAK ETHYLENE INSENSITIVE8 gene product that encodes for a Trp aminotransferase activity necessary for auxin biosynthesis (Stepanova et al., 2008). ACC180 signatures show robust negative correlation with ZEA, as ethylene is known to be antagonistic to cytokinin (Kudryakova et al., 2001). For example, the homeobox gene KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 acts synergistically with cytokinins and antagonistically with ethylene (Hamant et al., 2002).
The analysis carried out here documents a high degree of overlap in transcriptional responses and is consistent with the conclusions drawn by Goda et al. (2008). The vector-based algorithm allows continuous quantitative monitoring of comparisons, yielding a robust and revealing picture of hormone interactions.
Hormone Signatures of Mutants in Hormone Pathways
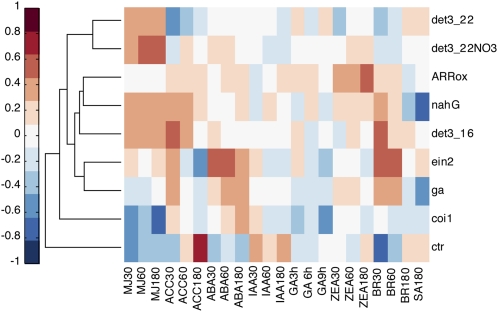
We further validated HORMONOMETER analysis by screening experimental data from hormone biosynthesis and signal transduction mutants. Figure 3 (for numerical values, see Supplemental Table S2) illustrates examples of such analysis for a series of mutants compared with their wild-type control.
Figure 3.
Clustergram representation of hierarchical tree groups of transcriptomes from experiments conducted in hormone biosynthesis or hormone signal transduction mutants. The following experiments were screened: det3_22, det3_22NO3, and det3_16 are det3 mutants at 22°C with or without nitrate and at 16°C; nahG, salicylate hydroxylase-expressing mutant; arrOX, ARR21C overexpression mutant; ein2, ethylene-insensitive mutant ein2; ga, GA biosynthetic mutant ga1; coi1, JA signaling mutant coi1; ctr1, constitutive ethylene response and wounding mutant ctr1. The hormone data and color-coded evaluation are as in Figure 2, while the sources of the particular experiments are tabulated in Supplemental Table S7.
CORONATINE INSENSITIVE1 (COI1) is required for JA-induced growth inhibition and encodes for an F-box protein (Xie et al., 1998). Its effect was previously examined in the context of JA responses, and 84% of JA-sensitive genes were found to be changed in the coi1 mutant (Devoto et al., 2005). Vector-based analysis of this mutant (grown under nonstressed conditions) shows consistent negative correlation of the MJ signature. The negative signature indicates that the stress-associated JA hormone exerts a significant effect during normal physiological growth. Interestingly, additional negative signatures appear, particularly for all GA signatures, while ABA (60 and 180) has a significant positive signature. GA and JA were shown to act synergistically on trichome induction (Traw and Bergelson, 2003). Apparently in seedlings, as revealed by HORMONOMETER analysis, additional cooperative interactions are present that are abrogated in coi1. ABA deficiency causes an increase in the basal levels of JA response genes (Anderson et al., 2004), and application of JA strongly down-regulates ABA biosynthetic genes (Jung et al., 2007). Thus, the lack of basal JA responses in the coi1 background appears to promote the appearance of an ABA signature.
JA and SA are known to exhibit antagonistic relationships (Koornneef and Pieterse, 2008). However, this is not strongly registered in the transcriptome of coi1, where the SA signature is slightly repressed. Nonetheless, that relationship is clearly seen in the analysis of NahG plants. NahG plants express salicylate hydroxylase and cannot accumulate SA (Delaney et al., 1994). The signature for SA is strongly negative, as expected, and strong positive values are registered for all MJ signatures due to the loss of basal-level suppression by the lack of endogenous SA (Fig. 3). Inspection of the coi1 hormonal signature exemplifies another striking property. With the exception of ABA responses, all other hormones are, to a greater or lesser degree, negatively correlated. This may have to do with the evidence that auxin, jasmonic acid, GA, and ethylene signaling are mediated by specific F-box protein-encoding genes. As the specific F-box proteins compete for binding to the SCF complex (for Skp1, cdc53/cullin, and F-box proteins), it may be that the coi1 mutation interferes with the control of individual hormone signals through this interaction. In this respect, ABA is exceptional, as it is not known to be associated with the SCF complex, and, as noted above, its positive signature in coi1 plants is evident (Yu et al., 2007).
Inspection of JA relationships among the recently described de-etiolated3 (det3) mutants of the vacuolar ATPase subunit C can provide insight about the quantitative nature of the hormone signatures. This mutant was originally identified as a negative regulator of photomorphogenesis (Schumacher et al., 1999). It shows a conditional defect in hypocotyl elongation in dark-grown seedlings (i.e. it exhibits either a reduced or a complete absence of phenotype at room temperature, a mild phenotype at 16°C, and a more severe phenotype in the presence of 5 mm nitrate). The manifestation of the unstable phenotype was shown to be correlated with JA accumulation and was abrogated in the double mutant det3/OPDA-reductase3 (Brux et al., 2008). The average correlation values (of the three time points) using the hormone index for the JA signature are 0.35, 0.37, and 0.47 for det3 at 22°C, 16°C, and 22°C + 5 mm nitrate, respectively (Fig. 3; Supplemental Table S2). The similarity of the phenotype gradations and the JA hormone signature values testify to the ability of the HORMONOMETER to detect fine differences in physiological states.
The ethylene insensitive2 (ein2) mutant shows the expected negative ACC signature, particularly at later times (i.e. ACC60 and ACC180). Interestingly, the ein2 mutant shows a consistent ABA-positive up-regulated signature. The relationship between ABA and ethylene is complex. In seed germination, ethylene has been shown to be a negative regulator of ABA, although in root growth, ein2 is required for ABA response (Beaudoin et al., 2000). ABA was also shown to inhibit ethylene-induced hyponastic (upward) leaf growth (Benschop et al., 2007), although, in all, this result is unexpected. The hormone signature of ethylene can also be seen in examination of the constitutive triple response1 (ctr1) mutant. CTR1 encodes for a Raf-like kinase, and the ctr1 mutant constitutively exhibits the phenotypes observed in plants treated with the plant hormone ethylene. The analysis shows a marked ethylene signature for ACC60 and particularly for ACC180, consistent with the constitutive nature of ethylene perception in the ctr1 mutant (Fig. 3). The reciprocal nature of ein2 and ctr1 behavior is notable across all signatures. Where ABA and BR signatures are prominent in ein2, they are negatively correlated in ctr1, lending credence to the significance of the signatures. The significant repression of the JA signature in ctr1 is less expected, as JA can show synergy with ethylene for the induction of plant defensins (Penninckx et al., 1998). However, in other cases, antagonistic relationships were shown. For example, the JASMONATE RESISTANT1-dependent JA pathway halts oxidative cell death promoted by ozone by directly suppressing ethylene signaling (Tuominen et al., 2004).
In GA requiring1 (ga1) mutants, GA biosynthesis is repressed due to the lack of ent-kaurene synthase A, which catalyzes the first committed step in the biosynthetic pathway of GA (Sun and Kamiya, 1994). Along with the expected absence of a GA signature, the complete hormonal signature response shows enhanced ABA and early BR signatures. The strong ABA signature is expected, as GA and ABA not only mutually inhibit each other's biosynthesis but also promote each other's catabolism. GA negatively regulated DELLA proteins, and they, in turn, targeted XERICO, which promotes the accumulation of ABA (Zentella et al., 2007). Strikingly, the repression of the SA signature in ga1 is even more negative than the missing cognate GA signatures. The reciprocal antagonistic relationship of SA and GA (note the result of NahG above) has not been noted before and needs to be further investigated.
ARABIDOPSIS RESPONSE REGULATOR21 (ARR21) is a representative of the type-B ARR transcription factors and positively regulates cytokinin responses. Overexpression of the constitutively active ARR21C protein results in abnormal development, with tissues resembling in vitro callus (Tajima et al., 2004). Analysis carried out by the FARO algorithm showed strong associations between ARR21C and ZEA treatments (Nielsen et al., 2007). That result is confirmed here (Fig. 3, ARRox) and further extended to show low-level but consistent ACC signatures. In summary, the results using mutants in hormone biosynthesis or signal transduction validate the vector-based HORMONOMETER strategy and show both expected and unexpected findings underlining the considerable complexity of hormone signatures.
Hormonal Signal Signatures during Plant-Insect and Plant-Pathogen Interactions
Plant surveillance and response systems have evolved to provide an answer to the diversity of pathogen lifestyles. Plant-pathogen or plant-pest interaction triggers the biosynthesis of SA, ethylene/ACC, and JA. The balance of these hormone systems determines resistance to particular pathogens and pests. Broadly put, SA has been implicated in local and systemic resistance to biotrophic pathogens (Glazebrook, 2005), hypersensitive responses are associated with ethylene production (Bouchez et al., 2007), and JA and ethylene contribute to resistance against necrotrophic pathogens or chewing insects (Glazebrook, 2005). The defense hormones show both antagonistic and cooperative actions that are further tempered by growth-stimulating phytohormones such as those controlling auxin, ABA, and GAs (Wang et al., 2007). The transcriptome of herbivory, pathogenesis, and environmental stress has been well articulated in relation to stress-hormone relationships. Less emphasis has been devoted to following concomitant growth hormone responses, although it is well appreciated that the cessation of growth plays an important role in the diversion of metabolic potential to defense. HORMONOMETER analysis of the hormone indexes is well suited for exemplifying these interactions, as shown below.
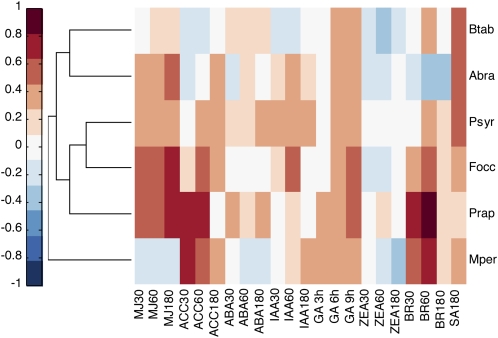
De Vos et al. (2005) monitored the dynamics of SA, JA, and ethylene accumulation and the concomitant transcriptome response due to attack by a range of plant pests. Among the microbial pathogens investigated were Pseudomonas syringae pv tomato, the causal agent of bacterial speck disease that will initiate systemic acquired resistance pathway responses when it contains avirulent genes, and Alternaria brassicicola, a necrotrophic plant pathogenic fungus that grows on the Arabidopsis phytoalexin deficient3 mutant, causing spreading necrotic lesions. Among the herbivorous insects were Pieris rapae and the thrip Frankliniella occidentalis. The former chews leaf parts, while the latter punctures the leaves and sucks up the exuding sap. In contrast, the aphid Myzus persicae passively feeds on sap of phloem vessels through a stylet. In addition to these data, a second type of phloem feeder is also examined here, the silverleaf whitefly (Bemisia tabaci type B; Kempema et al., 2007).
As shown in Figure 4 and Supplemental Table S3, the highest correlation values for the MJ signature are observed for the scraping-type herbivorous insects P. rapae (Prap) and the thrip F. occidentalis (Focc), while the lowest are for M. persicae (Mper) and B. tabaci (Btab). These results confirm and broaden the “attacker-specific” profile described for the MJ response (De Vos et al., 2005). Surprisingly, B. tabaci interaction leaves a stronger MJ footprint than M. persica, although the former is thought to cause less damage to epidermal or mesophyll cells prior to phloem puncture (Kempema et al., 2007).
Figure 4.
Clustergram representation of hierarchical tree groups of transcriptomes from plant-pathogen or plant-pest interactions. The indexes of hormone action appear on the abscissa, and the treatments appear on the ordinate. Treatments are as follows: Mper, M. persicae; Prap, P. rapae; Focc, F. occidentalis; Psyr, P. syringae pv tomato; Abra, A. brassicicola; Btab, B. tabaci. The hormone data and color-coded evaluation are as in Figure 2, while the sources of the particular experiments are tabulated in Supplemental Table S7.
P. syringae-infected leaves accumulated relatively high levels of SA (De Vos et al., 2005), and analysis of the transcriptome reveals a high SA signature correlation (0.54; Fig. 4). However, the other treatments also show a degree of SA-dependent signature (>0.26). These results could indicate a potentiation of sensitivity to SA rather than SA accumulation, as has been observed in other systems (Shirasu et al., 1997). Alternatively, herbivores like the aphid M. persicae and B. tabaci attempt to evade wound response by SA-induced repression of JA. This mechanism has been suggested for stylet feeders and particularly B. tabaci (Walling, 2008). Yet, inspection of Figure 4 shows that in the other herbivores, similar SA signature correlations are evident without accompanying repression of jasmonate-dependent responses. Indeed, in contrast to previous reports, in these experiments M. persicae appears to be more effective in the repression of global MJ signatures than B. tabaci (Kempema et al., 2007). These results likely indicate that additional mechanisms for wound stress avoidance in M. persicae-plant interactions come into play beyond direct repression of MJ responses through SA action.
Interestingly, most insect-plant interactions shown here appear to have a slight negative impact on ZEA signatures, which may reflect the decrease in growth rates brought about by the insect infestation. Another unexpected common feature is the increased 6-h and 9-h GA signatures (GA6h and GA9h). If this observed transciptome response is a direct result of GA biosynthesis, it may be related to a distinct phytochemical defense response. Indeed, GA has a specific negative effect on insect growth and is used as such in specific chemical applications (Alonso, 1971; Kaur and Rup, 2002; Uckan et al., 2008). Alternatively, changes common to all treatments may represent an age-dependent effect for these particular experiments, as pointed out in Figure 2A.
Actual increased production of the hormone ethylene was measured in P. syringae and A. brassicicola (De Vos et al., 2005). However, all other plant-pest interactions except B. tabaci show an unexpected early and sometimes late correlation with ethylene signatures. Aphids are known to induce ethylene production in cereals (Anderson and Peters, 1994; Argandona et al., 2001), but this has yet to be shown in dicots.
Remarkably, the vector-based analysis detects a significant auxin signature specific for P. syringae infection. The infections above were carried out using an avirulent strain that contains the avrRpt2 effector protein. It was recently shown that transgenic seedlings expressing avrRpt2 protein exhibit increased auxin sensitivity and increased auxin levels (Chen et al., 2007). As application of auxin can enhance disease symptoms, it was hypothesized that avrRpt2 may enhance the virulence of P. syringae by activating host auxin physiology. In this case, the higher SA levels/sensitivity that are evident may be part of a plant defense shift, as enhanced SA levels have been shown to decrease auxin sensitivity (Wang et al., 2007).
In diverse plant-pathogen interactions, exemplified by P. rapae, F. occidentalis, and M. persicae, a significant correlation signal is detected in early BR signatures (Fig. 4). The possibility that direct MJ applications induce such BR signatures as seen in Figure 2C does not explain the M. persicae BR responses, as it has no MJ signature. Alternatively, elevated BR levels/sensitivity have been shown to enhance tolerance in plants toward biotic and abiotic stresses (Dhaubhadel et al., 1999; Nakashita et al., 2003). A gene expression profile of lines defective in a step leading to BR biosynthesis shows lower transcript levels of stress-related genes (e.g. RAS-RELATED IN BRAIN18 and COLD REGULATED47), which indicates that brassinosteroids are required for these responses (Mussig et al., 2002). As a dual role for brassinosteroid receptors in development and pathogenesis has been reported (Montoya et al., 2002), it is not difficult to envision multiple inputs to this pathway. The results in Figure 4 exemplify the sensitivity of the vectorial approach to identify the underlying complexity of plant physiological responses related to hormones.
CONCLUSION
The HORMONOMETER application uses a vector-based correlation algorithm to compare transcriptomes with indexed data sets of hormone treatments. It was shown here to accurately discern gene signatures as related to plant hormone action. Importantly, the algorithm is not limited to this application and can readily be generalized for the treatment of other relevant biological circumstances by thoughtful selection and processing of select data sets. For example, it can be applied to discriminate the developmental stage of a leaf based on microarray data obtained from different ages of leaves or to discern the physiological state of a plant based on microarray data acquired from plants under different abiotic stresses. The HORMONOMETER application detailed here offers a novel portal by providing the user with calibrated correlation values that reflect a change in hormone-related transcript levels caused by external stimuli or mutation. The utility of HORMONOMETER is in providing a facile overview of hormone signatures that permits primary analysis of new mutants and novel environmental insults. HORMONOMETER can be accessed through user interface by submission of data in a “comma separated values” file format that includes fold change and the P value for each gene in the transcriptome to be examined. The user receives the computed correlation ranks for each of the hormones in a table and a clustergram that arranges the experiments according to their similarity to each other in terms of the ranks. The tool can be accessed at http://genome.weizmann.ac.il/hormonometer/.
MATERIALS AND METHODS
Microarray Experiments and Data Processing
CEL files for the Affymetrix ATH1 microarray data were downloaded from the following Web-available databases: The Arabidopsis Information Resource (Swarbreck et al., 2008) and Gene Expression Omnibus (Edgar et al., 2002). The experiments included in this work are summarized in Supplemental Table S7. The data arising from the microarray experiments were analyzed utilizing the Partek genomics solution (Downey, 2006). The processing of the data included quantile normalization according to the Robust Multiarray Analysis algorithm (Irizarry et al., 2003) and a one-way standard ANOVA model, treating each condition as a factor (seven treatments with three time points and one treatment with one time point = 22 conditions).
Building the Vector-Based Algorithm for the Hormone Response Index
In order to build the index, the results were first processed by quantile normalization as described above. The transcripts were then arranged by their decreasing absolute fold change values (i.e. up- and down-regulated transcripts). A Perl script was written to find for each condition the 1,000 transcripts with the most variable expression between the treatment and its control (i.e. the 1,000 highest absolute values of the fold change that have P < 0.05 from ANOVA modeling). The script utilizes fold change comparisons versus control samples to extract a gene expression index representing each experiment of individual hormone application. A summary of indexes generated for all hormone treatments is tabulated in Supplemental Table S4. The script uses an algebraic vector-based comparison to compare two experiments. When this is carried out over the 1,000 transcripts that constitute the index, they describe a Euclidean space of a multidimensional space.
Thus, for a particular signature:
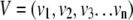 |
where v1, v2, v3, etc. represent the fold changes for each transcript in an index of size n. The list is used to define the vector in the particular experiment being scanned:
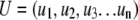 |
where u1, u2, etc. are conditionally included if their P value is <0.05. The cosine of the angle between the two vectors is then calculated:
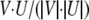 |
The resulting rank score is between −1 and 1. The closer the two vectors are, the higher the result. Two opposite vectors will result in a rank of −1. A simplified example and figure are provided in Supplemental Figure S1. A table of transcripts used for each hormone treatment is provided in Supplemental Table S4, and a Perl script of the algorithm will be provided on request.
FDR and Random Sampling Methods
When dealing with large numbers of individual measurements, the application of multiple hypothesis testing, FDR, can be appropriate to reduce the chance of false values (Benjamini and Hochberg, 1995). However, as detailed below, the vector-based algorithm used here can distinguish false values, as they have an equal chance of being positive or negative, effectively canceling each other out. Therefore, no further statistical processing is necessary.
We examined the appropriateness of further data processing by implementing FDR in the following manner. We first generated lists of 1,000 transcripts for each individual hormone treatment by applying a predefined fold induction criterion. For example, an index was generated for the MJ30 transcriptome by selecting 1,000 transcripts that were expressed at least 1.28-fold (absolute value) without considering their P values. The data sets to be screened by the index were then corrected by FDR for n = 1,000 and α = 0.05. The results are shown in Supplemental Table S5. When these results were compared with non-FDR processed correlation values (Supplemental Table S1), in which the selection criterion was noncorrected P < 0.05 (both to build the index and in the transcriptome data set screened), some differences were noted. As expected, the number of retrieved genes after FDR computation was much reduced. FDR correction tends to remove the lower fold induction values preferentially, as they tend to have lower P values. However, the low fold induction values are precisely those that are expected to be prominent when, for example, a hormone experiment is screened by its noncognate index. Furthermore, after FDR correction, only a few results are left, and the correlation values obtained by the vector analysis become erratic and can even change signs. For example, the results of the MJ30 index used to screen ZEA are as follows (−FDR/+FDR): ZEA30 (−0.01/−0.25), ZEA60 (0.25/0.23), and ZEA180 (0.19/−1; compare Supplemental Tables S1 and S5). On the other hand, without applying FDR prescreening, the numbers of transcripts scanned for the vector analysis is much larger and the correlation results are more consistent. That is because the vector-based algorithm easily distinguishes between signal and noise. “Noise” in the signature (i.e. false values) has an equal chance of being positive or negative, which would cancel each other out in vector analysis. This supposition is readily illustrated when the fold induction values and the P values are shuffled for a given index. For example, when the MJ180 transcriptome data were shuffled and then analyzed by the hormone index, a mean of 181 genes was scanned in each experiment but the absolute value of the correlation scores was near zero (0.05 ± 0.04; Supplemental Table S6). In the case of correction for FDR, the results are based on a lower number of genes scanned (mean = 48) but a much larger sd of the correlation scores 0.138 ± 0.1. Importantly, the vector algorithm can discern the low fold induction trends within the transcripts scanned if these are coherent with the signature. Hence, an increase in noise, the result of using larger indexes (i.e. uncorrected FDR input), will tend to reduce the correlation value but only slowly (Fig. 1). Thus, application of FDR, while perhaps providing a more secure database, also removes useful information that can be used safely by vector analysis. Due to these considerations, uncorrected P < 0.05 values were adopted throughout.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Example of the application of the vector-based correlation for comparison of the expression of three transcripts under two different conditions.
Supplemental Table S1. A, Table of correlation values for Figure 2A. B, Table of correlation values for Figure 2C.
Supplemental Table S2. Table of correlation values for Figure 3.
Supplemental Table S3. Table of correlation values for Figure 4.
Supplemental Table S4. Summary of indexes generated for all hormone treatments.
Supplemental Table S5. Correlation values obtained by correction for FDR (the FDR correction used was n = 1,000 and α = 0.05).
Supplemental Table S6. Correlation values and signature size for randomized MJ180 data.
Supplemental Table S7. Source of CEL files used in this work.
Supplementary Material
Acknowledgments
We thank Dr. Ester Feldmesser for assistance with data processing and Dr. Hillary Voet for consultations regarding statistics calculations. We also thank Drs. Yuval Eshed and David Weiss for critical reading of the manuscript.
This work was supported by the United States-Israel Binational Agricultural Research and Development Fund (grant no. IS–4141–08).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert Fluhr (robert.fluhr@weizmann.ac.il).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso C (1971) The effects of gibberellic acid upon developmental processes in Drosophila hydei. Entomol Exp Appl 14 73–82 [Google Scholar]
- Anderson JA, Peters DC (1994) Ethylene production from wheat seedlings infested with biotypes of Schizaphis graminum (Homoptera, Aphididae). Environ Entomol 23 992–998 [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argandona VH, Chaman M, Cardemil L, Munoz O, Zuniga GE, Corcuera LJ (2001) Ethylene production and peroxidase activity in aphid-infested barley. J Chem Ecol 27 53–68 [DOI] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methods 57 289–300 [Google Scholar]
- Benschop JJ, Millenaar FF, Smeets ME, van Zanten M, Voesenek LACJ, Peeters AJM (2007) Abscisic acid antagonizes ethylene-induced hyponastic growth in Arabidopsis. Plant Physiol 143 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez O, Huard C, Lorrain S, Roby D, Balague C (2007) Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1. Plant Physiol 145 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J (2001) Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol 127 450–458 [PMC free article] [PubMed] [Google Scholar]
- Brux A, Liu TY, Krebs M, Stierhof YD, Lohmann JU, Miersch O, Wastemack C, Schumacher K (2008) Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. Plant Cell 20 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Agnew JL, Cohen JD, He P, Shan LB, Sheen J, Kunkel BN (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32 D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grauwe L, Vandenbussche F, Tietz O, Palme K, Van Der Straeten D (2005) Auxin, ethylene and brassinosteroids: tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol 46 827–836 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gutrella M, Kessmann H, Ward E, et al (1994) A central role of salicylic acid in plant disease resistance. Science 266 1247–1250 [DOI] [PubMed] [Google Scholar]
- Deng ZP, Zhang X, Tang WQ, Oses-Prieto JA, Suzuki N, Gendron JM, Chen HJ, Guan SH, Chalkley RJ, Peterman TK, et al (2007) A proteomics study of brassinosteroid response in Arabidopsis. Mol Cell Proteomics 6 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux JP, Van Loon LC, Dicke M, et al (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18 923–937 [DOI] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58 497–513 [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, Chaudhary S, Dobinson KF, Krishna P (1999) Treatment with 24-epibrassinolide, a brassinosteroid, increases the basic thermotolerance of Brassica napus and tomato seedlings. Plant Mol Biol 40 333–342 [DOI] [PubMed] [Google Scholar]
- Downey T (2006) Analysis of a multifactor microarray study using Partek genomics solution. Methods Enzymol 411 256–270 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al (2008) The AtGenExpress hormone- and chemical-treatment data set: experimental design, data evaluation, model data analysis, and data access. Plant J 55 526–542 [DOI] [PubMed] [Google Scholar]
- Hamant O, Nogue F, Belles-Boix E, Jublot D, Grandjean O, Traas J, Pautot V (2002) The KNAT2 homeodomain protein interacts with ethylene and cytokinin signaling. Plant Physiol 130 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264 [DOI] [PubMed] [Google Scholar]
- Jung C, Lyou SH, Yeu S, Kim MA, Rhee S, Kim M, Lee JS, Do Choi Y, Cheong JJ (2007) Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana. Plant Cell Rep 26 1053–1063 [DOI] [PubMed] [Google Scholar]
- Kaur R, Rup PJ (2002) Evaluation of regulatory influence of four plant growth regulators on the reproductive potential and longevity of melon fruit fly (Bactrocera cucurbitae). Phytoparasitica 30 224–230 [Google Scholar]
- Kempema LA, Cui XP, Holzer FM, Walling LL (2007) Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs: similarities and distinctions in responses to aphids. Plant Physiol 143 849–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ (2008) Cross talk in defense signaling. Plant Physiol 146 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryakova NV, Burkhanova EA, Rakitin VY, Yakovleva LA, Smith A, Hall MA, Kulaeva ON (2001) Ethylene and cytokinin in the control of senescence in detached leaves of Arabidopsis thaliana eti-5 mutant and wild-type plants. Russ J Plant Physiol 48 624–627 [Google Scholar]
- Kuruvilla FG, Park PJ, Schreiber SL (2002) Vector algebra in the analysis of genome-wide expression data. Genome Biol 3 Research0011.1–Research0011.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al (2006) The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313 1929–1935 [DOI] [PubMed] [Google Scholar]
- Mandava NB (1988) Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39 23–52 [Google Scholar]
- Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts CJ, Stoughton R, Burchard J, Slade D, Dai HY, et al (1998) Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med 4 1293–1301 [DOI] [PubMed] [Google Scholar]
- Montoya T, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ (2002) Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14 3163–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussig C, Biesgen C, Lisso J, Uwer U, Weiler EW, Altmann T (2000) A novel stress-inducible 12-oxophytodienoate reductase from Arabidopsis thaliana provides a potential link between brassinosteroid action and jasmonic acid synthesis. J Plant Physiol 157 143–152 [Google Scholar]
- Mussig C, Fischer S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K, Takatsuto S, Yamaguchi I, Yoshida S (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J 33 887–898 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong FX, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2 1460–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HB, Mundy J, Willenbrock H (2007) Functional associations by response overlap (FARO), a functional genomics approach matching gene expression phenotypes. PLoS One 2 e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14 S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Metraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes Dev 13 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-P, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase-A of gibberellin biosynthesis. Plant Cell 6 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36 D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GTS, Sandberg G, Bhalerao R, Ljung K, Bennett MJ (2007) Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45 28–39 [DOI] [PubMed] [Google Scholar]
- Traw MB, Bergelson J (2003) Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol 133 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H, Overmyer K, Keinanen M, Kollist H, Kangasjarvi J (2004) Mutual antagonism of ethylene and jasmonic acid regulates ozone-induced spreading cell death in Arabidopsis. Plant J 39 59–69 [DOI] [PubMed] [Google Scholar]
- Uckan F, Tuven A, Er A, Ergin E (2008) Effects of gibberellic acid on biological parameters of the larval endoparasitoid Apanteles galleriae (Hymenoptera: Braconidae). Ann Entomol Soc Am 101 593–597 [Google Scholar]
- Walling LL (2008) Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol 146 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong XN (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17 1784–1790 [DOI] [PubMed] [Google Scholar]
- Weiss D, Ori N (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144 1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yu HC, Wu J, Xu NF, Peng M (2007) Roles of F-box proteins in plant hormone responses. Acta Biochim Biophys Sin (Shanghai) 39 915–922 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.