Abstract
Siderophores (ferric ion chelators) are secreted by organisms in response to iron deficiency. The pathogenic enterobacterium Erwinia chrysanthemi produces two siderophores, achromobactin and chrysobactin (CB), which are required for systemic dissemination in host plants. Previous studies have shown that CB is produced in planta and can trigger the up-regulation of the plant ferritin gene AtFER1. To further investigate the function of CB during pathogenesis, we analyzed its effect in Arabidopsis (Arabidopsis thaliana) plants following leaf infiltration. CB activates the salicylic acid (SA)-mediated signaling pathway, while the CB ferric complex is ineffective, suggesting that the elicitor activity of this siderophore is due to its iron-binding property. We confirmed this hypothesis by testing the effect of siderophores structurally unrelated to CB, including deferrioxamine. There was no activation of SA-dependent defense in plants grown under iron deficiency before CB treatment. Transcriptional analysis of the genes encoding the root ferrous ion transporter and ferric chelate reductase, and determination of the activity of this enzyme in response to CB or deferrioxamine, showed that these compounds induce a leaf-to-root iron deficiency signal. This root response as well as ferritin gene up-regulation in the leaf were not compromised in a SA-deficient mutant line. Using the Arabidopsis-E. chrysanthemi pathosystem, we have shown that CB promotes bacterial growth in planta and can modulate plant defenses through an antagonistic mechanism between SA and jasmonic acid signaling cascades. Collectively, these data reveal a new link between two processes mediated by SA and iron in response to microbial siderophores.
Iron is essential for most forms of life. It is required for the catalytic activity of proteins mediating electron transfer and redox reactions, such as those involved in respiration, photosynthesis, DNA synthesis, and defense against reactive oxygen species. However, it is often unavailable because it is present as insoluble ferric hydroxide complexes in aerobiosis and at neutral pH. In its ferrous form, iron is more soluble and catalyzes the Fenton reaction in the presence of hydrogen peroxide, which leads to the formation of hydroxyl radicals, resulting in protein denaturation, DNA breaks, and lipid peroxidation (Pierre and Fontecave, 1999). Therefore, iron acquisition, utilization, and storage are subject to different levels of homeostatic regulation.
In plants, iron is assimilated from the soil through the roots (Briat et al., 2007; Kim and Guerinot 2007). Under iron deficiency, Arabidopsis (Arabidopsis thaliana) activates processes described as strategy I based on the acidification of the soil by H+-ATPases, iron reduction by a ferric chelate reductase (FRO2; Robinson et al., 1999), and Fe2+ transport across the plasma membrane of root epidermal cells via the iron transporter IRT1 (Eide et al., 1996). Iron is then transported to plant organs essentially as citrate and nicotianamine complexes (Briat et al., 2007). Storage and buffering in dedicated compartments including apoplast and organelles (vacuole, plastids) avoid iron toxicity (Briat et al., 2007). In plastids, ferritins represent the major iron-containing proteins. In Arabidopsis, the ferritins AtFER1 to AtFER4 are mainly involved in buffering iron and protect the plant cells against oxidative stress (Ravet et al., 2009). Vacuolar iron stores can be mobilized to the cytosol via the divalent metal transporters AtNRAMP3 and AtNRAMP4 during seedling development (Lanquar et al., 2005).
Microorganisms have developed powerful iron acquisition systems based on the production of siderophores, which are selective ferric ion chelators secreted in response to iron deficiency (Andrews et al., 2003; Winkelmann, 2007). Siderophores have low molecular weights and very diverse chemical structures that can contain one or a combination of several types of iron-binding moieties: hydroxamate, catecholate, and hydroxycarboxylate. Once loaded with iron, siderophores are specifically transported through the bacterial envelope via protein transporters; in the cytosol, iron is reduced and distributed to iron-containing molecules. During microbial infection, a competition for iron between the host and the microorganism may take place. Phytopathogenic bacteria and fungi can use siderophores to multiply in the host and to promote infection (Expert, 1999; Haas et al., 2008). Oide et al. (2006) demonstrated that in four ascomycete species, Cochliobolus miyabeanus, Cochliobolus heterostrophus, Fusarium graminareum, and Alternaria brassicicola, siderophores are required for resistance to hydrogen peroxide and for full pathogenicity on their respective hosts maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum), and Arabidopsis. Likewise, the fire blight-causing agent Erwinia amylovora takes advantage of its siderophore deferrioxamine (DFO) for the infection of apple (Malus domestica) seedlings and flowers and for the resistance to hydrogen peroxide (Dellagi et al., 1998).
The importance of iron homeostasis in plant disease has been largely documented in the case of the bacterial pathogen Erwinia chrysanthemi (Expert, 1999). E. chrysanthemi is an enterobacterium causing soft rot on economically important crops including potato (Solanum tuberosum) and chicory (Cichorium intybus) and ornamentals like Saintpaulia plants (Pérombelon, 2002). The bacterial cells invade the intercellular spaces of parenchymatous tissues and secrete large quantities of plant cell wall-degrading enzymes, leading to tissue disorganization (Murdoch et al., 1999). Under iron deficiency, E. chrysanthemi releases two siderophores: the hydroxycarboxylate achromobactin, which is produced when iron becomes limiting (Münzinger et al., 2000), and the catecholate chrysobactin (CB; Persmark et al., 1989), which prevails under severe iron deficiency. CB and achromobactin production are required for the systemic progression of maceration symptoms on the hosts (Enard et al., 1988; Dellagi et al., 2005; Franza et al., 2005). Neema et al. (1993) showed that the production of CB enables bacterial cells to compete with plant cells for iron, preventing sequestration of this metal by the plant ferritins. Consistently, the low availability of iron in the apoplasm of infected Saintpaulia leaves induces the expression of the fct gene, encoding the bacterial outer membrane ferric-chrysobactin (Fe-CB) transporter, which is up-regulated under iron depletion (Masclaux and Expert, 1995). Unlike in mammals, where the iron-sequestrating proteins of the transferrin family are able to reduce extracellular iron availability upon infection (for review, see Schaible and Kaufmann, 2004; Weinberg, 2009), there is no evidence that these proteins exist in plants. In Arabidopsis, the genes encoding the ferritin AtFER1 and the vacuolar metal transporters AtNRAMP3 and AtNRAMP4 are involved in basal resistance to E. chrysanthemi, indicating that changes in plant iron trafficking occur during infection (Dellagi et al., 2005; Segond et al., 2009).
Interestingly, AtFER1 gene expression can be activated by the purified siderophore CB and not by Fe-CB in Arabidopsis leaves (Dellagi et al., 2005). This observation led us to suppose that the siderophore could act as a modulator of plant defense responses, since AtFER1 is part of the defense reactions triggered by E. chrysanthemi. Thus, we investigated the role of CB in the activation of Arabidopsis defense responses triggered by E. chrysanthemi upon infection, namely the salicylic acid (SA), the jasmonic acid (JA), and the ethylene (ET) pathways, which are three major signaling pathways involved in the plant's immune network (Glazebrook, 2005; Fagard et al., 2007).
In this work, we show that CB activates the SA-dependent defense pathway and that this process is dependent on iron availability in the plant. Not only CB but other types of siderophores could be elicitors, revealing a new function for these iron ligands in plant-microbe interactions. We also show that, when infiltrated into leaves, siderophores provoke iron deficiency in the roots. This work describes a new link between iron and immunity, which appears to be more complex than a simple nutritional competition.
RESULTS
CB Triggers the Signaling Cascade Mediated by SA
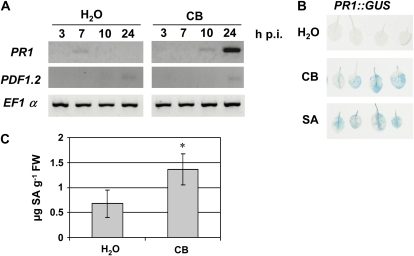
In Arabidopsis, E. chrysanthemi triggers defenses mediated by the major signaling molecules SA, JA, and ET, as revealed by marker gene expression analysis 24 h after bacterial inoculation (Fagard et al., 2007). We thus investigated whether the siderophore CB is able to activate similar responses. We monitored the expression of SA-, ET-, and JA-dependent defense genes by reverse transcription (RT)-PCR after water or CB infiltration in Arabidopsis leaves, using 0.25, 0.5, and 1 mm CB. All concentrations gave similar results (Supplemental Fig. S1), but the reproducibility of the data was best with 1 mm CB. Therefore, we used 1 mm CB in all the following experiments. We found that 24 h post infiltration (hpi), CB strongly activates the expression of the SA marker gene PR1 (Fig. 1A). We did not find any significant modification in the expression of PDF1.2, which is a good marker for the ET/JA pathway (Penninckx et al., 1998). Thus, we focused on the SA pathway. We then used Arabidopsis lines expressing GUS fusions to the PR1 promoter. We observed a strong GUS staining 24 h following infiltration of CB, which was not detected after water infiltration (Fig. 1B). The intensity of GUS staining in leaves treated with CB was similar to that observed in SA-treated leaves, used as positive controls. To determine whether the activation of PR1 expression correlated with an accumulation of SA, we measured the SA content by HPLC in Arabidopsis leaves 24 h after CB treatment. Figure 1C shows that siderophore treatment results in a 2- to 3-fold increase in SA content 24 hpi compared with control leaves. Altogether, these data show that CB triggers the SA defense pathway when infiltrated into Arabidopsis leaves.
Figure 1.
PR1 gene expression and SA production in Arabidopsis leaves following CB treatment. A, Expression patterns of PR1 (SA pathway) and PDF1.2 (ET/JA pathway) were monitored by RT-PCR using RNAs extracted from Col-0 leaves at the indicated times after infiltration of distilled water or CB. The constitutive EF1α gene was used as a control. B, GUS staining of leaves from transgenic PR1::GUS plants 24 h after infiltration of water, CB, or SA. C, Total SA content was measured by HPLC in Col-0 leaves 24 h after the indicated treatments (μg SA g−1 fresh weight [FW]). n = 6, error bars indicate sd, and the asterisk indicates a significant difference from the control by the Mann-Whitney test (P < 0.01).
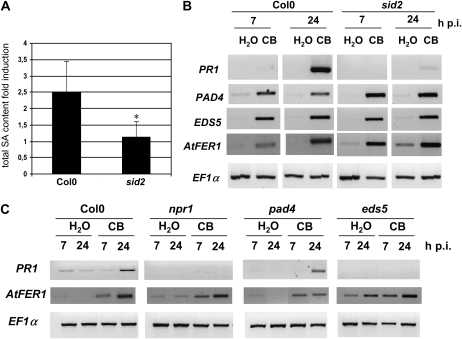
SA can be synthesized in Arabidopsis through two distinct pathways, involving either Phe ammonia-lyase or isochorismate synthase (ICS1/SID2). Because it was previously found that the ICS1/SID2 pathway is involved in the up-regulation of PR1 after E. chrysanthemi infection (Fagard et al., 2007), we proceeded on the hypothesis that this pathway could be also required for the CB-induced response. Therefore, we monitored the accumulation of SA in ecotype Columbia (Col-0) and in a sid2 mutant (Nawrath and Métraux, 1999). While CB infiltration resulted in a 2- to 3-fold accumulation of total SA in Col-0 leaves compared with control leaves 24 hpi, no significant accumulation of this hormone was observed in sid2 leaves (Fig. 2A). We can conclude that the SID2 gene is necessary for the biosynthesis of SA in response to CB.
Figure 2.
Roles of genes of the SA pathway in the signaling cascade triggered by CB. A, Total SA content (measured as in Fig. 1) fold induction (i.e. ratio of SA content in CB-treated leaves to SA content in water control). n = 6, error bars indicate sd, and the asterisk indicates a significant difference from Col-0 using the Mann-Whitney test (P < 0.01). B, RNAs from leaves were harvested after infiltration of water or CB (genotypes and times indicated). RT-PCR results with the indicated defense gene-specific primers are shown. The constitutive EF1α gene was used as a control. C, Expression profiles of AtFER1 and PR1 monitored by RT-PCR. RNAs were extracted from leaves of the indicated mutant genotypes harvested at the given times after the treatment indicated.
In order to check whether up-regulation of the PR1 gene in response to CB is dependent on SA biosynthesis, we monitored by RT-PCR the accumulation of its transcript in Col-0 and sid2 leaves treated with water or CB. PR1 expression was strongly activated by the siderophore 24 hpi in Col-0 leaves. By contrast, the presence of PR1 transcripts was hardly detected in sid2 leaves infiltrated with the siderophore (Fig. 2B). The up-regulation of PR1 by CB is thus dependent on the accumulation of SA in the leaves via the SID2 gene activity. We also monitored the expression of PAD4 and EDS5 genes known to act upstream of PR1 in the SA-mediated response. PAD4 encodes a protein similar to lipases and is required for resistance and accumulation of SA following infection with Pseudomonas syringae pv maculicola and Hyaloperonospora parasitica (Glazebrook et al., 1996, 1997; Zhou et al., 1998). EDS5 encodes a MATE-type multidrug efflux pump presumably involved in SA efflux from the chloroplast and is required for resistance to P. syringae and H. parasitica and the accumulation of SA in response to P. syringae (Nawrath and Métraux, 1999; Nawrath et al., 2002). We found that both PAD4 and EDS5 are up-regulated between 7 and 24 h after CB treatment (Fig. 2B). This response is independent of SA accumulation, since it was similar in Col-0 and in the sid2 deficient lines (Fig. 2B).
To determine whether the SA-mediated response activated by CB requires functional PAD4 and EDS5 genes, as well as the NPR1 gene encoding the SA sensor protein (Mou et al., 2003), CB or water was infiltrated onto the leaves of Col-0, eds5, pad4, and npr1 plants (Fig. 2C). No up-regulation of PR1 was observed in eds5 and npr1 mutants, indicating that the corresponding genes must be functional to mediate the response to CB. In the pad4 mutant, the expression of PR1 was still up-regulated. Collectively, these results indicate that CB activates a signaling pathway leading to PR1 up-regulation that is independent of PAD4 but dependent on SA production via SID2 and EDS5 and on the perception of this hormone via NPR1.
As CB infiltration in Arabidopsis leaves activates the expression of the ferritin-encoding gene AtFER1 (Dellagi et al., 2005), we asked whether this response requires the integrity of the SA pathway. We monitored the expression of this gene in Col-0, sid2, eds5, and npr1 leaves treated with CB. AtFER1 up-regulation was observed in all lines in response to the siderophore (Fig. 2, B and C). Similar results were obtained with the pad4 mutant line. These results indicate that AtFER1 up-regulation by CB is independent of the SA-mediated signaling pathway.
The Iron-Chelating Property of Siderophores Is Required for the Activation of the SA-Mediated Signaling Pathway
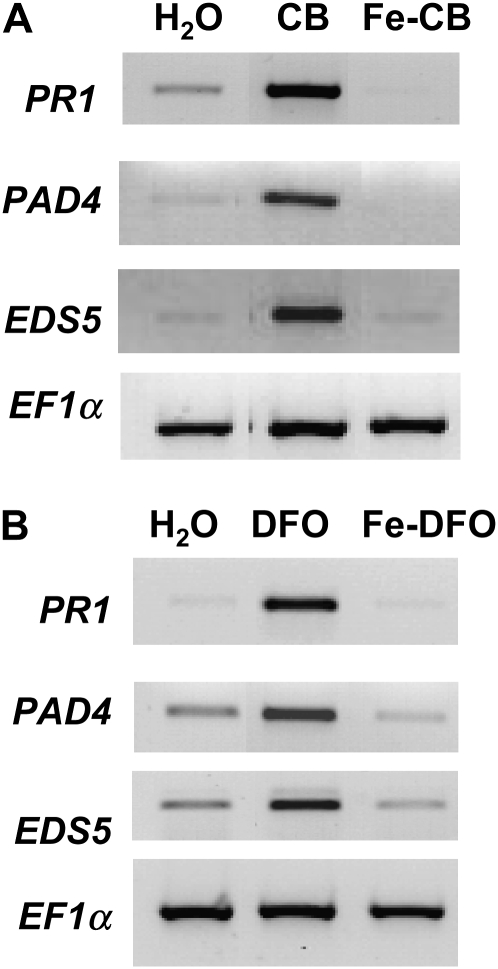
AtFER1 gene transcription is not activated by Fe-CB (Dellagi et al., 2005). Siderophore iron-binding activity is measured by calculating the pFe {defined as −log [Fe3+], where [Fe3+] = free [Fe3+] in solution calculated at determined concentrations of ligand and Fe(III) and pH; for CB, pFe = 14.5 [Tomisić et al., 2008]}. Thus, we analyzed the expression of the SA pathway in response to leaf infiltration with CB or Fe-CB (Fig. 3A). We found that the PR1, EDS5, and PAD4 genes were not up-regulated by Fe-CB. These results indicate that the elicitor activity of the siderophore is related to its chemical state.
Figure 3.
Importance of the chemical state of different siderophores in the activation of the SA pathway. Col-0 leaves were treated with water, CB, Fe-CB, DFO, or Fe-DFO as indicated. RNAs were extracted from leaves harvested 24 h after treatment, and RT-PCR with the indicated defense markers was performed. The constitutive EF1α gene was used as a control.
To determine whether the activation of the SA pathway is specific to CB, we tested the activity of a structurally unrelated siderophore, DFO. DFO is produced by the bacterial plant pathogen E. amylovora (Kachadourian et al., 1997) and is able to activate the transcription of AtFER1 in Arabidopsis leaves (Dellagi et al., 2005). Compounds of the DFO family harbor three hydroxamate groups that can bind Fe3+ very efficiently (pFe = 24.2; Tomisić et al., 2008). We found that, like CB, DFO infiltrated onto Arabidopsis leaves results in transcript accumulation of genes from the SA pathway (Fig. 3B). Ferrioxamine (Fe-DFO) did not induce this response. The same results were obtained with ferrichrome, another hydroxamate-type siderophore (data not shown).
Collectively, these results suggest that the presence of siderophores in intercellular spaces of Arabidopsis leaves, when they are iron free, induces an SA-mediated response similar to that activated by pathogens. This process is not specific to the siderophore structure, as it can be activated by either catecholates or hydroxamates.
Activation of the SA-Mediated Signaling Pathway by CB Depends on Iron Availability to the Plant
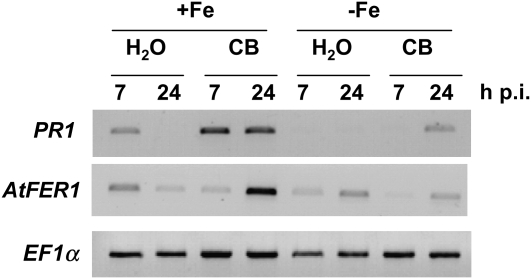
When present in the plant tissue, a siderophore should rapidly take up iron from iron-containing molecules, suggesting that this metal plays a critical role in activation of the SA-dependent process, depending on whether it is bound or not to the ligand. To check whether the nutritional iron status of the plant influences the SA-mediated response, we compared the effect of CB on plants grown under iron-sufficient and iron-deficient conditions. We used hydroponically grown plants for which nutritional iron was adjusted as described in “Materials and Methods” and analyzed the expression of PR1. The results (Fig. 4) indicate that PR1 is up-regulated in plants grown under iron sufficiency, while this was not the case in iron-deficient plants. We measured the amounts of SA in leaves treated with CB from plants grown under both conditions. In agreement with PR1 expression profiles, we found that iron-deficient plants do not accumulate significant amounts of SA (data not shown). These results show that iron present in the plant growth medium is necessary for up-regulation of the SA-mediated pathway in response to CB.
Figure 4.
Influence of plant iron nutrition on PR1 and AtFER1 gene expression levels following CB treatment. Hydroponically grown plants under 50 μm Fe-EDTA for approximately 6 weeks (+Fe) or under 50 μm Fe-EDTA for 5 weeks and then without iron for 5 d (−Fe) were infiltrated with water or CB (1 mm) as indicated. RT-PCR with the indicated gene-specific primers was performed with RNAs extracted from leaves harvested at the indicated times after treatment. The constitutive EF1α gene was used as a control.
Up-regulation of AtFER1 is observed in the presence of iron (Gaymard et al., 1996). As expected, the up-regulation of AtFER1 occurring in response to CB treatment (Dellagi et al., 2005) was not detected with plants grown under iron-deficient conditions (Fig. 4).
Siderophores Trigger an Iron Deficiency Response in the Roots
As the presence of siderophores in the plant leaves can lead to iron withholding, we investigated whether these ligands are able to trigger an iron deficiency reaction in the plant. We analyzed the expression of IRT1 and FRO2 genes, encoding the iron transporter IRT1 (Eide et al., 1996) and the ferric chelate reductase FRO2 (Robinson et al., 1999), known to respond to iron deficiency in the root. Both genes appeared to be up-regulated in roots 7 h after CB leaf treatment compared with control plants (Fig. 5A). Fe-CB infiltration in leaves did not activate the expression of these genes, as expected (Fig. 5A). We also observed a similar response to that observed with CB after infiltration of DFO (Fig. 5B).
Figure 5.
Iron deficiency root response caused by CB or DFO leaf infiltration. Col-0 plants were grown under hydroponic conditions with 50 μm Fe-EDTA. A and B, Leaves were infiltrated with water, CB, or DFO, and then RNAs were extracted from roots harvested at the indicated times after treatment. Expression patterns of IRT1 and FRO2 genes were analyzed by northern blots. Ethidium bromide staining is shown as a loading control. C, Ferric chelate reductase activity measured in roots at the indicated times after CB treatment. n = 6, error bars indicate sd, and the asterisk indicates a significant difference from the control using the Mann-Whitney test (P < 0.01). FW, Fresh weight.
We then determined the enzymatic activity of FRO2 in roots from plants treated with CB. Figure 5C shows that 24 h after siderophore infiltration in leaves of hydroponically grown plants, the FRO2 activity in roots was three times higher than in control plants. These data indicate that the presence of a siderophore in Arabidopsis leaves causes an iron deficiency in the roots, suggesting the propagation of a leaf-to-root signal.
The SA- and ET-Mediated Signaling Pathways Are Dispensable for IRT1 and FRO2 Up-Regulation by CB
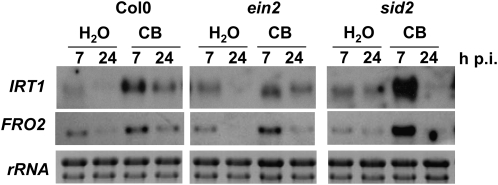
As the SA pathway is induced by CB and ET is involved in the up-regulation of FRO2 and IRT1 genes in response to iron deficiency (Lucena et al., 2006), we investigated the role of SA and ET in expression of the root response induced by CB. We used the sid2 and ein2 mutants, the latter being affected in ET perception (Alonso et al., 1999). CB infiltration in the leaves of these mutants led to the activation of IRT1 and FRO2, and notably, expression levels of these genes were higher in the sid2 mutant (Fig. 6). Thus, the SA and ET pathways are not required to mediate the iron deficiency root response induced by CB.
Figure 6.
IRT1 and FRO2 gene expression in Arabidopsis SA and ET mutants following CB leaf treatment. Plants were grown under hydroponic conditions with 50 μm Fe-EDTA. Expression patterns of IRT1 and FRO2 genes were analyzed by northern blots using RNA extracted from roots harvested from ET (ein2) and SA (sid2) mutants at the indicated times after treatment. Ethidium bromide staining is shown as a loading control.
CB Manipulates Plant Defenses and Promotes in Planta Bacterial Growth
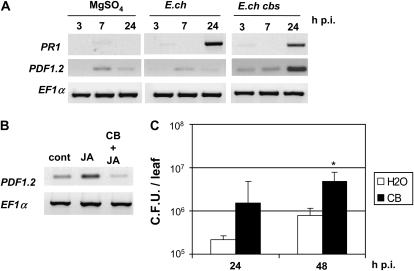
The data presented above indicate that siderophores act as elicitors of plant defense controlled by the SA hormone. As E. chrysanthemi triggers a set of defenses in Arabidopsis during infection, including the SA-mediated signaling pathway (Fagard et al., 2007), we wondered if the activation of this pathway was reduced after inoculation of a siderophore-deficient mutant compared with the wild-type strain. Thus, we used the E. chrysanthemi CB-deficient mutant affected in the cbsE gene (Franza et al., 2005). Expression of the PR1 gene was monitored 3, 7, and 24 h after infiltration of Arabidopsis leaves with wild-type cells or the siderophore-deficient mutant. As observed previously, the PR1 gene was strongly up-regulated by the wild-type bacteria compared with the control plants (Fig. 7A). Infection by the siderophore-deficient mutant resulted in reduced expression of this gene. This result indicates that CB, during bacterial infection, contributes to the activation of the SA pathway, although it is not the only elicitor of this process. Interestingly, expression of PDF1.2, the gene marker of the ET/JA pathway that is not activated by wild-type bacteria 24 h after infiltration, was strongly up-regulated in response to the siderophore-deficient mutant (Fig. 7A). This result suggests that CB represses the expression of PDF1.2. As PDF1.2 expression is known to be activated by JA, we analyzed the expression of PDF1.2 in leaves treated with JA or with both JA and CB (Fig. 7B). We observed an accumulation of PDF1.2 transcripts in response to JA, which was not detected after coinfiltration of CB and the hormone. This result confirms that CB can repress the expression of PDF1.2.
Figure 7.
Effects of CB on the expression of PR1 and PDF1.2 genes during E. chrysanthemi infection. A, Col-0 leaves were infiltrated with 10 mm MgSO4 or 107 colony-forming units mL−1 bacterial suspension of E. chrysanthemi wild type (E.ch) or CB negative mutant (E.ch cbs). Leaves were harvested at the indicated times after treatment. Expression patterns of PR1 (SA pathway) and PDF1.2 (ET/JA pathway) were monitored by RT-PCR. The constitutive EF1α gene was used as a control. B, RT-PCR using RNAs extracted from leaves 24 h after treatment with 0.05% (w/v) methanol (cont), JA, or JA + CB. C, Plants were infiltrated with water or CB 48 h before inoculation with a bacterial suspension of wild-type E. chrysanthemi cells. Leaves were harvested at the times indicated after bacterial infiltration, and then bacterial counts were performed as indicated in “Materials and Methods.” n = 6, error bars indicate sd, and the asterisk indicates a significant difference from the control using the Mann-Whitney test (P < 0.05).
Previous studies using bacterial mutants unable to produce CB or achromobactin have shown that these siderophores promote the infection process in host plants (Enard et al., 1988; Dellagi et al., 2005; Franza et al., 2005). We thus asked whether infiltration of CB onto Arabidopsis leaves prior to E. chrysanthemi inoculation could affect the bacterial growth. We infiltrated water or 1 mm CB onto Arabidopsis leaves 48 h before bacterial challenge. Bacterial populations were determined over 2 d post inoculation (Fig. 7C). In the control leaves preinfiltrated with water, E. chrysanthemi grew by less than 1 order of magnitude after 2 d of infection. In the leaves pretreated with CB, we observed a much faster growth and an increase in bacterial counts by 1 order of magnitude. These data indicate that preinfiltration of CB stimulates bacterial growth.
DISCUSSION
The plant pathogenic bacterium E. chrysanthemi requires the production of siderophores for systemic progression in host tissues (Enard et al., 1988; Dellagi et al., 2005; Franza et al., 2005). Production of siderophores and pectinases is controlled by iron availability, indicating that high-affinity iron uptake by this bacterium is a critical factor during pathogenesis (Franza et al., 2002). In order to know the role of siderophores in the infection process more precisely, we need to understand how these compounds are perceived in the host. In this work, we have investigated the plant's response to the siderophore CB. We found that two physiological functions are modulated by this molecule: plant defense and iron assimilation.
Role of CB in the Activation of the SA-Mediated Signaling Pathway
We found that CB in Arabidopsis activates the SA-mediated signaling pathway leading to PR1 gene expression. Our results using the sid2 mutant show that CB activates SA biosynthesis in Arabidopsis. The structural similarity between SA and CB allowed the hypothesis that CB or its potential degradation products could act as precursor(s) in SA biosynthesis. However, since other siderophores with no structural relationship to SA are also able to trigger the SA pathway, we excluded this hypothesis. CB also requires NPR1 to activate the expression of PR1. The NPR1 protein is an important player in SA signaling and in systemic acquired resistance (Dong, 2004). In the cytosol, it is present as disulfide-bound oligomers that monomerize following reduction consecutive to SA-controlled redox changes (Mou et al., 2003). The monomers are translocated to the nucleus, where they interact with TGA transcription factors that recognize cis elements in PR gene promoters (Johnson et al., 2003). This means that the response induced by the siderophore could also result in a cellular redox change involving SA and leading to the activation of the PR1 gene via NPR1. We also show that siderophore-mediated PR1 up-regulation does not require PAD4. PAD4 encodes a triacyl-glycerol lipase acting upstream of SA (Jirage et al., 1999) and is necessary for SA accumulation and amplification of SA-dependent defense responses (Zhou et al., 1998). It is not required for PR1 up-regulation during the hypersensitive response observed in an incompatible interaction involving resistant plants. However, PAD4 is required for full PR1 up-regulation in compatible interactions involving susceptible plants (Zhou et al., 1998). In light of these data, it is possible that the SA response triggered by a siderophore is strong enough and is comparable to an incompatible interaction, except that there is no reaction of cell death. Indeed, we never observed necrosis after siderophore treatment at the macroscopic level or at the microscopic level after trypan blue staining (data not shown).
Our results indicate that CB activates the AtFER1 gene independently of the SA-mediated signaling pathway (Fig. 8). Thus, it may be possible that regulation of the ferritin gene by CB takes place upstream of the SA response. It would be helpful to determine whether AtFER1 contributes to the responses induced by CB in an atfer1 mutant. As an iron-buffering molecule, ferritin could contribute to changes in the cellular iron status and activate downstream signals.
Figure 8.
Diagram showing the responses of Arabidopsis to microbial siderophores. Leaf infiltration of iron-free siderophores (CB or DFO) activates the SA-mediated signaling pathway leading to PR1 up-regulation, the basal defense marker PAD4, ferritin accumulation via AtFER1, and root iron uptake via IRT1 and FRO2. Up-regulation of IRT1 and FRO2 appeared to be partially repressed by SA. Activation of the SA pathway and AtFER1 up-regulation depends on iron availability to the plant (indicated with dashed arrows). Further details are discussed in the text.
Role of CB in the Activation of the Iron Deficiency Root Response
We recently reported that infection of Arabidopsis by E. chrysanthemi triggers an iron deficiency response in the roots (Segond et al., 2009). This work shows that this response is also induced by infiltration of a siderophore on the leaf, suggesting that this ligand is responsible for the root reaction when it is released by bacterial cells during infection.
The elicitor activity of siderophores is likely due to their strong iron-chelating capacity rather than to recognition by a plant receptor. Indeed, we found that the siderophores induce a reaction similar to iron deficiency consisting of IRT1 and FRO2 expression and production of the FRO2 enzymatic activity. It is tempting to speculate that the iron taken up by the roots is rapidly translocated to the leaves, a process that may cause an oxidative stress (Fig. 8). This oxidative stress could activate the SA pathway and AtFER1 gene expression, as these two responses are known to be inducible by reactive oxygen species (Leon et al. 1995; Gaymard et al., 1996; Petit et al., 2001). Two observations are in agreement with this hypothesis. First, under iron deficiency, CB treatment does not induce the up-regulation of PR1 or that of AtFER1, indicating that iron is required for activation of the SA response and confirming that this metal is essential to AtFER1 up-regulation. Second, the expression of IRT1 is rapid (7 hpi) but decreases between 7 and 24 h, indicating that the iron deficiency signal disappears during this period, likely because of a negative feedback due to iron uptake via IRT1. The timing of activation of the various SA marker genes (7–24 hpi) is compatible with this interpretation. In addition, the protein IRT1 can transport other cations than Fe2+, including Zn2+, Mn2+, and Cd2+ (Korshunova et al., 1999), and it is conceivable that some of these metals are taken up by the plant after IRT1 induction and contribute to the responses observed.
We investigated whether SA or ET is involved in the activation of the iron deficiency response by CB. Our data show that the ein2 mutation does not affect IRT1 and FRO2 up-regulation following CB treatment, suggesting that ET is not involved in this process. On the other hand, we found that IRT1 and FRO2 transcripts accumulate to higher levels in the sid2 mutant compared with the wild-type ecotype. This observation suggests that SA could exert a negative control of the iron deficiency response. This is consistent with the iron-binding capacity of SA (pFe = 12.1; Nurchi et al., 2009), a property that might confer onto this molecule a cellular iron-sensing function, as suggested in bacteria (Adilakshmi et al., 2000).
Role of CB in the Control of E. chrysanthemi Pathogenesis
CB pretreatment enhances the multiplication of E. chrysanthemi cells in the leaf (Fig. 7), which is in agreement with the fact that siderophore-deficient mutants are affected in their aggressiveness (Enard et al., 1988; Dellagi et al., 2005; Franza et al., 2005). The weaker activation of PR1 after inoculation of the CB-deficient mutant compared with the wild-type strain indicates that CB produced during infection is likely responsible for activation of the SA pathway. However, under this condition, the PR1 gene is still expressed, indicating the existence of other elicitors of the SA pathway. Achromobactin, the second E. chrysanthemi siderophore, could contribute to this response, and oligogalacturonides generated by pectinases are likely to be involved (Fagard et al., 2007). We also found that, unlike wild-type bacteria, the CB-deficient mutant activates the expression of a marker of the JA/ET pathway, PDF1.2, 24 hpi. This suggests that CB represses this pathway that is involved in Arabidopsis resistance to E. chrysanthemi (Fagard et al., 2007). By activating the biosynthesis of SA via CB, the bacteria modulate the plant defense responses and take advantage of the antagonism between the SA and JA pathways. Furthermore, as siderophores activate iron uptake in the roots, the plant iron content must increase, thus explaining the beneficial effect of CB on E. chrysanthemi growth in the leaves.
Some siderophores secreted by soil-borne Pseudomonas species (pyoverdin and pyocyanin) can promote systemic plant protection against soil and foliar pathogens, a phenomenon known as induced systemic resistance (Audenaert et al., 2002; Haas and Défago, 2005). Induced systemic resistance is known to be dependent on the ET and JA pathways and independent of the SA pathway (Pieterse et al., 1998). In this work, we show that the elicitor activity of the siderophore CB that we observed is SA dependent, indicating that this process is different from induced systemic resistance.
In conclusion, this work shows that microbial pathogens can modulate the activity of the plant iron acquisition system via the modulation of siderophore production during infection and that this process can lead to changes in the expression of plant immune responses. These changes may be to the advantage of the pathogen or may help the plant to resist the pathogen. This could explain why in a number of plant-pathogen interactions, no role for siderophores was found in virulence, while in others, siderophores are important pathogenicity factors. The future challenges now are to better understand the molecular mechanisms by which siderophores activate the SA pathway and the root iron deficiency response.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds from the Col-0 ecotype were obtained from the Institut National de la Recherche Agronomique Versailles collection. The sid2-1 mutant was kindly donated by J.-P. Métraux. Seeds of ein2-1 (Guzman and Ecker, 1990), npr1-1 (Cao et al., 1994), pad4-1 (Glazebrook et al., 1997), and eds5-1 (Glazebrook et al., 1996) were provided by the Nottingham Arabidopsis Stock Center (Scholl et al., 2000). The PR1::GUS lines were kindly provided by F. Ausubel. Plants were grown as described by Fagard et al. (2007). Hydroponic cultures were performed as described by Segond et al. (2009) and were used for all experiments where roots were used for RNA extraction or FRO2 activity monitoring. Experiments with iron-starved plants were performed as follows. Plants were first grown under the above-described conditions for 5 weeks and then transferred to iron-deficient medium after washing the roots for 5 min with medium containing the reductant sodium dithionite (5.7 mm) and the chelator bathophenanthrolinedisulfonic acid (0.3 mm), both from Sigma. The plants were kept under high-moisture conditions during the experiments.
Chemical Preparations
Leaf treatment with the following chemicals was performed by infiltration of the solutions at the indicated concentrations in the intercellular leaf space using a syringe without a needle. CB was a gift from J. Buyer (Lu et al., 1996), and Fe-CB was prepared as described by Rauscher et al. (2002). DFO and Fe-DFO (a gift from R. Kachadourian) were prepared as described by Kachadourian et al. (1997). JA and SA were purchased from Sigma-Aldrich. Siderophores and SA were used at a concentration of 1 mm diluted in distilled water. JA was used at 0.1 mm in 0.05% (w/v) methanol.
Bacterial Strains and Culture Conditions
The wild-type strain, Erwinia chrysanthemi 3937 (our collection), was isolated from Saintpaulia ionantha (African violet). The CB-deficient mutant PPV11 is derived from strain 3937 that contains an insertional element inactivating the biosynthetic CB gene cbsE (cbs-E1::Ω; Franza et al., 2005). Growth conditions were as described by Dellagi et al. (2005).
Plant Inoculations and Determination of Bacterial Growth
To monitor bacterial growth after siderophore treatment, we first infiltrated water or CB on the entire leaf. Forty-eight hours later, a small hole was made with a needle within the leaf, and then 5 μL of a bacterial suspension at a density of 5 × 107 colony-forming units mL−1 made up in 50 mm potassium phosphate buffer (pH 7) was spotted on the hole. Leaves were harvested in 0.9% NaCl and ground using a pestle and sterile sand. The resulting suspensions were used for serial dilutions followed by plating on an appropriate medium. For RNA extractions and GUS fusions, we used a syringe without a needle to infiltrate the entire leaf or a portion of the leaf with SA, siderophore solution, or bacterial suspensions at 5 × 107 colony-forming units mL−1 in 10 mm MgSO4 (half a leaf was infiltrated for GUS staining).
RNA Extraction, Northern Blotting, and RT-PCR
Northern-blot hybridization was carried out as described by Dellagi et al. (2005). IRT1 and FRO2 probes were prepared as described by Segond et al. (2009). For RT-PCR analysis, reverse transcription was performed as described by Fagard et al. (2007). PCR runs were of 94°C for 4 min, 26 to 30 cycles, and each cycle consisting of 94°C for 30 s, 54°C to 58°C for 30 s, and 72°C for 1 min, with a final step of 72°C for 10 min to complete polymerization. Primers for EF1α, PR1, PAD4, CHIB, and EDS5 were described by Fagard et al. (2007). The other gene-specific primers were as follows: PDF1.2-F (AT5G44420; 5′-TCATGGCTAAGTTTGCTTCCATCATCACCC-3′) and PDF1.2-R (5′-GTAGATTTAACATGGGAC-3′). Equal cDNA amounts were checked by performing different PCR cycles with EF1α primers (Supplemental Fig. S2). Experiments were repeated at least three times. Representative data are shown.
In Planta GUS Expression Detection
In planta GUS expression detection was performed as described by Dellagi et al. (2005). Experiments were repeated three times with similar results.
Root FRO Assays
Root FRO activity was performed as described by Yi and Guerinot (1996). Briefly, roots from control plants or plants treated with the siderophore were incubated in a solution containing 0.1 mm Fe(III)-EDTA and 0.3 mm ferrozine in distilled water in the dark. After 20 min, the absorbance of the solution was measured at 562 nm, using the same solution without roots as a control.
SA
Treated leaves were harvested and then weighed before freezing in liquid nitrogen. They were ground in a frozen state in Eppendorf tubes using TissueLyser II (Qiagen). [7-14C]SA (1 nCi, 54 mCi mmol−1; New England Nuclear) was used for recovery determination. Total SA was extracted and analyzed as described by Baillieul et al. (1995) with a Nova-Pak 4-μm C-18 column (150 × 3.9 mm; Waters) as part of the Waters system (1525 Binary HPLC Pump, 2475 Multi λ Fluorescence Detector, 2996 Photodiode Array Detector, 717 Autosampler; Waters). Data were analyzed using Empower Pro Software (Waters).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Defense gene expression in Arabidopsis leaves infiltrated with CB.
Supplemental Figure S2. Validation of the RT-PCR approach using EF1α as a constitutively expressed gene.
Supplementary Material
Acknowledgments
We thank Floriant Belvert for help with SA quantification. We thank J.-P. Métraux for kindly providing the seeds of sid2 mutants and F. Ausubel for seeds of the PR1::GUS line. We are grateful to S. Davenport for English editing of the manuscript.
This work was supported by the Institut National de la Recherche Agronomique, the Centre National de la Recherche Scientifique (D.E.), and the Ministère de l'Enseignement Supérieur et de la Recherche (D.S. and C.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alia Dellagi (dellagi@agroparistech.fr).
The online version of this article contains Web-only data.
References
- Adilakshmi T, Ayling PD, Ratledge C (2000) Mutational analysis of a role for salicylic acid in iron metabolism of Mycobacterium smegmatis. J Bacteriol 182 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152 [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodríguez-Quiñones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27 215–237 [DOI] [PubMed] [Google Scholar]
- Audenaert K, Pattery T, Cornelis P, Höfte M (2002) Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol Plant Microbe Interact 15 1147–1156 [DOI] [PubMed] [Google Scholar]
- Baillieul F, Genetet I, Kopp M, Saindrenan P, Fritig B, Kauffmann S (1995) A new elicitor of the hypersensitive response in tobacco: a fungal glycoprotein elicits cell death, expression of defence genes, production of salicylic acid, and induction of systemic acquired resistance. Plant J 4 551–560 [DOI] [PubMed] [Google Scholar]
- Briat JF, Curie C, Gaymard F (2007) Iron utilization and metabolism in plants. Curr Opin Plant Biol 10 276–282 [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 11 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellagi A, Brisset MN, Paulin JP, Expert D (1998) Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol Plant Microbe Interact 8 734–742 [DOI] [PubMed] [Google Scholar]
- Dellagi A, Rigault M, Segond D, Roux C, Kraepiel Y, Cellier F, Briat JF, Gaymard F, Expert D (2005) Siderophore-mediated upregulation of Arabidopsis ferritin expression in response to Erwinia chrysanthemi infection. Plant J 43 262–272 [DOI] [PubMed] [Google Scholar]
- Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7 547–552 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard C, Diolez A, Expert D (1988) Systemic virulence of Erwinia chrysanthemi 3937 requires a functional iron assimilation system. J Bacteriol 170 2419–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expert D (1999) Withholding and exchanging iron: interactions between Erwinia spp. and their plant hosts. Annu Rev Phytopathol 37 307–334 [DOI] [PubMed] [Google Scholar]
- Fagard M, Dellagi A, Roux C, Périno C, Rigault M, Boucher V, Shevchik VE, Expert D (2007) Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi. Mol Plant Microbe Interact 20 794–805 [DOI] [PubMed] [Google Scholar]
- Franza T, Mahé B, Expert D (2005) Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol Microbiol 55 261–275 [DOI] [PubMed] [Google Scholar]
- Franza T, Michaud-Soret I, Piquerel P, Expert D (2002) Coupling of iron assimilation and pectinolysis in Erwinia chrysanthemi 3937. Mol Plant Microbe Interact 15 1181–1191 [DOI] [PubMed] [Google Scholar]
- Gaymard F, Boucherez J, Briat JF (1996) Characterization of a ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem J 318 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Zook M, Mert F, Kagan I, Rogers EE, Crute IR, Holub EB, Hammerschmidt R, Ausubel FM (1997) Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3 307–319 [DOI] [PubMed] [Google Scholar]
- Haas H, Eisendle M, Turgeon BG (2008) Siderophores in fungal physiology and virulence. Annu Rev Phytopathol 46 149–187 [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J (2003) Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15 1846–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachadourian R, Chuilon S, Merienne C, Kunesch G, Deroussent A (1997) A new total synthesis of ferrioxamine E through metal-templated cyclic trimerization. Supramolec Chem 8 301–308 [Google Scholar]
- Kim SA, Guerinot ML (2007) Mining iron: iron uptake and transport in plants. FEBS Lett 581 2273–2280 [DOI] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40 37–44 [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D, Vansuyt G, Curie C, Schröder A, Krämer U, et al (2005) Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J 23 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon J, Lawton MA, Raskin I (1995) Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol 108 1673–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Buyer JS, Okonya JF, Miller MJ (1996) Synthesis of optically pure chrysobactin and immunoassay development. Biometals 9 377–383 [DOI] [PubMed] [Google Scholar]
- Lucena C, Waters BM, Romera FJ, García MJ, Morales M, Alcántara E, Pérez-Vicente R (2006) Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J Exp Bot 57 4145–4154 [DOI] [PubMed] [Google Scholar]
- Masclaux C, Expert D (1995) Signalling potential of iron in plant-microbe interactions: the pathogenic switch of iron transport in Erwinia chrysanthemi. Plant J 7 121–128 [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944 [DOI] [PubMed] [Google Scholar]
- Münzinger M, Budzikiewicz H, Expert D, Enard C, Meyer JM (2000) Achromobactin, a new citrate siderophore of Erwinia chrysanthemi. Z Naturforsch [C] 55 328–332 [DOI] [PubMed] [Google Scholar]
- Murdoch L, Corbel JC, Reis D, Bertheau Y, Vian B (1999) Differential cell wall degradation by Erwinia chrysanthemi in petiole of Saintpaulia ionantha. Protoplasma 210 59–74 [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neema C, Laulhere JP, Expert D (1993) Iron deficiency induced by chrysobactin in Saintpaulia leaves inoculated with Erwinia chrysanthemi. Plant Physiol 102 967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurchi VM, Pivetta T, Lachowicz JI, Crisponi V (2009) Effect of substituents on complex stability aimed at designing new iron (III) and aluminium (III) chelators. J Inorg Biochem 103 227–236 [DOI] [PubMed] [Google Scholar]
- Oide S, Moeder W, Krasnoff S, Gibson D, Haas H, Yoshioka K, Turgeon BG (2006) NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 10 2836–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérombelon MCM (2002) Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol 51 1–12 [Google Scholar]
- Persmark M, Expert D, Neilands JB (1989) Isolation, characterization, and synthesis of chrysobactin, a compound with siderophore activity from Erwinia chrysanthemi. J Biol Chem 264 3187–3193 [PubMed] [Google Scholar]
- Petit JM, van Wuytswinkel O, Briat JF, Lobréaux S (2001) Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. J Biol Chem 276 5584–5590 [DOI] [PubMed] [Google Scholar]
- Pierre JL, Fontecave M (1999) Iron and activated oxygen species in biology: the basic chemistry. Biometals 3 195–199 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, van Wees SC, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 9 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher L, Expert D, Matzanke BF, Trautwein AX (2002) Chrysobactin-dependent iron acquisition in Erwinia chrysanthemi: functional study of a homolog of the Escherichia coli ferric enterobactin esterase. J Biol Chem 277 2385–2395 [DOI] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57 400–412 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397 694–697 [DOI] [PubMed] [Google Scholar]
- Schaible UE, Kaufmann SH (2004) Iron and microbial infection. Nat Rev Microbiol 12 946–953 [DOI] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segond D, Dellagi A, Lanquar V, Rigault M, Patrit O, Thomine S, Expert D (2009) NRAMP genes function in Arabidopsis thaliana resistance to Erwinia chrysanthemi infection. Plant J 58 195–207 [DOI] [PubMed] [Google Scholar]
- Tomisić V, Blanc S, Elhabiri M, Expert D, Albrecht-Gary AM (2008) Iron(III) uptake and release by chrysobactin, a siderophore of the phytopathogenic bacterium Erwinia chrysanthemi. Inorg Chem 47 9419–9430 [DOI] [PubMed] [Google Scholar]
- Weinberg ED (2009) Iron availability and infection. Biochim Biophys Acta 1790 600–605 [DOI] [PubMed] [Google Scholar]
- Winkelmann G (2007) Ecology of siderophores with special reference to the fungi. Biometals 20 379–392 [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10 835–844 [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.