Abstract
Eukaryotic initiation factor (eIF) 4B is known to interact with multiple initiation factors, mRNA, rRNA, and poly(A) binding protein (PABP). To gain a better understanding of the function of eIF4B, the two isoforms from Arabidopsis (Arabidopsis thaliana) were expressed and analyzed using biophysical and biochemical methods. Plant eIF4B was found by ultracentrifugation and light scattering analysis to most likely be a monomer with an extended structure. An extended structure would facilitate the multiple interactions of eIF4B with mRNA as well as other initiation factors (eIF4A, eIF4G, PABP, and eIF3). Eight mRNAs, barley (Hordeum vulgare) α-amylase mRNA, rabbit β-hemoglobin mRNA, Arabidopsis heat shock protein 21 (HSP21) mRNA, oat (Avena sativa) globulin, wheat (Triticum aestivum) germin, maize (Zea mays) alcohol dehydrogenase, satellite tobacco necrosis virus RNA, and alfalfa mosaic virus (AMV) 4, were used in wheat germ in vitro translation assays to measure their dependence on eIF4B and eIF4F isoforms. The two Arabidopsis eIF4B isoforms, as well as native and recombinant wheat eIF4B, showed similar responses in the translation assay. AMV RNA 4 and Arabidopsis HSP21 showed only a slight dependence on the presence of eIF4B isoforms, whereas rabbit β-hemoglobin mRNA and wheat germin mRNA showed modest dependence. Barley α-amylase, oat globulin, and satellite tobacco necrosis virus RNA displayed the strongest dependence on eIF4B. These results suggest that eIF4B has some effects on mRNA discrimination during initiation of translation. Barley α-amylase, oat globulin, and rabbit β-hemoglobin mRNA showed the highest activity with eIF4F, whereas Arabidopsis HSP21 and AMV RNA 4 used both eIF4F and eIF(iso)4F equally well. These results suggest that differential or optimal translation of mRNAs may require initiation complexes composed of specific isoforms of initiation factor gene products. Thus, individual mRNAs or classes of mRNAs may respond to the relative abundance of a particular initiation factor(s), which in turn may affect the amount of protein translated. It is likely that optimal multifactor initiation complexes exist that allow for optimal translation of mRNAs under a variety of cellular conditions.
Initiation of eukaryotic protein synthesis requires numerous factors to correctly bind and position mRNA on the 40S ribosome. Before the 40S ribosome and mRNA interact, the eukaryotic initiation factor 4 (eIF4) group of initiation factors, eIF4A, eIF4B, and eIF4F, function in the ATP-dependent unwinding of secondary structure in the 5′ untranslated region (UTR) of the mRNA (for review, see Dever, 2002; Mathews, 2002; Sonenberg and Dever, 2003; Pestova et al., 2007; Gallie, 2007). eIF4A, a single polypeptide (approximately 50 kD), is the prototype DEAD/H helicase and has ATP binding and helicase motifs (for review, see Rogers et al., 2002). eIF4B is an RNA-binding protein (approximately 59 kD) that functions to enhance the helicase activity of eIF4A and eIF4F (Bi et al., 2000; Rogers et al., 2001). The cap binding complex, eIF4F, is composed of two subunits, eIF4E (approximately 24 kD) and eIF4G (approximately 200 kD). eIF4E is the cap binding protein, and eIF4G interacts with several components of the translational machinery, including eIF3, eIF4A, and poly(A) binding protein (PABP; for review, see Prévôt et al., 2003). Plants have a second form of eIF4F, eIF(iso)4F, which has similar activities to eIF4F but is composed of the genetically distinct subunits eIF(iso)4G (approximately 86 kD) and eIF(iso)4E (approximately 24 kD; Browning, 1996).
eIF4B has been isolated from rabbit reticulocytes (Trachsel et al., 1977; Benne and Hershey, 1978), yeast (Saccharomyces cerevisiae; Altmann et al., 1993; Coppolecchia et al., 1993), Drosophila melanogaster (Hernandez et al., 2004), and plants (Browning et al., 1989; Metz et al., 1999). It stimulates mRNA translation, ATP hydrolysis, and unwinding activities of eIF4F and eIF4A (Grifo et al., 1983, 1984; Ray et al., 1985; Abramson et al., 1987, 1988a, 1988b; Browning et al., 1989; Rozen et al., 1990; Lorsch and Herschlag, 1998; Bi et al., 2000; Rogers, et al., 2001; Khan and Goss, 2005). eIF4B also has a role in picornaviral internal ribosome entry site-mediated translation (Meyer et al., 1995; Ochs et al., 1999, 2002; Rust et al., 1999), the shutdown of translation of herpes simplex host mRNA (Doepker et al., 2004), and reinitiation of polycistronic RNA of Cauliflower mosaic virus (Park et al., 2004). eIF4B is a target for degradation during apoptosis (Bushell et al., 2000). Such reports suggest an important role for eIF4B in the initiation of translation. However, disruption of the single gene for yeast eIF4B is not lethal. It results in a phenotype that is cold sensitive and that grows slowly (Altmann et al., 1993; Coppolecchia et al., 1993). Similarly, RNA interference silencing of the single Drosophila eIF4B gene suggests that eIF4B is needed during starvation but is not essential for survival under optimal conditions (Hernandez et al., 2004). Unlike other initiation factors, eIF4B is poorly conserved in plants, animals, and fungi at the level of amino acid sequence. Thus, eIF4B is most likely conserved at the level of structure and/or function.
Mammalian eIF4B contains one RNA binding region in the N-terminal domain (Milburn et al., 1990), while a second binding region is located within the C-terminal domain (Méthot et al., 1994; Naranda et al., 1994). The RNA recognition motif (RRM) portion of human eIF4B and other RRMs are structurally similar, but they differ in the way that they bind RNA (Fleming et al., 2003). The presence of two RNA binding regions suggests that during initiation, mammalian eIF4B may interact concurrently with two different RNA molecules, mRNA and 18S rRNA (Methot et al., 1996a, 1996b; Cheng and Gallie, 2006). Mammalian eIF4B contains a central DRYG-rich domain (Milburn et al., 1990) that purportedly mediates eIF4B self-association and the direct association of eIF4B with the eIF3a (p170) subunit of eIF3 (Methot et al., 1996b). The self-association conveyed by the DRYG region in mammalian eIF4B is unlikely to function in RNA binding or helicase stimulation (Methot et al., 1996b). Regions rich in acidic and basic amino acid repeats are present in yeast (Altmann et al., 1993; Coppolecchia et al., 1993) and Drosophila eIF4B (Hernandez et al., 2004). In wheat (Triticum aestivum) and Arabidopsis (Arabidopsis thaliana), the comparable DRYG region of plant eIF4B has repeat regions of GGGFRESS and four to nine Gly residues, respectively (Metz et al., 1999). Wheat eIF4B has several RNA binding domains and two adjacent PABP/eIF4A binding sites in the C-terminal half of the protein (Cheng and Gallie, 2006). An N-terminal binding site for eIF(iso)4G overlaps with part of an RNA binding domain (Cheng and Gallie, 2006). These multiple protein interactions with other initiation factors suggest a role for eIF4B in regulation or coordination of the events of initiation.
The region responsible for low level in vitro dimerization of wheat eIF4B and RNA binding is located in the C terminus of the protein and connects the two tandem eIF4A and PABP interaction sites (Cheng et al., 2008). The partial dimerization of wheat eIF4B in vitro is stimulated in the presence of zinc (Cheng et al., 2008).
Both mammalian and wheat eIF4B are highly phosphorylated (Duncan and Hershey, 1985, 1989; Manzella et al., 1991; Gallie et al., 1997; Le et al., 1998, 2000), suggesting a role in regulation of protein synthesis. S6 kinase and other members of the AGC kinase family phosphorylate Ser-405 and/or Ser-422, two serum-responsive sites in mammalian eIF4B (Raught et al., 2004; Shahbazian et al., 2006; van Gorp et al., 2009). Whether comparable sites exist in plant eIF4B is not known at this time. In plants, eIF4B interacts with PABP (Le et al., 1997, 2000; Bi and Goss, 2000; Luo and Goss, 2001; Khan and Goss, 2005; Cheng and Gallie, 2007), and this interaction is stimulated by the presence of zinc. Zinc also confers specificity of PABP binding to eIF4B over eIF(iso)4G (Cheng et al., 2008). The phosphorylation state of both eIF4B and PABP are important for their interaction (Le et al., 2000). Plant eIF4B shows higher binding for polypurines, although it will bind polypyrimidines (Gallie and Tanguay, 1994; Cheng and Gallie, 2006). The interaction of eIF4B and PABP increases the ATPase and RNA helicase activity of the eIF4A/eIF4B/eIF(iso)4F complex. This suggests that PABP could stimulate mRNA scanning by the eIF4 complex and thereby increase the rate of initiation of translation (Bi and Goss, 2000). The cap-binding affinity of eIF(iso)4F and the poly(A)-binding affinity of PABP are also enhanced by these interactions (Le et al., 1997; Wei et al., 1998; Khan and Goss, 2005). Thus, interactions among PABP, eIF4B, and eIF(iso)4F could provide some capped and polyadenylated mRNAs with a competitive translational advantage (Bi and Goss, 2000; Khan and Goss, 2005).
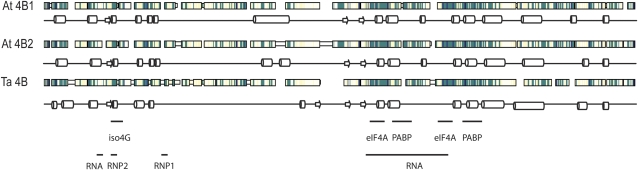
Arabidopsis has two genes for eIF4B. Based on EST libraries, both forms are highly expressed. The similarity between the two Arabidopsis isoforms is approximately 84% compared to approximately 60% between wheat and Arabidopsis eIF4B isoforms. The predicted secondary structures of Arabidopsis and wheat eIF4B show a high degree of conservation of two apparently folded domains linked by a region that is either unstructured or whose structure cannot be predicted by the current in silico methods (Fig. 1). Interestingly, no obvious βαββαβ RRM motif was predicted for the plant eIF4Bs, although plant eIF4B does interact with RNA (Browning et al., 1989; Le et al., 1997; Metz et al., 1999; Cheng and Gallie, 2006). The lack of a consensus RRM motif suggests that plant eIF4B may have a novel RNA binding fold. Similarity of plant eIF4B to other eukaryotic eIF4Bs (human, Drosophila, S. cerevisiae, and Schizosaccharomyces pombe) dropped dramatically to <40%, and finding regions of primary sequence that were common to all species was difficult (Metz et al., 1999). The large variation among primary sequences suggests that eIF4B has diverged more than other eIFs. Presumably, the structure of plant eIF4B must have retained some structural similarity or evolved new structural motifs to enable the interaction with RNA and the more conserved members of the translational machinery.
Figure 1.
Protein sequence alignment of Arabidopsis and wheat eIF4B. The protein sequences of the two eIF4B isoforms of Arabidopsis (At) were aligned with the wheat (Ta) eIF4B using CLUSTAL (Jeanmougin et al., 1998) and visualized graphically in MACAW v.2.0.5. The darkest blue regions indicate the greatest amino acid sequence similarity, and the lightest regions indicate no similarity. Large boxes indicate regions of high similarity between all three forms of eIF4B; small boxes indicate that there is no similarity. Gaps are introduced to maximize the alignments. The regions of α-helices (cylinders) and β-sheets (arrows) were predicted by PSIPRED (Bryson et al., 2005). RNA, eIF4A, eIF(iso)4G, and PABP binding domains were identified by Cheng and Gallie (2006).
The presence of multiple genes, as well as multiple phosphorylation states, suggests that plants may use eIF4B, as well as PABP (Belostotsky, 2003), to regulate translation of specific mRNAs. In order to understand how the isoforms of plant eIF4B might contribute to the regulation of translation, the two forms from Arabidopsis were overexpressed in Escherichia coli, purified, and compared to native and recombinant wheat eIF4B by biophysical and biochemical methods. The results suggest that purified plant eIF4B is a monomer with an extended shape. The abilities of the two Arabidopsis eIF4B isoforms and native and recombinant forms of wheat eIF4B to participate in the in vitro initiation of translation of several types of mRNAs in the presence of eIF4F or eIF(iso)4F cap-binding complexes were measured. The results suggest that eIF(iso)4F, eIF4F, and eIF4B isoforms all may contribute to differential translation of plant mRNAs.
RESULTS
Cloning and Expression of Arabidopsis eIF4B1 and eIF4B2
cDNAs for the two isoforms of eIF4B from Arabidopsis were obtained by reverse transcription of mRNA and DNA amplification. The cDNAs were cloned into pET22b for overexpression in E. coli. The complete cDNA sequences of the cloned isoform inserts were determined and compared to their respective chromosomal sequences (eIF4B1, At3g26400; eIF4B2, At1g13020). The alignment of each cloned isoform, originating from mRNA, with its chromosomal sequence yielded a perfect match and confirmed the three exons and two introns that were predicted from the genomic sequences of AteIF4B1 and AteIF4B2 genes (Metz et al., 1999).
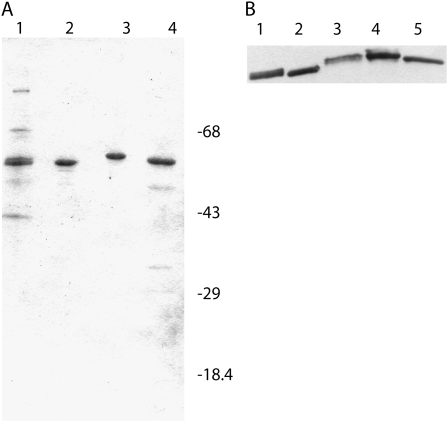
The two Arabidopsis eIF4B isoforms were expressed and purified by conventional ion-exchange chromatography. In SDS-PAGE, the migration of the purified products was similar to the migration of native and recombinant wheat eIF4B (Fig. 2A). A degradation product of AteIF4B1 was distinct and had a molecular mass of approximately 30 kD (data not shown). N-terminal sequencing of this fragment yielded the amino acid sequence AEKGLDXKKIDSEIE, which corresponded to amino acids 293 to 307 of AteIF4B1. A recombinant form of this fragment did not retain any biological activity in in vitro translation assays (data not shown). The presence of this fragment suggested a flexible region creating an exposed site within eIF4B1 that is susceptible to protease degradation.
Figure 2.
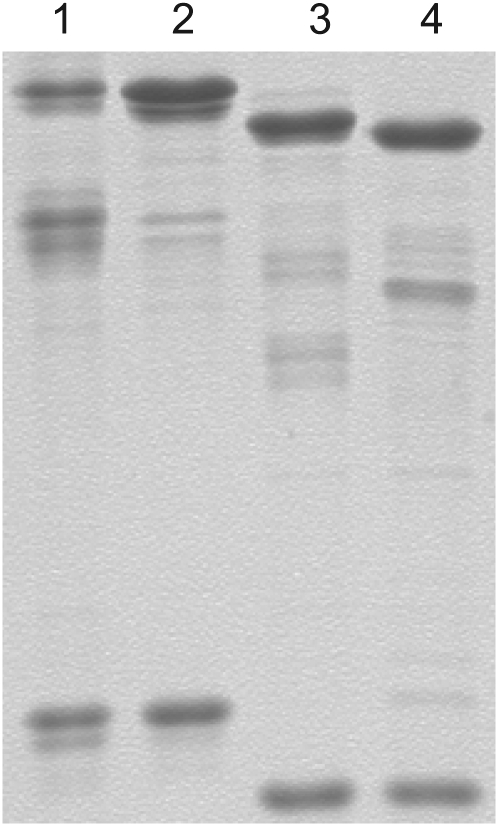
SDS-PAGE and western analysis of eIF4B preparations used in in vitro translation assays. A, Each lane contains approximately 2 μg of protein. Lane 1, Native wheat eIF4B; lane 2, recombinant wheat eIF4B; lane 3, recombinant Arabidopsis eIF4B2; lane 4, recombinant Arabidopsis eIF4B1. Molecular weight markers are as indicated. The gel was stained with Coomassie Brilliant Blue. B, The polyvinylidene fluoride blot was incubated with a 1/1,000 dilution of affinity-purified Arabidopsis eIF4B2 rabbit antibodies overnight at 4°C. A 1/25,000 dilution of goat-anti-rabbit horseradish peroxidase second antibody (Kirkegaard and Perry Laboratories) was incubated for 2 h at room temperature (Browning et al., 1990). The antibody reactive bands were visualized by chemiluminescence (SuperSignal; Pierce) and exposed to film. Lane 1, Native wheat eIF4B (0.9 μg); lane 2, recombinant wheat eIF4B (0.3 μg); lane 3, native Arabidopsis eIF4B (5 μg); lane 4, recombinant Arabidopsis eIF4B2 (0.3 μg); lane 5, recombinant Arabidopsis eIF4B1 (0.3 μg).
Antibodies to Arabidopsis eIF4B
Antibodies to Arabidopsis eIF4B2 were raised in a rabbit. The antisera cross-reacted with native AteIF4B, recombinant AteIF4B1, and with both native and recombinant wheat eIF4B (Fig. 2B). The eIF4B2 antisera also cross-reacted with the C-terminal degradation fragment of AteIF4B1 (data not shown).
Gel Filtration Analysis of Wheat and Arabidopsis eIF4B
Previous studies using gel filtration indicated that mammalian (Methot et al., 1996b) and wheat eIF4B (Metz et al., 1999) may exist as homodimers. To determine the native Mrs in low (0.1 m KCl) and high salt (0.5 m KCl), FPLC gel filtration was performed using a Sephacryl S-200HR column (separation range 5,000–250,000 molecular weight) for native and recombinant wheat eIF4B, as well as Arabidopsis eIF4B isoforms. We found no evidence that the elution volume differed between the low and high salt conditions for any of the eIF4B isoforms (Table I). In addition, all isoforms eluted to within 1.0 to 1.5 mL of each other, which suggested that they had similar Mrs. The differences in the apparent native molecular masses (approximately 100–120 kD) and the true molecular mass (approximately 59 kD) suggested that eIF4B forms dimers as previously reported (Metz et al., 1999) or does not have a globular shape and moves aberrantly in the gel filtration media.
Table I.
FPLC Sephacryl S-200 gel filtration analysis of wheat and Arabidopsis eIF4B isoforms
| Samplea | Native MMb | S200 Elution Volume
|
|
|---|---|---|---|
| 0.1 m KCl | 0.5 m KCl | ||
| kD | mL | ||
| Ferritinc | 440 | 37.0 | 39.8 |
| Bovine serum albumin | 67 | 53.9 | 54.8 |
| Ovalbumin | 43 | 59.7 | 60.5 |
| Chymotrypsin | 25 | 73.0 | 71.8 |
| nTaeIF4B | 45.8 | 46.8 | |
| rTaeIF4B | 45.8 | 46.1 | |
| rAteIF4B1 | 47.9 | 46.8 | |
| rAteIF4B2 | 47.6 | 46.5 | |
Protein samples were 1 to 2 mg/mL.
MM, Molecular mass.
Ferritin elutes at the void volume.
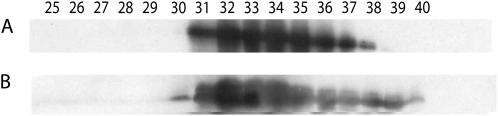
A Superose 6 column with a higher separation range (5,000–5,000,000 molecular weight) was used to compare native and recombinant wheat eIF4B to determine if the native protein potentially contained modifications that promoted formation of multimers that may be absent in the recombinant eIF4B. Native and recombinant wheat eIF4B eluted with similar patterns (Fig. 3), which indicated that the apparent native Mrs of the two protein preparations did not differ.
Figure 3.
Western-blot analysis of wheat eIF4B elution from Superose 6. Equal amounts (15 μg) of native and recombinant wheat eIF4B (predicted Mr of 56,800) were loaded on the Superose 6 column, and fractions of 50 μL were collected. An aliquot of 20 μL of each fraction was separated by SDS-PAGE, blotted to polyvinylidene fluoride, and incubated with a 1/1,000 dilution of rabbit antisera to wheat eIF4B and further processed as described in Figure 2. A, Native wheat eIF4B fractions. B, Recombinant wheat eIF4B fractions. The peaks of the elution of the Mr standards, bovine γ-globulin (158,000) and chicken ovalbumin (44,000), were in fractions 32 and 38, respectively.
Determination of Molecular Weight by Light Scattering
Recombinant AteIF4B1, AteIF4B2, and recombinant wheat eIF4B were analyzed by multiangle light scattering and refraction after passage through a Bio-Rad Bio-Gel SEC 40XL gel filtration column. This method is able to determine the Mr and purity of polymers (Wyatt, 1993). The average molar mass of the AteIF4B1 was approximately 47,000 D, and the purity is >95% (Table II). The lower-than-expected molecular mass (the calculated molecular mass was 57,727 D) was due to degradation of the AteIF4B1 into the approximately 30,000-D fragment, which lowered the average molecular mass. The average molar mass of AteIF4B2 was approximately 54,000, which was consistent with the calculated molecular mass of 59,332 D. The AteIF4B2 preparation (>95% purity) showed the least amount of degradation or aggregation. The recombinant wheat eIF4B (>94% purity) has an estimated molecular mass of 68,000 D in contrast with the calculated molecular mass of 56,835 D. The higher estimated molecular mass was due to the presence of protein aggregate (<6%) in the wheat recombinant eIF4B, which increased the average molar mass. This amount of aggregate is likely due to the low level of dimerization observed (<1%) in the absence of zinc by Cheng et al. (2008). These results were all consistent with purified eIF4B behaving predominantly as a monomer in solution. Note that the concentration of the protein preparations used for these experiments was in the 4 to 8 mg/mL range. If significant self-dimerization had occurred, it should have been obvious at this protein concentration. The results obtained by light scattering suggested that purified plant eIF4B in solution is a monomer of nonspherical shape. Although the light scattering experiments were carried out in the absence of zinc, the most self-association observed in the presence of zinc by Cheng et al. (2008) was approximately 28%, suggesting that the predominant form of purified eIF4B is still monomeric. It should be noted that only the wheat eIF4B displayed any evidence in light scattering of aggregates; neither preparation of AteIF4B appeared to have any aggregates.
Table II.
Weight average mass of peak for eIF4B determined by light scattering
| Samplea | Calculated MMb | Estimated MM by LSc | Estimated Protein Purity by LS |
|---|---|---|---|
| kD | kD | % | |
| rTaeIF4B | 56.8 | Approximately 68 | >94 |
| rAteIF4B1 | 57.7 | Approximately 47 | >95 |
| rAteIF4B2 | 59.3 | Approximately 54 | >95 |
Protein concentrations were 4 to 8 mg/mL.
MM, Molecular mass.
LS, Light scattering.
Analysis of eIF4B by Analytical Ultracentrifugation
Analytical ultracentrifugation confirmed that purified eIF4B was a nonspherical monomer. AteIF4B2 was used for analysis because each of the three preparations of recombinant plant eIF4B showed similar characteristics in light scattering. The three protein concentrations of AteIF4B2 used for analysis gave a calculated Mr of 55,500 to 64,000 (Table III), which indicated that plant eIF4B is a monomer in solution. The calculated frictional ratio (f/f0) was approximately 2.0. Frictional ratios for globular proteins typically are in the approximately 1.2 to 1.4 range, whereas asymmetrically shaped proteins have ratios >2 (Schuck, 2000). These results confirm that plant eIF4B is likely a monomer with an extended or asymmetric structure.
Table III.
Analytical ultracentrifugation analysis of AteIF4B2
| AteIF4B2 (mg/mL) | s (Svedbergs) (±95% Confidence Limits) | MMa (kD) (±95% Confidence Limits) | Root Mean Square Deviations |
|---|---|---|---|
| mg/mL | |||
| 0.046 | 1.8 (1.784, 1.817) | 64.0 (55.0, 73.4) | 0.0018 |
| 0.160 | 1.8 (1.790, 1.800) | 58.1 (55.3, 60.6) | 0.0019 |
| 0.570 | 1.8 (1.813, 1.817) | 55.5 (54.7, 56.5) | 0.0024 |
MM, Molecular mass.
Activity of Arabidopsis eIF4B1 and eIF4B2 in Protein Synthesis Assays
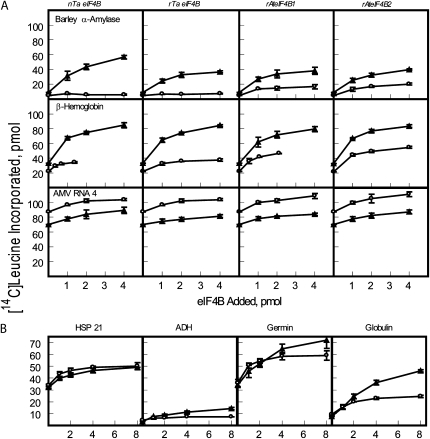
We evaluated three mRNAs: capped barley (Hordeum vulgare) α-amylase (a plant cellular mRNA), capped rabbit β-hemoglobin (a common mRNA used in in vitro translation), and capped alfalfa mosaic virus (AMV) RNA 4 (a plant viral RNA). The assays were carried out with single in vitro transcribed and capped mRNAs so that the observations were not complicated due to competition by a mixture of mRNAs for initiation factors. All of the eIF4B preparations (native and recombinant wheat eIF4B, AteIF4B1, and AteIF4B2) were analyzed by SDS-PAGE, and the protein concentrations were determined in the same assay to ensure that equivalent eIF4B protein levels were tested in the in vitro translation assays. The effects of various levels of the eIF4B isoforms on the translation of these mRNAs in the presence of eIF4F and eIF(iso)4F are shown in Figure 4A and summarized in Table IV. eIF4F was much more efficient than eIF(iso)4F in stimulating translation of the two cellular mRNAs, barley α-amylase and rabbit β-hemoglobin, in the presence of native and recombinant wheat eIF4B (Fig. 4A). There was an approximately 22-fold increase in the amount of protein synthesized in the presence of eIF4F over eIF(iso)4F for capped barley α-amylase mRNA or approximately 3-fold increase for capped rabbit β-hemoglobin mRNA in the presence of eIF4B isoforms (data in Fig. 4A is summarized in Table IV for 2 pmol of eIF4B). A slightly higher activity of native wheat eIF4B compared to recombinant wheat eIF4B with barley α-amylase mRNA was observed. This could be due to posttranslational modifications in the native protein, the presence of contaminating stimulatory proteins in the preparation of native protein (only approximately 80% pure; see Fig. 2), or folding issues with the recombinant protein. Mass spectrometry analysis of native eIF4B did not reveal any posttranslational modifications in any of the observed peptides. Mass spectrometry did recover peptides to additional proteins [eIF(iso)4G and a DEAD box helicase] that might account for the slight stimulation (data not shown). However, a comparable difference was not seen with rabbit β-hemoglobin mRNA or the other mRNAs, suggesting that the stimulation was specific to the barley α-amylase mRNA, which would seem to preclude contaminants as the source of the difference.
Figure 4.
eIF4B translation assays with wheat eIF4F and wheat eIF(iso)4F. A, Each 100-μL reaction contained 5 pmol of the indicated capped mRNA (barley α-amylase, rabbit β-hemoglobin, or AMV RNA 4) and 7.5 pmol recombinant wheat eIF4F (triangles) or 7.5 pmol recombinant wheat eIF(iso)4F (circles). Various eIF4B preparations were added as indicated. The left panels show native wheat eIF4B (nTa eIF4B) or recombinant wheat eIF4B (rTa eIF4B), and the right panels show recombinant Arabidopsis eIF4B1 (rAt eIF4B1) or recombinant Arabidopsis eIF4B2 (rAt eIF4B2). Each assay was performed in triplicate and the results averaged. The amount of [14C]Leu incorporated in the absence of additional eIF4B (not subtracted) was as follows: capped barley α-amylase mRNA [eIF4F, 9 pmol; eIF(iso)4F, 5 pmol]; capped rabbit β-hemoglobin [eIF4F, 33 pmol; eIF(iso)4F, 22 pmol]; capped AMV RNA 4 [eIF4F, 70 pmol; eIF(iso)4F, 87 pmol]. B, Each 100-μL reaction contained 5 pmol of the indicated capped mRNA (AtHSP21, maize ADH, wheat germin, or oat globulin) and 7.5 pmol recombinant wheat eIF4F (triangles) or 7.5 pmol recombinant wheat eIF(iso)4F (circles). Recombinant wheat eIF4B was added as indicated. The amount of [14C]Leu incorporated in the absence of additional eIF4B (not subtracted) was as follows: capped AtHSP21 [eIF4F, 32 pmol; eIF(iso)4F, 34 pmol]; capped maize ADH [eIF4F, 3 pmol; eIF(iso)4F, 4 pmol]; capped wheat germin [eIF4F, 34 pmol; eIF(iso)4F, 36 pmol]; capped oat globulin [eIF4F, 8 pmol; eIF(iso)4F, 9 pmol].
Table IV.
Summary of eIF4B stimulation in the presence of wheat eIF4F or eIF(iso)4F with different mRNAs
| TaeIF4Fa
|
TaeIF(iso)4Fa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| nTaeIF4B | rTaeIF4B | AteIF4B1 | AteIF4B2 | nTaeIF4B | rTaeIF4B | AteIF4B1 | AteIF4B2 | |
| [14C]Leu incorporated, pmolbc | ||||||||
| Barley α-amylased | 34.7 ± 3.7 | 23.6 ± 3.6 | 24.9 ± 5.4 | 23.6 ± 2.7 | 1.5 ± 0.4 | 1.1 ± 0.6 | 9.8 ± 2.4 | 11.8 ± 1.1 |
| Rabbit β-Hb2 | 41.8 ± 1.4 | 41.4 ± 1.1 | 38.7 ± 4.7 | 44.2 ± 2.1 | 12.0 ± 1.1 | 13.6 ± 0.2 | 19.6 ± 0.4 | 27.6 ± 1.1 |
| AMV RNA 42 | 13.8 ± 6.0 | 7.2 ± 2.5 | 11.1 ± 0.5 | 12.2 ± 3.4 | 15.1 ± 2.4 | 11.4 ± 2.0 | 15.2 ± 2.2 | 18.3 ± 5.8 |
Recombinant wheat eIF4F (7.5 pmol) or eIF(iso)4F (7.5 pmol).
The amount of eIF4B added was 2 pmol. Data are from Figure 4.
[14C]Leu incorporation in the absence of exogenous eIF4B was subtracted as follows: capped barley α-amylase mRNA [eIF4F, 9 pmol; eIF(iso)4F, 5 pmol]; capped rabbit β-hemoglobin [eIF4F, 33 pmol; eIF(iso)4F, 22 pmol]; capped AMV RNA 4 [eIF4F, 70 pmol; eIF(iso)4F, 87 pmol].
The amount of template mRNA added was 5 pmol.
In the presence of wheat eIF(iso)4F, both barley α-amylase mRNA and rabbit β-hemoglobin mRNA had a small advantage when AteIF4B1 or 2 was present (Table IV) over wheat eIF4B. This demonstrates that translational activities of different mRNAs may differ subtly, depending on the combinations of factors present.
In contrast with barley α-amylase mRNA or rabbit β-hemoglobin mRNA, the capped AMV RNA 4 showed modestly higher activity (approximately 20%) for eIF(iso)4F versus eIF4F. However, the dependence of AMV RNA 4 on eIF4B during initiation was quite low, as evidenced by the high background (see Fig. 4A) in the absence of eIF4B and the modest stimulation in the presence of all forms of eIF4B.
Additional experiments with four more capped plant mRNAs, Arabidopsis heat shock protein 21 (HSP21), maize (Zea mays) alcohol dehydrogenase (ADH), wheat germin, and oat (Avena sativa) globulin were carried out to determine if there were differences in the requirement for eIF4B. As shown in Figure 4B, these plant mRNAs also displayed varying degrees of dependence on recombinant wheat eIF4B. HSP21 and wheat germin were similar to AMV RNA 4 in having a low requirement for eIF4B, whereas globulin seems to have a high requirement for eIF4B similar to barley α-amylase. HSP21 mRNA shows similar responses to eIF4F and eIF(iso)4F, germin mRNA shows slightly higher activity with for eIF4F, and globulin mRNA shows much higher activity with eIF4F similar to that observed for barley α-amylase. ADH mRNA does not perform well in the fractionated assay; however, in wheat germ extracts, it is somewhat more active (data not shown). This suggests that ADH mRNA may have additional factor requirements that are not fully met in a fractionated assay. These additional mRNAs confirm that translational activities of different mRNAs may differ subtly, depending on the combinations of factors present.
Activity of AteIF(iso)4F1 or AteIF(iso)4F2 with Wheat eIF4B
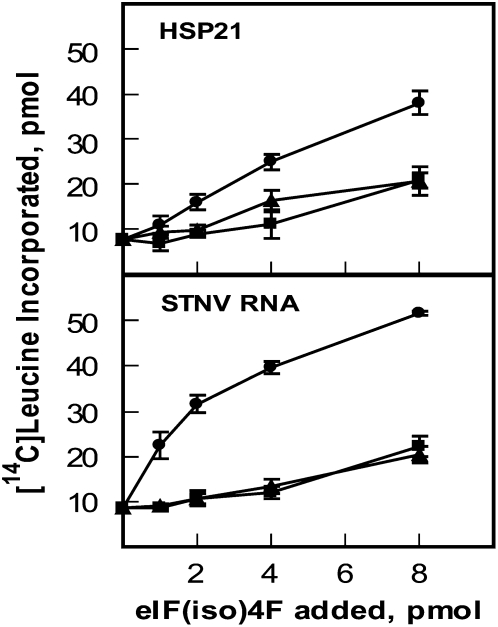
Arabidopsis has two genes for the large subunit of eIF(iso)4F, eIF(iso)4G1 (At5g57870) and eIF(iso)4G2 (At2g24050). However, there is only one gene for the cap-binding subunit, eIF(iso)4E (At5g35620). Recombinant Arabidopsis eIF(iso)4F1 [eIF(iso)4G1 and eIF(iso)4E] and eIF(iso)4F2 [eIF(iso)4G2 and eIF(iso)4E] complexes were expressed and purified from E. coli (Fig. 5). The Arabidopsis eIF(iso)4F forms were assayed in the presence of recombinant wheat eIF4B and were compared to recombinant wheat eIF(iso)4F to determine if there was specificity or discrimination by the different eIF(iso)4G forms. Because we had shown previously (Fig. 4) that AMV RNA 4 had a low requirement for eIF4B and barley α-amylase did not use eIF(iso)4F effectively (Fig. 4), these mRNAs were not used. Instead, another capped plant mRNA, Arabidopsis HSP21, and satellite tobacco necrosis virus (STNV; an uncapped plant viral RNA) were utilized as mRNAs in these assays. AtHSP21 mRNA showed similar activity for wheat eIF(iso)4F or AteIF(iso)4F1 or 2 (Fig. 6). This result was somewhat surprising because the HSP21 mRNA was from Arabidopsis. However, the eIF4B, as well as other components of these assays, was from wheat. Consequently, there may have been subtle differences in the optimal interactions of the translational machinery between monocot and dicot species that are independent of the mRNA source. Wheat eIF(iso)4F was more active than the AteIF(iso)4F isoforms in the presence of STNV RNA and wheat eIF4B.
Figure 5.
Purified wheat eIF(iso)4F and Arabidopsis eIF(iso)4F1 and eIF(iso)4F2. SDS-PAGE (12.5%) of purified complexes. Lane 1, 2.8 μg of native wheat eIF(iso)4F; lane 2, 2.8 μg of recombinant wheat eIF(iso)4F; lane 3, 2.8 μg of recombinant Arabidopsis eIF(iso)4F1; and lane 4, 2.8 μg of recombinant Arabidopsis eIF(iso)4F2. The gel was stained with Coomassie Brilliant Blue.
Figure 6.
Translation assay with wheat eIF4B and wheat eIF(iso)4F, AteIF(iso)4F1, or AteIF(iso)4F2. Each 100-μL reaction contained 5 pmol of capped AtHSP21 mRNA or uncapped STNV RNA and 10.5 pmol recombinant wheat eIF4B. Recombinant wheat (iso)4F (circles), recombinant AteIF(iso)4F1 (triangles), or recombinant AteIF(iso)4F2 (squares) was added as indicated. Each assay was performed in triplicate and the results averaged. The amount of [14C]Leu incorporated in the absence of additional elF(iso)4F (not subtracted) was as follows: 7.1 pmol, AtHSP21; 8.7 pmol, STNV RNA.
Activity of AteIF(iso)4F1 or AteIF(iso)4F2 with AteIF4B1 or AteIF4B2
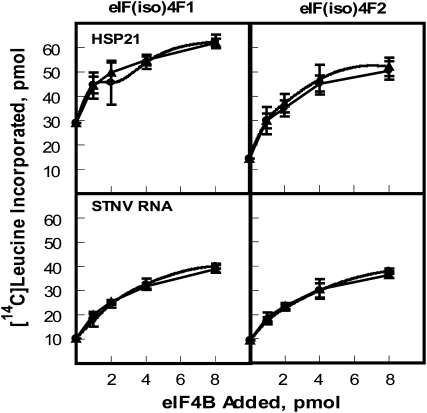
To determine if there are any differences in the interaction between isoforms of eIF4B and eIF(iso)4F, AteIF(iso)4F1 and AteIF(iso)4F2 were assayed with AteIF4B1 and 2 using AtHSP21 and STNV RNA mRNAs (Fig. 7). Interestingly, the only difference noted was an approximately 2-fold higher activity for AteIF(iso)4F1 versus AteIF(iso)4F2 by AtHSP21 mRNA in the absence of eIF4B. In the presence of AteIF4B1 or 2, AtHSP21 continued to show a slight increase in activity for AteIF(iso)4F1. STNV RNA showed similar activity with AteIF(iso)4F or AteIF4B forms.
Figure 7.
AteIF4B translation assays with AteIF(iso)4F1 and eIF(iso)4F2. Each 100-μL reaction contained 5 pmol of the indicated mRNA (capped AtHSP21 or uncapped STNV RNA) and 7.7 pmol recombinant AteIF(iso)4F1 or AteIF(iso)4F2. AteIF4B preparations were added as indicated, recombinant Arabidopsis eIF4B1(circles) or recombinant Arabidopsis eIF4B2 (triangles). Each assay was performed in triplicate and the results averaged. The amount of [14C]Leu incorporated in the absence of additional eIF4B (not subtracted) was as follows: AtHSP21, 29 pmol for eIF(iso)4F1 and 14.3 pmol for eIF(iso)4F2; STNV RNA, 10.3 pmol for eIF(iso)4F1 and 9.3 pmol for eIF(iso)4F2.
DISCUSSION
eIF4B is the most enigmatic of the translation initiation factors. It is poorly conserved on an amino acid sequence level, and it is known to interact with several other initiation factors (e.g. eIF4G, eIF3, eIF4A, and PABP). eIF4B could be required for the initiation of translation; alternatively, it could function as an enhancer of a regulatory protein or it may facilitate translation of specific mRNAs during development or environmental stress. Two observations support the latter hypothesis. First, eIF4B is not essential in yeast or Drosophila except under conditions of stress (Altmann et al., 1993; Coppolecchia et al., 1993; Hernandez et al., 2004). Second, the primary sequence of eIF4B has diverged considerably among plants, fungi, and animals. However, the exact function of eIF4B remains unclear. To gain a better understanding of plant eIF4B, its structure was analyzed biophysically and its ability to stimulate translation of several different mRNAs in the presence of plant eIF4F or eIF(iso)4F was analyzed biochemically.
Analysis of Arabidopsis eIF4B by light scattering and analytical ultracentrifugation showed that purified eIF4B exists in solution predominantly as a monomer that is nonspherical or elongated. These results contrast with those previously reported for mammalian eIF4B using far western analysis and the yeast two-hybrid system or gel filtration (Grifo et al., 1983; Methot et al., 1996b). Indeed, both wheat and Arabidopsis eIF4B show aberrant Mrs using gel filtration. Recombinant forms of wheat eIF4B were reported to dimerize at low levels (approximately 1%), and dimerization was stimulated (approximately 28%) in the presence of zinc (Cheng et al., 2008); however, the majority of protein appears to be monomeric in vitro. eIF4B contacts several other protein components of the translational machinery, including PABP, eIF3, and eIF4F. In addition, three RNA binding sites, two adjacent eIF4A/PABP binding domains in the C terminus and a binding site for eIF(iso)4G in the N terminus that overlaps the RNP2 motif, were identified for wheat eIF4B (Cheng and Gallie, 2006). The ability of wheat eIF4B to bind RNA was compared between GST-tagged eIF4B and His10-tagged eIF4B (Cheng and Gallie, 2006). Interestingly, only the GST-tagged eIF4B was able to robustly bind RNA in in vitro assays. Because GST is known to promote dimerization (Trakshel and Maines, 1988), a dimerized form of eIF4B may bind to RNA more effectively (Cheng and Gallie, 2006). Although pure, full-length, untagged eIF4B does not appear to form dimers in significant amounts in solution (this work), other factors may promote eIF4B dimerization in vivo. Alternatively, binding of RNA may have an allosteric effect such that eIF4B binding affinity for itself and/or RNA is enhanced. As shown in Figure 1, the secondary structure predictions show a potentially unstructured region between two domains that appear to be folded. NMR analysis of AteIF4B2 (D. Hoffman, data not shown) indicated that all the methyl groups were in a similar environment consistent with the prediction that the protein is of an extended shape in which two folded domains are connected by an extended, flexible linker. In addition, the linker region in AteIF4B1 appeared to be susceptible to degradation, as evidenced by the formation of a specific degradation product (data not shown). The apparent flexible, elongated shape of eIF4B may allow it to make multiple contacts with several translation factor components [e.g. PABP, eIF4G, eIF(iso)4G, eIF4A, and mRNA] simultaneously during initiation of translation. The flexibility of its shape and the divergence of the sequence of eIF4B in contrast to other initiation factors suggest that its protein and/or mRNA contacts are highly variable and that the potential exists for mRNA-specific interactions.
Although there were modest differences in the activity of the eIF4B isoforms with various mRNAs tested, there were certain combinations of eIF4B and eIF(iso)4F or eIF4F that were more active in the presence of some mRNAs. Barley α-amylase mRNA, rabbit β-hemoglobin mRNA, globulin mRNA, and STNV RNA translation were all stimulated by eIF4B, whereas AMV RNA 4, HSP21 mRNA, and germin mRNA showed little or modest stimulation by eIF4B. Phosphorylation of eIF4B and PABP dramatically affect the binding to each other and to eIF4F (Le et al., 2000). The phosphorylation state of eIF4B and PABP in vivo, combined with differences in the interactions between various mRNAs, may provide a potent mechanism for discriminating among mRNA species if those factors are limited during stress or other metabolic processes.
The in vitro translation assays also clearly show that mRNAs can use eIF4F and eIF(iso)4F differentially. There was higher activity for barley α-amylase mRNA, oat globulin, and rabbit β-hemoglobin with eIF4F versus eIF(iso)4F. We have previously shown that barley α-amylase mRNA showed a much higher requirement for eIF4F or eIF4B than AMV RNA 4 and that the 5′ UTR was not solely responsible for the amount of eIF4F required (Browning et al., 1988). Interestingly, AMV RNA 4 showed slightly higher activity for eIF(iso)4F versus eIF4F. These results suggest that the availability of eIF4F and eIF(iso)4F may determine which mRNAs are translated the most efficiently.
Plant initiation factor complexes could number in the thousands when the number of genes for each initiation factor and their variable phosphorylation states are taken into consideration. Distinct eIF3 complexes have been reported in S. pombe (Zhou et al., 2005) and Arabidopsis (Kim et al., 2004, 2007) to be associated with specific mRNAs, providing further evidence that initiation complexes may be optimized for some mRNAs leading to another layer of posttranscriptional regulation of gene expression. Further work will be necessary to identify classes of mRNAs that interact with these various isoforms of initiation factors in order to better understand their role in mRNA selection and/or discrimination in vivo.
MATERIALS AND METHODS
Cloning of Arabidopsis eIF4B1 and eIF4B2
Reverse transcription reactions were performed using Arabidopsis (Arabidopsis thaliana; Columbia-0) poly(A)-enriched mRNA as template and primers designed for the 3′ UTR of eIF4B1 (At3g26400) or C-terminal end of the coding region for eIF4B2 (At1g13020). Primers specific for the 5′ and 3′ coding regions of eIF4B1 or eIF4B2 were used for second-strand synthesis and DNA amplification. The primers contained appropriate restriction sites for cloning into pET22b (Novagen). The AteIF4B1 PCR product was digested with NdeI and SstI and inserted into pET22b. The AteIF4B2 PCR product was digested with NdeI and SacI and inserted into pET22b. All constructs were confirmed by DNA sequencing (DNA Core Facility, Institute for Cell and Molecular Biology, University of Texas).
Expression and Purification of AteIF4B1 and AteIF4B2
Recombinant Arabidopsis eIF4B isoforms were purified without the use of affinity tags using standard ion-exchange methods similar to those used to purify recombinant wheat (Triticum aestivum) eIF4B (Mayberry et al., 2007).
Affinity Purification of Antibodies to AteIF4B2 from Rabbit Serum
Rabbit antiserum to AteIF4B2 was prepared at the University of Texas, MD Anderson Cancer Center, Department of Veterinary Sciences (Bastrop, TX). Affinity-purified antibodies to AteIF4B2 were prepared as previously described (Browning et al., 1990).
Partial Purification of Native Arabidopsis eIF4B
Approximately 200 g (wet weight) of Arabidopsis cells from suspension cell cultures (culture generously provided by A.S.N. Reddy, Colorado State University) were rinsed with 20 mm HEPES-KOH, pH 7.6, containing 3% Suc. Portions of cells (100 g) were blended for 2 to 4 min with 100 mL of ice-cold extraction buffer (10% glycerol, 20 mm HEPES-KOH, pH 7.6, 5 mm MgCl2, 1 mm dithiothreitol (DTT), and 0.2 m KCl). Immediately prior to use, 1.0 mL of 100 mm phenylmethylsulfonyl fluoride (in isopropanol) was added to the extraction buffer. The extracts were centrifuged for 20 min at 29,000g at 4°C. The supernatants were pooled and then centrifuged at 218,000g at 4°C for 3 h. The supernatant was diluted to 0.08 m KCl with N1 buffer (20 mm HEPES-KOH, pH 7.6, 10% glycerol, 1 mm DTT, and 0.1 mm EDTA) containing no KCl and loaded onto an 80-mL DEAE column equilibrated in N1 buffer containing 0.08 m KCl. The column was washed with N1 containing 0.08 m KCl. The effluent, which contained eIF4B, was collected from the column and precipitated with 80% ammonium sulfate. The ammonium sulfate pellets were dissolved in N1 buffer containing 0.08 m KCl and dialyzed against the same buffer overnight at 4°C.
The dialyzed sample was loaded onto a 10-mL SP Sepharose column equilibrated in N1 buffer containing 0.08 m KCl. The column was washed with 10 to 20 mL of N1 buffer containing 0.1 m KCl and eluted with a 50-mL linear gradient (0.1–0.3 m KCl in N1 buffer). Fractions (1 mL) were collected and assayed for eIF4B activity. The fractions with the highest level of eIF4B activity were used for western-blot analysis.
Gel Filtration Analysis
An FPLC HiPrep 16/60 Sephacryl S-200 HR column with a bed volume of 126 mL was equilibrated with N1 buffer containing 0.1 or 0.5 m KCl. Molecular weight standards were from GE-Pharmacia (ferritin, 440,000; bovine serum albumin, 67,000; ovalbumin, 43,000; chymotrypsinogen A, 25,000). The elution volumes were determined by A280. Approximately 150 μg of native or recombinant wheat eIF4B or recombinant Arabidopsis eIF4B1 or eIF4B2 in 100 μL was applied to the standardized column, and the elution volumes were determined.
Additional gel filtration analysis was carried out using a 2.4-mL Sepharose 6 PC 3.2/30 column on the SMART system (GE-Pharmacia). Approximately 15 μg of native or recombinant wheat eIF4B in 50 μL was applied to the column equilibrated in N1 buffer containing 0.1 m KCl, and fractions were collected for further analysis by western blot.
Light Scattering Analysis
Recombinant AteIF4B1, AteIF4B2, or wheat eIF4B (4–8 mg/mL) were characterized by multiangle light scattering after passage through a Bio-Rad Bio-Gel SEC 40XL (G3000) column (300 × 7.8 mm) equilibrated with 20 mm HEPES-KOH, pH 7.6, 1 mm DTT, 0.1 mm EDTA, and 0.25 m KCl (passed through a 0.2-μm filter). The column was coupled to an 18-angle, light scattering photometer (Dawn EOS, Wyatt Technology) with a 30-mW gallium arsenide linear polarized laser (685 nm). Data were collected at 1.0-s intervals at ambient temperature (25°C) for light scattering and maintained at 25°C for the refraction measurements. Data were analyzed with ASTRA v4.90.08 software (Wyatt Technology) to calculate the molar mass and molar mass distribution across the peak.
Sedimentation Velocity Analysis
The ultracentrifugation analysis was carried out at the National Ultracentrifugation Facility located at the University of Connecticut, Storrs (Dr. James Cole, Director). Sedimentation velocity of eIF4B2 was measured at concentrations of approximately 0.1 to 1.0 mg/mL. Synthetic boundary cells were loaded with 420 μL of buffer and 410 μL of eIF4B2 (as indicated in Table III). The cells were placed in the rotor and accelerated to 12,000 rpm. The run was stopped and the rotor was gently inverted to mix the contents of the cells. The rotor was then equilibrated in vacuum at 20°C and accelerated to 50,000 rpm. Interference data were acquired at 1-min intervals for approximately 7 h. The data for each loading concentration was analyzed using the program Sedfit (version 8.5) and the model of a single, noninteracting species (Schuck, 2000).
Preparation of Recombinant Wheat eIF4F, Wheat eIF(iso)4F, and Arabidopsis eIF(iso)4F1 and eIF(iso)4F2
To facilitate the preparation of Arabidopsis eIF(iso)4F complexes, both the eIF(iso)4E and eIF(iso)4G subunits of the desired complex were cloned into the same pET22b plasmid to make a dicistronic construct. Briefly, the coding regions were amplified from Arabidopsis ESTs obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH for eIF(iso)4G1 (GenBank N38234), eIF(iso)4G2 (GenBank BE525240), and eIF(iso)4E (GenBank AA041060). Primers were designed for the 5′ and 3′ ends of the coding regions of eIF(iso)4G1 and eIF(iso)4G2 and included appropriate restriction enzyme sites. The amplified products for eIF(iso)4G1 (NdeI/SalI) and eIF(iso)4G2 (NdeI/BamHI) were digested as indicated and cloned into appropriate pET22b restriction sites. The eIF(iso)4E subunit was amplified with appropriate primers, and the forward primer included a ribosome binding site. The eIF(iso)4E amplification product was cloned into pCR-Blunt II TOPO vector (Invitrogen). The eIF(iso)4E coding sequence and ribosome binding site were removed from the plasmid with either NotI for cloning into eIF(iso)4G1/pET22b or BamHI for cloning into eIF(iso)4G2/pET22b. The resulting plasmids contained dicistronic constructs to make eIF(iso)4F1 or eIF(iso)4F2 complexes in Escherichia coli. The dicistronic complex constructs were sequenced to confirm that no mutations were introduced during the cloning process. AteIF(iso)4F1 or AteIF(iso)4F2 were expressed and purified as described for recombinant wheat eIF(iso)4F (Mayberry et al., 2007).
In Vitro Transcription
RNA was transcribed from linearized plasmid using mMessage mMachine (capped mRNA) or MegaScript (uncapped mRNA) T7 kits (Ambion) according to the manufacturer's instructions. Transcription reactions (0.5–1.0 mL reactions) were purified on sterile, 20-mL Sephadex G100 columns preequilibrated with sterile buffer (20 mm HEPES-KOH, pH 7.5, 0.1 mm EDTA, and 0.15 m KCl). Peak fractions were pooled, phenol extracted, and ethanol precipitated. The RNA pellet was rinsed with ethanol, dried, and resuspended in sterile water, and the concentration was determined from the A260.
Fractionated in Vitro Translation Assay
eIF4B was assayed for the ability to stimulate translation in a partially purified wheat germ system as previously described (Lax et al., 1986). The amounts of factors added were optimized for the in vitro reaction. Briefly, 100-μL reactions included 5 pmol mRNA template (as indicated), eIF4B (as indicated), 7.5 pmol recombinant eIF4F or eIF(iso)4F (wheat or Arabidopsis as indicated), 4 μg (5 pmol) native wheat eIF3, 11 μg (220 pmol) recombinant wheat eIF4A, 24 mm HEPES-KOH, pH 7.6, 2.9 mm MgAc2, 100 mm KAc, 30 mm KCl, 2.4 mm DTT, 0.1 mm spermine, 1 mm ATP, 0.2 mm GTP, 34 μm [14C]Leu, 50 μm 19 amino acids, 7.8 μm creatine phosphate, 3 μg creatine kinase, 0.75 A260 units of yeast tRNA, 1 to 2 A260 units of 1× washed wheat ribosomes, and 200 μg wheat germ 40% to 70% ammonium sulfate fraction. Incubation was for 30 min at 27°C, and the amount of [14C]Leu incorporated into protein was determined as previously described (Lax et al., 1986). The wheat germ 40% to 70% ammonium sulfate fraction contains the tRNA synthetases and elongation factors (Lax et al., 1986).
Acknowledgments
The authors thank Dr. Austen Riggs and Claire Riggs (Section of Neurobiology, University of Texas) for their expert help in obtaining the light scattering data. The light scattering instrumentation was obtained with aid from National Science Foundation Grant MCB 951179 (to A. Riggs). The authors thank Dr. Tanya Paull and Ben Hopkins for their assistance and use of the SMART system, Dr. Elizabeth Burks for purification of the native Arabidopsis eIF4B, Dr. E. Vierling (University of Arizona) for the AtHSP21 plasmid, Dr. J. Bailey-Serres (University of California-Riverside) for the maize ADH plasmid, Dr. B. Lane (University of Toronto) for the wheat germin plasmid, and Dr. B. Larkins (University of Arizona) for the oat globulin plasmid. Ultracentrifugation analysis was carried out at the National Ultracentrifugation Facility at the University of Connecticut at Storrs (Dr. James Cole, Director). The authors thank Dr. A.S.N. Reddy (Department of Biology, Colorado State University) for his gift of the Arabidopsis cell suspension culture. The University of Texas MD Anderson Cancer Center, Department of Veterinary Sciences (Bastrop, TX) is supported by Grant NIH-NCI CA-16672.
This work was supported by grants from the Department of Energy (DE–FG02–04ER15575), the National Science Foundation (MCB0214996), and The Welch Foundation (F1339) to K.S.B.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Karen S. Browning (kbrowning@mail.utexas.edu).
Open access articles can be viewed online without a subscription.
References
- Abramson RD, Browning KS, Dever TE, Lawson TG, Thach RE, Ravel JM, Merrick WC (1988. a) Initiation factors that bind mRNA: a comparison of mammalian factors with wheat germ factors. J Biol Chem 263 5462–5467 [PubMed] [Google Scholar]
- Abramson RD, Dever TE, Lawson TG, Ray BK, Thach RE, Merrick WC (1987) The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J Biol Chem 262 3826–3832 [PubMed] [Google Scholar]
- Abramson RD, Dever TE, Merrick WC (1988. b) Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J Biol Chem 263 6016–6019 [PubMed] [Google Scholar]
- Altmann M, Müller PP, Wittmer B, Ruchti F, Lanker S, Trachsel H (1993) A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J 12 3997–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky DA (2003) Unexpected complexity of poly(A)-binding protein gene families in flowering plants: three conserved lineages that are at least 200 million years old and possible auto- and cross-regulation. Genetics 163 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Hershey JWB (1978) The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem 253 3078–3087 [PubMed] [Google Scholar]
- Bi XP, Goss DJ (2000) Wheat germ poly(A)-binding protein increases the ATPase and the RNA helicase activity of translation initiation factors eIF4A, eIF4B, and eIF-iso4F. J Biol Chem 275 17740–17746 [DOI] [PubMed] [Google Scholar]
- Bi XP, Ren JH, Goss DJ (2000) Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 39 5758–5765 [DOI] [PubMed] [Google Scholar]
- Browning KS (1996) The plant translational apparatus. Plant Mol Biol 32 107–144 [DOI] [PubMed] [Google Scholar]
- Browning KS, Fletcher L, Lax SR, Ravel JM (1989) Evidence that the 59-kDa protein synthesis initiation factor from wheat germ is functionally similar to the 80-kDa initiation factor 4B from mammalian cells. J Biol Chem 264 8491–8494 [PubMed] [Google Scholar]
- Browning KS, Humphreys J, Hobbs W, Smith GB, Ravel JM (1990) Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J Biol Chem 265 17967–17973 [PubMed] [Google Scholar]
- Browning KS, Lax SR, Humphreys J, Ravel JM, Jobling SA, Gehrke L (1988) Evidence that the 5′-untranslated leader of mRNA affects the requirement for wheat germ initiation factors 4A, 4F and 4G. J Biol Chem 263 9630–9634 [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT (2005) Protein structure prediction servers at University College London. Nucleic Acids Res 33 W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Wood W, Clemens MJ, Morley SJ (2000) Changes in integrity and association of eukaryotic protein synthesis initiation factors during apoptosis. Eur J Biochem 267 1083–1091 [DOI] [PubMed] [Google Scholar]
- Cheng S, Gallie DR (2006) Wheat eukaryotic initiation factor 4B organizes assembly of RNA and eIFiso4G, eIF4A, and PABP. J Biol Chem 281 24351–24364 [DOI] [PubMed] [Google Scholar]
- Cheng S, Gallie DR (2007) eIF4G, eIFiso4G, and eIF4B bind the poly(A)-binding protein through overlapping sites within the RNA recognition motif domains. J Biol Chem 282 25247–25258 [DOI] [PubMed] [Google Scholar]
- Cheng S, Sultana S, Goss DJ, Gallie DR (2008) Translation initiation factor 4B homodimerization, RNA binding, and interaction with poly(A)-binding protein are enhanced by zinc. J Biol Chem 283 36140–36153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppolecchia R, Buser P, Stotz A, Linder P (1993) A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J 12 4005–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE (2002) Gene-specific regulation by general translation factors. Cell 108 545–556 [DOI] [PubMed] [Google Scholar]
- Doepker RC, Hsu WL, Saffran HA, Smiley JR (2004) Herpes simplex virus virion host shutoff protein is stimulated by translation initiation factors eIF4B and eIF4H. J Virol 78 4684–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan RF, Hershey JWB (1985) Regulation of initiation factors during translational repression caused by serum depletion. J Biol Chem 260 5493–5497 [PubMed] [Google Scholar]
- Duncan RF, Hershey JWB (1989) Protein synthesis and protein phosphorylation during heat stress, recovery, and adaptation. J Cell Biol 109 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming K, Ghuman J, Yuan X, Simpson P, Szendroi A, Matthews S, Curry S (2003) Solution structure and RNA interactions of the RNA recognition motif from eukaryotic translation initiation factor 4B. Biochemistry 42 8966–8975 [DOI] [PubMed] [Google Scholar]
- Gallie DR (2007) Translational control in plants and chloroplasts. In MB Mathews, N Sonenberg, JWB Hershey, eds, Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 747–774
- Gallie DR, Le H, Caldwell C, Tanguay RL, Hoang NX, Browning KS (1997) The phosphorylation state of translation initiation factors is regulated developmentally and following heat shock in wheat. J Biol Chem 272 1046–1053 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Tanguay R (1994) Poly(A) binds to initiation factors and increases cap-dependent translation in vitro. J Biol Chem 269 17166–17173 [PubMed] [Google Scholar]
- Grifo JA, Abramson RD, Satler CA, Merrick WC (1984) RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem 259 8648–8654 [PubMed] [Google Scholar]
- Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC (1983) New initiation factor activity required for globin mRNA translation. J Biol Chem 258 5804–5810 [PubMed] [Google Scholar]
- Hernandez G, Vazquez-Pianzola P, Zurbriggen A, Altmann M, Sierra JM, Rivera-Pomar R (2004) Two functionally redundant isoforms of Drosophila melanogaster eukaryotic initiation factor 4B are involved in cap-dependent translation, cell survival, and proliferation. Eur J Biochem 271 2923–2936 [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23 403–405 [DOI] [PubMed] [Google Scholar]
- Khan MA, Goss DJ (2005) Translation initiation factor (eIF) 4B affects the rates of binding of the mRNA m7G cap analogue to wheat germ eIFiso4F and eIFiso4F.PABP. Biochemistry 44 4510–4516 [DOI] [PubMed] [Google Scholar]
- Kim BH, Cai X, Vaughn JN, Von Arnim AG (2007) On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol 8 R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kim BH, Yahalom A, Chamovitz DA, Von Arnim AG (2004) Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16 3341–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax SR, Lauer SJ, Browning KS, Ravel JM (1986) Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol 118 109–128 [DOI] [PubMed] [Google Scholar]
- Le H, Browning KS, Gallie DR (1998) The phosphorylation state of the wheat translation initiation factors eIF4B, eIF4A, and eIF2 is differentially regulated during seed development and germination. J Biol Chem 273 20084–20089 [DOI] [PubMed] [Google Scholar]
- Le H, Browning KS, Gallie DR (2000) The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J Biol Chem 275 17452–17462 [DOI] [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR (1997) Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem 272 16247–16255 [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D (1998) The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 37 2180–2193 [DOI] [PubMed] [Google Scholar]
- Luo Y, Goss DJ (2001) Homeostasis in mRNA initiation: wheat germ poly(A)-binding protein lowers the activation energy barrier to initiation complex formation. J Biol Chem 276 43083–43086 [DOI] [PubMed] [Google Scholar]
- Manzella JM, Rychlik W, Rhoads RE, Hershey JWB, Blackshear PJ (1991) Insulin induction of ornithine decarboxylase. Importance of mRNA secondary structure and phosphorylation of eucaryotic initiation factors eIF-4B and eIF-4E. J Biol Chem 266 2383–2389 [PubMed] [Google Scholar]
- Mathews MB (2002) Lost in translation. Trends Biochem Sci 27 267–269 [DOI] [PubMed] [Google Scholar]
- Mayberry LK, Dennis MD, Allen ML, Nitka KA, Murphy PA, Campbell L, Browning KS (2007) Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol 430 397–408 [DOI] [PubMed] [Google Scholar]
- Méthot N, Pause A, Hershey JWB, Sonenberg N (1994) The translation initiation factor eIF-4B contains an RNA- binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol Cell Biol 14 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N, Pickett G, Keene JD, Sonenberg N (1996. a) In vitro RNA selection identified RNA ligands that specifically bind to eukaryotic translation initiation factor 4B; The role of the RNA recognition motif. RNA 2 38–50 [PMC free article] [PubMed] [Google Scholar]
- Methot N, Song MS, Sonenberg N (1996. b) A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol 16 5328–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz AM, Wong KCH, Malmström SA, Browning KS (1999) Eukaryotic initiation factor 4B from wheat and Arabidopsis thaliana is a member of a multigene family. Biochem Biophys Res Commun 266 314–321 [DOI] [PubMed] [Google Scholar]
- Meyer K, Petersen A, Niepmann M, Beck E (1995) Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J Virol 69 2819–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn SC, Hershey JWB, Davies MV, Kelleher K, Kaufman RJ (1990) Cloning and expression of eukaryotic initiation factor 4B cDNA: Sequence determination identifies a common RNA recognition motif. EMBO J 9 2783–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranda T, Strong WB, Menaya J, Fabbri BJ, Hershey JWB (1994) Two structural domains of initiation factor eIF-4B are involved in binding to RNA. J Biol Chem 269 14465–14472 [PubMed] [Google Scholar]
- Ochs K, Rust RC, Niepmann M (1999) Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S initiation complexes independently of initiator AUG location. J Virol 73 7505–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs K, Saleh L, Bassili G, Sonntag VH, Zeller A, Niepmann M (2002) Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J Virol 76 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Browning KS, Hohn T, Ryabova LA (2004) Eucaryotic initiation factor 4B controls eIF3-mediated ribosomal entry of viral reinitiation factor. EMBO J 23 1381–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT (2007) The mechanism of translation initiation in eukaryotes. In MB Mathews, N Sonenberg, JWB Hershey, eds, Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 87–128
- Prévôt D, Darlix JL, Ohlmann T (2003) Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell 95 141–156 [DOI] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW (2004) Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23 1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B, Lawson T, Kramer J, Cladaras M, Grifo J, Abramson R, Merrick W, Thach R (1985) ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem 260 7651–7658 [PubMed] [Google Scholar]
- Rogers GW Jr, Komar AA, Merrick WC (2002) eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 72 307–331 [DOI] [PubMed] [Google Scholar]
- Rogers GW Jr, Richter NJ, Lima WF, Merrick WC (2001) Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 276 30914–30922 [DOI] [PubMed] [Google Scholar]
- Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N (1990) Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol 10 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust RC, Ochs K, Meyer K, Beck E, Niepmann M (1999) Interaction of eukaryotic initiation factor eIF4B with the internal ribosome entry site of foot-and-mouth disease virus is independent of the polypyrimidine tract-binding protein. J Virol 73 6111–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 78 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N (2006) The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J 25 2781–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Dever TE (2003) Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol 13 56–63 [DOI] [PubMed] [Google Scholar]
- Trachsel H, Erni B, Schreier MH, Staehelin T (1977) Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J Mol Biol 116 755–767 [DOI] [PubMed] [Google Scholar]
- Trakshel GM, Maines MD (1988) Characterization of glutathione S-transferases in rat kidney. Alteration of composition by cis-platinum. Biochem J 252 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gorp AG, van der Vos KE, Brenkman AB, Bremer A, van den Broek N, Zwartkruis F, Hershey JW, Burgering BM, Calkhoven CF, Coffer PJ (2009) AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene 28 95–106 [DOI] [PubMed] [Google Scholar]
- Wei CC, Balasta ML, Ren JH, Goss DJ (1998) Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37 1910–1916 [DOI] [PubMed] [Google Scholar]
- Wyatt PJ (1993) Light scattering and the absolute characterization of macromolecules. Anal Chim Acta 272 1–40 [Google Scholar]
- Zhou C, Arslan F, Wee S, Krishnan S, Ivanov A, Oliva A, Leatherwood J, Wolf D (2005) PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol 3 14. [DOI] [PMC free article] [PubMed] [Google Scholar]