Abstract
In this article, we demonstrate the connection between intracellular iron storage and oxidative stress response in cyanobacteria. Iron is essential for the survival of all organisms. However, the redox properties that make iron a valuable cofactor also lead to oxidative interactions, resulting in the formation of harmful radicals. Therefore, iron accumulation in cells should be tightly regulated, a process in which ferritin family proteins play an important role. Synechocystis sp. PCC 6803 contains two ferritin-type storage complexes, bacterioferritin and MrgA. Previous studies demonstrated the role of bacterioferritin and MrgA in iron storage. In addition, MrgA was found to play a key role in oxidative stress response. Here, we examined the dual role of the ferritin family proteins using physiological and transcriptomic approaches. Microarray analysis of iron-limited wild-type and ΔmrgA cultures revealed a substantial up-regulation of oxidative stress-related genes in mutant cells. The PerR regulator was found to play an important role in that process. Furthermore, we were able to demonstrate the connection between internal iron quota, the presence of the two storage complexes, and the sensitivity to externally applied oxidative stress. These data suggest a pivotal role for the ferritin-type proteins of Synechocystis sp. PCC 6803 in coordinating iron homeostasis and in oxidative stress response. The combined action of the two complexes allows for the safe accumulation and release of iron from storage by minimizing damage resulting from interactions between reduced iron and the oxygen radicals that are produced in abundance by the photosynthetic apparatus.
In the oxidative environment of Earth, organisms must contend with the problem of transporting, storing, and assembling iron into active cofactors and, at the same time, protect themselves against oxidative damage due to the interactions of iron with dioxygen and reactive oxygen species. The problem of balancing iron homeostasis and oxidative stress is most acute in photosynthetic organisms (Shcolnick and Keren, 2006). On the one hand, the photosynthetic electron transfer chain utilizes radicals and reduced metal species as part of its normal catalysis, all prone to cause oxidative damage if not handled properly. On the other hand, the photosynthetic apparatus imposes an iron requirement that far exceeds that of nonphotosynthetic organisms. The iron quota of the cyanobacterium Synechocystis sp. PCC 6803 (Synechocystis 6803 hereafter) is in the 106 atoms per cell range (Keren et al., 2004), 1 order of magnitude higher than that of the similarly sized nonphotosynthetic Escherichia coli (Finney and O'Halloran, 2003). Iron bioavailability in water bodies is limited by the tendency of Fe3+ to form insoluble ferric oxide crystals. Fe2+ is soluble and easy to transport and utilize for the construction of enzymatic cofactors but prone to interactions with reactive oxygen species and rapidly oxidized. The large demand for iron in photosynthetic organisms and its low bioavailability result in a severe limitation of primary productivity in many large water bodies (Morel and Price, 2003). Furthermore, iron supply is often intermittent, being high for short periods following aeolian dust deposition events (Morel and Price, 2003). Under these conditions, iron should be transported and stored as rapidly as possible, in preparation for long periods of iron scarcity.
A number of response mechanisms are induced upon the onset of iron limitation in different cyanobacterial species. Examples include the isiAB operon, coding for the CP43′ antennae protein and for a flavodoxin (Laudenbach et al., 1988; Burnap et al., 1993), the IdiA, IdiB, and IdiC proteins (Michel et al., 1996, 2001; Pietsch et al., 2007; Nodop et al., 2008), and the iron-sulfur cluster assembly proteins encoded by suf operon genes (Wang et al., 2004). Analysis of cyanobacterial sequences indicated a certain degree of variability in the complement of iron stress-responsive genes coded in the genomes of different species (Bibby et al., 2009; Rivers et al., 2009). In this study, we focus on Synechocystis 6803, for which a considerable amount of information on iron transport and iron stress response is already available (Katoh et al., 2001; Singh et al., 2003; Singh and Sherman, 2007). A major plasma membrane transporter in this organism is the FutABC protein complex. Inactivation mutants in genes encoding components of this transporter can only survive on high iron concentrations (Katoh et al., 2001). A number of studies indicated that the two periplasmic binding proteins of this transporter, FutA1 and FutA2, chelate Fe3+ exclusively (Waldron et al., 2007; Badarau et al., 2008). However, Fe2+ binding to one of these proteins was also demonstrated (Koropatkin et al., 2007).
Once transported into the cell, iron can be stored in ferritin complexes (Lewin et al., 2005). Synechocystis 6803 contains two ferritin-type storage complexes, bacterioferritin and MrgA, a member of the DPS (for DNA-binding proteins from starved cells) family. Bacterioferritins function as ferroxidases, oxidizing Fe2+ to Fe3+ while generating hydrogen peroxide. Fe3+ is stored as iron oxide in the cavity at the center of their 24-mer ultrastructure (Lewin et al., 2005). Synechocystis 6803 bfr genes belong to a subfamily of bacterioferritin genes in which one gene codes for a protein with a conserved heme ligand and the other codes for a protein with conserved di-iron center ligands (Keren et al., 2004). Inactivation mutants lacking either of the two proteins exhibited a loss of approximately 50% of the cellular iron quota and the induction of the iron stress response pathway even under iron-replete growth conditions (Keren et al., 2004). The double insertional mutant does not display a more severe phenotype, indicating that both proteins are required for effective iron storage. DPS proteins are members of the ferritin family that lack the fifth C-terminal helix present in other ferritins (Zeth et al., 2004; Lewin et al., 2005).
Evolutionarily, DPS proteins represent a more diverse group than the other ferritin families, with members functioning as iron storage proteins, DNA-binding proteins protecting against oxidative stress, cold shock proteins, neutrophile activators, or pili components (Zeth et al., 2004). The BFR ferroxidase activity generates hydrogen peroxide, whereas DPS functions as a catalase donating the electron from Fe2+ to hydrogen peroxides (Wiedenheft et al., 2005). A study of MrgA in Synechocystis demonstrated a reduction in the ability of ΔmrgA cells to withstand exposure to hydrogen peroxide (Li et al., 2004). In addition, MrgA was found to have an important role in iron homeostasis. ΔmrgA cells grew much slower than wild-type cells when transferred from iron-sufficient to iron-deficient conditions, like the BFR mutants (Shcolnick et al., 2007). However, unlike the BFR mutants, the internal iron quota of ΔmrgA cells grown on sufficient iron was similar to that of wild-type cells (Shcolnick et al., 2007). Based on these results, we suggest that MrgA functions downstream of BFR in the intracellular iron storage pathway, not affecting the extent of storage but limiting iron mobilization from BFRs under limiting conditions.
In this study, we examined the dual role of the ferritin family proteins using physiological and transcriptomic approaches. Our results uncovered an interrelation between iron homeostasis and oxidative stress response in which ferritin complexes and the oxidative stress response regulator PerR play an important role.
RESULTS
The Response of Wild-Type and ΔmrgA Cells to Iron Limitation Induced by Chelator Treatment
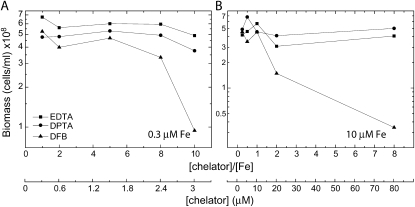
In preparation for a transcriptomic analysis of the effect of MrgA on iron mobilization, we fine-tuned our method for limiting iron bioavailability. Traditionally, iron limitation of cyanobacterial cultures was achieved by repeated washing in iron-free medium (Riethman and Sherman, 1988; Singh et al., 2003). This method suffers from the tendency of Fe3+ to form insoluble iron oxide crystals that adhere to the solid surfaces of glassware and cell walls. This renders the wash protocol only partially effective and requires extensive washing of glassware. As a result, the transition of cyanobacterial cultures into an iron-limited state is slow. An alternative to the wash procedure is the use of iron chelators (Castruita et al., 2007; Shcolnick et al., 2007). The effect of metal chelators is 2-fold: they prevent the formation of insoluble iron oxide crystals, and they compete with the cyanobacterial cells for iron binding. For example, EDTA (binding constant, 1024) successfully inhibits iron acquisition by eukaryotic phytoplankton, causing a reduction in growth rates (Morel et al., 2008). However, in Synechocystis 6803 cultures grown in BG11 medium, EDTA enhanced rather than inhibited growth (Shcolnick et al., 2007). In this study, we examined the effect of two additional chelators, diethylenetriaminopentaacetic acid (binding constant, 1026) and deferoxamine B (DFB; binding constant, 1030). Cells grown on iron-limited EDTA-amended BG11 medium (YBG11 containing 0.3 μm Fe; Shcolnick et al., 2007) were exposed to the chelators in limiting or in sufficient medium (0.3 or 10 μm Fe; Fig. 1, A and B, respectively). Biomass accumulation was not affected by EDTA or diethylenetriaminopentaacetic acid in either of the growth conditions. DFB restricted the growth of iron-limited cultures at DFB-iron ratios higher than 4 (Fig. 1A) and in iron-sufficient conditions at ratios higher than 1 (Fig. 1B). The stronger effect observed in the sufficient culture, as compared with the limited culture, is a result of the higher free DFB concentrations with which cells needed to compete in order to acquire iron.
Figure 1.
The effect of different chelators on biomass accumulation of wild-type cells. Biomass of wild-type cells after 90 h of growth in YBG11 containing 0.3 μm iron (A) and 10 μm iron (B) and amended with 0.3 to 3.0 μm (A) or 10 to 80 μm (B) chelator. Starter cultures were all grown on YGB11 containing 0.3 μm iron. DPTA, Diethylenetriaminopentaacetic acid.
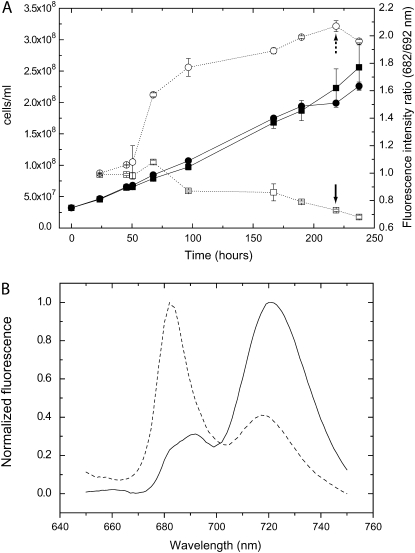
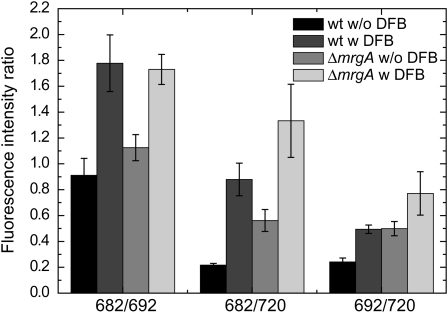
Using both EDTA and DFB, we designed a method for inducing iron limitation. Cultures were grown in an iron-sufficient YBG11 medium allowing for the buildup of intracellular iron storage. Iron limitation was achieved by addition of DFB, forcing the cells to utilize stored iron instead of acquiring it from the medium. The results of such an experiment are presented in Figure 2. Cultures were grown in shaking flasks, conditions that are easily reproduced and that allow for the generation of the large biomass required for the extraction of mRNA for the transcriptomic analysis. The addition of DFB at time zero had little effect on the growth rate of the culture (Fig. 2A). Although growth rates were not affected, the physiological state of DFB-treated cells, as detected by 77 K chlorophyll fluorescence spectroscopy, changed markedly (Fig. 2B). The increase observed in the 682-nm fluorescence band on day 3 is indicative of the accumulation of the iron stress response CP43′ chlorophyll-binding protein (Burnap et al., 1993). The effect of DFB on the chlorophyll fluorescence spectra was quantified as the ratio of the fluorescence emission intensities at 682 and 692 nm (Fig. 2A). According to this parameter, the transition into iron limitation started approximately 48 h after the addition of DFB. As a test for the robustness of this experimental procedure, we evaluated the reproducibility of the physiological response of wild-type and ΔmrgA cells to the DFB treatment (Fig. 3). Two separate experiments (with three repeats each) yielded very reproducible results in terms of fluorescence peak ratios. Interestingly, the relative fluorescence intensity of ΔmrgA cells at 682 nm, grown on sufficient iron, was higher than that observed in wild-type cells.
Figure 2.
Development of iron limitation in DFB-treated cells. A, Cell density (black symbols) and 77 K chlorophyll fluorescence emission spectra peak ratios (white symbols) of cultures grown in YBG11 containing 10 μm iron in the presence (circles) or absence (squares) of 50 μm DFB. Starter cultures were grown on YBG11 containing 10 μm iron. The sd was calculated from two repeats. B, Sample fluorescence spectra (collected at 218 h; marked by arrows in A). The fluorescence at 682 nm is mainly a result of the CP43′ antenna complex emission, 692 nm of PSII reaction center core emission, and 720 nm of PSI emission.
Figure 3.
77 K chlorophyll fluorescence emission spectra of wild-type (wt) and ΔmrgA cells. Chlorophyll fluorescence spectra at 77 K of wild-type cells and ΔmrgA grown for 190 to 210 h in YBG11 medium containing 10 μm iron with (w) or without (w/o) the addition of 50 μm DFB were measured. The mean fluorescence peak ratios of the six biological repeats are shown.
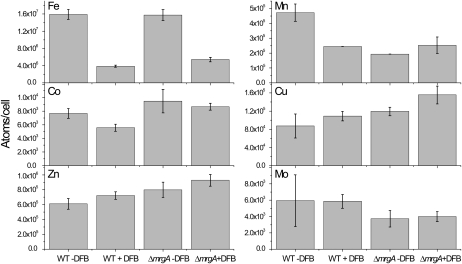
In addition to fluorescence spectroscopy, we measured the cellular quota of a number of biologically relevant transition metals (Fig. 4). As expected, the iron quota of DFB-treated Synechocystis 6803 cells dropped to approximately 26% of the untreated cell value. The iron quota of untreated ΔmrgA was similar to that of untreated wild-type cells, as reported (Shcolnick et al., 2007). In the DFB-treated ΔmrgA culture, the cellular quota dropped to approximately 34%, a statistically significant difference compared with wild-type cells. This result is in agreement with the proposed role of MrgA in mobilizing iron from intracellular storage (Shcolnick et al., 2007). In addition, we found that the manganese quota is affected by the addition of DFB to wild-type cells. Since DFB does not form stable complexes with Mn(II) (Duckworth and Sposito, 2005), the reduction in cellular quota cannot be attributed to lower bioavailability. Therefore, we can only assume that the iron content affects manganese uptake or accumulation in the cells. ΔmrgA cells contain less manganese than wild-type cells under iron-sufficient conditions. However, it is important to note that the reduction in manganese was not severe enough to affect photosynthesis, as the Vmax for oxygen evolution in ΔmrgA cultures was approximately 96% that of wild-type cultures (299 versus 282 μmol oxygen mg−1 chlorophyll h−1, respectively). Under iron-limiting conditions, the manganese quota of the mutant cells was similar to that of wild-type cells. The cellular quotas of cobalt, copper, zinc, and molybdenum did not change significantly as a result of the DFB treatment or mrgA disruption (Fig. 4).
Figure 4.
Internal transition metal ion quotas of wild-type and ΔmrgA cells. Cells were grown in 10 μm iron with or without DFB. At the time of harvest, the peak ratios at 682 nm to 692 nm observed in 77 K chlorophyll fluorescence spectra were as follows: 1.03 ± 0.05 (wild type [WT] − DFB), 1.97 ± 0.05 (WT + DFB), 1.16 ± 0.10 (ΔmrgA − DFB), and 1.79 ± 0.11 (ΔmrgA + DFB). Error bars represent sd, based on two to three different cultures.
Microarray Analysis of Iron-Sufficient Versus Iron-Deficient Wild-Type and ΔmrgA Cells
Microarray analysis was performed using a loop design comparing two strains (wild type and ΔmrgA), two conditions (wild type + DFB and ΔmrgA + DFB), and two dye swaps, as described by Singh and coworkers (2003). Differential transcript abundance, in units of fold change, is reported in Supplemental File S1, which includes data on 1,803 genes that passed a minimum statistical threshold. A breakdown of the affected transcripts into functional categories is presented in Table I for each of the comparisons that were made from the loop design as depicted in Supplemental File S1.
Table I.
Differentially regulated genes in response to the DFB treatment or to deletion of mrgA, according to their functional categories
Genes were considered differentially regulated in conditions of fold change > 1.4 and P < 0.05. The number of transcripts that were down-regulated by the treatment (DFB, ΔmrgA, or both) is marked by ↓, and the number of up-regulated genes is marked by ↑. Categories discussed in this article appear in boldface type. WT, Wild type.
| General Pathway | No. of Genes | Differentially Regulated Genes
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔmrgA/WT | WT DFB/WT | ΔmrgA DFB/ΔmrgA | ΔmrgA DFB/WT DFB | ||||||
| Amino acid biosynthesis | 83 | 1↓ | 2↑ | 4↓ | 1↑ | 16↓ | 14↑ | 16↓ | 6↑ |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 116 | 3↓ | 5↑ | 5↓ | 8↑ | 10↓ | 25↑ | 23↓ | 18↑ |
| Cell envelope | 63 | 0↓ | 8↑ | 3↓ | 1↑ | 12↓ | 5↑ | 9↓ | 4↑ |
| Cellular processes | 61 | 3↓ | 3↑ | 4↓ | 1↑ | 8↓ | 19↑ | 3↓ | 13↑ |
| Central intermediary metabolism | 31 | 2↓ | 1↑ | 0↓ | 2↑ | 3↓ | 3↑ | 4↓ | 1↑ |
| DNA replication, restriction, modification, recombination, and repair | 51 | 3↓ | 1↑ | 2↓ | 0↑ | 5↓ | 10↑ | 8↓ | 6↑ |
| Energy metabolism | 86 | 1↓ | 3↑ | 0↓ | 4↑ | 10↓ | 13↑ | 21↓ | 8↑ |
| Fatty acid, phospholipid, and sterol metabolism | 34 | 2↓ | 1↑ | 2↓ | 1↑ | 7↓ | 5↑ | 10↓ | 3↑ |
| Hypothetical | 449 | 41↓ | 45↑ | 26↓ | 23↑ | 81↓ | 200↑ | 118↓ | 112↑ |
| Other categories | 258 | 13↓ | 9↑ | 6↓ | 7↑ | 21↓ | 93↑ | 28↓ | 36↑ |
| Photosynthesis and respiration | 129 | 3↓ | 5↑ | 15↓ | 5↑ | 54↓ | 10↑ | 47↓ | 4↑ |
| Purines, pyrimidines, nucleosides, and nucleotides | 39 | 0↓ | 0↑ | 0↓ | 0↑ | 1↓ | 2↑ | 3↓ | 2↑ |
| Regulatory functions | 156 | 7↓ | 8↑ | 5↓ | 6↑ | 22↓ | 29↑ | 25↓ | 16↑ |
| Transcription | 27 | 3↓ | 2↑ | 0↓ | 2↑ | 3↓ | 5↑ | 8↓ | 3↑ |
| Translation | 146 | 17↓ | 4↑ | 8↓ | 3↑ | 33↓ | 15↑ | 56↓ | 11↑ |
| Transport and binding proteins | 169 | 9↓ | 6↑ | 7↓ | 17↑ | 18↓ | 38↑ | 34↓ | 14↑ |
| Unknown | 1,267 | 19↓ | 25↑ | 10↓ | 11↑ | 38↓ | 86↑ | 45↓ | 64↑ |
| Total | 3,165 | 127↓ | 128↑ | 97↓ | 92↑ | 342↓ | 572↑ | 458↓ | 321↑ |
It was interesting to compare the results obtained for the DFB treatment of wild-type cells with those previously obtained for iron depletion in the same organism (Singh et al., 2003). Overall, the change in transcript levels observed in this study was considerably smaller than that previously observed (312 differently expressed genes as compared with 866, using a 1.25-fold difference selection criteria as described by Singh et al. [2003]). The important differences between the treatments used in the two experiments were responsible for these results. In the previous experiment, cells were grown in iron-depleted medium until a strong iron-depleted phenotype was observed. Then, iron was added and the time course of release from iron limitation was studied. In this experiment, we examined the transition into iron limitation rather than from iron limitation. In addition, the DFB treatment lowered iron bioavailability considerably, but not completely, and did not affect the growth rate (Fig. 2). This treatment reduced the general stress response of the cells. For example, in wild-type cells, three chaperone transcripts were slightly down-regulated (sll1514, sll0416, and sll0430) as a result of the DFB treatment (Table II). Severe iron deficiency, on the other hand, resulted in the up-regulation of six chaperone genes (Singh et al., 2003). Energy metabolism genes (pfkA, pgi, and fda), whose transcripts were strongly down-regulated in the iron starvation experiment (Singh et al., 2003), were not affected by the DFB treatment. The effect on photosynthesis and respiration was also much smaller, with a total of 25 differentially expressed genes (Table I) as compared with 70 under iron starvation. At the same time, specific iron response transcripts, like the hallmark iron stress response operon isiAB, were strongly up-regulated by the DFB treatment (sll0247-8; Table II). Apart from isiAB, we observed an increase in the transcript levels of a number of iron transport system components. These include the futC and feoB genes, for which experimental data showing involvement in iron transport exist (Katoh et al., 2001), the putative outer membrane transporter slr1406, and the entire slr1316 to slr1319 region, exhibiting similarity to an ABC-type iron transport system from E. coli (Katoh et al., 2001). A complete list of the responses of all putative iron transporters is provided in Supplemental File S1 under the iron transport tab.
Table II.
Microarray results for open reading frames that are discussed in this article
Changes in transcript abundance between the different treatments are presented in units of fold change. Statistical analysis (P treatment values) was performed as described by Singh and coworkers (2003). WT, Wild type.
| Open Reading Frame | Gene Function | 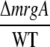 |
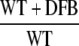 |
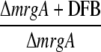 |
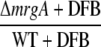 |
P |
|---|---|---|---|---|---|---|
| sll1514 | 16.6-kD small heat shock protein | −1.5 | −1.9 | 2.5 | 3.2 | 6.1 × 10−10 |
| sll0416 | 60-kD chaperonin 2, GroEL2 | −1.1 | −1.5 | 1.9 | 2.4 | 1.4 × 10−6 |
| sll0430 | HtpG, heat shock protein 90 | −1.1 | −1.6 | 1.9 | 2.8 | 5.5 × 10−4 |
| sll0247 | Iron-stress chlorophyll-binding protein CP43′ | 1.4 | 11.4 | 7.9 | −1.0 | 1.5 × 10−13 |
| sll0248 | Flavodoxin | 1.5 | 5.3 | 2.8 | −1.2 | 4.7 × 10−5 |
| slr0984 | CDP-Glc 4,6-dehydratase | 2.0 | 1.4 | −1.6 | −1.1 | 1.1 × 10−5 |
| slr1072 | GDP-d-Man dehydratase | 2.9 | −2.8 | −2.4 | 3.5 | 1.3 × 10−4 |
| ssl2542 | High-light-inducible polypeptide HliA | −3.2 | 1.6 | 4.4 | −1.2 | 2.4 × 10−11 |
| ssr2595 | High-light-inducible polypeptide HliB | −2.1 | 2.2 | 1.0 | −4.5 | 4.3 × 10−9 |
| slr0898 | Ferredoxin-nitrite reductase | −1.3 | 1.4 | −2.8 | −4.8 | 1.7 × 10−9 |
| sll1453 | Nitrate/nitrite transport ATP-binding protein | 1.1 | 1.6 | −3.2 | −4.5 | 2.7 × 10−12 |
| slr1756 | Glu-ammonia ligase | −1.0 | −1.1 | −2.0 | −2.0 | 7.1 × 10−7 |
| sll1502 | Glu synthase large subunit | −1.0 | −2.2 | −4.0 | −1.9 | 5.6 × 10−6 |
| sll1450 | Nitrate/nitrite transport substrate-binding protein | 1.1 | 1.6 | −1.9 | −2.8 | 5.6 × 10−7 |
| sll1451 | Nitrate/nitrite transport system permease protein | 1.1 | 1.5 | −2.2 | −2.9 | 6.8 × 10−7 |
| sll1452 | Nitrate/nitrite transport ATP-binding protein | 1.1 | 1.3 | −3.5 | −4.3 | 1.5 × 10−10 |
| sll1987 | Catalase peroxidase | −1.0 | 1.0 | 2.0 | 1.9 | 1.2 × 10−6 |
| slr1516 | Superoxide dismutase | −1.1 | −1.8 | 1.3 | 2.2 | 2.8 × 10−3 |
| sll0184 | Group 2 RNA polymerase sigma factor sigC | −1.2 | 1.1 | 3.2 | 2.4 | 3.5 × 10−9 |
| sll0856 | RNA polymerase (group 3) sigma E factors sigH | −1.5 | 1.7 | 8.4 | 3.3 | 1.3 × 10−10 |
| slr0854 | DNA photolyase phrA | 1.1 | 1.0 | 3.2 | 3.5 | 9 × 10−5 |
| slr1667 | Hypothetical protein (target gene of Sycrp1) | 1.6 | 1.3 | 13.4 | 17.1 | 2.3 × 10−7 |
| slr1668 | Periplasmic protein (target gene of Sycrp1) | −1.3 | 1.1 | 7.9 | 5.8 | 1.4 × 10−11 |
| slr1991 | Adenylate cyclase | 1.6 | 3.3 | 1.8 | −1.1 | 5.5 × 10−10 |
| sll0517 | Putative RNA-binding protein | 1.1 | 1.1 | −1.4 | −1.4 | 2.2 × 10−6 |
| sll1937 | Ferric uptake regulation protein | 1.1 | 1.0 | −2.4 | −2.4 | 8.1 × 10−7 |
| slr1738 | Transcription regulator Fur family | −1.2 | 1.3 | 13.3 | 8.5 | 1.5 × 10−16 |
| slr1740 | Oligopeptide-binding protein of ABC transporter | 1.7 | −4.8 | 3.2 | 25.4 | 1.4 × 10−3 |
| sll0312 | Oligopeptide ABC transporter permease protein | −1.4 | 1.3 | 2.0 | 1.2 | 5.4 × 10−5 |
| sll0833 | Oligopeptide ABC transporter permease protein | 1.0 | −1.1 | −1.4 | −1.2 | 2.3 × 10−4 |
| sll1927 | ABC transporter ATP-binding protein | −1.0 | 1.2 | 1.5 | 1.2 | 9 × 10−3 |
The inactivation of the mrgA gene resulted in significant changes in the expression pattern of 255 genes, about half of which were up-regulated and half down-regulated (Table I). When examining the differential regulation of these genes, a strong positive effect on cell envelope-related genes could be observed (Table I). The largest differences were found for genes involved in polysaccharide metabolism, such as CDP-Glc 4,6-dehydratase (slr0984) and GDP-d-Man dehydratase (slr1072; Table II). The top three up-regulated genes, as a result of the mrgA inactivation, were all hypothetical (Table II). One of the most down-regulated genes was HliA (ssl2542), with HliB (ssr2595) exhibiting a similar response, albeit to a smaller extent (Table II). These genes code for proteins from the HLIP CAB-like family, involved in the stabilization of PSI under high light intensities (Wang et al., 2008). The isiA transcript exhibited a small increase that correlated well with the small increase in 682-nm chlorophyll fluorescence observed in Figure 3.
The DFB-induced transcriptional perturbation in ΔmrgA cells was much stronger than in wild-type cells. The difference was greater than we would have predicted from analyzing the relationship of the wild type to ΔmrgA under Fe-sufficient conditions. Overall, the transcripts of 914 genes were differentially affected by the treatment, 4.8 times higher than in the wild type (Table I). Direct comparison of DFB-treated wild-type and ΔmrgA cells yielded 779 differentially regulated genes.
One important category in the comparison of the effect of DFB on ΔmrgA versus the wild type included the photosynthesis and respiration genes, with about 10 times more genes being down-regulated than up-regulated. The changes were not random but affected genes for specific functions. The addition of DFB to the medium led to a decrease in all of the genes encoding the ATP synthase and the cytochrome oxidase, NADH dehydrogenase genes associated with both low-affinity and high-affinity CO2 uptake mechanisms (18 of 20 genes), the flavoproteins sll0217 and sll0219, and many important phycobiliproteins. A decline in phycobilisomes is typical of iron deficiency, but the decrease in transcript level in the ΔmrgA strain is greater than that seen in the wild type (Supplemental File S1, phycobiliproteins tab). Iron-sufficient ΔmrgA cultures often appear more blue in color, and our results indicated higher transcript levels of phycobilisome genes under these conditions. However, the decline in ΔmrgA under iron deficiency is more precipitous, and the overall decrease is more than 2-fold greater than in the wild type. The transcript level of sll0247 (isiA) was up-regulated some 8-fold, and all of the genes in this operon were similarly up-regulated (Fig. 5A). Once again, our results demonstrated that there was more IsiA transcript under iron-sufficient conditions in ΔmrgA than in the wild type and that the enhancement was not as great in ΔmrgA (8-fold) relative to the wild type (11.4-fold). Therefore, the transcription results are consistent with those observed in the 77 K chlorophyll fluorescence (Fig. 3).
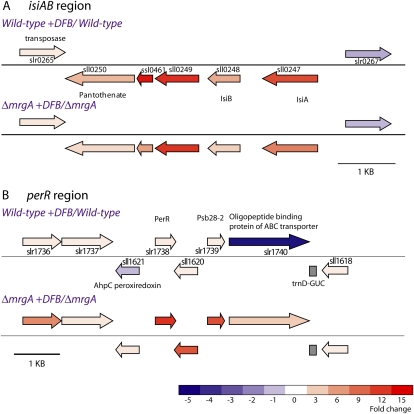
Figure 5.
A map of the iron and oxidative stress-responsive regions in the Synechocystis 6803 genome. The genomic regions surrounding the isiA (A) and perR (B) genes are depicted. The fold change in transcript level, recorded in the microarray experiment, is represented by a color scale. Results for both wild type + DFB/wild type and ΔmrgA + DFB/ΔmrgA are presented. The color scale represents fold change in transcript levels between ΔmrgA + DFB and wild type + DFB. A complete listing of the results for these open reading frames can be found in Table II and Supplemental File S1.
An additional category that exhibited substantial up-regulation in DFB-treated ΔmrgA cultures was the detoxification-related genes. Transcript levels of all five genes in this category, including catalase (sll1987) and superoxide dismutase (slr1516), were increased by an average of 2-fold. In DFB-treated wild-type cultures, the transcripts of genes in this category were hardly affected (Table II).
The largest differential down-regulation in the ΔmrgA strain compared with the wild type in the presence of DFB was recorded for the transcript of a ferredoxin-nitrite reductase (nir, slr0898), followed by a nitrate/nitrite transporter subunit (nrtD, sll1453). Other members of the nitrogen assimilation pathway, such as Gln synthase (glnA, slr1756), the Glu synthase large subunit (gltB, sll1502), and the entire nrt nitrate/nitrite transporter system (sll1450–sll1453), were all significantly down-regulated (Table II).
In addition, there were significant transcriptional alterations in regulatory genes. In most cases, changes in the wild type were minor, again indicating the need for ΔmrgA to establish a different homeostasis. Two sigma factors were up-regulated in ΔmrgA (+DFB). The gene encoding the group 2 sigma factor SigC was enhanced some 3-fold in ΔmrgA, whereas the sigH gene was strongly up-regulated (sll0856; 8.4-fold) in ΔmrgA (but only 1.7-fold in the wild type). All of the genes in the sigH region, including phrA (slr0854), which encodes a DNA photolyase (Ng and Pakrasi, 2001), were also induced from 2- to 6-fold. These results suggest that sigH may be an important transcriptional factor under iron deficiency (as suggested by Foster et al., 2007), but especially in the ΔmrgA mutant. It is also possible that the increase in the DNA photolyase transcript level is of physiological significance. DPS proteins were originally identified as DNA-binding proteins in starved E. coli cells (Almiron et al., 1992). Additional differences between the wild type and ΔmrgA upon the addition of DFB were observed for slr1667 and slr1668, which were strongly up-regulated in ΔmrgA (8- to 13-fold) versus no change in the wild type. These genes are known to be the targets of adenylyl cyclase (encoded by the cya gene), which produces cAMP (Katayama et al., 1995). The cya gene itself (slr1991) is up-regulated in both the wild type and ΔmrgA under iron-deficient conditions to a similar extent, but its slr1667/68 targets are highly up-regulated only in the mutant. Both proteins appear to have one transmembrane helix with most of the protein mass in the periplasm.
In Synechocystis 6803, three proteins are annotated as Fur-like proteins (sll0517, sll1937, and slr1738). Only slr1738, encoding the perR gene, was up-regulated in ΔmrgA upon the addition of DFB, whereas transcript levels of genes for the other two proteins were either unchanged or slightly decreased upon DFB addition. The perR gene showed a significant increase (13.3-fold; Fig. 5B) in ΔmrgA compared with virtually no change in the wild type, a result that highlights an important difference in ΔmrgA under iron deficiency relative to the wild type. The perR gene and protein have been studied by a number of groups and have been shown by Li et al. (2004) to control a small regulon. The increased transcription of perR in ΔmrgA in the presence of DFB led to an induction of virtually all of the genes in the PerR regulon (Supplemental File S1, PerR regulon tab). Of course, this includes isiA but also futA2 (slr0513). These results strongly demonstrated a relationship between iron deficiency and oxidative stress.
Comparison of transcript levels in the ΔmrgA strain (+DFB) with the wild type (+DFB) revealed that the gene with the highest differential up-regulation encodes for a putative ABC transporter solute-binding protein (slr1740; Table II). This protein exhibits considerable similarity to the Bacillus subtilis AppA protein involved in peptide transport. The APP transporter is part of a system controlling sporulation in this organism (Levdikov et al., 2005). The transcript levels of other putative subunits of the same transporter (appB sll0312, appC sll0833, and appD/F sll1927) were not affected. The PhrA peptide, which is the substrate of the APP transporter, and the RapA phosphatase, which it inhibits, have no orthologs in the Synechocystis 6803 genome.
The transcript levels of four additional genes in this region (Fig. 5B) also were strongly up-regulated in DFB-treated ΔmrgA cells (slr1739, slr1738, sll1620, and sll1621). The slr1739 transcript codes for the protein formerly known as PsbW. Currently, this protein is annotated as Psb28-2, not to be confused with Psb28, which is coded by sll1398 (Kashino et al., 2002). In cyanobacteria, the function of this protein is still unknown. slr1738 codes for PerR, which is discussed above. The coregulated genes on the complementary strand of this region code for a hypothetical protein (sll1620) and a peroxiredoxin (ahpC and sll1621). ahpC shares a divergent promoter with perR (Li et al., 2004). Disruption of this gene reduced the ability to grow under low light intensities (Hosoya-Matsuda et al., 2005).
The Effect of Iron Homeostasis on Oxidative Stress Response and Vice Versa
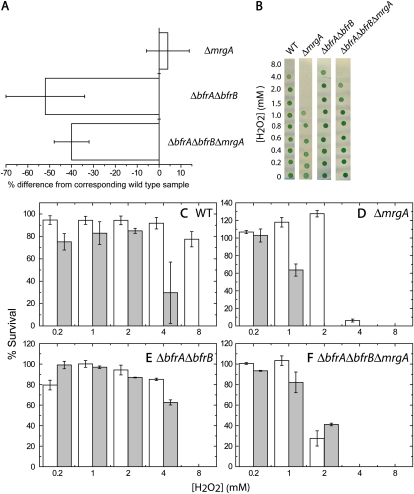
The transcriptomic data pointed to an interrelationship between iron homeostasis and the oxidative stress response. To probe these interactions, we tested the susceptibility of cells containing different iron quotas (Fig. 6A) to the oxidative stress imposed by exposure to hydrogen peroxide. We used the wild type and three disruption strains: ΔmrgA, the bacterioferritin mutant ΔbfrAΔbfrB, and ΔbfrAΔbfrBΔmrgA. The triple mutant exhibited a reduction in the iron quota similar to that observed in the bacterioferritin mutant (Fig. 6A). The four strains were grown on two different iron concentrations: 10 μm, which, in an EDTA-amended growth medium, is more than sufficient, and 0.3 μm, which is limiting (Shcolnick et al., 2007). Cells were washed free of external iron and resuspended in an iron-free YBG11 medium to avoid the effect of extracellular Fenton reactions and to force the cells to utilize stored iron. Exposure to 0 to 8 mm hydrogen peroxide was performed in the iron-free medium. Following the treatment, samples from each culture were spotted on solid medium to determine viability (Fig. 6B).
Figure 6.
Survival of cells after exposure to hydrogen peroxide (H2O2). Wild-type (WT), ΔbfrAΔbfrB, ΔmrgA, and ΔbfrAΔbfrBΔmrgA cells were grown for 7 d in YBG11 medium containing either 0.3 or 10 μm iron, washed, and adjusted to a final concentration of 7 × 107 cells mL−1 in iron-free YBG11. The adjusted cultures were incubated for 20 h in the presence of 0 to 8 mm H2O2 in darkness. In order to assess viability after the H2O2 treatment, 2 μL from each treatment was spotted onto BG11 plates. The results were documented by photography, and brightness was quantified. Results are presented as percentages of the 0 mm H2O2 control. A, The relative iron quota of the mutant cells grown on sufficient iron (data for ΔbfrAΔbfrB were extracted from Keren et al., 2004). B, Samples of spot assays are shown (from cultures grown in 10 μm iron). C to F, The responses of the wild type and mutant strains to H2O2 treatment after growth in 0.3 μm (white bars) or 10 μm iron (gray bars). The data for 0.4 to 0.8 mm H2O2 were omitted for clarity. sd was derived from two biological repeats. [See online article for color version of this figure.]
Iron-deficient wild-type cells could withstand exposure of up to 8 mm hydrogen peroxide, 2-fold higher than iron-sufficient cells (Fig. 6C). ΔmrgA cells were much more sensitive, as observed before (Li et al., 2004). Nevertheless, the 2-fold improvement in survival of iron-limited cells was conserved (Fig. 6D). ΔbfrAΔbfrB cells withstood exposure of up to 4 mm hydrogen peroxide and were not affected by iron availability (Fig. 6E). ΔbfrAΔbfrBΔmrgA cells were clearly more sensitive than either ΔbfrAΔbfrB or wild-type cells (Fig. 6F). The differential effect imposed by iron bioavailability, observed in wild-type and ΔmrgA cultures was not observed in the triple mutant cultures (Fig. 6F).
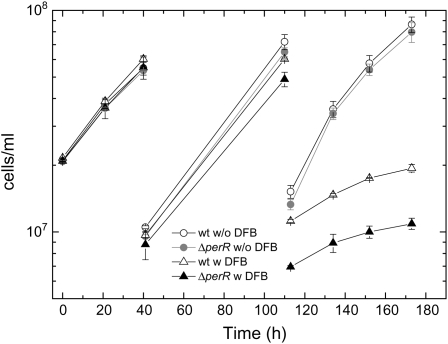
An additional aspect of the connection between iron homeostasis and oxidative stress response exposed in the microarray analysis was the strong induction of the perR transcription in response to iron limitation of ΔmrgA cells. To examine the role of PerR under iron limitation, we tested the effect of DFB on the growth of ΔperR as compared with wild-type cells (Fig. 7). In order to enhance the effect of DFB on the growth rate, we periodically diluted the cultures 5-fold. The dilutions kept the cultures in a logarithmic stage, replenished the medium, and reduced the effects of self-shading (Takahashi et al., 2008), resulting in a faster growth rate than that observed in Figure 2. In the absence of DFB, the growth rates of wild-type and ΔperR cultures were comparable. In the presence of DFB, ΔperR cultures grew considerably slower than wild-type cells.
Figure 7.
Response of ΔperR cells to iron limitation. Wild-type (wt) and ΔperR cells were grown for 4 d in YBG11 + 10 μm iron. At time zero, each culture was split into four subcultures, two of which were amended with 50 μm DFB. Cells were diluted 5-fold every 2 d and amended with DFB to maintain the initial concentration. Culture density was measured as optical density at 730 nm. Following the second dilution, the growth rate of ΔperR cultures was 93% ± 13% of wild-type cultures in the absence of DFB but only 48% ± 8% in the presence of DFB (1.2 × 106 versus 1.1 × 106 and 1.4 × 105 versus 6.6 × 104 cells mL−1 h−1, respectively). w, With DFB; w/o, without DFB.
DISCUSSION
The data presented here indicate a connection between oxidative stress response and iron homeostasis in the cyanobacterium Synechocystis 6803. Furthermore, our results suggest a role for ferritin-type proteins in both processes. In the following section, we will attempt to present a model for the nature of these connections, based on the data presented here.
Bioavailable iron, in oxygenated aqueous solutions, exists mainly in the form of Fe3+. A significant proportion of the bioavailable iron is chelated by organic acids (Morel et al., 2008). Our data demonstrate a remarkable ability of Synechocystis 6803 to utilize this iron source (Fig. 1). For comparison, growth of eukaryotic phytoplankton can be inhibited by EDTA (Hudson and Morel, 1990), while the growth of Synechocystis 6803 is affected only by DFB, a chelator 106 times more efficient. This higher uptake efficiency observed in cyanobacteria is a significant advantage in an iron-limited environment.
The FutABC transporter appears to be a major uptake route into Synechocystis 6803. However, as FutABC disruption mutants are viable (Katoh et al., 2001), it is clear that additional transporters exist. Based on our transcriptomic data, we can suggest the slr1316 to slr1319 proteins and the putative outer membrane slr1406 protein as likely candidates (Table II). It is important to note that disruption mutants in the genes coding for these proteins failed to display a significant phenotype (Katoh et al., 2001). However, it is possible that these transporters function under iron limitation and, therefore, that their effect was not exposed under iron starvation conditions.
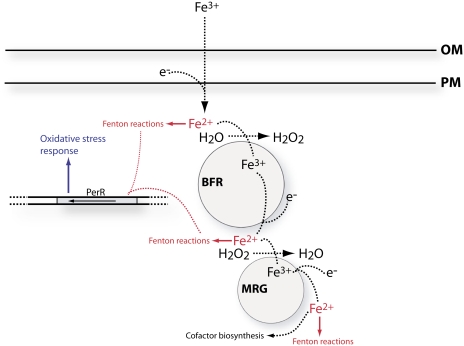
The valency of iron, transported through the plasma membrane, is still under debate (Koropatkin et al., 2007; Badarau et al., 2008). However, regardless of the mode of transport, bacterioferritins catalyze the conversion Fe2+ to Fe3+ oxide (Lewin et al., 2005), which necessitates a reduction of Fe3+ on its way to storage. BFRs form massive (approximately 400 kD; Laulhere et al., 1992) complexes that are slightly hydrophobic (Keren et al., 2004). The level of bfr mRNA does not fluctuate much with changes in iron availability (Supplemental File S1; Singh et al., 2003) or oxidative stress (Li et al., 2004). Similar results were obtained for BFR proteins (Laulhere et al., 1992). These results suggest a role for BFR in bulk stationary storage, probably in association with membranes. Utilization of the sorted iron requires a more nimble entity. The MrgA protein fits this description well. It forms smaller complexes than BFR and catalyzes a similar reaction, the conversion Fe2+ to Fe3+ oxide, reducing hydrogen peroxide instead of water (Lewin et al., 2005). Disruption of mrgA results in a limited ability to utilize stored iron, supporting a function downstream of BFR (Shcolnick and Keren, 2006). Fe3+ can be released directly from the stable Fe3+ oxide crystal formed at the cavity of the BFR (Laulhere et al., 1992). Such a mechanism is expected to be kinetically limited because of the low solubility of Fe3+. A much faster way for dissolving Fe3+ oxides is to reduce them. A reductive pathway fits well with the enzymatic activity of MrgA, which is designed to handle Fe2+ rather than Fe3+. Taking these considerations into account, we are able to suggest a mechanism for the intracellular iron storage pathway in Synechocystis 6803 (Fig. 8). According to this mechanism, iron enters the storage pathway as Fe2+ and leaves as Fe2+, a net redox change of zero. The hydrogen peroxide produced by BFR can be consumed by MrgA. Nevertheless, along the pathway, there are three junctions at which Fe2+ is produced. Fe2+ can interact with hydrogen peroxide to form hydroxyl radicals (Fenton reaction; Shcolnick and Keren, 2006), and the resulting Fe3+ can be reduced by superoxide, forming a positive feedback loop.
Figure 8.
A working model for the mechanism of the intracellular iron storage pathway in Synechocystis 6803. The pathway of iron flow is represented by dashed black arrows. Fenton reactions, in red, are the hydrogen peroxide-driven oxidation of iron (H2O2 + Fe2+ → Fe3+ + OH− + OH·); Fe2+ can be regenerated in a reaction with superoxide (O2− + Fe3+ → Fe2+ + O2). Protective mechanisms are represented in blue. Circles stand for bacterioferritin (BFR) and MrgA (MRG). OM, Outer membrane; PM, plasma membrane. [See online article for color version of this figure.]
Under iron-sufficient conditions, Fe2+ is transported and stored in BFRs. Upon transfer to the iron-free medium, BFR turns from a sink into a source and MrgA functions as a shuttle between BFR and the location of cofactor biogenesis. In the process, two additional Fe2+-producing reactions occur, increasing the potential for oxidative damage (Fig. 8). Under iron-limiting conditions, storage in BFRs is limited and the oxidative danger arising from a massive release of iron is smaller. Consequently, iron-limited cells stand a better chance of surviving the externally applied oxidative stress (Fig. 6C). The reduction in the intracellular manganese quota under iron limitation (Fig. 4) may also have a protective effect, since Mn2+ can participate in Fenton-type reactions much like Fe2+.
The ΔmrgA cultures exhibited an increased sensitivity to externally applied oxidative stress (Li et al., 2004; Foster et al., 2009; Fig. 6D) as well as to internally applied oxidative stress (Foster et al., 2009). However, it is hard to explain the sensitivity of the mutant in terms of MrgA's catalse activity alone. DPS-type proteins, like MrgA, have only a weak catalase activity (Pena and Bullerjahn, 1995). An alternative to a direct role in detoxification is an indirect effect through its function in iron homeostasis (Fig. 8). In the absence of MrgA, Fe2+ that leaves the BFR complex is left unattended and free to interact with reactive oxygen species. This can explain the induction of transcription of genes involved in the oxidative stress response in DFB-treated ΔmrgA cultures. Diverting energetic resources to detoxification can also explain the depression in the transcription of genes involved in energy-consuming reactions such as nitrogen assimilation. According to our scheme, while the lack of MrgA determined the overall sensitivity to Fe2+-enhanced oxidative stress, the concentration of released Fe2+ was still modulated by storage in BFR complexes, being lower in iron-limited cells (Figs. 4 and 6).
Additional support for this theory can be gained from the response of ΔbfrAΔbfrB and ΔbfrAΔbfrBΔmrgA cultures to oxidative stress. The internal iron quota of both mutants is considerably smaller than that of either wild-type or ΔmrgA cells (Fig. 6A), and the differential effect of growth on sufficient or limiting iron is lost in both (Fig. 6, E and F). However, the hydrogen peroxide sensitivity of ΔbfrAΔbfrB is similar to that of wild-type cells, whereas the sensitivity of ΔbfrAΔbfrBΔmrgA is similar to that of ΔmrgA cells. In ΔbfrAΔbfrB cells, Fe2+ transported into the cell can be scavenged by MrgA. In ΔbfrAΔbfrBΔmrgA cells, Fe2+ cannot be stored in BFR complexes and cannot be scavenged by MrgA.
It is important to note that the connection between oxidative stress and iron homeostasis is bidirectional. Large iron quotas expose the cells to oxidative stress, and the deletion of perR resulted in a reduced ability to grow under iron-limiting conditions (Fig. 7). The effect of PerR on iron homeostasis can be indirect through its function in regulating oxidative stress response. However, as PerR is a member of the FUR family of transcriptional regulators and, like many other proteins in this family, may directly bind metal ions (Lee and Helmann, 2007), we cannot exclude a direct role in regulating iron homeostasis. DpsA is a member of the PerR regulon in Synechocystis 6803 (Li et al., 2004). A similar situation was observed in Anabaena sp. PCC7120, where a FUR family protein was found to regulate the expression of a DpsA homolog (Hernandez et al., 2007).
The components of the system suggested here, ferritin family proteins and FUR regulators, are ubiquitous among cyanobacterial species. A simple bioinformatic survey detected ferritin family genes in all of the fully sequenced cyanobacterial genomes (data not shown). It is also interesting to compare the functions of ferritins in cyanobacteria and in eukaryotic photosynthetic organisms. In the green algae Chlamydomonas reinhardtii, two chloroplast-localized ferritin complexes were identified, one of which is constitutive while the other accumulates under iron-limiting conditions (Long et al., 2008). It was proposed that one plays a role in iron storage while the other plays a role in iron reallocating under limiting conditions. In Arabidopsis (Arabidopsis thaliana), iron storage in a triple ferritin disruption strain was not affected, thus indicating the existence of additional storage compartments. Nevertheless, the triple disruption did result in an enhanced oxidative stress in vegetative tissues (Ravet et al., 2009).
In conclusion, the data presented here suggest a pivotal role for the two ferritin-type protein complexes of Synechocystis 6803 cells in coordinating iron homeostasis and the oxidative stress response. The combined action of the two complexes allows for the safe accumulation and release of iron from storage by minimizing damage caused by the interaction of reduced iron and oxygen radicals, which are abundant in cyanobacterial cells due to the function of the photosynthetic apparatus.
MATERIALS AND METHODS
Spectroscopy
Cultures were prepared for metal ion quota analysis as described by Shcolnick et al. (2007). Metal ion concentrations were determined using an inductively coupled plasma mass spectrometer (Agilent 7500). Chlorophyll fluorescence spectroscopy at 77 K was measured as described by Shcolnick et al. (2007) using a Fluoromax-3 spectrofluorometer (Jobin Ivon). The excitation wavelength was set at 420 ± 5 nm, and the emission window was 5 nm. Each spectrum was internally normalized to its minimum and maximum values, as shown in Figure 2B.
Strains and Growth Conditions
Cells were grown in YBG11 medium (Shcolnick et al., 2007) at 30°C with constant shaking. Light intensity was set at 60 μmol photons m−2 s−1. Mutant strains used in this project included ΔmrgA (Li et al., 2004), ΔperR (Li et al., 2004), and ΔbfrAΔbfrB (Keren et al., 2004). The triple ΔbfrAΔbfrBΔmrgA strain was constructed by transformation of the ΔbfrAΔbfrB strain with the mrgA disruption construct. Culture density was measured at 730 nm using a Carry 300 spectrophotometer (Varian). The optical density was calibrated to cells per milliliter values counted using a hemocytometer (optical density at 730 nm −0.032/1.3 × 10−8). Different chelators were applied as described in the figure legends. Chelators were obtained from Sigma-Aldrich. Cultures for inductively coupled plasma mass spectrometry measurements (100-mL cultures in 500-mL flasks), chlorophyll fluorescence spectroscopy (Figs. 3 and 4), and microarray analysis were grown in 10 μm iron with or without 50 μm DFB.
RNA Isolation and Microarray Analysis
Culture used for microarray analysis was centrifuged at 4°C and 7,000 rpm and flash frozen in liquid N2 for later use. RNA was extracted and purified using phenol-chloroform extraction and CsCl2 gradient purification as described previously (Reddy et al., 1990; Sen et al., 2000). A complete description of array construction has been provided previously by Postier et al. (2003). cDNA labeling, glass treatment, prehybridization, and hybridization protocols have been described in detail by Singh et al. (2003). The experimental loop design used for probing the microarray in this experiment is presented in Supplemental File S1. Biological variation was sampled by extracting RNA from three biological repeats and pooling the RNA prior to hybridization. Data acquisition and statistical analysis were performed as described previously (Li et al., 2004). The approximately 1,800 genes represented in Supplemental File S1 each have a P treatment of <0.01 (P treatment is the P value from the ANOVA model that is most representative of the complete loop design), and this should be a highly reliable data set. This microarray platform has been used for many experiments, and validation using reverse transcription-PCR and RNA gel blots have resulted in only one gene out of over 100 tested that did not show qualitative agreement with the microarray results (Singh et al., 2003; Li et al., 2004; Summerfield and Sherman, 2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental File S1. Microarray results.
Supplementary Material
This work was supported by the Israeli Science Foundation (grant no. 1168/07) and by the United States-Israel Binational Science Foundation (grant no. 205196).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nir Keren (nirkeren@vms.huji.ac.il).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Almiron M, Link AJ, Furlong D, Kolter R (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6 2646–2654 [DOI] [PubMed] [Google Scholar]
- Badarau A, Firbank SJ, Waldron KJ, Yanagisawa S, Robinson NJ, Banfield MJ, Dennison C (2008) FutA2 is a ferric binding protein from Synechocystis PCC 6803. J Biol Chem 283 12520–12527 [DOI] [PubMed] [Google Scholar]
- Bibby TS, Zhang Y, Chen M (2009) Biogeography of photosynthetic light-harvesting genes in marine phytoplankton. PLoS One 4 e4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnap RL, Troyan T, Sherman LA (1993) The highly abundant chlorophyll-protein complex of iron-deficient Synechococcus sp. PCC7942 (CP43') is encoded by the isiA gene. Plant Physiol 103 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castruita M, Elmegreen LA, Shaked Y, Stiefel EI, Morel FMM (2007) Comparison of the kinetics of iron release from a marine (Trichodesmium erythraeum) Dps protein and mammalian ferritin in the presence and absence of ligands. J Inorg Biochem 101 1686–1691 [DOI] [PubMed] [Google Scholar]
- Duckworth OW, Sposito G (2005) Siderophore-manganese(III) interactions. I. Air-oxidation of manganese(II) promoted by desferrioxamine B. Environ Sci Technol 39 6037–6044 [DOI] [PubMed] [Google Scholar]
- Finney LA, O'Halloran TV (2003) Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300 931–936 [DOI] [PubMed] [Google Scholar]
- Foster JS, Havemann SA, Singh AK, Sherman LA (2009) Role of mrgA in peroxide and light stress in the cyanobacterium Synechocystis sp PCC 6803. FEMS Microbiol Lett 293 298–304 [DOI] [PubMed] [Google Scholar]
- Foster JS, Singh AK, Rothschild LJ, Sherman LA (2007) Growth-phase dependent differential gene expression in Synechocystis sp. strain PCC 6803 and regulation by a group 2 sigma factor. Arch Microbiol 187 265–279 [DOI] [PubMed] [Google Scholar]
- Hernandez JA, Pellicer S, Huang L, Peleato ML, Fillat MF (2007) FurA modulates gene expression of alr3808, a DpsA homologue in Nostoc (Anabaena) sp. PCC7120. FEBS Lett 581 1351–1356 [DOI] [PubMed] [Google Scholar]
- Hosoya-Matsuda N, Motohashi K, Yoshimura H, Nozaki A, Inoue K, Ohmori M, Hisabori T (2005) Anti-oxidative stress system in cyanobacteria: significance of type II peroxiredoxin and the role of 1-Cys peroxiredoxin in Synechocystis sp. strain PCC 6803. J Biol Chem 280 840–846 [DOI] [PubMed] [Google Scholar]
- Hudson RJM, Morel FMM (1990) Iron transport in marine-phytoplankton: kinetics of cellular and medium coordination reactions. Limnol Oceanogr 35 1002–1020 [Google Scholar]
- Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh J, Satoh K, Pakrasi HB (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41 8004–8012 [DOI] [PubMed] [Google Scholar]
- Katayama M, Wada Y, Ohmori M (1995) Molecular cloning of the cyanobacterial adenylate cyclase gene from the filamentous cyanobacterium Anabaena cylindrica. J Bacteriol 177 3873–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Hagino N, Grossman AR, Ogawa T (2001) Genes essential to iron transport in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 183 2779–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Aurora R, Pakrasi HB (2004) Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol 135 1666–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin N, Randich AM, Bhattacharyya-Pakrasi M, Pakrasi HB, Smith TJ (2007) The structure of the iron-binding protein, FutA1, from Synechocystis 6803. J Biol Chem 282 27468–27477 [DOI] [PubMed] [Google Scholar]
- Laudenbach DE, Reith ME, Straus NA (1988) Isolation, sequence analysis, and transcriptional studies of the flavodoxin gene from Anacystis nidulans R2. J Bacteriol 170 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulhere JP, Laboure AM, Vanwuytswinkel O, Gagnon J, Briat JF (1992) Purification, characterization and function of bacterioferritin from the cyanobacterium Synechocystis PCC-6803. Biochem J 281 785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD (2007) Functional specialization within the Fur family of metalloregulators. Biometals 20 485–499 [DOI] [PubMed] [Google Scholar]
- Levdikov VM, Blagova EV, Brannigan JA, Wright L, Vagin AA, Wilkinson AJ (2005) The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J Mol Biol 345 879–892 [DOI] [PubMed] [Google Scholar]
- Lewin A, Moore GR, Le Brun NE (2005) Formation of protein-coated iron minerals. Dalton Trans 3597–3610 [DOI] [PubMed]
- Li H, Singh AK, McIntyre LM, Sherman LA (2004) Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J Bacteriol 186 3331–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Sommer F, Allen MD, Lu SF, Merchant SS (2008) FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron. Genetics 179 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel KP, Pistorius EK, Golden SS (2001) Unusual regulatory elements for iron deficiency induction of the idiA gene of Synechococcus elongatus PCC 7942. J Bacteriol 183 5015–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel KP, Thole HH, Pistorius EK (1996) IdiA, a 34 kDa protein in the cyanobacteria Synechococcus sp. strains PCC 6301 and PCC 7942, is required for growth under iron and manganese limitations. Microbiology 142 2635–2645 [DOI] [PubMed] [Google Scholar]
- Morel FMM, Kustka AB, Shaked Y (2008) The role of unchelated Fe in the iron nutrition of phytoplankton. Limnol Oceanogr 53 400–404 [Google Scholar]
- Morel FMM, Price NM (2003) The biogeochemical cycles of trace metals in the oceans. Science 300 944–947 [DOI] [PubMed] [Google Scholar]
- Ng WO, Pakrasi HB (2001) DNA photolyase homologs are the major UV resistance factors in the cyanobacterium Synechocystis sp. PCC 6803. Mol Gen Genet 264 924–930 [DOI] [PubMed] [Google Scholar]
- Nodop A, Pietsch D, Hocker R, Becker A, Pistorius EK, Forchhammer K, Michel KP (2008) Transcript profiling reveals new insights into the acclimation of the mesophilic fresh-water cyanobacterium Synechococcus elongatus PCC 7942 to iron starvation. Plant Physiol 147 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena MM, Bullerjahn GS (1995) The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein: linkage of the Dps and bacterioferritin protein families. J Biol Chem 270 22478–22482 [DOI] [PubMed] [Google Scholar]
- Pietsch D, Staiger D, Pistorius EK, Michel KP (2007) Characterization of the putative iron sulfur protein IdiC (ORF5) in Synechococcus elongatus PCC 7942. Photosynth Res 94 91–108 [DOI] [PubMed] [Google Scholar]
- Postier BL, Wang HL, Singh A, Impson L, Andrews HL, Klahn J, Li H, Risinger G, Pesta D, Deyholos M, et al (2003) The construction and use of bacterial DNA microarrays based on an optimized two-stage PCR strategy. BMC Genomics 4 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57 400–412 [DOI] [PubMed] [Google Scholar]
- Reddy KJ, Webb R, Sherman LA (1990) Bacterial RNA isolation with one hour centrifugation in a table-top ultracentrifuge. Biotechniques 8 250–251 [PubMed] [Google Scholar]
- Riethman HC, Sherman LA (1988) Purification and characterization of an iron stress-induced chlorophyll-protein from the cyanobacterium Anacystis nidulans R2. Biochim Biophys Acta 935 141–151 [DOI] [PubMed] [Google Scholar]
- Rivers AR, Jakuba RW, Webb EA (2009) Iron stress genes in marine Synechococcus and the development of a flow cytometric iron stress assay. Environ Microbiol 11 382–396 [DOI] [PubMed] [Google Scholar]
- Sen A, Dwivedi K, Rice KA, Bullerjahn GS (2000) Growth phase and metal-dependent regulation of the dpsA gene in Synechococcus sp strain PCC 7942. Arch Microbiol 173 352–357 [DOI] [PubMed] [Google Scholar]
- Shcolnick S, Keren N (2006) Metal homeostasis in cyanobacteria and chloroplasts: balancing benefits and risks to the photosynthetic apparatus. Plant Physiol 141 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcolnick S, Shaked Y, Keren N (2007) A role for mrgA, a DPS family protein, in the internal transport of Fe in the cyanobacterium Synechocystis sp. PCC6803. Biochim Biophys Acta 1767 814–819 [DOI] [PubMed] [Google Scholar]
- Singh AK, McIntyre LM, Sherman LA (2003) Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 132 1825–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Sherman LA (2007) Reflections on the function of IsiA, a cyanobacterial stress-inducible, Chl-binding protein. Photosynth Res 93 17–25 [DOI] [PubMed] [Google Scholar]
- Summerfield TC, Sherman LA (2008) Global transcriptional response of the alkali-tolerant cyanobacterium Synechocystis sp strain PCC 6803 to a pH 10 environment. Appl Environ Microbiol 74 5276–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Uchimiya H, Hihara Y (2008) Difference in metabolite levels between photoautotrophic and photomixotrophic cultures of Synechocystis sp. PCC 6803 examined by capillary electrophoresis electrospray ionization mass spectrometry. J Exp Bot 59 3009–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron KJ, Tottey S, Yanagisawa S, Dennison C, Robinson NJ (2007) A periplasmic iron-binding protein contributes toward inward copper supply. J Biol Chem 282 3837–3846 [DOI] [PubMed] [Google Scholar]
- Wang Q, Jantaro S, Lu B, Majeed W, Bailey M, He Q (2008) The high light-inducible polypeptides stabilize trimeric photosystem I complex under high light conditions in Synechocystis PCC 6803. Plant Physiol 147 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Shen G, Balasubramanian R, McIntosh L, Bryant DA, Golbeck JH (2004) The sufR gene (sll0088 in Synechocystis sp. strain PCC 6803) functions as a repressor of the sufBCDS operon in iron-sulfur cluster biogenesis in cyanobacteria. J Bacteriol 186 956–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Mosolf J, Willits D, Yeager M, Dryden KA, Young M, Douglas T (2005) An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci USA 102 10551–10556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K, Offermann S, Essen LO, Oesterhelt D (2004) Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states. Proc Natl Acad Sci USA 101 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.