Abstract
Red clover (Trifolium pratense) leaves accumulate several μmol g−1 fresh weight of phaselic acid [2-O-(caffeoyl)-l-malate]. Postharvest oxidation of such o-diphenols to o-quinones by endogenous polyphenol oxidases prevents breakdown of forage protein during storage. Forage crops like alfalfa (Medicago sativa) lack both polyphenol oxidase and o-diphenols, and breakdown of their protein upon harvest and storage results in economic losses and release of excess nitrogen into the environment. Understanding how red clover synthesizes o-diphenols such as phaselic acid will help in the development of forage crops utilizing this natural system of protein protection. A possible pathway for phaselic acid biosynthesis predicts a hydroxycinnamoyl transferase (HCT) capable of forming caffeoyl and/or p-coumaroyl esters with malate. Genes encoding two distinct HCTs were identified in red clover. HCT1 shares more than 75% amino acid identity with a number of well-characterized shikimate O-HCTs implicated in monolignol biosynthesis. HCT2 shares only 34% amino acid sequence identity with HCT1 and has limited sequence identity to any previously identified HCT. Expression analyses indicate that HCT1 mRNA accumulates to 4-fold higher levels in stems than in leaves, whereas HCT2 mRNA accumulates to 10-fold higher levels in leaves than in stems. Activity assays of HCT1 and HCT2 proteins expressed in Escherichia coli indicate that HCT1 transfers caffeoyl or p-coumaroyl moieties from a coenzyme A-thiolester to shikimate but not malate, whereas HCT2 transfers caffeoyl or p-coumaroyl moieties from a coenzyme A-thiolester to malate but not shikimate. Together, these results indicate that HCT1 is involved in monolignol biosynthesis and HCT2 is a novel transferase likely involved in phaselic acid biosynthesis.
In contrast to many other forage legumes (e.g. alfalfa [Medicago sativa]; Jones et al., 1995), red clover (Trifolium pratense) accumulates relatively high levels of the phenylpropanoid o-diphenol phaselic acid [2-O-(caffeoyl)-l-malic acid; hereafter referred to as caffeoyl-malate or phaselic acid] in its leaves (Hatfield and Muck, 1999; Winters et al., 2008). In red clover, upon cellular disruption, phaselic acid and other o-diphenols are readily oxidized by a soluble polyphenol oxidase (PPO) to produce their corresponding o-quinones (Hatfield and Muck, 1999; Sullivan et al., 2004). The formation of such o-quinones by PPO, and the subsequent secondary reactions of these quinones, are most often associated with browning of fresh fruits and vegetables (Steffens et al., 1994), which has a negative impact on perceived quality. When preserved by ensiling, however, oxidation of o-diphenols by PPO in red clover prevents degradation of protein during storage (Sullivan et al., 2004; Sullivan and Hatfield, 2006). Although alfalfa lacks significant levels of both PPO activity and o-diphenol compounds in its leaves, red clover's natural system of protein protection has been transferred to this forage legume by expressing a red clover PPO transgene in alfalfa and exogenously adding o-diphenol PPO substrates to the resulting tissues or tissue extracts (Sullivan et al., 2004; Sullivan and Hatfield, 2006). Because ruminant animals poorly utilize degraded protein, adaptation of the PPO system to alfalfa and other forage crops would have substantial positive economic and environmental impacts (Sullivan and Hatfield, 2006). Unfortunately, lack of system components in these forage crops, especially the o-diphenol PPO substrates, presents a challenge to practical adaptation of this natural system of protein preservation. Consequently, understanding how red clover is able to accumulate o-diphenols such as phaselic acid will be a key step to adapt the PPO/o-diphenol system to a wide range of economically important forage crops.
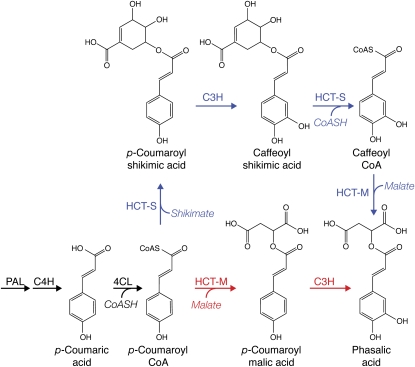
The biosynthetic pathways whereby red clover synthesizes and accumulates phaselic acid and other o-diphenols have not been defined. However, in the Brassicaceae, hydroxycinnamoyl esters with malic acid can be made via the action of sinapoyl-Glc:malate sinapoyltransferase (SMT; EC 2.3.1), which is capable of transferring a hydroxycinnamoyl moiety from a hydroxycinnamoyl-Glc ester to a malic acid acceptor. In Arabidopsis (Arabidopsis thaliana), SNG1 (for sinapoylglucose accumulator 1), which encodes the enzyme, has been shown to be responsible for the accumulation of sinapoylmalate in seeds and leaves (Lehfeldt et al., 2000). An SMT from radish (Raphanus sativus), presumably the homolog of the Arabidopsis SNG1 gene product, has been purified to apparent homogeneity and characterized (Grawe et al., 1992). The purified enzyme is capable of utilizing sinapoyl-, feruloyl-, caffeoyl-, and to a lesser extent p-coumaroyl-Glc esters to form the corresponding malic acid esters, suggesting that it is responsible for the accumulation of these esters in vivo. In contrast, in many plants, formation of certain hydroxycinnamoyl esters is often mediated by a member of the BAHD transferase family (D'Auria, 2006) that utilize a CoA thiolester hydroxycinnamoyl donor. Some of the best characterized of these hydroxycinnamoyl transferases (HCTs) are those associated with the biosynthesis of monolignols (Hoffmann et al., 2003, 2004; Shadle et al., 2007). These are capable of transferring p-coumaroyl or caffeoyl moieties from the respective CoA thiolesters to form 5-O-esters with shikimic acid or, to a lesser extent, 3-O-esters with quinic acid. Separable enzymatic activities capable of transferring a p-coumaroyl moiety to either shikimate/quinate or to 4′-hydroxyphenyllactate in basil (Ocimum basilicum) peltate gland extracts have been identified, although genes encoding these activities have not been cloned (Gang et al., 2002). Niggeweg et al. (2004) used gene-silencing experiments to definitively demonstrate that a hydroxycinnamoyl-CoA:quinate hydroxycinnamoyl transferase (HQT) is responsible for chlorogenic acid accumulation in the Solanaceae. Although phaselic acid biosynthesis in red clover could be via a pathway utilizing SMT, lack of an apparent SNG1 homolog in a collection of red clover EST sequences derived from leaves and young plants suggests otherwise (see “Discussion”). Therefore, pathways in red clover for the biosynthesis of phaselic acid utilizing one or more BAHD family transferase (Fig. 1) should be considered. In these proposed pathways, Phe would be converted to p-coumaroyl-CoA by the sequential action of Phe ammonia lyase (PAL), cinnamate-4-hydroxylase (C4H), and 4-coumarate:CoA ligase (4CL). The action of one or more specific HCTs and one or more p-coumarate 3′-hydroxylases (C3Hs) would then result in the formation of phaselic acid.
Figure 1.
Possible pathways for phaselic acid biosynthesis in red clover. Proposed pathway enzymes for the production of phaselic acid include PAL, 4CL, hydroxycinnamoyl:shikimate transferase (HCT-S), hydroxycinnamoyl:malate transferase (HCT-M), and C3H. The branch point at p-coumaroyl-CoA represents two alternative pathways. For simplicity, not all reactants and products are shown.
Existing literature suggests that C3H enzymes, which are cytochrome P450 enzymes (CYP98A subfamily), do not directly hydroxylate p-coumaric acid to caffeic acid but rather act on p-coumaroyl ester derivatives. For example, the enzyme from Arabidopsis hydroxylates shikimic and quinic acid esters of p-coumaric acid but only poorly or not at all p-coumaric acid or its Glc or CoA esters (Schoch et al., 2001; Franke et al., 2002). Thus, one model of phaselic acid biosynthesis is the formation of 2-O-(p-coumaroyl)-l-malic acid (hereafter referred to as p-coumaroyl-malate) by a HCT and its subsequent hydroxylation by a C3H enzyme capable of utilizing the malic acid ester as a substrate (Fig. 1, bottom, red pathway). An alternative model would require at least two HCT activities for phaselic acid biosynthesis (Fig. 1, top, blue pathway). The first activity would form a substrate suitable for hydroxylation (e.g. p-coumaroyl-shikimate, since several characterized C3H enzymes appear to favor this substrate [Schoch et al., 2001; Franke et al., 2002; Gang et al., 2002; Morant et al., 2007]). Following hydroxylation to the caffeoyl derivative by a C3H, the first HCT activity could synthesize caffeoyl-CoA via its reverse reaction (Hoffmann et al., 2003; Niggeweg et al., 2004). A second HCT activity would then transfer the caffeoyl moiety to malic acid to form phaselic acid. Both pathways predict a transferase capable of transferring a hydroxycinnamoyl moiety (either p-coumaroyl or caffeoyl) to malic acid. Also, these pathways are consistent with the observation that, at least in vitro, several characterized HCT enzymes are capable of transfer reactions utilizing either p-coumaroyl- or caffeoyl-CoA (Hoffmann et al., 2003; Niggeweg et al., 2004). The identification and characterization of two distinct HCTs from red clover, one of which has properties consistent with a role in phaselic acid biosynthesis, are reported here.
RESULTS
Phaselic Acid and p-Coumaroyl-Malate Content of Red Clover Tissues
To confirm and extend previous studies estimating the phaselic acid (caffeoyl-malate) content of red clover leaves (Hatfield and Muck, 1999; Sullivan and Hatfield, 2006; Winters et al., 2008), we measured extractable phaselic acid and p-coumaroyl-malate from red clover leaves, stems, and flowers. Phaselic acid content was detected in all tissues examined but was present at 6- to 10-fold higher levels in leaves compared with stems and flowers, respectively (Table I). p-Coumaroyl-malate, a potential precursor to phaselic acid, was detected in leaves, but at lower levels than the caffeoyl ester, and in flowers at levels similar to that seen for the caffeoyl ester. p-Coumaroyl-malate was not detected in stems. The phaselic acid content of red clover leaves measured here (4 μmol g−1 fresh weight) is in good agreement with our previous estimate based on enzymatic browning (10–15 μmol g−1 fresh weight; Sullivan and Hatfield, 2006) and that measured by Winters et al. (2008) for field-grown red clover (5–20 μmol g−1 fresh weight). p-Coumaroyl- and caffeoyl-shikimate esters, the products of several characterized HCTs (Hoffmann et al., 2003, 2004; Shadle et al., 2007), were not detected in the red clover tissues, even though the methods employed would allow their detection.
Table I.
Hydroxycinnamoyl-malate content (μmol g−1 fresh weight) of red clover tissues
| Tissue | p-Coumaroyl-Malate | Phaselic Acid |
|---|---|---|
| Leaves | 1.3 | 4.0 |
| Stems | NDa | 0.4 |
| Flowers | 0.4 | 0.6 |
ND, Not detected.
Identification and Cloning of HCT Genes from Red Clover
To identify HCT sequences from red clover, degenerate oligonucleotide primers were designed based on hydroxycinnamoyl-CoA transferase sequences from several species encoding proteins reported to be capable of transferring hydroxycinnamic acids to shikimic or quinic acids. These sequences included those derived from tomato (Solanum lycopersicum; GenBank accession no. AJ582652 and The Gene Index Project [TGI; compbio.dfci.Harvard.edu/tgi] no. TC138039), Medicago truncatula (TGI no. TC114220), and Arabidopsis (GenBank accession no. NM_124270). The primers were used in PCR with cDNA derived from young red clover leaves, and the resulting approximately 880-bp DNA fragments were cloned. cDNA from young leaves was used as the PCR template, since this tissue contains abundant o-diphenols, but should not be highly lignified. Several plasmids carrying the 880-bp fragment were sequenced. All were nearly identical to each other and highly similar to the M. truncatula and Arabidopsis sequences (92% and 70% nucleotide sequence identity, respectively) on which the primer design was based.
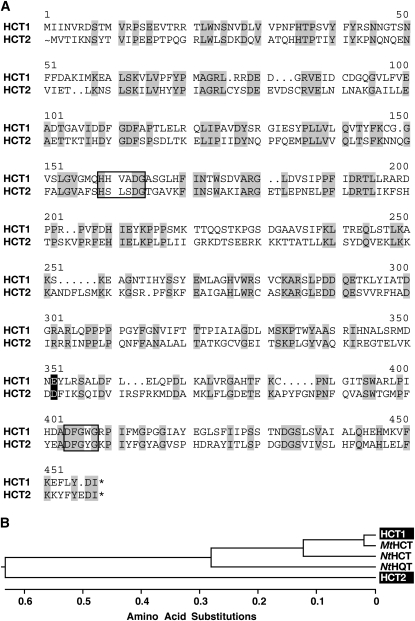
To obtain full-length cDNAs corresponding to red clover HCT genes, a young leaf cDNA library was screened by hybridization under moderate stringency (0.8 m NaCl, 55°C) using the 880-bp HCT fragment obtained above as the hybridization probe. Using this approach, six cDNA clones were identified that correspond to two unique sequences. These sequences, designated HCT1A and HCT1B, are nearly identical to each other and the 880-bp hybridization probe used to isolate them and share 91% nucleotide sequence identity with the M. truncatula TGI sequence TC114220. Coding regions of HCT1A and HCT1B, predicted to encode 434-amino acid (48.3-kD) proteins, have seven nucleotide differences, only one of which alters the amino acid sequence resulting in substitution of Asp (HCT1B) for Glu (HCT1A) at position 337 (Fig. 2). The encoded proteins contain conserved motifs characteristic of BAHD acyltransferases (D'Auria, 2006) and are highly similar (96% identity) to the M. truncatula protein predicted to be encoded by TC114220. The predicted HCT1 proteins are 78% and 58% identical to well-characterized Nicotiana tabacum HCT (GenBank accession no. AJ507825; Hoffmann et al., 2003) and HQT (GenBank accession no. AJ582651; Niggeweg et al., 2004) that mediate the transfer of hydroxycinnamoyl moieties from CoA derivatives to shikimic and quinic acids, respectively.
Figure 2.
Amino acid sequence similarity of HCTs. A, Amino acid sequence comparison of red clover HCT1A and HCT2. Identities are shaded in gray. The amino acid at the position shaded in black corresponds to the single substitution observed for HCT1B (D for E). Conserved BAHD acyltransferase domains are boxed. B, Phenogram of amino acid sequences of HCT1 and HCT2 from red clover (shaded in black), MtHCT (TGI no. TC114220) from M. truncatula, and NtHCT and NtHQT (GenBank accession nos. AJ507825 and AJ582651, respectively) from N. tabacum. The phenogram was generated by the neighbor-joining method following alignment by the ClustalW algorithm using Lasergene MegAlign software.
To identify additional HCT sequences in red clover that might lack sufficient homology to the 880-bp HCT gene fragment to be detected in a library screen, a bioinformatics approach was used. The amino acid sequence predicted to be encoded by HCT1A was used as the query in a tBLASTn search of a collection of EST sequences derived from red clover leaves and 3-week-old whole plants (Sato et al., 2005). In addition to two ESTs encoding proteins nearly identical to those encoded by the already cloned HCT1A and HCT1B sequences (GenBank accession nos. BB911266 and BB903226), this search identified nine overlapping ESTs having amino acid sequence similarity (approximately 35% identity) to the C-terminal half of the predicted HCT1 proteins. These nine ESTs are nearly identical to each other (more than 99% nucleotide sequence identity), with GenBank accession BB926056 being the most 5′ EST identified. No additional overlapping ESTs were identified using BB926056 as the query in Megablast searches of the red clover EST collection. For this reason, a 5′ RACE PCR approach using cDNA from unexpanded red clover leaves as template was used to obtain a full-length cDNA sequence corresponding to this new HCT gene, designated HCT2 (Fig. 2). The HCT2 coding region shares 53% nucleotide sequence identity with that of HCT1 and is predicted to encode a 454-amino acid (51.5-kD) protein that is only 35% identical to red clover HCT1 and 37% identical to N. tabacum HCT and HQT proteins. Like HCT1, HCT2 contains motifs conserved among BAHD acyltransferases (D'Auria, 2006). The sequences most closely related to HCT2 identified in BLASTn and tBLASTn searches of GenBank were M. truncatula genomic sequences CU468290 (71% amino acid identity) and CT954272 (65% amino acid identity) and M. truncatula EST BG454551 (60% amino acid identity). This level of sequence identity is relatively low compared with that typically observed (more than 90%) for functional homologs from red clover and M. truncatula (Sullivan and Thoma, 2006). Other legume EST sequences, including those from Phaseolus vulgaris, Glycine max, and Vigna unguiculata, are predicted to encode proteins with only 50% to 60% amino acid sequence identity to HCT2. Together, these results suggest that red clover HCT2 may be a unique transferase, even among legumes.
Red Clover HCT1 and HCT2 Are Differentially Expressed
The expression patterns of red clover HCT1 and HCT2 were examined using quantitative real-time PCR (q-rtPCR). cDNA made from RNA prepared from various red clover tissues and primers designed to detect HCT1 or HCT2 were used in these experiments. Specificity of the HCT1 and HCT2 primers was validated using dilutions of the cloned genes as the amplification target in rtPCR. These validation experiments showed that the HCT1 primer pair recognized HCT1A and HCT1B with similar efficiencies but failed to detect HCT2, whereas the HCT2 primer pair detected HCT2 but failed to detect either of the HCT1 genes (data not shown), and that Ct (for cycle threshold) varied with DNA concentration as expected (i.e. 1ΔCt corresponds to a 2-fold change in target DNA concentration). Additionally, mRNA levels of the housekeeping gene EIF4A (for eukaryotic initiation factor 4A) determined by q-rtPCR were used to normalize expression data, as has been done previously in expression analyses in other plant species, including Arabidopsis (Taylor et al., 1993), tomato (Howe et al., 2000), and Lotus corniculatus (Chen et al., 1998).
As shown in Figure 3, both HCT1 and HCT2 mRNAs are abundant in flowers. For HCT1, mRNA abundance in stems is similar to that observed in flowers and 4- to 5-fold higher than that in unexpanded or mature leaves. In contrast, HCT2 mRNA abundance in unexpanded or mature leaves is about half that observed in flowers but more than 10-fold higher than that observed in stems. The relatively high-level expression of HCT1 in lignifying stems is consistent with a role in monolignol biosynthesis, whereas the relatively high level of expression of HCT2 in leaf tissue is consistent with it having a role in secondary metabolite biosynthesis in this tissue. That both HCT1 and HCT2 show high levels of expression in flowers suggests both play roles in biosynthesis of secondary metabolites in floral tissues.
Figure 3.
Expression of HCT1 and HCT2 in various red clover tissues. mRNA abundance in unexpanded leaves (UL), mature leaves (ML), stems (S), and flowers (F) was determined by q-rtPCR. Expression levels for the various tissues were normalized to the expression of EIF4A and presented relative to that of flowers, which had the highest level expression for both HCT genes. Data are averages of four biological replicates, and error bars represent se.
Expression of Red Clover HCT1 and HCT2 Proteins in Escherichia coli and Evaluation of Their Enzymatic Activities in Vitro
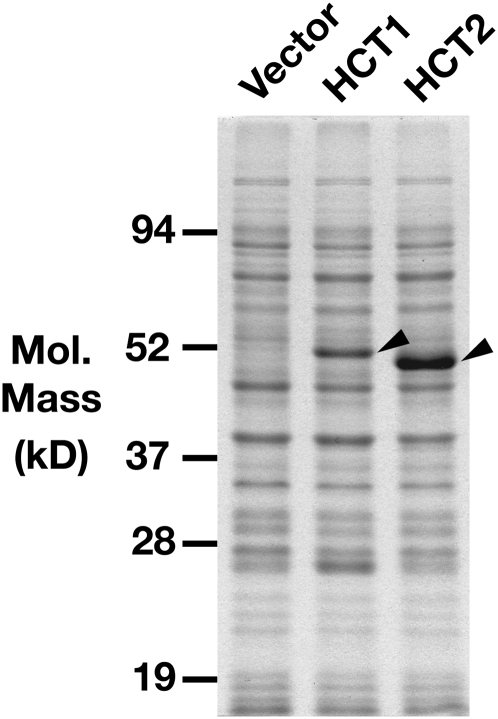
To study the enzymatic activities of the red clover HCT1 and HCT2 proteins, their entire coding regions were cloned into pET vectors for expression in E. coli. Because HCT1A and HCT1B encode nearly identical proteins, only HCT1A was evaluated in this study. The resulting constructs were designed to express full-length native versions of the proteins and had no introduced mutations that would change the amino acid sequence of the encoded proteins. Following transformation of the expression constructs into the E. coli host strain BL21(DE3)RIL, induction with isopropyl-β-d-thiogalactoside (IPTG) resulted in accumulation of nearly all of the HCT protein as insoluble inclusions when cultures were grown at 37°C (data not shown). When bacterial cultures were grown at 22°C following induction, however, approximately 50% of both HCT1 and HCT2 protein accumulated as soluble protein (Fig. 4; data not shown). Both proteins were easily detectible on SDS-PAGE gels following staining, although HCT2 accumulated to higher levels than HCT1. Relative mobility of HCT2 was slightly higher than that of HCT1 despite the slightly higher predicted molecular mass of HCT2. This likely represents anomalous migration in the SDS-PAGE gel system, since sequencing the entire coding regions and cloning junctions of both expression constructs showed them to be free of errors.
Figure 4.
Expression of HCT1 and HCT2 proteins in E. coli. Soluble extracts (5 μg of protein) of E. coli transformed with the pET28 vector (Vector) or HCT1 or HCT2 expression constructs were resolved on a 10% SDS-PAGE gel. Arrowheads indicate protein bands derived from the HCT1 and HCT2 expression constructs.
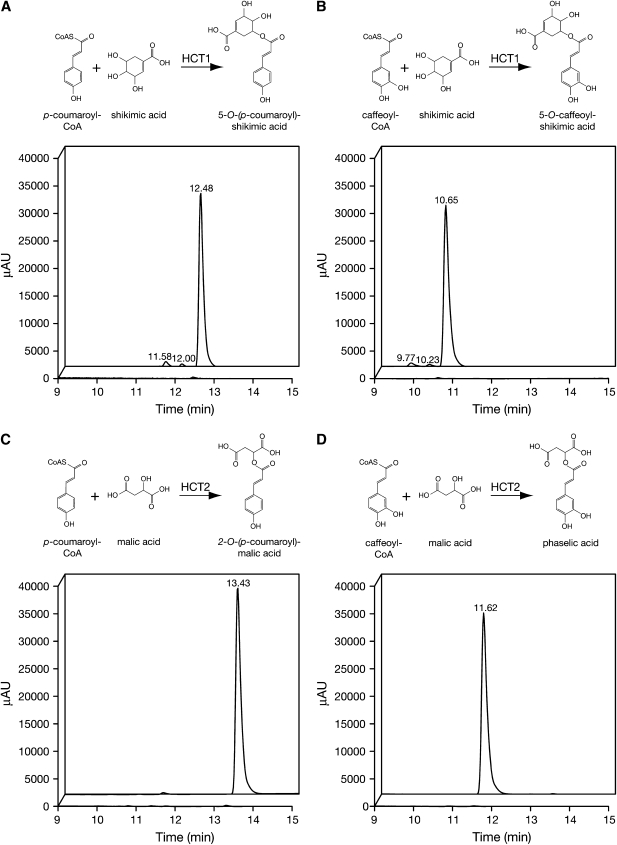
To examine enzymatic activities of the proteins, extracts of E. coli transformed with HCT1 or HCT2 expression constructs or a control extract from E. coli transformed with the empty pET28 vector were incubated with p-coumaroyl- or caffeoyl-CoA and an acceptor for the transferase reaction. Acceptors included shikimic acid, since it has been shown to be a preferred acceptor for many characterized HCTs implicated in monolignol biosynthesis (Hoffmann et al., 2003, 2004; Shadle et al., 2007); malic acid, since transfer to this moiety would be expected if either of the enzymes was directly involved in phaselic acid biosynthesis; quinic acid, Glc, and 3,4-dihydroxy-l-Phe (l-DOPA). Following incubation at 30°C for 60 min, reactions were quenched with acid and resolved by reverse-phase HPLC. For HCT1, incubation with either hydroxycinnamoyl-CoA derivative and shikimic acid resulted in the appearance of reaction products whose elution times and absorption spectra were indistinguishable from those of authentic 5-O-(p-coumaroyl)-shikimate or 5-O-caffeoyl-shikimate (Fig. 5, A and B; data not shown). Mass spectral analysis of the HCT1 reaction products gave mass-to-charge ratios of 319.09 and 335.08, in good agreement with the expected values of 319.08 and 335.08 for the p-coumaroyl-shikimate and caffeoyl-shikimate ions. No reaction products were detected when HCT1 and either hydroxycinnamoyl-CoA derivative were incubated with malic acid, Glc, or l-DOPA (data not shown). Reaction products were detected when HCT1 and either hydroxycinnamoyl-CoA derivative were incubated with quinic acid, although the amount of product formed was less than 2% of that formed when shikimic acid served as the acceptor (data not shown). Although mass spectral analysis was not carried out on these quinic acid reaction products, they are likely 3-O-(p-coumaroyl)- and 3-O-caffeoyl-quinate (chlorogenic acid) based on their UV absorption spectra and elution profiles. For HCT2, incubation with either p-coumaroyl- or caffeoyl-CoA and malic acid resulted in the appearance of reaction products (Fig. 5, C and D). Elution time and absorption spectrum of the product of the reaction of p-coumaroyl-CoA and malic acid were indistinguishable from those of authentic 2-O-(p-coumaroyl)-l-malate (data not shown). While no authentic phaselic acid standard was available for this analysis, elution of the product of the reaction of caffeoyl-CoA and malic acid is distinct from that of pure caffeic acid (11.62 versus 8.27 min), ruling out the possibility of simple hydrolysis of the caffeoyl-CoA reactant, and its UV absorption spectrum is similar to, but distinct from, that of caffeic acid (data not shown). Mass spectral analysis of the HCT2 reaction products gave mass-to-charge ratios of 279.05 and 295.05, in agreement with the expected values of 279.05 and 295.05 for the p-coumaroyl-malate and caffeoyl-malate ions. No reaction products were detected when HCT2 and either hydroxycinnamoyl-CoA derivative were incubated with shikimic acid, quinic acid, Glc, or l-DOPA (data not shown). Furthermore, no reaction products were detected upon incubation of either of the hydroxycinnamoyl-CoA derivatives and any of the acceptors with control extract from E. coli transformed with the empty pET28 vector (data not shown).
Figure 5.
Analysis of HCT1 and HCT2 reaction products by reverse-phase HPLC. Extracts of E. coli expressing HCT1 or HCT2 were incubated with hydroxycinnamoyl-CoA derivatives and shikimic or malic acid as indicated. Reaction samples at 0 and 60 min (foreground and background of each panel, respectively) were resolved by reverse-phase HPLC, and elution was monitored using a UV/Visible photodiode array detector (250–500 nm). Small peaks eluting just before the main 5-O-(p-coumaroyl)- and 5-O-(caffeoyl)-shikimic acid esters (A and B) are likely due to isomerization of the 5-O- products to 3-O- and 4-O- products that is known to occur at physiological pH (Kuhnl et al., 1987). AU, Absorbance units.
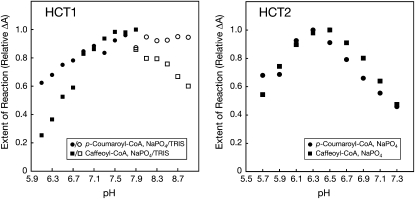
To optimize reaction conditions, the effect of pH on reaction rate was assessed by measuring loss of p-coumaroyl- or caffeoyl-CoA spectrophotometrically (Fig. 6). Reaction conditions were selected such that the extent of reaction was less than 30% of the maximum possible observed, so that the change in A would be an accurate reflection of reaction rate. For HCT1 with shikimic acid as the acceptor, highest reaction rates were observed for both hydroxycinnamoyl-CoA derivatives between pH 7.5 and 7.9 using sodium phosphate buffer, the upper range of pH that can be achieved using this buffer system. Although HCT1 activity remained high through pH 8.9 using Tris buffer, the measured activity was lower than that observed using phosphate buffer. For HCT2 with malic acid as the acceptor, highest reaction rates were observed for both hydroxycinnamoyl-CoA derivatives between pH 6.1 and 6.7. Because of these results, subsequent enzyme activity experiments were carried out using pH-7.9 and -6.5 sodium phosphate buffer for HCT1 and HCT2, respectively.
Figure 6.
Effect of pH on HCT1 and HCT2 activity. HCT1 or HCT2 was incubated with p-coumaroyl-CoA (circles) or caffeoyl-CoA (squares) as indicated and either shikimic (HCT1) or malic (HCT2) acid in sodium phosphate (black symbols) or Tris (white symbols) buffers adjusted to the indicated pH. The extent of the reaction was assessed by measuring the loss of hydroxycinnamoyl-CoA thiolesters spectrophotometrically as described in “Materials and Methods” and is expressed as the change in A (at λmax for each thiolester) relative to the maximum change observed. Reactions were carried out using conditions (incubation time and amount of enzyme) that resulted in less than approximately 30% maximum hydroxycinnamoyl-CoA substrate utilization to ensure that any differences in reaction rates would be detectable.
Observations during the course of the above experiment suggested that reaction rates varied considerably for the different enzyme and substrate combinations. Therefore, reaction rates for transfer of p-coumaroyl or caffeoyl moieties from their CoA derivatives to shikimic acid by HCT1 or malic acid by HCT2 were determined by measuring product formation over time. For HCT1, hydroxycinnamoyl transfer to shikimic acid from p-coumaroyl-CoA was only slightly (1.2 times) faster than from caffeoyl-CoA (specific activities of 1,200 and 980 nkat mg−1 for p-coumaroyl- and caffeoyl-CoA, respectively). In contrast, for HCT2, hydroxycinnamoyl transfer to malic acid from p-coumaroyl-CoA was substantially (6.7 times) faster than from caffeoyl-CoA (specific activities of 34 and 5.1 nkat mg−1 for p-coumaroyl- and caffeoyl-CoA, respectively). Although the specific activities measured in this experiment are substantially higher for HCT1 than for HCT2, whether this difference is meaningful is unclear. This may simply be a reflection of a higher proportion of E. coli-expressed HCT1 being correctly folded and active or reaction conditions being more optimal for HCT1 than for HCT2. Together, these data indicate that HCT1 and HCT2 are capable of transferring p-coumaroyl or caffeoyl moieties from their respective CoA thiolesters to shikimic (HCT1) or malic (HCT2) acid acceptors. In the case of HCT2, p-coumaroyl-CoA appears to be the preferred substrate, at least under the reaction conditions tested.
Measurement of p-Coumaroyl-CoA Transferase Activity in Red Clover Tissues
If red clover HCT2 plays a role in phaselic acid biosynthesis in red clover tissues, especially leaves, the corresponding enzyme activity should be present in those tissues. Extracts were prepared from red clover leaves, stems, and flowers and depleted of endogenous HCT substrates and products by gel filtration. The extracts were then used in transferase reactions with p-coumaroyl-CoA and either malic or shikimic acid as acceptor. p-Coumaroyl-CoA was used as the hydroxycinnamoyl donor in these experiments because the higher rate of reaction seen for HCT2 using this substrate would allow more sensitive detection of its activity. Control reactions with no acceptor were also run to assess the level of thiolesterase activity present in the extracts. Reaction mixtures were analyzed by HPLC, and reaction products were quantified. As shown in Table II, p-coumaroyl-CoA:malate p-coumaroyl transferase activity was readily detected in red clover leaves and stems, with activity in leaves being more than 3-fold higher than in stems. This higher transferase activity in leaves is consistent with the higher phaselic acid content and level of HCT2 mRNA accumulation measured in this tissue. Substantial levels of p-coumaroyl-CoA:shikimate p-coumaroyl transferase activity were detected in both leaves and stems, with stems having approximately 10-fold more activity than leaves, consistent with the higher HCT1 mRNA accumulation measured in stems. That relatively high levels of p-coumaroyl-CoA:shikimate p-coumaroyl transferase activity observed in leaves as well as stems suggests that the HCT1 gene product could have roles not only in the biosynthesis of monolignols in stems but also in other phenylpropanoid secondary metabolites in leaves. Both tissues also contain substantial levels of thiolesterase activity that hydrolyzed the p-coumaroyl-CoA substrate to yield p-coumaric acid. The resulting loss of the CoA thiolester substrate may have resulted in an underestimation of transferase activity. No p-coumaroyl:malate transferase, p-coumaroyl:shikimate transferase, or thiolesterase activity was detected in flowers, but this might have been due to the extremely low protein content of the extracts prepared, which, even after concentration, had 40- and 150-fold less protein than the stem and leaf extracts, respectively.
Table II.
p-Coumaroyl-CoA transferase with shikimate and malate acceptors and thiolesterase activities (pkat mg−1 protein) in red clover tissues
| Tissue |
p-Coumaroyl-CoA Transferase
|
Thiolesterase | |
|---|---|---|---|
| Shikimate | Malate | ||
| Leaves | 16.0 | 2.0 | 17.0 |
| Stems | 150.0 | 0.6 | 23.0 |
| Flowers | NDa | ND | ND |
ND, Not detected.
DISCUSSION
In this report, two classes of HCTs were identified in red clover that differ substantially in sequence, expression pattern, and enzymatic activities. The first class, represented by HCT1A and HCT1B, have amino acid sequences highly similar to those of HCTs in M. truncatula, N. tabacum, and Arabidopsis (96%, 78%, and 77% identity, respectively) that have been implicated in monolignol biosynthesis (Hoffmann et al., 2003, 2004; Shadle et al., 2007). Near identity with the M. truncatula enzyme (96%) suggests that red clover HCT1 is its functional homolog. Supporting this, HCT1 mRNA accumulates to approximately 5-fold higher levels in stems than in leaves, and its gene product is capable of transferring p-coumaroyl moieties from the corresponding CoA derivative to shikimic acid, an activity that has been shown to be important in the biosynthesis of monolignol lignin precursors (Hoffmann et al., 2004; Shadle et al., 2007). Mirroring the mRNA accumulation results, p-coumaroyl-CoA:shikimate p-coumaroyl transferase activity levels are nearly 10-fold higher in red clover stems than in red clover leaves, although substantial transferase activity was detected in leaves. Despite the high levels of transferase activity in red clover leaves and stems, p-coumaroyl- and caffeoyl-shikimate were not detected in these tissues, presumably because they are intermediate compounds that fail to accumulate to measurable levels.
The second class of red clover HCT, represented by HCT2, appears to be unique. Searches of the GenBank database failed to identify any highly similar sequences, even among other legumes. Although amino acid sequences derived from M. truncatula genomic clones show the highest degree of sequence similarity to HCT2 (approximately 70%), this level of identity is far below the more than 90% identity usually observed for homologous gene products from these two species (Sullivan and Thoma, 2006). The expression pattern of HCT2 is distinct from that of HCT1, with HCT2 mRNA accumulating to approximately 10-fold higher levels in leaves than in stems. Finally, HCT2 is capable of transferring hydroxycinnamoyl moieties (p-coumaroyl and caffeoyl) from the corresponding CoA derivatives to malic acid but not to shikimic acid. This activity has not been previously described for any transferase enzymes but would be predicted for the phaselic acid biosynthetic pathways depicted in Figure 1. Although the specific activity of HCT2 expressed in E. coli is substantially lower than that of HCT1, this finding may have little significance. It could be that only a relatively small fraction of the soluble HCT2 produced in E. coli is correctly folded and active, since solubility alone is not indicative of correct folding (Novagen, 2005). Also, the reaction conditions used here could have been more optimal for the HCT1 reactions than those of HCT2. Consistent with the mRNA accumulation results, p-coumaroyl-CoA:malate p-coumaroyl transferase activity levels are higher in red clover leaves than in red clover stems. The expected p-coumaroyl- and caffeoyl-malate reaction products accumulate in leaves to levels similar to those estimated (Sullivan and Hatfield, 2006) or measured (Winters et al., 2008) previously. These hydroxycinnamoyl esters accumulated to a lesser extent in stems and flowers. Despite high HCT2 mRNA levels in flowers, accumulation of p-coumaroyl- and caffeoyl-malate was low relative to leaves on a fresh weight basis, raising the possibility that in flowers these hydroxycinnamoyl esters are metabolic intermediates or that limited flux through the biosynthetic pathway limits their accumulation. However, given the low protein concentration of flower extracts and the lack of detectable enzyme activity, for this tissue the accumulation of these hydroxycinnamoyl-malate esters observed in this study may be substantial.
Although additional experiments will be required to definitively implicate the HCT2 gene product in phaselic acid biosynthesis, the findings presented here (lack of a functional homolog in M. truncatula, a closely related legume species that does not accumulate phaselic acid; foliar expression pattern of its mRNA; and enzyme activity capable of transferring hydroxycinnamic moieties to malic acid) are consistent with a role for HCT2 in a red clover biosynthetic pathway for phaselic acid. More detailed analyses of HCT2 enzyme activity (e.g. determination of kinetic parameters for various donor and acceptor substrates for the forward reaction [formation of hydroxycinnamic esters] and determination of the potential for the reverse reaction [formation of hydroxycinnamoyl-CoA from hydroxycinnamic esters]) should provide additional insights into the in vivo functions of the enzyme. Down-regulation of HCT2 in red clover will provide the strongest evidence of the role of HCT2 in phaselic acid biosynthesis. Although we cannot rule out the possibility that in red clover phaselic acid is synthesized via the action of a sinapoyl-Glc:malate sinapoyltransferase, as appears to be the case for the biosynthesis of hydroxycinnamoyl-malic acid esters in the Brassicaceae, this seems unlikely given the lack of an apparent homolog to SNG1, the Arabidopsis gene encoding this enzyme. In tBLASTn searches of red clover ESTs, the best matches with Arabidopsis SNG1 shared only about 40% amino acid sequence identity, far below what is seen for several other enzymes involved in phenylpropanoid biosynthesis. For example, Arabidopsis genes encoding PAL, C4H, 4CL1, and HCT all have corresponding red clover ESTs whose predicted proteins share 70% to 90% identity with their Arabidopsis counterparts. Nevertheless, my laboratory is currently determining whether red clover leaf extracts contain SMT activity so that this alternative pathway can be either further investigated or ruled out.
Although the preliminary analyses presented here indicate that HCT2 more readily utilizes p-coumaroyl-CoA than caffeoyl-CoA, more detailed analyses of HCT2 enzyme activity should aid in determining the likelihood of phaselic acid formation directly from caffeoyl moieties in vivo (i.e. the blue pathway Fig. 1, top). If transfer from p-coumaroyl moieties is favored over caffeoyl moieties and the red pathway in Figure 1 (bottom) is favored in vivo, a red clover C3H capable of hydroxylating p-coumaroyl-malate must exist. My laboratory has identified a single red clover C3H (CYP98A44) through a conventional library screen and BLAST searches of available red clover ESTs. We are currently characterizing the red clover C3H enzyme to determine whether it is capable of hydroxylating p-coumaroyl-malate and how likely this reaction is to occur in vivo. Even if the red clover C3H is in-capable of hydroxylating p-coumaroyl-malate, it seems likely it will be capable of hydroxylating p-coumaroyl-shikimate like many of the already characterized C3H enzymes (Schoch et al., 2001; Franke et al., 2002; Gang et al., 2002; Morant et al., 2007). Because red clover leaves appear to have sufficiently high levels of p-coumaroyl-CoA:shikimate p-coumaroyl transferase activity to provide p-coumaroyl-shikimate as a hydroxylation substrate, the blue pathway in Figure 1 (top) remains plausible.
In the course of identifying HCT2, red clover genes encoding at least six additional related HCTs were also identified. These HCTs share 65% to 95% amino acid sequence identity with HCT2 but only approximately 35% amino acid sequence identity with HCT1. Given the limited sequence identity of their protein products with HCT1, it seems unlikely that HCT2 or any of these additional red clover HCT genes would have been identified using hybridization-based library screens, underscoring the utility of genomics-based approaches. The additional red clover HCT genes and corresponding proteins are currently being characterized to determine in vivo function. One or more of these HCTs could play a role in the biosynthesis of clovamide [N-(caffeoyl)-(3,4-dihydroxy-l-Phe)], another major o-diphenol accumulating in red clover leaves (Hatfield and Muck, 1999; Winters et al., 2008).
Phaselic acid and other o-diphenols along with PPO constitute a natural system of protein protection in red clover (Lee et al., 2004; Sullivan et al., 2004; Sullivan and Hatfield, 2006; Winters et al., 2008). Unfortunately, aerial tissues of alfalfa and other important forage crops lack both PPO and the o-diphenol substrates that lead to protection of protein in harvested forage crops. Because ruminant animals poorly utilize degraded protein, it is estimated that adaptation of the PPO/o-diphenol system to alfalfa alone could save U.S. farmers more than $100 million annually and prevent the release of excess nitrogen into the environment (Sullivan and Hatfield, 2006). There are also indications that milk and meat products with healthier lipid profiles result when ruminant animals are fed diets high in PPO- and o-diphenol-containing forage crops (Lee et al., 2004, 2006). Besides roles in protein preservation and healthier lipid profiles for meat and milk, o-diphenols are natural antioxidants; consequently, they have potential to be used in human and animal nutrition (Niggeweg et al., 2004, and refs. therein). Identification of red clover HCT2 may be a crucial first step in understanding o-diphenol biosynthetic pathways in red clover and in recreating them in alfalfa and other important forage crops. Knowledge of the pathway may also allow it to be controlled in red clover and other crops in situations where undesirable browning occurs due to the oxidation of abundant o-diphenols.
MATERIALS AND METHODS
Reagents
All purchased reagents were of molecular biology or higher grade. 5-O-(p-Coumaroyl)-shikimate, 2-O-(p-coumaroyl)-l-malate, and 5-O-caffeoyl-shikimate standards were prepared by the method described by Hemmerle et al. (1997). The NMR spectra of the standards were consistent with the assigned structures. 2-O-Caffeoyl-l-malate was so readily oxidized during its chemical synthesis that the authentic standard prepared in this manner was unavailable for this study. 2-O-Caffeoyl-l-malate prepared enzymatically using HCT2 and whose identity was confirmed by UV spectroscopy and mass spectrometry (described below and in “Results”) was used as a standard for experiments examining the hydroxycinnamoyl-l-malate content of red clover (Trifolium pratense) tissues.
Caffeoyl- and p-coumaroyl-CoA thiolesters were prepared using recombinant Arabidopsis (Arabidopsis thaliana) 4CL1 protein (Lee et al., 1995) produced in Escherichia coli using the pET30 expression vector (Novagen). The thiolesters were synthesized in 200-mL reactions consisting of 400 mm Tris (pH 7.8), 0.5 mm CoA, 5 mm ATP, 5 mm MgCl2, 1 mm caffeic or p-coumaric acid, and 0.5 mL of E. coli extract (equivalent to a 5-mL culture induced with IPTG and grown overnight at 16°C). The reaction mixture was incubated at 37°C and monitored by measuring the A346 (for caffeoyl-CoA) or A333 (for p-coumaroyl-CoA). When no additional change in A at λmax was observed (approximately 2 h), the entire reaction mixture was applied to a 20-mL ENVI-18 solid-phase extraction column (Supelco) preequilibrated with 3 × 20 mL of methanol and 3 × 20 mL of 50 mm Tris, pH 7.8. The column was washed with 3 × 20 mL of 50 mm Tris, pH 7.8, and 20 mL of water. The CoA thiolesters were eluted with 20 mL of methanol, the methanol was removed by evaporation, and the thiolesters were dissolved in approximately 4 mL of 25 mm MOPS buffer, pH 7.5. Concentration of the thiolesters was determined spectrophotometrically using extinction coefficients of 21 and 18 mm−1 cm−1 for p-coumaroyl-CoA (λmax = 333 nm) and caffeoyl-CoA (λmax = 346 nm; Stoeckigt and Zenk, 1975), respectively.
Plant Materials
A red clover genotype (lab designation “PPO”; Sullivan et al., 2004) selected from a population of WI-2 germplasm (Smith and Maxwell, 1980) was the source material for the isolation of HCT genes. This same plant, and three additional genotypes derived from the WI-2 germplasm, were used for expression and other analyses. The plants were clonally propagated from crown pieces and grown in a greenhouse with temperatures maintained between 20°C and 30°C and light intensities between 400 and 1,000 μmol m−2 s−1. Supplemental lighting was used when daylength was less than 13 h per day. Plants were fertilized weekly with Peter's soluble 20-10-20 (Scott's).
HPLC Analysis and Quantitation of Hydroxycinnamoyl Esters
Aliquots of samples of interest were transferred to 0.3-mL autosample vials and analyzed by HPLC on a Gemini 5-μm C6-Phenyl 110Å column (Phenomenex; 250 × 3.0 mm × 5 μm) using a two-solvent system (solvent A, deionized water with 0.0125% [v/v] trifluoroacetic acid; solvent B, acetonitrile with 0.0125% [v/v] trifluoroacetic acid) at a flow rate of 1 mL min−1. For analysis of hydroxycinnamoyl esters from red clover tissues, the HPLC conditions were 5 min of isocratic 10% solvent B, 30-min gradient to 25% solvent B, 3 min of isocratic 25% solvent B, 1-min gradient to 100% solvent B, 4 min of isocratic 100% solvent B, 5-min gradient to 10% solvent B, and 5 min of isocratic reequilibration at 10% solvent B. For analysis of HCT reactions, HPLC conditions were 5 min of isocratic 10% solvent B, 8-min gradient to 34% solvent B, 3 min of isocratic 34% solvent B, 1-min gradient to 100% solvent B, 4 min of isocratic 100% solvent B, 5-min gradient to 10% solvent B, and 5 min of isocratic reequilibration at 10% solvent B. Compound elution was monitored (250–500 nm) with a UV/Visible photodiode array detector. Peaks of interest were quantified with Xcaliber software (Thermo Scientific) using p-coumaric and caffeic acids as standards as described by Nielsen et al. (1984).
Analysis of Soluble Hydroxycinnamoyl Ester Content of Red Clover Tissues
Red clover young fully expanded leaves, stems, or flowers were powdered with a mortar and pestle in liquid nitrogen and extracted with 5 mL g−1 fresh weight of 50 mm MOPS and 50 mm ascorbic acid, pH 7.5, and the resulting slurry was filtered through Miracloth (Calbiochem). The filtrate was acidified by the addition of 0.125 volume of 1 n HCl and centrifuged for 10 min at 22,000g. A portion (1 mL) of the acidified clarified extract was applied to a 1-mL ENVI-18 solid-phase extraction column preequilibrated with 3 × 1 mL of methanol and 3 × 1 mL of 0.1% acetic acid in water, pH adjusted to 2.75 with HCl. The column was washed with 6 × 1 mL of 0.1% acetic acid in water (pH 2.75 with HCl) and eluted with 1 mL of methanol. The eluate was analyzed by HPLC, and hydroxycinnamoyl esters were quantified as described above.
Nucleic Acid Methodologies
Preparation of RNA and cDNA, Plasmid Preparation, and Sequence Analysis
Total RNA was prepared from plant tissues by the method of Chang et al. (1993). Oligo(dT)-primed cDNA was prepared using SuperScript III reverse transcriptase according to the manufacturer's protocol (Invitrogen) from DNase I-treated total RNA. Plasmid DNA was prepared using the QIAprep Spin Miniprep Kit (Qiagen). DNA sequence was determined by cycle sequencing using Big Dye version 3.1 (Applied Biosystems) and run on ABI automated sequencers by the University of Wisconsin Biotechnology Center. Sequence analyses were carried out using the Wisconsin Package Version 10 (Accelrys), Lasergene Version 8 (DNAStar), and BLAST programs using the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) and TGI (compbio.dfci.Harvard.edu/tgi) Web sites.
Generation of a Red Clover HCT Gene Fragment by PCR and cDNA Library Screening
Oligo(dT)-primed cDNA from unexpanded red clover leaves corresponding to 25 ng of total RNA was used as a template with degenerate primers ms248 and ms252 (for a list of all primers used in these studies, see Table III) in PCR using JumpStart KlenTaq LA polymerase (Sigma) using 40 cycles of 30 s at 94°C, 30 s at 50°C, and 90 s at 68°C. The resulting DNA fragment was purified from an agarose gel using QIAEX II resin (Qiagen) and cloned into pGEM-T Easy (Promega) according to the manufacturer's instructions. Approximately 106 recombinant phage from a λ ZAP II (Stratagene) red clover young leaf cDNA library were screened essentially as described previously (Sullivan et al., 2004) using an 880-bp PCR-derived red clover HCT1 gene fragment (described above) as the hybridization probe. Phage clones of interest were rescued to pBluescript II SK− phagemids according to the manufacturer's protocol.
Table III.
Oligonucleotide primers for cloning and expression analysis
| Name | +/−a | Sequence (5′ to 3′) | Purpose |
|---|---|---|---|
| ms248 | + | GATGATTTTGGNGAYTTTRCWCC | HCT cloning |
| ms252 | − | AATNCMRSCWGGTCCCAT | |
| SMART | + | TACGGCTGCGAGAAGACGACAGAAGGG | HCT2 RACE |
| 5′ primer | + | TACGGCTGCGAGAAGACGACAGAA | |
| ms492 | − | CCCTTATCTTTTGTGCTACATAG | |
| ms493 | − | CCTAAAGGCTTTAATGTGATTTCTC | |
| ms521 | + | GATGTTAACTTAAACCATAATACC | HCT2 cloning |
| ms490 | − | GTTTGATAAATGCAAAGAACTAAACTTT | |
| ms407 | + | GCGCGCCATATGATCATAAACGTGAGAGAT | HCT1 expression |
| ms408 | − | CGCGCGCTCGAGTTATCAGATATCATACAAGAATTCC | |
| ms522 | + | GCCATGGTTACCATTAAAAATTCTTAC | HCT2 expression |
| ms523 | − | GCTCGAGTCATATATCCTCATAGAAGTAC | |
| ms381 | + | CGCGAATGGACAACGAGTATT | HCT1 q-rtPCR |
| ms382 | − | TACCAAGATTCGGGCACTTGA | |
| ms595 | + | ACACCCTCGAAAGTACCACGTT | HCT2 q-rtPCR |
| ms596 | − | TCTTCACTTGTGTCTTTTCTTCCTATAATG | |
| ms597 | + | GACATGGACCAAAATACTCGTGATA | EIF4A q-rtPCRb |
| ms598 | − | CAGTTGTTATGAGCACTCGTGAAGA |
Orientation: +, sense; −, antisense.
Based on GenBank accession BB919542.
5′ RACE and Cloning of Full-Length HCT2
A SMART oligonucleotide and 5′ primer combination (Clontech Laboratories) in conjunction with nested antisense primers (Table III; ms492 and ms493) specific for the HCT2 coding region based on the GenBank EST BB926056 were used for 5′ RACE. cDNA synthesis from mature red clover leaf RNA was carried out using the distal antisense primer (ms492) and SMART oligonucleotide essentially as described by Clontech using SuperScript III reverse transcriptase (Invitrogen). The resulting cDNA was used in nested PCR using first the distal antisense (ms492)/5′ primer combination (30 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 2 min) followed by the proximal antisense (ms493)/5′ primer combination (30 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 2 min). The major PCR product of these reactions was cloned into pGEM-T Easy and sequenced as described above. The 5′ HCT2 sequence information from this clone was used to design an oligonucleotide primer 5′ of the HCT2 start codon (ms521) that was used along with a primer immediately following the HCT2 coding region as predicted from BB926056 (ms490) to generate a full-length HCT2 cDNA fragment by PCR using oligo(dT)-primed mature red clover leaf cDNA prepared as described above. The resulting coding region fragments from multiple independent PCRs were cloned into pGEM-T Easy and sequenced as described above. Comparison of sequences from clones obtained from three independent PCRs allowed the identification of an error-free HCT2 cDNA clone.
q-rtPCR
Primers for q-rtPCR (Table III) were designed using Primer Express software (Applied Biosystems) using the default parameters for the Taqman protocol (melting temperature = 59°C ± 1°C; 30%–80% GC; length of nine to 40 nucleotides, optimal length = 20). q-rtPCR was carried out using SYBR Green PCR Master Mix (Applied Biosystems) in 25-μL reactions containing cDNA equivalent to 0.025 ng of total RNA and 50 nm each primer, with triplicate reactions run for each cDNA sample for a given primer pair. Reactions were run in an ABI Prism 7000 Sequence Detection System. Initial denaturation was 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 58°C for 1.5 min. Ct values were determined with auto Ct and auto baseline functions of the system, and the three experimental replicates were averaged. Relative mRNA levels were calculated assuming that 1ΔCt corresponds to a 2-fold change in mRNA level and normalized using expression levels of the housekeeping gene EIF4A (Taylor et al., 1993). HCT expression levels were averaged for four biological replicates, normalized to those of the tissue with the highest level of expression, and are reported with se.
Construction of Plasmids for Expression of Red Clover HCTs in E. coli
For all gene constructions, standard molecular biology techniques were used (Sambrook et al., 1989; Ausubel et al., 1998). When fragments for cloning were generated via PCR, the cloned insert was sequenced to ensure that no mutations were introduced that would alter the sequence of the translated protein.
Plasmids containing full-length red clover HCT1A or HCT2 coding regions were used as templates in PCRs with primers designed to introduce an NdeI (HCT1) or NcoI (HCT2) restriction site at the start codons and an XhoI site immediately following the stop codon of each open reading frame (ms407/ms408 and ms522/ms523 for HCT1 and HCT2, respectively; Table III). The resulting PCR products were digested with NdeI and XhoI (HCT1) or NcoI and XhoI (HCT2) and inserted into pET28a digested with NdeI and XhoI or pET42 (Novagen) digested with NcoI and XhoI, respectively.
Expression of Red Clover HCT Proteins in E. coli
pET28 derivatives containing HCT1 or HCT2 coding regions (detailed above) or pET28 (as a negative control) were transformed into BL21(DE3)RIL Codon Plus E. coli (Stratagene). Cultures of E. coli harboring HCT-containing or control empty vector plasmids were grown at 37°C with shaking (225 rpm) in Luria-Bertani medium supplemented with 50 μg mL−1 kanamycin to an optical density at 600 nm of approximately 0.5. Cultures were cooled on ice to approximately 22°C, induced by the addition of IPTG to 1 mm, and incubated with shaking for an additional 18 h at approximately 22°C. Cultures were lysed using BugBuster reagent (Novagen) according to the manufacturer's suggested procedures and fractionated into soluble and insoluble portions. Total protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories). SDS-PAGE (10% acrylamide; 37.5:1 acrylamide:bis-acrylamide) was carried out using standard methodologies (Harlow and Lane, 1988). Gels were stained using Gel Code Blue Stain Reagent (Pierce Biotechnology). Concentrations of soluble HCT1 and HCT2 in E. coli extracts were estimated by comparing staining intensity with known amounts of bovine serum albumin.
Analysis of Transferase Activity of Recombinant and Endogenous HCTs
HCT Enzyme Activity Assays
Initially, reaction mixtures for HCT activity contained 1 mm p-coumaroyl- or caffeoyl-CoA; 3 mm shikimic acid, malic acid, quinic acid, Glc, or l-DOPA; 25 mm ascorbate; and 0.125 mg mL−1 total E. coli extract in 50 mm MOPS buffer, pH 7.5. Reaction mixtures were incubated at 30°C for up to 60 min. Reactions were quenched by the addition of one-tenth volume of 10% (v/v) acetic acid in water (to give 1% [v/v] final concentration). Samples were diluted 5-fold with acidified water (pH 3.0 with acetic acid), passed through a 0.2-μm filter, and analyzed by HPLC as described above. For mass spectral analysis of reaction products, 1-mL reactions were carried out as described above and reaction progress was monitored spectrophotometrically. When no further changes were observed in the absorption spectra, the reaction mixtures for HCT1 with shikimic acid and p-coumaroyl- or caffeoyl-CoA and the reaction mixture for HCT2 with malic acid and p-coumaroyl-CoA were acidified by the addition of one-tenth volume of 10% (v/v) acetic acid in water and applied to a 1-mL ENVI-18 solid-phase extraction column preequilibrated with 3 × 1 mL of methanol and 3 × 1 mL of 0.1% acetic acid in water. The column was washed with 3 × 1 mL of 0.1% acetic acid in water, and the reaction product was eluted with 1 mL of methanol. For the reaction mixture for HCT2 with malic acid and caffeoyl-CoA, the solid-phase extraction procedure was essentially as described above, except that the reaction mixture was acidified by adding one-tenth volume of 1.2 m HCl and the column was equilibrated and washed using 0.1% acetic acid in water, pH adjusted to 2.75 with HCl. Following methanol elution, the sample was dried under a stream of nitrogen gas to remove residual HCl and then resuspended in methanol. These lower pH conditions were required for the product of this reaction (phaselic acid) to bind to the solid-phase extraction column matrix. A subsample of each purified reaction product was run on HPLC as described above to assess purity. The remaining sample was submitted to the University of Wisconsin Biotechnology Center Mass Spectrometry Facility for analysis on an Agilent liquid chromatography/mass selective detector time-of-flight instrument in negative ion mode.
Determination of pH Optima of Red Clover HCT1 and HCT2
Reaction mixtures (50 μL) containing 100 mm sodium phosphate (for pH 5.7–7.9) or Tris (for pH 7.9–8.9) buffer; 25 mm ascorbic acid; 3 mm shikimic acid (for HCT1) or malic acid (for HCT2); 1 mm p-coumaroyl- or caffeoyl-CoA; and 0.0125 μg of recombinant HCT1 or 0.5 or 5.0 μg of recombinant HCT2 (for p-coumaroyl- or caffeoyl-CoA substrate, respectively) were incubated at 30°C for 15 min. Reactions were stopped by adding 950 μL of 1% (v/v) acetic acid. The extent of the reaction was measured as the change in A333 or A346 (i.e. λmax for p-coumaroyl- or caffeoyl-CoA reactant, respectively) compared with an unincubated control. Reactions were carried out using conditions (incubation time and amount of enzyme) that resulted in less than approximately 30% maximum hydroxycinnamoyl substrate utilization to ensure that any differences in reaction rates would be detectable.
Determination of HCT1 and HCT2 Specific Activities
To determine reaction rates, reactions were run as described above except that the reaction buffer was 100 mm sodium phosphate, pH 7.9 (for HCT1) or pH 6.5 (for HCT2). Reactions contained 0.56 μg mL−1 HCT1 protein (for either CoA derivative) or 11.2 and 112 μg mL−1 HCT2 protein for reactions with p-coumaroyl- or caffeoyl-CoA, respectively. Following enzyme addition, samples were withdrawn at 0, 5, 10, 15, 30, and 60 min, quenched with acid, processed, and analyzed by HPLC as described above. Reaction rates were determined by plotting product formation versus time and calculating the slope from the initial linear portion of the curve.
Analysis of Endogenous HCT and Thiolesterase Activity in Red Clover Tissues
Red clover young fully expanded leaves, stems, or flowers were powdered in liquid nitrogen with a mortar and pestle and extracted with 1 mL g−1 fresh weight of 100 mm sodium phosphate and 50 mm ascorbic acid, pH 6.5. Protease inhibitor cocktail (P-9599; Sigma) was added to 1% (v/v) to the buffer immediately before extraction. The resulting slurry was filtered through Miracloth (Calbiochem), and the filtrate was centrifuged at 4°C for 5 min at 22,000g. Low Mr compounds, including endogenous phenylpropanoids, were removed from the clarified extracts essentially as described previously (Sullivan and Hatfield, 2006) using Sephadex G-25 (GE Healthcare Biosciences) spin columns equilibrated with 100 mm sodium phosphate buffer, pH 6.5. Protease inhibitor cocktail (0.5%, v/v) was added to the column eluate. Flower extract was concentrated 10-fold using a Microcon YM-10 centrifugal concentrator (Millipore). Protein concentrations of the extracts were determined as described above. To assess HCT activity in the extract, 250-μL reactions containing 177.25 μL of red clover tissue extract, 26 μL of p-coumaroyl-CoA solution (2 mm final), 31.25 μL of 200 mm ascorbic acid solution (pH 6.5 with NaOH; 25 mm final), and 15 μL of 100 mm malic or shikimic acid solution in water (6 mm final) or 15 μL of water as a no-acceptor control (to measure thiolesterase activity [release of free p-coumaric acid from p-coumaroyl-CoA]) were incubated at 30°C. Following extract addition, 50-μL samples were withdrawn at 0, 60, 180, and 360 min and processed and analyzed by HPLC as described above.
Sequences of red clover HCT1A, HCT1B, and HCT2 have been deposited in GenBank under accession numbers EU861218, FJ151489, and EU861219, respectively.
Acknowledgments
I thank Sara Zerbel and Lisa Koch for excellent technical assistance; Paul Schatz and John Ralph for providing hydroxycinnamoyl ester standards; Clint Chapple, Jo Cusumano, and Nick Bonawitz for providing the 4CL1 expression construct and helpful advice; and Jane Marita and Ron Hatfield for technical advice and helpful discussions and comments on the manuscript.
This work was supported by the U.S. Department of Agriculture-Cooperative State Research, Education, and Extension Service-National Research Initiative Competitive Grants Program (grant no. 2009–35318–05048).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michael L. Sullivan (michael.sullivan@ars.usda.gov).
Open Access articles can be viewed online without a subscription.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Deidman JG, Smith JA, Struhl K, editors (1998) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- Chen R, Silver DL, de Bruijn FJ (1998) Nodule parenchyma-specific expression of the Sesbania rostrata early nodulin gene SrEnod2 is mediated by its 3′ untranslated region. Plant Cell 10 1585–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria JC (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9 331–340 [DOI] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30 33–45 [DOI] [PubMed] [Google Scholar]
- Gang DR, Beuerle T, Ullmann P, Werck-Reichart D, Pichersky E (2002) Differential production of meta-hydroxylated phenlypropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant Physiol 130 1536–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawe W, Bachhuber P, Mock HP, Strack D (1992) Purification and characterization of sinapoylglucose-malate sinapoyltransferase from Raphanus sativus L. Planta 187 236–241 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, p 508
- Hatfield RD, Muck R (1999) Characterizing proteolytic inhibition in red clover silage. In XIIth International Silage Conference. Swedish University of Agriculture and Sciences, Uppsala, Sweden, pp 147–148
- Hemmerle H, Burger HJ, Below P, Schubert G, Rippel R, Schindler PW, Paulus E, Herling AW (1997) Chlorogenic acid and synthetic chlorogenic acid derivatives: novel inhibitors of hepatic glucose-6-phosphate translocase. J Med Chem 40 137–145 [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278 95–103 [DOI] [PubMed] [Google Scholar]
- Howe GA, Lee GI, Itoh A, Li L, DeRocher AE (2000) Cytochrome P450-dependent metabolism of oxylipins in tomato: cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol 23 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BA, Hatfield RD, Muck RE (1995) Screening legume forages for soluble phenols, polyphenol oxidase and extract browning. J Sci Food Agric 67 109–112 [Google Scholar]
- Kuhnl T, Koch U, Heller W, Wellmann E (1987) Chlorogenic acid biosynthesis: characterization of a light-induced microsomal 5-O-(4-coumaroyl)-d-quinate/shikimate 3′-hydroxylase from carrot (Daucus carota L.) cell suspension cultures. Arch Biochem Biophys 258 226–232 [DOI] [PubMed] [Google Scholar]
- Lee D, Ellard M, Wanner LA, Davis KR, Douglas CJ (1995) The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol Biol 28 871–884 [DOI] [PubMed] [Google Scholar]
- Lee MRF, Parfitt LJ, Minchin FR (2006) Lipolysis of red clover with differing polyphenol oxidase activities in batch culture. J Anim Sci 84 101–101 [Google Scholar]
- Lee MRF, Winters AL, Scollan ND, Dewhurst RJ, Theodorou MK, Minchin FR (2004) Plant-mediated lipolysis and proteolysis in red clover with different polyphenol oxidase activities. J Sci Food Agric 84 1639–1645 [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M, Schoch GA, Ullmann P, Ertunc T, Little D, Olsen CE, Petersen M, Negrel J, Werck-Reichhart D (2007) Catalytic activity, duplication and evolution of the CYP98 cytochrome P450 family in wheat. Plant Mol Biol 63 1–19 [DOI] [PubMed] [Google Scholar]
- Nielsen JK, Olsen O, Pedersen LH, Sorensen H (1984) 2-O-(p-Coumaroyl)-L-malate, 2-O-caffeoyl-L-malate and 2-O-feruloyl-L-malate in Raphanus sativus. Phytochemistry 23 1741–1743 [Google Scholar]
- Niggeweg R, Michael AJ, Martin C (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22 746–754 [DOI] [PubMed] [Google Scholar]
- Novagen (2005) pET System Manual, Ed 11. Novagen-EMD Biosciences, Madison, WI
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sato S, Isobe S, Asamizu E, Ohmido N, Kataoka R, Nakamura Y, Kaneko T, Sakurai N, Okumura K, Klimenko I, et al (2005) Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.). DNA Res 12 301–364 [DOI] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276 36566–36574 [DOI] [PubMed] [Google Scholar]
- Shadle G, Chen F, Reddy MSS, Jackson L, Nakashima J, Dixon RA (2007) Down-regulation of hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68 1521–1529 [DOI] [PubMed] [Google Scholar]
- Smith RR, Maxwell DP (1980) Registration of WI-1 and WI-2 red clover. Crop Sci 20 831 [Google Scholar]
- Steffens JC, Harel E, Hunt MD (1994) Polyphenol oxidase. In BE Ellis, GW Kuroki, HA Stafford, eds, Genetic Engineering of Plant Secondary Metabolism, Vol 28. Plenum Press, New York, pp 275–312
- Stoeckigt J, Zenk MH (1975) Chemical synthesis and properties of hydroxycinnamoyl-coenzyme A derivatives. Z Naturforsch [C] 30 352–358 [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Hatfield RD (2006) Polyphenol oxidase and o-diphenols inhibit postharvest proteolysis in red clover and alfalfa. Crop Sci 46 662–670 [Google Scholar]
- Sullivan ML, Hatfield RD, Thoma SL, Samac DA (2004) Cloning and characterization of red clover polyphenol oxidase cDNAs and expression of active protein in Escherichia coli and transgenic alfalfa. Plant Physiol 136 3234–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan ML, Thoma SL (2006) Cloning, molecular characterization, and expression analysis of several red clover cDNAs. Can J Plant Sci 86 465–468 [Google Scholar]
- Taylor CB, Bariola PA, Delcardayre SB, Raines RT, Green PJ (1993) Rns2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA 90 5118–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters AL, Minchin FR, Michaelson-Yeates TPT, Lee MRF, Morris P (2008) Latent and active polyphenol oxidase (PPO) in red clover (Trifolium pratense) and use of a low PPO mutant to study the role of PPO in proteolysis reduction. J Agric Food Chem 56 2817–2824 [DOI] [PubMed] [Google Scholar]