Abstract
Bacterial plant pathogens manipulate their hosts by injection of numerous effector proteins into host cells via type III secretion systems. Recognition of these effectors by the host plant leads to the induction of a defense reaction that often culminates in a hypersensitive response manifested as cell death. Genes encoding effector proteins can be exchanged between different strains of bacteria via horizontal transfer, and often individual strains are capable of infecting multiple hosts. Host plant species express diverse repertoires of resistance proteins that mediate direct or indirect recognition of bacterial effectors. As a result, plants and their bacterial pathogens should be considered as two extensive coevolving groups rather than as individual host species coevolving with single pathovars. To dissect the complexity of this coevolution, we cloned 171 effector-encoding genes from several pathovars of Pseudomonas and Ralstonia. We used Agrobacterium tumefaciens-mediated transient assays to test the ability of each effector to induce a necrotic phenotype on 59 plant genotypes belonging to four plant families, including numerous diverse accessions of lettuce (Lactuca sativa) and tomato (Solanum lycopersicum). Known defense-inducing effectors (avirulence factors) and their homologs commonly induced extensive necrosis in many different plant species. Nonhost species reacted to multiple effector proteins from an individual pathovar more frequently and more intensely than host species. Both homologous and sequence-unrelated effectors could elicit necrosis in a similar spectrum of plants, suggesting common effector targets or targeting of the same pathways in the plant cell.
Plants and potential pathogens are locked in continual antagonism involving alternating cycles of selection to increase resistance and virulence, respectively. There are many biochemical exchanges between plants and pathogens, and selection can act at multiple points in the host-pathogen interaction. Several overlapping mechanisms of resistance in plants and strategies of pathogens to interdict these resistance responses are being elucidated (Jones and Dangl, 2006). The combined abilities of a pathogen to overcome host resistance mechanisms are major determinants of its host range and success as a pathogen.
Plants possess elaborate mechanisms for detecting the presence of potential pathogens. Basal defenses are triggered by microbe (or pathogen)-associated molecular patterns (MAMPs), ubiquitous components of microbes such as flagellin, lipopolysaccharide, and bacterial translation factor EF-Tu (for review, see Grant et al., 2006; Jones and Dangl, 2006). Detection of MAMPs by extracellular receptor-like kinases results in signal transduction cascades and the elicitation of basal defense or MAMP-triggered immunity, which includes production of active oxygen species and antimicrobial compounds as well as modification of cell walls, including callose deposition (for review, see Bent and Mackey, 2007; McDowell and Simon, 2008).
Pathogens have evolved effector molecules that are translocated into their hosts and often interfere with one or more steps in the induction of resistance (Alfano and Collmer, 2004; Grant et al., 2006; Gohre and Robatzek, 2008; Hogenhout et al., 2009). This has been best characterized for effectors from gram-negative bacterial pathogens. Pseudomonas, Xanthomonas, and Ralstonia species and other bacterial pathogens cause disease by injecting 20 or more effector proteins into host cells via the type III secretion system (Lindeberg et al., 2006; for review, see Mudgett, 2005). Multiple effector proteins have been shown to function as virulence factors in the absence of the cognate resistance (R) protein; the type III secretion system is essential for pathogenicity, and strains defective in the secretion apparatus are nonpathogenic (Lindgren et al., 1986, 1988; Ashfield et al., 1995). While the precise activity of the majority of secreted proteins is currently unknown, there is increasing evidence that the function of many, but not all, effectors is to inhibit plant defenses, including MAMP-triggered immunity (DebRoy et al., 2004; Block et al., 2008; Boller and He, 2009; Cunnac et al., 2009; Jeong et al., 2009).
Plants have countered the pathogen's virulence actions by evolving the ability to directly or indirectly detect the activities of effectors (for review, see Chisholm et al., 2006; Bent and Mackey, 2007). Such recognition results in effector-triggered immunity (ETI) and is mediated predominantly by intracellular nucleotide-binding site-Leu-rich repeat (NBS-LRR) proteins that have often been identified as monogenic R genes. ETI involves signal transduction and a resistance response that overlaps qualitatively with basal defenses but differs in its timing and amplitude, often resulting in a more extreme resistance phenotype. R genes correspond in a gene-for-gene manner to genetically defined avirulence (avr) genes in pathogens (Flor, 1971). In addition to inhibiting basal defense responses, pathogens have in turn evolved effectors that abrogate ETI and that plants have subsequently evolved to detect (Jones and Dangl, 2006). The cycle of plants developing novel recognition specificities and pathogens evolving to overcome them is a continuous process the molecular diversity of which remains to be resolved. Most known gene-for-gene interactions have been characterized in fairly homogenous germplasm of cultivated crop species or in Arabidopsis (Arabidopsis thaliana). There are few data currently available that document resistance mediated by recognition of individual effectors among diverse plant species (e.g. Ashfield et al., 2004; Kuang et al., 2006).
The recognition of effectors by the plant's receptor proteins may occur through direct or indirect interaction (for review, see Jones and Dangl, 2006; Bent and Mackey, 2007). Direct recognition of effectors involves protein-protein interactions between the R protein and the effector. This has been demonstrated for the Avr-Pita/Pi-ta, PopP2-RRS1, and AvrL576-L pairs of avirulence factors and corresponding R proteins (Jia et al., 2000; Deslandes et al., 2003; Dodds et al., 2006). Indirect recognition involves R proteins acting as “guards” that monitor the status of plant proteins targeted by pathogen effectors (Dangl and Jones, 2001). The best understood example of the “target and guard” situation is recognition by the R proteins RPM1 and RPS2 of the effects of three effectors from Pseudomonas syringae (Ps), AvrRpm1, AvrB, and AvrRpt2, on the RIN4 protein in Arabidopsis (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003).
Therefore, there are at least three broad classes of coevolving molecules involved in plant-bacterial pathogen interactions: the pathogen effectors, the plant targets of these effectors (either as the virulence target or decoys), and the plant receptor proteins that recognize the presence of effectors on their targets (Jones and Dangl, 2006; Van der Hoorn and Kamoun, 2008; Caldwell and Michelmore, 2009). Bacterial pathogens contain clusters of genes encoding effector proteins that exhibit elevated levels of sequence variation consistent with diversifying selection acting on these genes (Rohmer et al., 2004; Ma et al., 2006). Because of the selective advantage conferred by such genes, there has been extensive horizontal transfer among microbial pathogens of genes that encode effectors (Lindgren et al., 1988; Hueck, 1998; Deng et al., 2003; Rohmer et al., 2004; Ma et al., 2006; Lindeberg et al., 2008). As a result, diverse pathogens express overlapping sets of effectors (Lindeberg et al., 2006; Sarkar et al., 2006; Stavrinides et al., 2006). On the plant side, R genes, particularly those encoding NBS-LRR proteins and receptor-like kinases, are among the largest and most diverse classes of plant proteins (Clark et al., 2007). Genes encoding R proteins are often clustered in plant genomes, which helps to maintain existing resistance specificities and generate new ones (Halterman et al., 2001; Meyers et al., 2003; Kuang et al., 2004; McHale et al., 2006).
There are several implications arising from the coevolution of these three classes of molecules. Pathogens, particularly bacterial species that infect multiple hosts, should be considered as coevolving with a broad range of plant species rather than individual pathovars coevolving with a limited number of plant hosts. This paradigm leads to several testable hypotheses. One is that plant populations have been exposed to overlapping subsets of pathogen effectors and, consequently, individual plant species have evolved the ability to recognize numerous effectors. Second, nonhosts will react to effectors from nonpathogens either as a consequence of direct recognition or due to the detection of similar effector activities. Recognition of some effectors will be fixed in the species, while recognition of others will display intraspecific variation, especially if there is a fitness cost associated with the expression of the cognate resistance gene (Tian et al., 2003; Bomblies et al., 2007). Screens of plant germplasm, therefore, will reveal intraspecific variation for the reaction to effectors, and the detection of orthologous effectors will vary within and between species. Furthermore, there may be a limited number of points of vulnerability in plants that are targeted by multiple effectors from diverse pathogens. Consequently, host and nonhost plants may exhibit parallel reactions to nonhomologous effectors.
To test these hypotheses, we conducted a large-scale comparative analysis of the reactions of germplasm of several crop species to a library of effector proteins representing nearly the entire secretomes of five bacterial plant pathogens. The reactions of 59 plant genotypes representing 13 species from four dicotyledonous families were tested for reactions to over 171 bacterial effector proteins. Agrobacterium tumefaciens-mediated transient expression was used to provide isogenic delivery of individual effectors into a broad range of species to avoid the confounding effects of multiple effectors secreted by a pathogen and to overcome the limited host ranges of individual pathogens. Variation in effector-elicited chlorotic and necrotic phenotypes that resulted from cell death was observed both within and between species. Several lines of evidence indicated that necrotic phenotypes were often the result of effector recognition rather than the result of their overexpression or enzymatic activity related to their virulence function. Effectors from incompatible pathovars induced necrosis substantially more frequently in nonhosts compared with host species. Twenty-seven known avirulence factors and their homologs frequently induced a necrotic phenotype in multiple taxonomically unrelated species. An additional 32 novel putative avirulence determinants were identified. Common patterns of reaction were identified for homologous as well as sequence-unrelated effectors, implying that multiple effectors targeted the same host proteins or pathways. Finally, we identified several potential new sources of resistance to bacterial plant pathogens.
RESULTS
Confirmed and Putative Effector Proteins Induced Necrotic Responses across Diverse Plant Species
Reactions to 171 effector and other pathogenicity-related proteins were tested in 59 plant accessions to assay interspecific and intraspecific diversity for the elicitation of a phenotypic response, particularly necrosis. We used previously published data and our own sequence searches to identify genes encoding confirmed and putative effectors representing nearly the entire secretomes of four pathovars of Ps and one strain of Ralstonia solanacearum (Rs; Supplemental Table S1). Specifically, we cloned 42 of 54 genes encoding effectors and related proteins from Ps pv tomato DC3000 (Pto DC3000), 24 of 36 from Ps pv phaseolicola 1448A (Pph 1448A), and 22 of 27 from Ps pv syringae B728a (Psy B728a; Table I; Guttman et al., 2002; Greenberg and Vinatzer, 2003; Chang et al., 2004; Schechter et al., 2004, 2006; Joardar et al., 2005; Vinatzer et al., 2005, 2006; Lindeberg et al., 2006; http://pseudomonas-syringae.org/). The sequence of the Ps pv maculicola strain ES4326 (Pma ES4326) genome was not available to us when we initiated these studies; therefore, we could not determine the exact number of effector genes in this strain. Nevertheless, genes encoding 14 of 16 effectors previously identified in Pma ES4226 (Guttman et al., 2002; Vinatzer et al., 2005) were cloned and utilized (Table I). The genome of Rs strain BS048 also had not been sequenced, but it is known to be similar to the recently sequenced Rs strain UW551 (Gabriel et al., 2006; Castillo and Greenberg, 2007); 41 putative effector genes were successfully amplified from Rs BS048 using primers based on the genomic sequence of Rs UW551. In addition to the 143 effector-encoding and pathogenicity-related genes from the five pathogens mentioned above, we cloned genes encoding several effectors from other bacterial strains, including five from the biocontrol Pseudomonas fluorescens strain SBW25 (Rainey, 1999), seven from Xanthomonas campestris strain ATCC33913, and five from Rs strain GMI1000. We also cloned a limited number of genes encoding confirmed and putative effector proteins from other strains of Pseudomonas and Ralstonia (Table I; Supplemental Table S1). Results are presented for putative effectors and their homologs as well as for so-called helper proteins with predicted activity outside the plant cell, regardless of whether secretion or translocation of each protein into or out of the plant cell had been previously confirmed (Table I; Supplemental Table S1; Lindeberg et al., 2006; http://pseudomonas-syringae.org/).
Table I.
Genes encoding effectors and related proteins assayed for their ability to elicit a macroscopic reaction in planta following A. tumefaciens-mediated transient expression
Cloned and tested effectors are indicated in boldface. Superior letters are defined as follows: a, known avirulence factor; h, identifier of the closest known homolog in Ps or other Rs strain; t, truncated; ta, truncated to the first 2,000 bp of the N terminus to make amplification possible; tr, transposon. *, Identifiers of effectors from Rs BS048 were created by adding “BS0” to the last four digits of identifiers assigned to putative effectors identified in Rs strain UW551 by Gabriel et al. (2006).
| Pathogen | Effector (Locus) | ||
|---|---|---|---|
| P. fluorescens SBW25 | avrA | OspC2 | ExoY |
| Ipa | PopC | ||
| Pto DC3000 | avrE1t (PSPTO_1377) | hopP1 (PSPTO_2678) | hopAG∷ISPssytr (PSPTO_0901) |
| avrPto1 (PSPTO_4001) | hopQ1-1a (PSPTO_0877) | hopAH1 (PSPTO_0905) | |
| hopA1 (PSPTO_5354) | hopQ1-2 (PSPTO_4732) | hopAH2-1 (PSPTO_3292) | |
| hopB1 (PSPTO_1406) | hopR1t (PSPTO_0883) | hopAH2-2 (PSPTO_3293) | |
| hopC1 (PSPTO_0589) | hopS1∷ISPssytr (PSPTO_4597) | hopAI1 (PSPTO_0906) | |
| hopD1 (PSPTO_0876) | hopS2 (PSPTO_4588) | hopAJ1 (PSPTO_0852) | |
| hopE1 (PSPTO_4331) | hopT1-1 (PSPTO_A0019) | hopAK1 (PSPTO_4101) | |
| hopF2 (PSPTO_0502) | hopT1-2 (PSPTO_4593) | hopAM1-1 (PSPTO_1022) | |
| hopG1 (PSPTO_4727) | hopT2 (PSPTO_4590) | hopAN1 (PSPTO_5061) | |
| hopH1 (PSPTO_0588) | hopU1 (PSPTO_0501) | hopAO1 (PSPTO_4722) | |
| hopI1 (PSPTO_4776) | hopV1 (PSPTO_4720) | hopAQ1 (PSPTO_4703) | |
| hopJ1 (PSPTO_1179) | hopX1 (PSPTO_A0012) | hopAS1 (PSPTO_0474) | |
| hopK1 (PSPTO_0044) | hopY1 (PSPTO_0061) | hopAT1' (PSPTO_5618) | |
| hopL1 (PSPTO_2872) | hopAA1-1 (PSPTO_1372) | hrpA1 (PSPTO_1381) | |
| hopM1 (PSPTO_1375) | hopAA1-2 (PSPTO_4718) | hrpK1 (PSPTO_1405) | |
| hopN1 (PSPTO_1370) | hopAB2a (PSPTO_3087) | hrpW1 (PSPTO_1373) | |
| hopO1-1 (PSPTO_A0018) | hopAD1 (PSPTO_4691) | hrpZ1 (PSPTO_1382) | |
| hopO1-2 (PSPTO_4594) | hopAF1 (PSPTO_1568) | ||
| hopO1-3 (PSPTO_4592) | |||
| Ps pv tomato JL1065 | avrPto1a | avrRpt2a | |
| Ps pv tomato T1 | hopAB2 | ||
| Pma ES4326 | avrE1 | hopX1 | hopW1-1a |
| hopAA1 | hopX2 | hopO1-1 | |
| hopAA1-2 | hopZ1 | hopT1-1 | |
| hopAB3 | hopAJ1 | hrpW1 | |
| hopAK1 | hopAL1 | ||
| hopJ1 | hopI1 | ||
| Ps pv maculicola M2 | avrRpm1a | ||
| Psy B728a | avrB3a (Psyr_1219) | hopX1 (Psyr_1220) | hopAI1 (Psyr_0780) |
| avrE1 (Psyr_1188) | hopZ3a (Psyr_1224) | hopAJ2 (Psyr_4357) | |
| avrPto1 (Psyr_4919) | hopAA1a (Psyr_1183) | hopAK1 (Psyr_3839) | |
| avrRpm1 (Psyr_0738) | hopAB1a (Psyr_4659) | hopAN1 (Psyr_0465) | |
| hopH1 (Psyr_1889) | hopAE1 (Psyr_4269) | hopAP1 (Psyr_1890) | |
| hopI1 (Psyr_4326) | hopAF1 (Psyr_3813) | hrpA2 (Psyr_1192) | |
| hopJ1(Psyr_1017) | hopAG1 (Psyr_0778) | hrpK1 (Psyr_1218) | |
| hopL1 (Psyr_2631) | hopAH1 (Psyr_0779) | hrpW1 (Psyr_1184) | |
| hopM1 (Psyr_1186) | hopAH2 (Psyr_3123) | hrpZ1 (Psyr_1193) | |
| Ps pv syringae Cit7 | hopAE1 | hopI1 | |
| Pph 1448A | avrB2 (PSPPH_A0120) | hopQ1-1 (PSPPH_A0012) | hopAJ1 (PSPPH_0763) |
| avrB4-1 (PSPPH_3028) | hopR1 (PSPPH_0171) | hopAJ2 (PSPPH_4398) | |
| avrB4-2 (PSPPH_0784) | hopV1 (PSPPH_2351) | hopAK1 (PSPPH_1424) | |
| avrD1 (PSPPH_A0113) | hopW1-1 (PSPPH_A0009) | hopAN1 (PSPPH_0456) | |
| avrE1t (PSPPH_1268) | hopW1-2 (PSPPH_A075) | hopAS1 (PSPPH_4736) | |
| avrRps4 (PSPPH_A0087) | hopX1 (PSPPH_1296) | hopAU1 (PSPPH_A0031) | |
| hopD1 (PSPPH_A0010) | hopAA1 (PSPPH_1263) | hopAV1t (PSPPH_A0056) | |
| hopF3 (PSPPH_3498) | hopAB1 (PSPPH_A0127) | hopAW1 (PSPPH_A0122) | |
| hopG1 (PSPPH_0767) | hopAB3' (PSPPH_2294) | hrpA2 (PSPPH_1272) | |
| hopI1 (PSPPH_4366) | hopAE1t (PSPPH_4326) | hrpK (PSPPH_1295) | |
| hopJ1 (PSPPH_1068) | hopAF1 (PSPPH_1443) | hrpW1 (PSPPH_1264) | |
| hopM1 (PSPPH_1266) | hopAH2 (PSPPH_3036) | hrpZ1 (PSPPH_1273) | |
| Ps pv phaseolicola | hopAR1a | ||
| Ps pv pisi | avrRps4a | hopAB1 | |
| Ps pv glycinea race 4 | avrB1a | ||
| Ps pv glycinea 49a/90 | hopAB1 | ||
| Rs BS048* | BS00326 | BS01019 | BS03105 |
| BS00508 [hrpY]h | BS01066 | BS03109 | |
| BS00515 [hpaP]h | BS01071ta | BS03113 | |
| BS00531 | BS01260 | BS03375ta [hopR1]h | |
| BS00532 [Gala 4]h | BS01554 | BS03418ta | |
| BS00546ta | BS01561 [Gala 4]h | BS03559 | |
| BS00571 | BS01562 [Gala 5]h | BS03923 [hopX1]h | |
| BS00576 | BS01581 [AvrA]h | BS04655 | |
| BS00703 | BS02213 [hopAV1]h | BS04736 | |
| BS00752 [Gala 3]h | BS02264ta [Gala]h | BS04744 [hopAJ1]h | |
| BS00852 [hopX1]h | BS02442 [popB]h | BS04764 [popF1]h | |
| BS00926 [hopG1]h | BS02443 [popA]h | BS10001 | |
| BS00947 [hopD1]h | BS02573 | BS10010 [hopH1]h | |
| BS01016 [popC]h | BS02682 | ||
| Rs GMI1000 | AvrPphF | AvrD | ORF5CEL |
| RS04833 (RSc0608) | hopQ1-1 | ||
| X. campestris ATCC33913 | hopB | XopD | hopH1 |
| Hpa2 | hopG1 | hopQ1-1 | |
| HrpB2 | |||
To provide transient expression of effectors in planta, genes encoding putative effectors were cloned behind the cauliflower mosaic virus 35S promoter in the binary vector pBAV139 (Vinatzer et al., 2006) and subsequently transformed into two strains of A. tumefaciens. Transient expression experiments were performed by infiltrating suspensions of A. tumefaciens into leaves as described previously (Wroblewski et al., 2005). A wide range of interaction phenotypes were observed that varied from no visible macroscopic symptoms through various degrees of chlorosis to extensive tissue damage and cell death in the infiltrated area, as evidenced by extensive macroscopic necrosis (Fig. 1). Infiltration with A. tumefaciens harboring a vector expressing GFP produced no reaction or, occasionally, mild chlorosis. For several effectors, the reaction was weak and manifested as slight discoloration or chlorosis of the infiltrated area. The most severe phenotypes involved extensive cell death accompanied by a brown or black color and collapse of the leaf tissue. The severity of the reactions increased with time, so the reactions were recorded at multiple time points; phenotypes scored 4 to 5 d post infiltration (dpi) were usually more definitive compared with the first scorings at 2 to 3 dpi. Occasionally, necrosis occurred slowly and was only observed at the second scoring (4–5 dpi) with no reaction visible earlier. We generated over 25,000 data points, including replications, that have been archived in a publicly accessible database (http://charge.ucdavis.edu). The consensus result for each interaction was calculated automatically as described in “Materials and Methods” and displayed using a color code to facilitate the visual identification of trends and patterns. Although chlorotic phenotypes were often highly reproducible, we primarily focused on the phenotypes exhibiting stronger reactions involving at least some level of macroscopic necrosis (Fig. 1).
Figure 1.
Interaction phenotypes resulting from transient expression of effectors at 4 to 5 dpi. Each interaction phenotype was assigned a color to represent the reactions in the database. Panels 1 to 8, Lettuce cv Ninja: 1, HopAB2PtoDC3000; 2, HopM1PtoDC3000; 3, AvrRpt2PtoJL1065; 4, AvrB4-2Pph1448A; 5, HopM1PsyB728a; 6, HopZ3PsyB728a; 7, HopT1-1PmaES4326; 8, vector control. Panels 9 to 16, Tomato cv Mocimor: 9, HopAA1PsyB728A; 10, HopAR1Pph; 11, HopW1-1PmaES4326; 12, HopG1PtoDC3000; 13, HopM1PtoDC3000; 14, HopZ3PsyB728a; 15, HopT1-1PtoDC3000; 16, vector control. Panels 17 to 24, Pepper line RNaKy: 17, HopH1PsyB728a; 18, HopW1-1PmaES4326; 19, AvrB2Pph1448A; 20, AvrPto1PtoJL1065; 21, AvrB3PsyB728A; 22, HopAE1PsyCit7; 23, AvrRps4Pph1448A; 24, vector control. Panels 25 to 32, N. benthamiana: 25, HopT1-1PtoDC3000; 26, AvrB1Pgyrace4; 27, HopQ1-1PtoDC3000; 28, HopQ1-1RalGMI1000; 29, HopC1PtoDC3000; 30, HopAA1PsyB728A; 31, HopX1PmaM2; 32, vector control. Enlarged, high-resolution versions of all images in Figure 1 can be found at http://charge.ucdavis.edu/supdata/effectors_in_planta_2009.html.
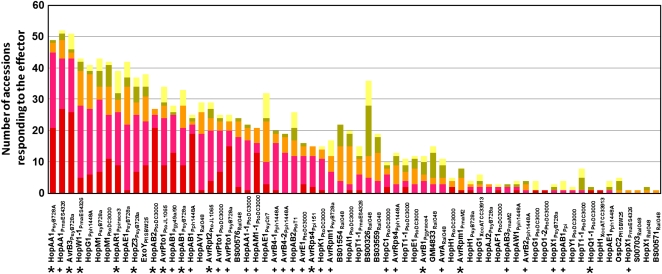
No effector induced a response in all plants tested, but many induced responses in multiple accessions. More than one-third (66 of 171 tested) of the effectors elicited a reaction (chlorosis or necrosis) in at least one genotype (Fig. 2). Four of 14 effectors from Pma ES4326, nine of 24 from Pph 1448A, 11 of 22 from Psy B728a, 19 of 42 from Pto DC3000, and 11 of 41 from Rs BS048 induced a reaction in at least one accession. Of the 22 effectors from other strains of Pseudomonas, Ralstonia, and Xanthomonas, 15 induced reactions on at least one plant genotype. All of the known avirulence determinants tested induced a necrotic response in at least one genotype, and most of them elicited a reaction in many accessions. Several homologs of known avirulence determinants also induced necrotic phenotypes in one or more accessions (Fig. 2).
Figure 2.
Frequency of chlorotic or necrotic phenotypes of reaction to 66 effectors in 59 plant accessions and genotypes. Color coding corresponds to the severity of the different reactions as shown in Figure 1. Known avirulence determinants are indicated by asterisks; homologs of known avirulence determinants are indicated by plus signs.
All accessions reacted to multiple effectors from multiple pathogens. The average number of reactions per accession was 19. Lettuce (Lactuca sativa), tomato (Solanum lycopersicum), and pepper (Capsicum annuum), each represented by multiple accessions, were able to react to at least one effector from each of the five major bacterial pathogens. The severity of the reactions was highly variable but consistent across the plant accessions tested. Variability in the severity of reactions to individual effectors was observed at the family, genus, and species levels.
The Frequency of the Determinants of the Interaction Phenotypes Varied within Cultivated Lettuce and Tomato
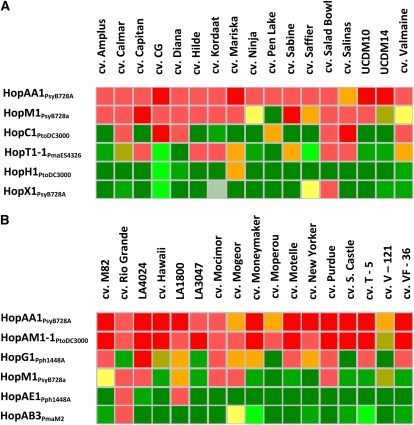
We sampled the diversity of interaction phenotypes within cultivated germplasm of lettuce and tomato. Nineteen cultivars of lettuce and 19 cultivars of tomato were selected as representing a large number of known resistance specificities, many of which had been introgressed from wild species (Farrara et al., 1987; Laterrot, 1987; Williams and St. Clair, 1993; Grube et al., 2000; Van Deynze et al., 2007; Michelmore and Wong, 2008). These accessions, therefore, were expected to express diverse clusters of R genes. Fifty-four and 32 effectors induced strong reactions in at least one genotype of lettuce and tomato, respectively. Some effectors (16 in lettuce and nine in tomato) elicited a necrotic reaction in most or even all genotypes tested (Fig. 3). For example, HopAA1PsyB728A induced a strong reaction in all of the lettuce and tomato genotypes tested. Expression of HopM1PsyB728a caused strong reaction in most of the lettuce genotypes tested, and expression of HopAM1-1PtoDC3000 caused strong necrosis in all tomato accessions (Fig. 3). Other effectors, 23 in lettuce and seven in tomato, elicited reactions in only a subset of the accessions tested (e.g. HopC1PtoDC3000 and HopT1-1PmaES4326 in lettuce and HopG1Pph1448A and HopM1PsyB728a in tomato; Fig. 3). Yet other effectors, five in lettuce and 10 in tomato, rarely induced necrosis (i.e. reactions were observed in three or fewer cultivars within a species). For example, only lettuce cultivars Mariska and Salad Bowl reacted to HopH1PtoDC3000 and HopX1PsyB728A, respectively (Fig. 3). Among the tomato accessions tested, only cv Rio Grande and LA1800 responded with necrosis to HopAE1Pph1448A; cv Rio Grande was also the sole tomato accession showing a necrotic reaction to HopAB3PmaM2 (Fig. 3). These rare reactions to effectors were distributed among various cultivars rather than being restricted to one or a few specific accessions. Therefore, the frequency of plant determinants responsible for reactions to effectors varied considerably within cultivated lettuce and tomato. Some were present in multiple or even all accessions, several had intermediate frequency, while others were rare and present in only one or a few accessions.
Figure 3.
Examples of different patterns of interaction phenotypes observed among cultivars of lettuce (A) and tomato (B). Color coding is as described in Figure 1; gray indicates inconsistent reaction.
Reactions to Individual Effectors Were Similar among, But Highly Polymorphic within, Taxonomically Distinct Groups of Plants
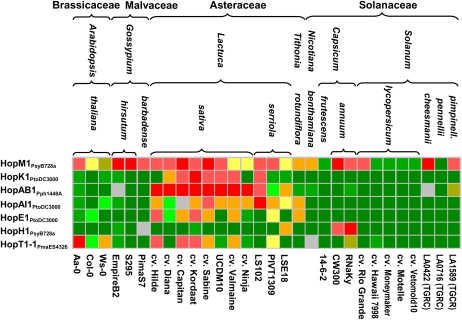
The 59 plant accessions tested represented four families, seven genera, and 14 species (Fig. 4). This allowed us to investigate the specificity of interaction phenotypes relative to different taxonomic groups. Within the Compositae, in addition to the 19 accessions of cultivated L. sativa, we tested five lines of L. serriola (the wild progenitor of L. sativa) and one line each of L. saligna and the more distantly related Mexican sunflower (Tithonia rotundiflora). Within the Solanaceae, in addition to the 19 cultivars of tomato, we tested three species of wild tomato (S. pimpinellifolium, S. cheesmanii, and S. pennellii), four lines of pepper from three different species (C. annuum, C. chinense, and C. frutescens), as well as one line of Nicotiana benthamiana. Within the Malvaceae, we tested three lines of cotton from two different species (Gossypium hirsutum and G. barbadense). The Brassicaceae were represented by three accessions of Arabidopsis.
Figure 4.
Interspecific differences in reactions to transient expression of effectors in 13 species from seven genera belonging to four plant families: Brassicaceae, Malvaceae, Asteraceae (referred to as Compositae in the text), and Solanaceae. Color coding is as described in Figure 1; gray indicates inconsistent reaction.
Thirty-nine of the 66 effectors capable of eliciting necrotic reactions listed in Figure 3 did so in more than one family. Three effectors, HopM1PsyB728a (Fig. 4), HopAA1PmaM2, and HopAE1PsyCit7, induced necrotic reactions across all four families. None of the effectors induced necrosis in all of the accessions tested; however, three effectors, AvrB3PsyB728A and two HopAA1 homologs from Pma ES4326 and Psy B728a, elicited necrosis in more than 90% of the genotypes tested.
Generally, there was no clear relationship between the distribution of necrotic reactions and the taxonomic affinity of the plant genotypes tested. For example, most of the 23 effectors that induced necrosis in lettuce but not in tomato were able to elicit reactions in pepper or N. benthamiana, indicating that these responses were not specific to lettuce or the Compositae family. Similarly, T. rotundifolia did not respond to 12 effectors that elicited strong reactions in most lettuce accessions; however, five of those induced necrosis in tomato. A few reactions were specific to particular taxonomic groups and present in all or nearly all accessions within that group. For example, multiple accessions of L. sativa and L. serriola reacted to AvrRps4 homologs, including HopK1PtoDC3000 or HopAB1Pph1448A (Fig. 4). However, none of the other plants tested (including T. rotundiflora, another member of the Compositae) responded with necrosis to these effectors, making the reaction specific to the genus Lactuca (Fig. 4). Reactions to HopAI1PtoDC3000 and HopE1PtoDC3000 were observed exclusively among several Lactuca lines and in T. rotundiflora and, therefore, were family specific among the accessions tested (Fig. 4). In contrast, determinants of responses to HopH1PsyB728a were specific to L. serriola (LSE18) and two accessions of pepper (CW300 and RNaKy). Similarly, determinants of responses to HopT1-1PmaES4326 were present among accessions of lettuce and Arabidopsis belonging to the Compositae and Brassicaceae families, respectively. Substantially fewer effectors (13) induced reactions in T. rotundiflora as compared with lettuce, in which each accession responded to an average of 29 effectors, ranging from 19 in cv Pennlake to 33 in cv Salad Bowl. The only effector that elicited a reaction in T. rotundiflora but not in lettuce was HopQ1-1PtoDC3000 (data not shown). Within the Solanaceae, tomato, pepper, and N. benthamiana genotypes responded to similar numbers of effectors on average (12, 12, and 13, respectively); even though the subset of effectors that induced necrosis in each genus overlapped, several effectors only induced necrosis in accessions from one or two genera and elicited no responses in the other(s). After several repetitions using all 171 effectors, we were able to identify 12 effectors that induced necrosis at least once. Despite efforts to optimize assays in Arabidopsis (Wroblewski et al., 2005), this species remained recalcitrant to reliable A. tumefaciens-mediated transient assays; therefore, while observations of necrotic reactions are informative, the lack of a reaction was not.
Overall, almost two-thirds of effectors that induced a response in at least one accession were able to elicit a reaction in more than one family.
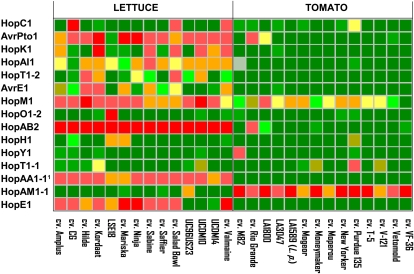
Effectors from Pto DC3000 and Rs BS048 Elicited More Frequent and Stronger Responses in Lettuce Than in Tomato
Lettuce is a nonhost for Pto DC3000 and Rs BS048, while tomato is a good host for both of these pathogens. Of 42 effectors from Pto DC3000, 13 elicited necrosis in one or more accessions of lettuce but only six did so in tomato (Fig. 5). Furthermore, six of these effectors (AvrPto1, HopAI1, HopM1, HopAB2, HopAA1-1, and HopE1) elicited reactions in all or nearly all of the lettuce genotypes tested, whereas only two effectors (HopM1 and HopAM1-1) elicited reactions in more than one accession of tomato, and only one of them (HopAM1-1) elicited a reaction in multiple tomato lines (Fig. 5). Interestingly, the avirulence determinants detected by the product of the Pto gene in tomato and that provide resistance against Pto DC3000, AvrPto1, and HopAB2 rapidly elicited severe necrosis in nearly all of the lettuce genotypes tested as well as in L. serriola UC96US23 (Fig. 5). Of 41 effectors from Rs BS048 (Table I), seven induced necrotic reactions in at least one lettuce genotype and six did so in tomato accessions (Supplemental Fig. S1). Similar to the effectors from Pto DC3000, the reactions to the Rs BS048 effectors in lettuce were stronger and more frequent than those observed among the tomato genotypes. For example, Ral028 and Ral033 induced necrosis in all lettuce accessions tested. In contrast, responses in tomato were rare: five effectors elicited necrosis in only one or two accessions, and a sixth effector, Ral040, elicited necrotic reactions in approximately half of the genotypes tested (Supplemental Fig. S1). Therefore, effectors from Pto DC3000 and Rs BS048 elicited stronger responses more frequently in the nonhost lettuce as compared with their tomato host.
Figure 5.
The differences between lettuce and tomato in their reactions to the effector repertoire from Pto DC3000. Only a representative subset of plants and the effectors that elicited reactions are shown. 1 The reaction of tomato to HopAA1-1 differs from results reported by Munkvold et al. (2008). Color coding corresponds to that in Figure 1; gray indicates inconsistent reaction.
Effectors Known to be Avirulence Determinants Induced Necrosis at High Frequency, and Several New Putative Avirulence Determinants Were Identified
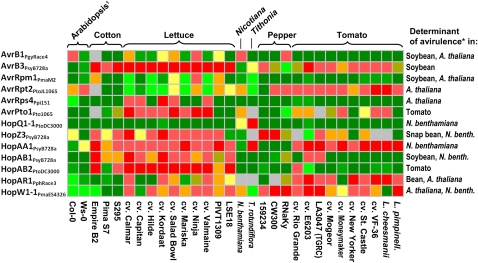
Several effector proteins were historically identified through their avirulence activities and are known to trigger necrotic responses when the corresponding R gene is present in the host plant (Yu et al., 1993; Van den Ackerveken et al., 1996). Our library of effectors contained 13 genes that had been previously described as determining avirulence in various pathosystems (Fig. 6). All of them induced necrosis in at least one among the plants tested, and notably, many did so in species in addition to the ones in which their avirulence function had been initially identified (Fig. 6). Therefore, effectors originally identified as avirulence factors seem to maintain this activity across multiple plant species. The products of most of the known avirulence genes induced strong necrotic reactions in at least one lettuce accession, even though none of the genes had been isolated from a bacterial strain that is pathogenic on this species.
Figure 6.
Reactions of different plant accessions to known avirulence determinants. Avirulence activity of AvrB1Pgyrace4 was reported by Tamaki et al. (1988) and Wanner et al. (1993), AvrB3PsyB728a by Alfano et al. (2000), AvrRpm1PmaM2 by Ashfield et al. (1995) and Ritter and Dangl (1995), AvrRpt2PtoJL1065 by Innes et al. (1993), AvrRps4Ppi151 by Hinsch and Staskawicz (1996), AvrPto1Pto1065 by Ronald et al. (1992), HopQ1-1PtoDC3000 by Wei et al. (2007), HopZ3PsyB728a and HopAA1PsyB728a by Vinatzer et al. (2006), HopAB1PsyB728a by Jackson et al. (1999) and Vinatzer et al. (2006), HopAB2PtoDC3000 by Kim et al. (2002), HopAR1Pphrace3 by Puri et al. (1997) and Simonich and Innes (1995), and HopW1-1PmaES4326 by Lee et al. (2008). Color coding corresponds to that in Figure 1; gray indicates inconsistent reaction. 1, Ecotype Columbia (Col-0) did not respond to AvrRpm1PmaM2 and ecotype Wassilewskija (Ws-0) did not respond to AvrRps4Ppi151, probably due to insufficient effector expression. *, The pathosystem in which the avirulence activity for a particular effector was determined.
In addition to the known avirulence genes, our library contained 39 homologs of known avirulence genes. These homologs induced necrosis more frequently than did other putative or confirmed effectors, and 21 of them induced a necrotic response in at least one genotype (Fig. 2); 63% of the known avirulence factors and their homologs induced necrotic response in at least one genotype tested. Conversely, among the 105 effectors and related proteins that did not elicit a reaction, only 18 were homologs of known avirulence determinants. The fraction of known avirulence factors and their homologs that elicited necrosis in at least one accession (63%) was significantly higher (P < 0.05) than the 22% of all the effectors tested that showed such activity. Therefore, the ability to elicit necrotic response was strongly biased toward effectors with known avirulence activity and their homologs.
As a result of this study, we identified 32 effectors capable of eliciting necrosis that, to our knowledge, had not previously been reported to be avirulence factors. These effectors, including HopG1Pph1448A, HopM1 homologs from Psy B728a and Pto DC3000, ExoYPflSBW25, HopAE1PsyCit7, HopAV1Ral048, and several other effectors from Rs, were capable of eliciting strong reactions in multiple accessions (Fig. 2). Therefore, these 32 effectors could potentially be novel avirulence determinants (see “Discussion”).
Homologous as Well as Sequence-Unrelated Effectors Elicited Similar Patterns of Reactions
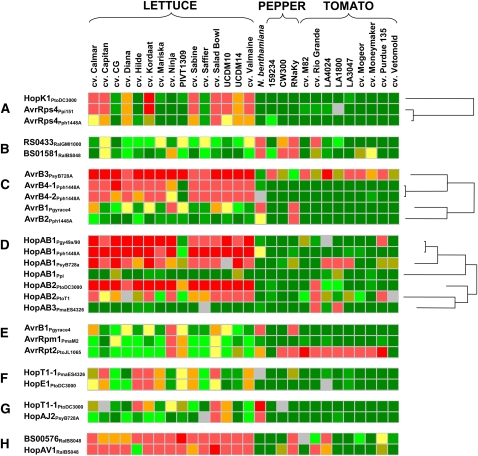
To search for similarities in patterns of reactions produced by homologous and sequence-unrelated effectors across our plant collection, we performed extensive visual inspection as well as cluster analysis of the entire database or of data for subsets of accessions (Supplemental Fig. S2). Our library of effectors included 33 series of homologs containing two or more paralogs from the same pathogen or more often orthologs from different pathogens (Supplemental Table S2). Some of these series, such as those constituting AvrB, HopAB1, or HopAH1, could be divided into subsets based on sequence similarities (Lindeberg et al., 2005; Fig. 7; Supplemental Table S2). For 12 of the 33 series, no reaction to any member was observed among the plants tested. For 21 series, at least one member induced a necrotic reaction in at least one plant genotype; 10 of these reactions were elicited in similar groups of plants by at least some, but not all, of the homologs in the series. For example, the pattern of reactions produced by three AvrRps4 homologs (HopK1DC3000, AvrRps4Ppi151, and AvrRps4Pph1448A) were nearly identical across all accessions tested (Fig. 7); HopK1DC3000 homology (84% similarity) is restricted to the first two-thirds of the protein and, therefore, is considered to be a chimera. For this reason, the N-terminal region of the protein is most likely responsible for triggering plant response. The patterns produced by AvrB4-1Pph1448A and AvrB4-2Pph1448A paralogs (99% protein identity) were also nearly identical; however, the reaction elicited by the other AvrB paralog (29% and 47% protein identity and similarity, respectively), AvrB2Pph1448A, was very different (Fig. 7). Patterns of reaction to RS04833RalGMI1000 and BS01581RalBS048 were similar among tomato and pepper accessions, but among lettuce accessions cv Ninja responded only to BS01581RalBS048 (Fig. 7) but not to RS04833RalGMI1000. Three HopAB1 homologs from Ps pv glycinea 49a/90, Pph 1448A, and Psy B728a elicited similar patterns of reactions among lettuce accessions but different ones among tomato accessions (Fig. 7). In summary, although there were several examples of differences in the patterns of reactions produced by homologous effectors, the majority of homologs elicited similar reaction patterns among the accessions tested.
Figure 7.
Common patterns induced among tested plant accessions by homologous (A–D) and sequence-unrelated (E–H) effectors. Homologous effectors often produced identical or nearly identical patterns (A and B), although differences within orthologous series were also observed (C and D). The patterns of reaction produced by three sequence-unrelated effectors, AvrB1Pgyrace4, AvrRpm1PmaM2, and AvrRpt2PtoJL1065 (E), were similar among lettuce accessions but different among other plants tested. Similarly, the patterns of reaction elicited by other sequence-unrelated effectors, HopT1-1PmaES4326 and HopE1PtoDC3000 (F), HopT1-1PtoDC3000 and HopAJ2PsyB728a (G), and BS00756RalBS048 and HopAV1RalBS048 (H), were common at least in the subset of accessions tested and detectable by cluster analysis (Supplemental Fig. S2).
Cluster analysis using just lettuce accessions identified some similarities between patterns elicited by sequence-unrelated effectors (Supplemental Fig. S2). AvrB1Pgyrace4, AvrRpm1PmaM2, and AvrRpt2PtoJL1065, each of which targets the RIN4 protein in Arabidopsis, all induced strong reactions in lettuce cv Ninja, milder reactions in cv Salad Bowl and PIVT1309, but no reaction in any other lettuce accession tested. Two other sequence-unrelated effectors, HopT1-1PmaES4326 and HopE1PtoDC3000, showed similar patterns of necrotic elicitation across all of the lettuce accessions. Two accessions of lettuce, cv Salad Bowl and line UCDM10, and N. benthamiana displayed parallel reactions to HopT1-1PtoDC3000 and HopAJ2PsyB728A. Finally, the reactions to BS00576RalBS048 and HopAV1RalBS048 were similar among lettuce, pepper, and tomato accessions (Fig. 7). These parallel patterns suggest that these effectors may be interacting with the same plant target(s) or affecting the same plant pathway(s).
The Genetic Determinants of Necrotic Reactions Mapped to R Gene Candidates in Lettuce
Intraspecific polymorphism in the reactions elicited by several effectors allowed the determinants of the necrotic response to be mapped in lettuce. The determinants of the reactions to six effectors, AvrRps4Ppi151, HopK1PtoDC3000, AvrRpm1PmaM2, AvrB1Pgyrace4, AvrRpt2PtoJL1065, and HopC1PtoDC3000, were mapped using 106 F2 individuals from a cross between L. sativa ‘Valmaine’ and L. sativa ‘Ninja’, and the reactions to AvrRps4Ppi151, AvrRps4Pph1448A, and HopK1PtoDC3000 were mapped using 113 recombinant inbred lines derived from L. sativa ‘Salinas’ × L. serriola UC96US23 (Supplemental Table S3). In addition, the determinants of the reaction to AvrPto1PtoJL1065 and AvrRps4Ppi151 were mapped using 107 F3 families derived from a cross between L. sativa ‘Valmaine’ and L. serriola LSE18. The determinants of the reaction to each effector segregated as a single dominant locus (Supplemental Table S3).
Determinants of the reaction to two homologous effectors, AvrRps4Ppi151 and HopK1PtoDC3000, cosegregated and mapped in each of three populations to the same locus on linkage group 8 that contained multiple NBS-LRR-encoding RGC4 (for Resistance Gene Candidate4) genes. No disease resistance phenotypes have been mapped to this locus to date, although based on the number of NBS-LRR-encoding sequences present at the locus, it is one of the larger RGC-encoding loci in lettuce (McHale et al., 2009).
The determinants of the reactions to all three of the sequence-unrelated effectors, AvrRpm1PmaM2, AvrB1Pgyrace4, and AvrRpt2PtoJL1065, that target RIN4 in Arabidopsis cosegregated in lettuce and mapped to a region coincident with many RGC genes and resistance phenotypes on linkage group 1, including race-specific resistances to lettuce downy mildew caused by Bremia lactucae as well as Turnip mosaic virus (McHale et al., 2009). This locus, referred to as the Dm5/8 locus, is unlinked to LsRIN4 and the closest homologs of the Arabidopsis RPS2 and RPM1 genes in lettuce (McHale et al., 2009).
The determinant of the responses to HopC1PtoDC3000 mapped to the terminal region of linkage group 8, a region that contains three NBS-LRR-encoding sequences and a gene that confers race-specific resistance to anthracnose, Ant3. Finally, the determinant of reactions to AvrPto1PtoJL1065 mapped to the bottom of linkage group 9 in the region containing a single NBS-LRR-encoding RGC sequence.
Overall, these data indicate that the plant determinants of the necrotic responses to these bacterial effectors are genetically linked to confirmed and putative disease resistance genes. This is consistent with the necrotic reaction elicited by transient expression of effectors being analogous to the hypersensitive response (HR) elicited in gene-for-gene interactions following pathogen challenge.
DISCUSSION
Bacterial plant pathogens such as Pseudomonas and Ralstonia species secrete repertoires of effector proteins into the extracellular spaces and cells of their hosts (Hueck, 1998). Extensive research is revealing the roles of an increasing number of these effectors in pathogen virulence and/or avirulence (for review, see Grant et al., 2006; Jeong et al., 2009). Multiple effectors have been shown to interfere with plant defense responses in compatible interactions (for review, see Jeong et al., 2009) and to elicit an HR when a cognate gene is present (Van den Ackerveken et al., 1996). Much of our current understanding has been generated using somewhat artificial yet highly informative experimental systems, particularly Arabidopsis and AvrB1Pgyrace4, AvrRps4Ppi151, or AvrPphB (HopAR1Pph). These effectors originated from the pathogens of legumes, Ps pv glycinea, pisi, and phaseolicola, respectively, and were introduced to pathogens of Arabidopsis, Pma ES4326 and Pto DC3000, to study their activity in plant cells. It should be noted that Pma ES4326 and Pto DC3000 do not even contain homologs of AvrB1.
This paper describes a comparative approach to assess natural variation in reactions to bacterial effectors by host and nonhost species. We used A. tumefaciens to deliver genes encoding effector proteins into plant cells to overcome several constraints associated with using the donor pathogens, Pseudomonas, Xanthomonas, and Ralstonia. This strategy allowed isogenic assays of a large number of effectors in a wide range of plants. Restrictions associated with the specificity of individual pathovars to particular hosts were avoided, and each effector could be assessed without the confounding effects of the activities of other secreted effectors. Even though A. tumefaciens-mediated transient expression of an effector inside of the host cell may result in protein levels different from those occurring during infection and infiltration with A. tumefaciens is not completely benign (it may trigger ETI or interfere with salicylic acid-mediated defense signaling; Zipfel et al., 2006; Rico and Preston, 2008), our observations recapitulated previously reported phenotypes for known avirulence proteins and therefore is an informative method of analysis.
We observed a wide range of macroscopic phenotypes, from mild chlorosis to severe necrosis, in response to transient expression of various effectors. Necrotic phenotypes can be triggered in plant cells in response to a variety of signals, including molecular components of pathogens. In some cases, cell death may be triggered by activity of an effector that is associated with its virulence function, particularly if the level of expression after agroinfiltration was substantially higher than following infection with Pseudomonas. However, the best characterized necrosis is programmed cell death resulting in the HR elicited by avirulence factors and mediated by intracellular NBS-LRR proteins, several of which have been identified as the products of R genes. The HR is thought to constrain pathogen proliferation, particularly in the case of biotrophs (Goodman and Novacky, 1994; Greenberg and Yao, 2004). Partial inhibition of the HR can result in increased growth of Ps (Yao and Greenberg, 2006). In several cases, HR has been shown to be sufficient, although not always required, for resistance (Yu et al., 1998; Gassmann et al., 1999; Gassmann, 2005; Römer et al., 2007). Several lines of evidence suggested that many, although not necessarily all, of the necrotic responses elicited by transient expression of effectors reported here reflect the elicitation of the HR mediated by R genes. First, approximately 80% of the necrotic reactions were induced by previously identified avirulence factors or their homologs; the remaining 20% of reactions were induced by effectors whose avirulence status has not been determined. Second, reaction to the effectors was frequent and often stronger in nonhost compared with host species, consistent with some effectors being involved in nonhost resistance. Third, the determinants of reactions to all seven effectors mapped to clusters of NBS-LRR-encoding genes in lettuce, which is a nonhost for all of the bacterial strains used as sources of effector genes in this study. Fourth, the reaction was polymorphic even in otherwise relatively monomorphic germplasm of cultivated species; this is consistent with the selection of the lines screened for their diversity of R genes, many of which had been introgressed as part of breeding programs. Finally, necrosis caused by two effectors for which the avirulence functions had not been determined, HopAE1PsyB728a and HopM1PsyB728a, was SGT1 dependent in N. benthamiana (Vinatzer et al., 2006); SGT1 is a well-characterized mediator of R-triggered defense responses (Azevedo et al., 2006).
The ability to elicit necrosis in our transient assays does not preclude a role for an effector in virulence when secreted by a pathogen. In some cases, we observed a necrotic reaction elicited by effectors from virulent pathogens. For example, HopAM1-1PtoDC3000 elicited necrosis in all genotypes of tomato tested, which is a good host for Pto DC3000. There are several potential explanations of this result. First, other effectors secreted by the pathogen may block recognition or the subsequent elicitation of HR (Abramovitch et al., 2003; Kim et al., 2005; Cunnac et al., 2009). Second, not all cases of the elicitation of necrosis may be detrimental to the pathogen; in some cases, it may be critical to the pathogenesis and triggering cell death may actually benefit the pathogen. Some necrotrophs have host-specific toxins that enhance virulence by inducing programmed cell death in host cells (Sweat and Wolpert, 2007; Sweat et al., 2008). Similarly, some hemibiotrophic oomycetes such as Phytophthora benefit from killing host cells via the activity of necrosis-inducing proteins such as Nep1-like proteins (Pemberton and Salmond, 2004) at the later stages of infection (Kanneganti et al., 2006). Interestingly, these necrotrophic or hemibiotrophic pathogens induce programmed cell death by triggering the plant's defense response (Kanneganti et al., 2006; Sweat and Wolpert, 2007; Sweat et al., 2008). Necrosis may also help to release nutrients or aid in the liberation and dissemination of biotrophic bacteria from infected tissue (Liang et al., 2003; Abramovitch and Martin, 2004; Greenberg and Yao, 2004), or may simply contribute to disease symptoms (Badel et al., 2003, 2005). Alternatively, effectors capable of inducing HR may not be secreted from compatible strains or do not accumulate to a level required to trigger necrosis. HopAJ2PsyB728a, for example, induces a necrotic response in multiple plant accessions (Fig. 7) but lacks a Hrp box and is not translocated from its source strain (Lindeberg et al., 2005; Oh et al., 2007); however, they may be secreted from other strains, and it is interesting that these genes still encode proteins that elicit necrosis in multiple plant genotypes. The presence of these proteins in the effector repertoires of their source strains may be a relic of virulence on other hosts, and the fact that they are not secreted may be the result of adaptation to species that can react to them.
The significance of the chlorotic phenotypes is less obvious than the consequences of necrosis. The degree of chlorosis varied from barely discernible to extensive and accompanied by limited necrosis, particularly several days after infiltration. The chlorosis observed in these studies possibly had a variety of causes. In some cases, it may have been symptomatic of a partial resistance response, as was recently described for AvrB1Pgyrace4, which elicits TAO1-dependent chlorosis in plants lacking RPM1 (Eitas et al., 2008). Therefore, TAO1, a NBS-LRR protein, is a second, minor recognition determinant of AvrB1Pgyrace4 in addition to RPM1. The appearance of minor recognition determinants or quantitative avirulence factors has previously been reported for a few other effectors (Chang et al., 2002; Vinatzer et al., 2006). In other cases, chlorosis may have resulted from the activity of the effector in the plant cell; however, there is no published evidence clearly correlating chlorosis with an effector's virulence-enhancing activity.
The taxonomic distribution of necrotic reactions to individual avirulence factors among the plant genotypes tested is consistent with either convergent evolution or the maintenance of ancient recognition specificities. Known avirulence determinants often retain their activity in other pathosystems (e.g. AvrRps4Ppi151, AvrB1Pgyrace4, and HopAR1Pph in Arabidopsis and HopZ3PsyB728a in N. benthamiana; Wanner et al., 1993; Simonich and Innes, 1995; Hinsch and Staskawicz, 1996; Vinatzer et al., 2006). However, even if the determinants of a reaction were present in all or nearly all of the accessions belonging to a particular genus in our study, they were often absent in related species in the same family. Nonetheless, they were frequently present in more distantly related taxa. This is consistent with convergent evolution of recognition specificities in different families or loss during the evolution of particular species. Convergent evolution of resistance specificities has been reported for the NBS-LRR-encoding genes determining recognition of AvrB1Pgyrace4 and AvrRpm1 in soybean (Glycine max) and Arabidopsis (Grant et al., 1995; Ashfield et al., 2004). Similarly, the closest homologs of RPM1 and RPS2 in lettuce map to a different position than the Dm5/8 locus that determines the reaction to AvrRpm1PmaM2, AvrB1Pgyrace4, and AvrRpt2PtoJL1065, and none of the NBS-LRR-encoding genes at the Dm5/8 locus are similar in sequence to RPM1 or RPS2. Not all of the taxonomically widespread recognition specificities observed in this study may be the result of convergent evolution; resistance genes exhibit heterogeneous rates of evolution, and some resistance specificities may persist over long periods of evolutionary time (Kuang et al., 2004). The loss of resistance specificities in a particular lineage could be the consequence of a lack of selection due to the absence of pathogens with cognate avirulence genes or a selective disadvantage due to the presence of a resistance gene, possibly due to the presence of necrotrophic pathogens that exploit the resistance specificity to elicit the HR to their benefit (Govrin and Levine, 2000; Kanneganti et al., 2006; Sweat and Wolpert, 2007; Sweat et al., 2008).
The repertoire of effectors in bacterial pathogens is a major factor determining host specificity. This could be due to either host-specific virulence activities or the detection of effectors by the plant and elicitation of the resistance response. A previous study failed to identify host-specific virulence determinants (Sarkar et al., 2006). In our study, nonhosts responded to more effectors than hosts. Therefore, host specificity may be determined by lack of (many) avirulence determinants and nonhost resistance may be, at least in part, the result of the recognition of multiple effectors. Of the homologous effectors that elicited necrosis in one or more accessions, approximately half exhibited patterns of reactions similar to those of another homolog. However, patterns of reactions for homologous effectors were rarely identical among accessions. This implies extensive coevolution of effectors and their plant targets and/or the corresponding plant resistance genes. This conclusion is consistent with convergent evolution of resistance specificities, particularly when patterns of recognition were similar in one family but different in others.
Patterns of reaction to sequence-related effectors do not imply similarities or differences in their virulence functions or targets. Sequence-related effectors are presumably functional orthologs and have similar targets, although this has to be verified. The variation in reactions to effectors observed in this study, therefore, probably reflects evolution in pathogens to avoid recognition. It has yet to be demonstrated that the effectors that did not elicit a reaction in this study retain virulence activity; however, they originated from virulent pathogens and often elicited a reaction in some accessions (Fig. 7). Therefore, particularly if recognition is indirect, they must have some activity. Because the number of points of vulnerability in a host may be limited (Caldwell and Michelmore, 2009), sequence-unrelated effectors may have redundant functions or common targets; for example, RIN4 in Arabidopsis is targeted by at least three effectors, AvrRpm1PmaM2, AvrB1Pgyrace4, and AvrRpt2PtoJL1065 (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003). This may be recapitulated in lettuce; the determinant of reactions to the same three effectors maps to one locus. In addition, similarities in the patterns of reactions to three other pairs of sequence-unrelated effectors suggest that they may modify the same target or pathway (Fig. 7). In future studies, we will investigate the virulence activities associated with various alleles encoding several effectors as well as the ability of those effectors to interact with target proteins using the yeast two-hybrid system. The necrotic and chlorotic phenotypes associated with the expression of bacterial effector proteins may enable the cloning of the cognate determinants of the reaction from multiple plant species. Such determinants could be useful as disease resistance genes. This strategy had been successfully employed using effectors of the oomycete pathogen Phythophthora infestans and potato (Solanum tuberosum; Vleeshouwers et al., 2008).
In summary, our data are consistent with the hypotheses proposed at the outset of this paper. Individual plant species have evolved the ability to recognize, either directly or indirectly, numerous effectors, suggesting that most plants have been exposed to overlapping subsets of pathogen effectors. Nonhosts react to effectors from nonpathogens either as a consequence of direct recognition or due to similarity of the effector's activity to the activity of an analogous effector in a pathogen of the plant. Screens of germplasm revealed intraspecific variation in reactions to individual effectors, while recognition of other effectors was constant within a species. Reaction to orthologous effectors varied within and between species. Host and nonhost plants sometimes exhibited parallel reactions to nonhomologous effectors, indicating that there may be a limited number of points of vulnerability in plants that can be targeted by multiple effectors from diverse pathogens. Therefore, bacterial pathogens should be considered as coevolving with a broad range of potential plant hosts rather than as individual pathogen species coevolving with a limited number of plant species. It will be interesting to see to what extent this paradigm can be extended to other classes of pathogens, such as oomycetes and fungi, that do not have such well-documented evidence for horizontal transfer of virulence genes.
MATERIALS AND METHODS
Identification and Cloning of Confirmed and Putative Effector-Encoding Genes
A three-tiered approach was used to mine the genomic sequence of Pseudomonas syringae pv phaseolicola 1448A. First, BLASTX analysis was used to identify homologs of previously identified effectors. Second, all open reading frames located within 300 bp downstream of sequences resembling the conserved hrp-promoter element were translated and analyzed for characteristics common to known effectors, including a high Ser or Pro bias in the first 50 amino acids, an aliphatic amino acid at the third or fourth position, and the lack of negatively charged residues in the first 12 positions (Guttman et al., 2002; Petnicki-Ocwieja et al., 2002; Schechter et al., 2004). Finally, open reading frames immediately downstream of putative effector genes identified above were also analyzed because genes encoding effectors are often clustered in the genome and can exist in operons.
Sequences encoding known and putative effectors were amplified by PCR and cloned into Gateway shuttle vector pDONR207 (Invitrogen; http://www.invitrogen.com). For five large effector-encoding genes (those encoding AvrE1PtoDC3000, HopR1PtoDC3000, AvrE1Pph1448A, HopAE1Pph1448A, and HopAV1Pph1448A), only the N-terminal portion of the gene was cloned. After sequence validation, genes were transferred to the Gateway-compatible binary vector pBAV139 containing in its T-DNA the cauliflower mosaic virus 35S promoter to drive their expression in planta and a His tag to produce a C-terminal fusion with the protein as it is expressed (Vinatzer et al., 2006). Details of all effectors and clones are available from http://charge.ucdavis.edu.
Plant Material and Growth Conditions
All lettuce (Lactuca sativa) accessions used in these experiments were obtained from the collection at the University of California, Davis. Most of them had been used previously in genetic studies of resistance to lettuce downy mildew and contain diverse resistance loci introgressed from wild species (Farrara et al., 1987; Ilott et al., 1987; Maisonneuve et al., 1994). Tomato (Solanum lycopersicum) cultivars and genetic stocks were similarly selected to ensure high diversity in resistance to various pathogens and were obtained from the Tomato Genetic Resource Center (http://tgrc.ucdavis.edu). Pepper (Capsicum annuum) and cotton (Gossypium hirsutum) accessions had been used in previous studies involving genetic mapping (Paran et al., 2004; Gingle et al., 2006). Tithonia rotundiflora was selected as an additional member of the Compositae family (in addition to lettuce) because of its high compatibility with Agrobacterium tumefaciens and strong transient expression following agroinfiltration. Similarly, the Arabidopsis (Arabidopsis thaliana) ecotypes used were selected based on high expression following agroinfiltration as reported previously (Wroblewski et al., 2005). Lettuce, tomato, T. rotundiflora, Nicotiana benthamiana, and Arabidopsis plants were grown under conditions described previously (Wroblewski et al., 2005). Cotton plants were grown in the growth chamber at 26°C during the day and 22°C during the night, with a 14-h-light/10-h-dark photoperiod, and light intensity of approximately 300 μE generated by high-pressure sodium lamps.
Transient Expression Assay and Scoring Criteria
Transient expression assays were performed as described previously (Schob et al., 1997) by infusing a suspension of A. tumefaciens in water (optical density at 600 nm = 0.4–0.5) into the leaf lamina using a blunt-ended 1-mL syringe. To facilitate high-throughput analyses, our collection of effector genes in A. tumefaciens was cultured and manipulated using 48-cell deep-well polypropylene plates. To induce transient expression in tomato and pepper, and to avoid nonspecific necrosis, A. tumefaciens strain 1D1249 was used (Wroblewski et al., 2005); for all other plants tested, A. tumefaciens strain C58 was used. The consistency in reactions induced by effectors delivered using strains 1D1249 and C58 was confirmed in N. benthamiana. The onset of reaction varied among the plants and was usually quicker in lettuce and T. rotundiflora as compared with the other species tested. Reactions were scored two and 4 dpi in lettuce and T. rotundiflora and 3 and 5 dpi in all other species. Eight types of reaction phenotypes could be distinguished in the infiltrated areas (Fig. 1), and they were assigned numeric values for computational analyses and colors for visualization as follows: 0 (dark green), no reaction or minor discoloration comparable to “no-effector” control; 1 (green), mild discoloration, a reaction often observed as a result of plant response to the presence of A. tumefaciens even if no effector was delivered; 2 (light green), chlorosis manifested as discoloration of the infiltrated area; 3 (yellow), extensive chlorosis manifested as severe discoloration of the infiltrated area but not accompanied by necrosis; 4 (orange), chlorosis accompanied by necrosis, a reaction similar to chlorosis but with stronger discoloration and some browning; 5 (brown-green), incipient necrosis, occasional small necrotic spots within the infiltrated area considered to be indicative of necrosis having been induced in only a subset of the plant cells (because this phenotype sometimes appeared to be the plant's response to clumps of A. tumefaciens cells, “potential necrosis” was considered a significant reaction only if the response was consistent from experiment to experiment); 6 (magenta), necrosis manifested as browning and often tissue collapse within the infiltrated tissue; and 7 (red), severe necrosis of the infiltrated area. In most cases, each effector was tested in each genotype three times. All numerical values, summary scores, and other supplementary information are available at http://charge.ucdavis.edu.
Data Management and in Silico Analyses
The Comparative Analyses of Resistance Gene Evolution database (http://charge.ucdavis.edu) was designed to implement, store, and display information about these pathogen-plant interactions. We developed a Web interface to progressively enter the data from different experiments in the form of the numerical values described directly above. All interaction data are stored in linear tables that contain the name of the effector, plant genotype, number of replications of the assay, and experimental conditions. A summary reaction was automatically generated for each interaction based on reproducibility of the replications. The summary scores are stored in both a linear table and a matrix table for easy query. The database is fully searchable using the Web interface by effector, pathogen, pathovar, plant species, or particular genotype. The cluster analysis to search for common patterns of reactions among the summarized reaction data was performed using a modified version of MadMapper (http://cgpdb.ucdavis.edu/XLinkage/MadMapper/). The grouping was performed using Python_UniCluster_V014.py, and the order was refined using Python_MadMapper_V248_XDELTA_119.py (http://cgpdb.ucdavis.edu/scripts_and_tools/). For further manual inspection, alignment was visualized using Pixelirator (http://cgpdb.ucdavis.edu/data_pixelirator/; Supplemental Fig. S2).
Genetic Analysis
Phenotypic data were collected for 106 F2 plants descendent from an L. sativa ‘Ninja’ × L. sativa ‘Valmaine’ cross segregating in response to AvrRps4Ppi151, HopK1PtoDC3000, AvrRpm1PmaM2, AvrB1Pgyrace4, AvrRpt2PtoJL1065, and HopC1PtoDC3000, for 113 recombinant inbred lines (F7 generation) descendent from an L. sativa ‘Salinas’ × L. serriola (UC96US23) cross segregating in response to AvrRps4Ppi151 and HopK1DC3000, and for 107 F3 families descendent from an L. sativa ‘Valmaine’ × L. serriola (LSE18) cross segregating in response to AvrPto1PtoJL1065. Initially, the mapping was done by bulk segregant analysis (Michelmore et al., 1991) using previously developed molecular markers derived from RGC sequences (http://cgpdb.ucdavis.edu; McHale et al., 2009). Ninety-six PCR-based markers were run on contrasting pairs of bulks of genomic DNA from 10 individuals that produced necrotic responses and from 10 individuals that did not show the reaction. Separate pairs of bulks were made for responses to AvrPto1PtoJL1065 and HopC1PtoDC3000. Additionally, a single pair of bulks each was made for the responses to AvrRps4Ppi151 and HopK1PtoDC3000 and to AvrRpm1PmaM2, AvrB1Pgyrace4, and AvrRpt2PtoJL1065, as these phenotypes cosegregated. Markers that distinguished the bulks as well as additional markers used for fine-mapping were then analyzed on all phenotyped individuals in the population (McHale et al., 2009). Genetic linkage was determined using MapMaker version 3.0 as described previously (Lander et al., 1987).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The differences between lettuce and tomato in their reactions to the effector repertoire from Rs BS048.
Supplemental Figure S2. Example of a two-dimensional diagonal CheckMatrix heat plot after cluster analysis of the reactions to effectors in the 59 plant accessions tested.
Supplemental Table S1. Genes encoding effectors and related proteins.
Supplemental Table S2. Orthologous series of effectors used in the experiments.
Supplemental Table S3. Segregation of the reaction determinants to seven effector proteins in three lettuce mapping populations.
Supplementary Material
Acknowledgments
We thank multiple laboratories for providing DNA of effector-encoding genes. We thank Pauline Sanders (University of California, Davis) for greenhouse assistance, Luis Williams (University of California, Davis) and Daniel Coerper (University of Chicago) for technical help, members of the Michelmore laboratory for helpful discussions, and Belinda Martineau (University of California, Davis) for editing the manuscript. Seeds of pepper, cotton, and T. rotundiflora were provided by M. Jahn (Cornell University), A. Paterson, and S. Knapp (both at University of Georgia), respectively. Seeds of tomato and Arabidopsis were obtained from the Tomato Genetic Resource Center (http://tgrc.ucdavis.edu) and the Arabidopsis Biological Resource Center (http://arabidopsis.org).
This work was supported by the National Science Foundation Plant Genome Program (grant no. 0211923 to R.W.M. and J.T.G.) and by a postdoctoral fellowship award from the National Institutes of Health (grant no. 1 F32 G066606–02 to B.A.V.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard W. Michelmore (rwmichelmore@ucdavis.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB (2003) Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 22 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch RB, Martin GB (2004) Strategies used by bacterial pathogens to suppress plant defenses. Curr Opin Plant Biol 7 356–364 [DOI] [PubMed] [Google Scholar]
- Alfano JR, Charkowski AO, Deng WL, Badel JL, Petnicki-Ocwieja T, van Dijk K, Collmer A (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci USA 97 4856–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Collmer A (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42 385–414 [DOI] [PubMed] [Google Scholar]
- Ashfield T, Keen NT, Buzzell RI, Innes RW (1995) Soybean resistance gene specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics 141 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noël L, Sadanandom A, Casais C, Parker J, Shirasu K (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25 2007–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel JL, Nomura K, Bandyopadhyay S, Shimizu R, Collmer A, He SY (2003) Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol Microbiol 49 1239–1251 [DOI] [PubMed] [Google Scholar]
- Badel JL, Shimizu R, Oh HS, Collmer A (2005) A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant Microbe Interact 19 99–111 [DOI] [PubMed] [Google Scholar]
- Bent A, Mackey D (2007) Elicitors, effectors and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45 17.1–17.38 [DOI] [PubMed] [Google Scholar]
- Block A, Li G, Fu ZQ, Alfano JR (2008) Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol 11 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 8 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D (2007) Autoimmune response as a mechanism for Bateson-Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5 e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell KS, Michelmore RW (2009) Arabidopsis thaliana genes encoding defense signaling and recognition proteins exhibit contrasting evolutionary dynamics. Genetics 181 671–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A, Greenberg JT (2007) Evolutionary dynamics of Ralstonia solanacearum. Appl Environ Microbiol 73 1225–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JH, Tai YS, Bernal AJ, Lavelle DT, Staskawicz BJ, Michelmore RW (2002) Functional analysis of the Pto resistance gene family in tomato. Mol Plant Microbe Interact 15 281–291 [DOI] [PubMed] [Google Scholar]
- Chang JH, Urbach JM, Law TF, Arnold LW, Hu A, Gombar S, Grant SR, Ausubel FM, Dangl JL (2004) A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc Natl Acad Sci USA 102 2549–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342 [DOI] [PubMed] [Google Scholar]
- Cunnac S, Lindeberg M, Collmer A (2009) Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr Opin Microbiol 12 53–60 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411 826–833 [DOI] [PubMed] [Google Scholar]
- DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY (2004) A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA 101 9927–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WL, Rehm A, Charkowski A, Rojas M, Collmer A (2003) Pseudomonas syringae exchangeable effector loci: sequence diversity in representative pathovars and virulence function in P. syringae pv. syringae B728a. J Bacteriol 185 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG (2006) Direct protein interaction underlines gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA 103 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas TK, Nimchuk ZL, Dangl JL (2008) Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc Natl Acad Sci USA 105 6475–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrara BF, Ilott TW, Michelmore RW (1987) Genetic analysis of factors for resistance to downy mildew (Bremia lactucae) in species of lettuce (Lactuca sativa and L. serriola). Plant Pathol 36 499–514 [Google Scholar]
- Flor HH (1971) Current status of gene-for-gene concept. Annu Rev Phytopathol 9 275–296 [Google Scholar]
- Gabriel DW, Allen C, Schell M, Denny TP, Greenberg JT, Duan YP, Flores-Cruz Z, Huang Q, Clifford JM, Presting G, et al (2006) Identification of open reading frames unique to select agent: Ralstonia solanacearum race 3 biovar 2. Mol Plant Microbe Interact 19 69–79 [DOI] [PubMed] [Google Scholar]
- Gassmann W (2005) Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol Plant Microbe Interact 18 1054–1060 [DOI] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20 265–277 [DOI] [PubMed] [Google Scholar]
- Gingle AR, Yang H, Chee PW, May OL, Rong J, Bowman DT, Lubbers EL, Day JL, Paterson AH (2006) An integrated Web resource for cotton. Crop Sci 46 1998–2007 [Google Scholar]
- Gohre V, Robatzek S (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46 189–215 [DOI] [PubMed] [Google Scholar]
- Goodman RN, Novacky AJ (1994) The Hypersensitive Reaction in Plants to Pathogens: a Resistance Phenomenon. American Phytopathological Society Press, St. Paul
- Govrin EM, Levine A (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10 751–757 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269 843–846 [DOI] [PubMed] [Google Scholar]
- Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL (2006) Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol 60 425–449 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Vinatzer BA (2003) Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr Opin Microbiol 6 20–28 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6 201–211 [DOI] [PubMed] [Google Scholar]
- Grube RC, Radwanski ER, Jahn M (2000) Comparative genetics of disease resistance within the Solanaceae. Genetics 155 873–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT (2002) A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295 1722–1726 [DOI] [PubMed] [Google Scholar]
- Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P (2001) The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J 25 335–348 [DOI] [PubMed] [Google Scholar]
- Hinsch M, Staskawicz B (1996) Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, AvrRps4, from Pseudomonas syringae pv. pisi. Mol Plant Microbe Interact 9 55–61 [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Van der Hoorn RAL, Terauchi R, Kamoun S (2009) Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22 115–122 [DOI] [PubMed] [Google Scholar]
- Hueck CJ (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilott TW, Durgan ME, Michelmore RW (1987) Genetics of virulence in California populations of Bremia lactucae (lettuce downy mildew). Phytopathology 77 1381–1386 [Google Scholar]
- Innes RW, Bent AF, Kunkel BN, Bisgrove SR, Staskawicz BJ (1993) Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J Bacteriol 175 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RW, Athanassopoulos E, Tsiamis G, Mansfield JW, Sesma A, Arnold DL, Gibbon MJ, Murillo J, Taylor JD, Vivian A (1999) Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc Natl Acad Sci USA 96 10875–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong BR, van Dijk K, Alfano JR (2009) Pseudomonas syringae type III-secreted proteins and their activities and effects on plant innate immunity. Annu Plant Rev 34 48–76 [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Harshey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joardar V, Lindeberg M, Jackson RW, Selengut J, Dodson R, Brinkac LM, Daugherty SC, Deboy R, Durkin AS, Giglio MG, et al (2005) Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol 187 6488–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Huitema E, Cakir C, Kamoun S (2006) Synergistic interaction of the plant cell death pathway induced by Phytophthora infestans Nep1-like protein piNPP1.1 and INF1 elicitin. Mol Plant Microbe Interact 19 854–863 [DOI] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL (2005) The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA 102 6496–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB (2002) Two highly distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109 589–598 [DOI] [PubMed] [Google Scholar]
- Kuang H, Ochoa OE, Nevo E, Michelmore RW (2006) The disease resistance gene Dm3 is infrequent in natural populations of Lactuca serriola due to deletions and frequent gene conversions. Plant J 47 38–48 [DOI] [PubMed] [Google Scholar]
- Kuang H, Woo SS, Meyers B, Nevo E, Michelmore RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster of disease resistance genes in lettuce. Plant Cell 16 2870–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Day MJ, Lincoln SE, Newberg L (1987) MapMaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genetics 121 174–181 [DOI] [PubMed] [Google Scholar]
- Laterrot H (1987) Near isogenic tomato line in Moneymaker type with different genes for disease resistances. Tomato Genetics Coop Rep 37 91 [Google Scholar]
- Lee MW, Jelenska J, Greenberg JT (2008) Arabidopsis proteins important for modulating defense responses to Pseudomonas syringae that secrete HopW1-1. Plant J 54 452–465 [DOI] [PubMed] [Google Scholar]
- Liang H, Yao N, Song JT, Luo S, Lu H, Greenberg JT (2003) Ceramides modulate programmed cell death in plants. Genes Dev 17 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A (2006) Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact 19 1151–1158 [DOI] [PubMed] [Google Scholar]
- Lindeberg M, Myers CR, Collmer A, Schneider DJ (2008) Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization. Mol Plant Microbe Interact 21 685–700 [DOI] [PubMed] [Google Scholar]
- Lindeberg M, Stavrinides J, Chang JH, Alfano JR, Collmer A, Dangl JL, Greenberg JT, Mansfield JW, Guttman DS (2005) Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III hop effector proteins in the plant pathogen Pseudomonas syringae. Mol Plant Microbe Interact 18 275–282 [DOI] [PubMed] [Google Scholar]
- Lindgren PB, Panopoulos NJ, Staskawicz BJ, Dahlbeck D (1988) Genes required for pathogenicity and hypersensitivity are conserved and interchangeable among pathovars of Pseudomonas syringae. Mol Gen Genet 211 520–5312835637 [Google Scholar]
- Lindgren PB, Peet RC, Panopoulos NJ (1986) Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of non-host plants. J Bacteriol 168 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Dong F, Stavrinides J, Guttman DS (2006) Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet 2 e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF III, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated disease resistance in Arabidopsis. Cell 108 743–754 [DOI] [PubMed] [Google Scholar]
- Maisonneuve B, Bellec Y, Anderson P, Michelmore RW (1994) Rapid mapping of two genes for resistance to downy mildew from Lactuca serriola to existing clusters of resistance genes. Theor Appl Genet 89 96–104 [DOI] [PubMed] [Google Scholar]
- McDowell JM, Simon SA (2008) Molecular diversity at the plant-pathogen interface. Dev Comp Immunol 32 736–744 [DOI] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale LK, Truco MJ, Kozik A, Wroblewski T, Ochoa OE, Lahre KA, Knapp SJ, Michelmore RW (2009) The genomic architecture of disease resistance in lettuce. Theor Appl Genet 118 565–580 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R, Wong J (2008) Classical and molecular genetics of Bremia lactucae, cause of lettuce downy mildew. Eur J Plant Pathol 122 19–30 [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol 56 509–531 [DOI] [PubMed] [Google Scholar]
- Munkvold KR, Martin ME, Bronstein PA, Collmer A (2008) A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae. Mol Plant Microbe Interact 21 490–502 [DOI] [PubMed] [Google Scholar]
- Oh HS, Kvitko BH, Morello JE, Collmer A (2007) Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effector proteins into plant cells. J Bacteriol 189 8277–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran I, van der Voort JR, Lefebvre V, Jahn M, Landry L, van Schriek M, Tanyolac B, Caranta C, Chaim AB, Livingstone K, et al (2004) An integrated genetic linkage map of pepper (Capsicum spp.). J Mol Breed 13 251–261 [Google Scholar]
- Pemberton CL, Salmond GPC (2004) The Nep1-like proteins: a growing family of microbial elicitors of plant necrosis. Mol Plant Pathol 5 353–359 [DOI] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Schneider DJ, Tam VC, Chancey ST, Shan L, Jamir Y, Schechter LM, Janes MD, Buell CR, Tang X, et al (2002) Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 99 7652–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri N, Jenner C, Bennett M, Stewart R, Mansfield J, Lyons N, Taylor J (1997) Expression of avrPphB, an avirulence gene from Pseudomonas syringae pv. phaseolicola, and the delivery of signal causing the hypersensitive reaction in bean. Mol Plant Microbe Interact 2 247–256 [DOI] [PubMed] [Google Scholar]
- Rainey PB (1999) Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ Microbiol 1 243–257 [DOI] [PubMed] [Google Scholar]
- Rico A, Preston GM (2008) Agrobacterium suppresses P. syringae-elicited salicylate production in Nicotiana tabacum leaves. In M Fatmi, A Collmer, NS Iacobellis, JW Mansfield, J Murillo, NW Schaad, M Ullrich, eds, Pseudomonas syringae Pathovars and Related Pathogens: Identification, Epidemiology and Genomics. Springer, Dordrecht, The Netherlands, pp 97–102
- Ritter C, Dangl JL (1995) The AvrRpm1 gene of Pseudomonas syringae pv. maculicola is required for avirulence on Arabidopsis. Mol Plant Microbe Interact 8 444–453 [DOI] [PubMed] [Google Scholar]
- Rohmer L, Guttman D, Dangl JL (2004) Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in plant pathogenic Pseudomonas syringae. Genetics 167 1341–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer P, Hahn S, Jordan T, Strauß T, Bonas U, Lahaye T (2007) Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318 645–648 [DOI] [PubMed] [Google Scholar]
- Ronald PC, Salmeron JM, Carland FM, Staskawicz BJ (1992) The cloned avirulence gene AvrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J Bacteriol 174 1604–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar SF, Gordon JS, Martin GB, Guttman DS (2006) The comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174 1041–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol 186 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Vencato M, Jordan KL, Schneider SE, Schneider DJ, Collmer A (2006) Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol Plant Microbe Interact 19 1180–1192 [DOI] [PubMed] [Google Scholar]
- Schob H, Kunz C, Meins F Jr (1997) Silencing of transgenes introduced into leaves by agroinfiltration: a simple, rapid method for investigation of sequence requirements for gene silencing. Mol Gen Genet 256 581–585 [DOI] [PubMed] [Google Scholar]
- Simonich MT, Innes RW (1995) A disease resistance gene in Arabidopsis with specificity for the AvrPph3 gene of Pseudomonas syringae pv. phaseolicola. Mol Plant Microbe Interact 8 637–640 [DOI] [PubMed] [Google Scholar]
- Stavrinides J, Ma W, Guttman DS (2006) Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathog 2 e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweat TA, Lorang JM, Bakker EG, Wolpert TJ (2008) Variation in the LOV1 gene: a CC-NBS-LRR gene conferring victorin sensitivity and disease susceptibility in Arabidopsis. Mol Plant Microbe Interact 21 7–19 [DOI] [PubMed] [Google Scholar]
- Sweat TA, Wolpert T (2007) Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis. Plant Cell 19 673–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Dahlbeck D, Staskawicz B, Keen NT (1988) Characterization and expression of two avirulence genes cloned from Pseudomonas syringae pv. glycinea. J Bacteriol 170 4846–4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423 74–77 [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken G, Maroi E, Bonas U (1996) Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87 1307–1316 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RAL, Kamoun S (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deynze A, Stoffel K, Buell RC, Kozik A, Liu J, van der Knaap E, Francis D (2007) Diversity in conserved genes in tomato. BMC Genomics 8 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer BA, Jelenska J, Greenberg JT (2005) Bioinformatics correctly identifies many type III secretion substrates in the plant pathogen Pseudomonas syringae and the biocontrol isolate P. fluorescens SBW25. Mol Plant Microbe Interact 18 877–888 [DOI] [PubMed] [Google Scholar]
- Vinatzer BA, Teitzel GM, Lee M-W, Jelenska J, Hotton S, Fairfax K, Jenrette J, Greenberg JT (2006) The type III effector repertoire of Pseudomonas syringae B728a and its role in survival and disease on host and non-host plants. Mol Microbiol 62 26–44 [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, Oh S-K, Wang M, Bouwmeester K, Vosman B, Visser RGF, et al (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One 3 e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner LA, Mittal S, Davis KR (1993) Recognition of the avirulence gene avrB from Pseudomonas syringae pv. glycinea by Arabidopsis thaliana. Mol Plant Microbe Interact 6 582–591 [DOI] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 51 32–46 [DOI] [PubMed] [Google Scholar]
- Williams EC, St Clair DA (1993) Phenetic relationships and levels of variability detected by restriction fragment length polymorphism and random amplified polymorphic DNA analysis of cultivated and wild accessions of Lycopersicon esculentum. Genome 36 619–630 [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore RW (2005) Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3 259–273 [DOI] [PubMed] [Google Scholar]
- Yao N, Greenberg JT (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GL, Katagiri F, Ausubel FM (1993) Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol Plant Microbe Interact 6 434–443 [DOI] [PubMed] [Google Scholar]
- Yu I, Parker J, Bent AF (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones J, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.