Abstract
Measuring the dynamics of plant growth is fundamental to the understanding of plant development processes. This paper describes a high-throughput, automatic method to trace Arabidopsis (Arabidopsis thaliana) seedling roots grown on agarose plates. From the trace, additional software can quantify length, curvature, and stimulus response parameters such as onset of gravitropism. The method combines a particle-filtering algorithm with a graph-based method to trace the center line of a root. This top-down approach is robust to a variety of noise effects and is reasonably flexible across different image sets. The resulting tool requires minimal interaction from the user and is able to process long time-lapse sequences with user interaction only required on the first frame. The tool is described first, followed by its use on two sample data sets, one measuring root length and the other additionally analyzing the gravitropic response and curvature. The tool, RootTrace, is open source; both the program and source code will be available online.
Plant development is a highly complex process involving many physiological changes. Quantification of these changes is important if the developmental process is to be understood (Chavarría-Krauser et al., 2007). The ability to monitor the growth of plant organs provides valuable information about how those organs respond in or adapt to different situations (Ishikawa and Evans, 1997; Walter et al., 2002).
Root development is a dynamic process in which several events, including cell elongation and cell division, regulate organ growth (Beemster and Baskin, 1998). Roots are constantly integrating hormonal, developmental, and environmental signals in order to control the dynamics of growth (Chavarría-Krauser et al., 2007). The majority of traditional root development bioassays, however, are based on end-point analysis or at best consider a small number of time points (Parry et al., 2001). These are informative but have the limitation of only examining long-term effects on root growth. Transient events and subtle temporal changes can be missed. Recent approaches have considered more frequent time points, allowing the evolution of growth processes and responses to be studied (Ishikawa and Evans, 1997; van der Weele et al., 2003; Chavarría-Krauser et al., 2007; Miller et al., 2007).
Image sequences provide a rich source of data on plant development. Each image contains a potentially rich description of a plant's state of development at the time of acquisition, and images can be captured at high speeds. Manual time-lapse photography was used as early as the 1930s (Van der Laan, 1934; Michener, 1938) to measure the heights of seedlings at various times after application of the phytohormone ethylene, providing important information about the timing of the effects of this hormone on growth regulation. Improvements in digital camera technology and a decrease in digital storage cost have meant it is now comparatively easy to automatically acquire large, high-quality, time-stamped digital image data sets detailing root growth. The ability to correlate the timing of root growth changes (extracted from the image data) with the effect of different signals (hormonal, developmental, or environmental) on processes such as cell division and cell expansion will provide valuable information on understanding root development.
Human analysis of image data, however, is time consuming, subjective, and prone to error. Analysis “by eye” can produce measurements that are difficult to replicate and may result in subtle phenotypes, such as a delayed response, being missed. Previous work has demonstrated the power of image-analysis techniques over manual methods. Examples include the Multi-ADAPT technique being used to examine gravitropic data (Rashotte et al., 2001) and the high-resolution analysis of the effect of pin-formed3 mutants on gravitropism (Chavarría-Krauser et al., 2007). As the potential role of digital image-acquisition technologies in the plant sciences increases, a corresponding demand has arisen for high-throughput software tools capable of producing objective, quantitative analyses of large image sets. In order to study the kinematics of root growth in Arabidopsis (Arabidopsis thaliana), we have developed such an approach, combining automated time-lapse photography with automatic analysis of the resulting images.
A number of software packages exist that aim to automate aspects of the kinematic analysis of growing roots. Many of these software tools attempt to measure growth parameters from high-resolution images. RootFlow (van der Weele et al., 2003) and relative elemental growth rate (REGR) analysis (Walter et al., 2002) have made use of optic flow-based techniques (Barron et al., 1994;Barron and Liptay, 1994) to recover the motion of texture features through a sequence of root images. Intensity features are identified and matched between frames, and corresponding velocity flow fields are calculated. From these vector fields, estimates of growth can be made across any part of the image, provided that reliable image features are available in the areas of interest. Other work has used cell-scale models of features to determine image-plane motion and hence estimate growth parameters (Roberts et al., 2006).
The work described here is motivated by the need to understand larger scale growth effects through a sizable population of roots, as there is considerable variability between seedlings. The approach does not require high-magnification images. Instead, standard digital images are taken of roots, growing on agarose on square plates, using standard consumer cameras (Fig. 1, A and B). Some software tools exist that can take measurements of roots from similar images; all have their strengths and weaknesses, and a selection of popular ones are highlighted below.
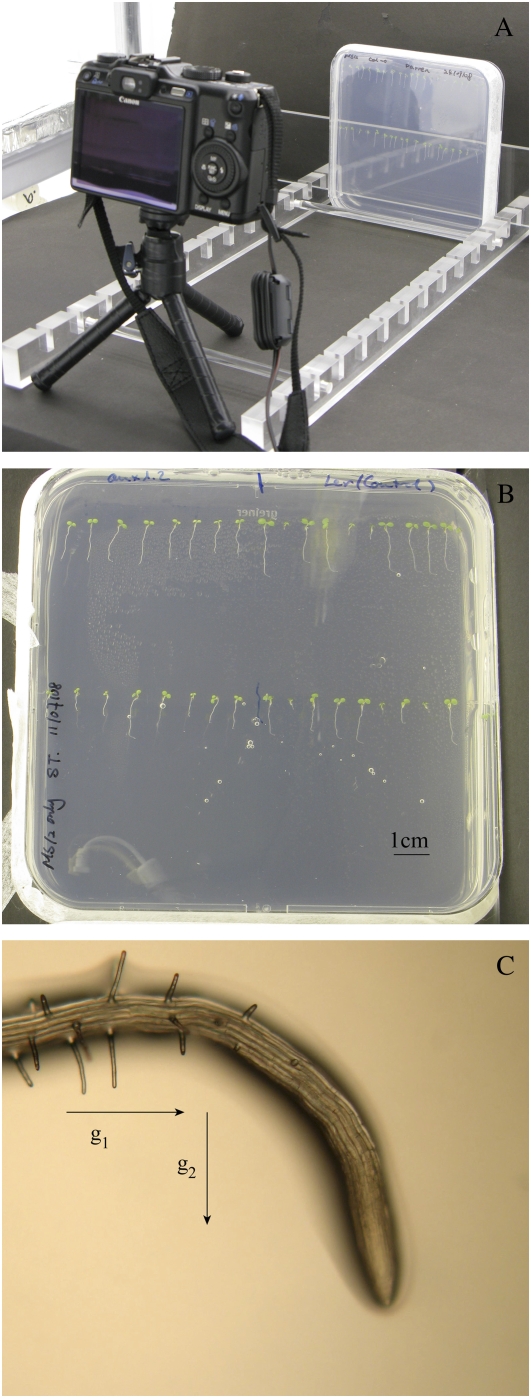
Figure 1.
A, Image of the setup in the growth room, illustrating how the camera (Canon G9) and the plate are arranged during a time-lapse experiment. B, Typical precropped image of a plate of Arabidopsis seedlings. C, Light microscope image of a root exhibiting a gravitropic response, taken approximately 6 h after the plate was rotated 90°. Arrows indicate the direction of the gravity stimulus before (g1) and after (g2) the rotation of the plate. [See online article for color version of this figure.]
KineRoot (Basu et al., 2007) requires the user to initially define a sufficiently large number of points per root and also requires a sufficiently textured root surface to allow reliable tracking of control patches. Although KineRoot was demonstrated on Arabidopsis roots, the images employed were microscope scale, reducing the number of roots that can be analyzed at each time step. The unmagnified images used were of physically larger roots than Arabidopsis, allowing more detail to be visible. Advantages of the tool include the ability to detect root edges and hence produce measures for the diameter of the root and the ability to measure curvature and elongation rates along the root.
SmartRoot (International Atomic Energy Agency, 2006; Draye, 2008) has been developed as a method of analyzing root architecture from scanned images of plant roots. The system provides a powerful vector representation of the root architecture, allowing comparison of architectures across plants and time points. However, it is designed with a relatively large amount of interaction from the user in mind; therefore, it is not currently suited to high-throughput analysis.
RootLM (Qi et al., 2007) can measure the lengths of segments of Arabidopsis roots, allowing the determination of per-day growth rates, for example. However, the tool requires the user to manually mark up the plate on which the roots sit with different colored marker pens to indicate the growth during different growth phases (e.g. the growth from day to day). The plates are then scanned in, and the images are manually touched up in photo-processing software, if required.
Multi-ADAPT (Ishikawa and Evans, 1997) can be used to measure root elongation and curvature from digitized microscope images of a root. It measures the angle (relative to the vertical) of each of a set of sequential root subsections from the root tip. Because it uses magnified images, Multi-ADAPT is able to measure elongation along both sides of a root independently. Due to the nature of the microscope imaging, however, the throughput of the system is limited by human interaction.
Phytomorph (Miller et al., 2007; Spalding, 2009) is a comprehensive project that includes a description of an automated robotic system for high-throughput image capture using backlit plates illuminated by infrared light-emitting diodes and high-end imaging equipment. The resulting images feature high-resolution seedling hypocotyls, which can be analyzed using the HYPOTrace software (Wang et al., 2009).
The method proposed by Chavarría-Krauser et al. (2007) measures REGRs and curvature distribution given microscope images of single roots captured using transmitted near-infrared light. A velocity field is constructed using the structure tensor method. The position of a point can be tracked in time; this was used to track the control points of a curve. The change in distance between control points gives rise to the REGR distribution and growth velocity. Curvature was determined as being the angle formed by the tip and the horizontal, measured using cross-correlation to fit a region of interest (the root tip) to a reference image and thus determine the new orientation.
The software tool described in this paper exploits visual tracking techniques developed within the computer vision community to provide a flexible, extensible root-measuring tool, RootTrace, which can automatically measure multiple growth parameters on multiple roots throughout large time series. User interaction is kept to a minimum in order to reduce the demand on the plant biologist's time. Key unique features of the tool are the high-throughput design and the flexibility of the top-down image analysis approach employed. It is designed to work with images of roots grown on agarose plates, imaged using standard digital cameras, without any special lighting (Fig. 1, A and B).
The primary output of RootTrace is a detailed description, in the form of a set of closely spaced points, of the center line of each root visible in each image of the input sequence. Center lines are associated between frames, so each root is represented as a time-ordered set of center lines whose starting points coincide. Once this data structure is available, it can be analyzed in a wide variety of ways. Root length can be measured and plotted over time, and further general properties (e.g. local orientation and/or curvature) can be estimated. This is of interest, for example, in gravitropic studies. Plant roots respond to gravity by utilizing gravity-sensing cells located within the columella and root cap (Blancaflor et al., 1998). When a root is treated with a gravity stimulus, the cells in the elongation zone on the lower side of the root expand at a much reduced rate compared with the cells on the upper side (Mullen et al., 1998). This differential growth pattern causes the root to curve downward (Fig. 1C).
Once the correct orientation is achieved, nondifferential expansion is resumed. The signal required to respond to gravity is the phytohormone auxin (Swarup et al., 2005). Plant lines defective in auxin biosynthesis, signaling, or transport typically show defects in their gravitropic responses (Leyser et al., 1993, 1996; Bennett et al., 1996; Chen et al., 1998; Luschnig et al., 1998; Muller et al., 1998; Rouse et al., 1998; Utsuno et al., 1998; Friml et al., 2002; Swarup et al., 2005; Abas et al., 2006). These agravitropic phenotypes have been identified by analyzing images of growing roots to estimate angles of root growth following a gravitropic stimulus (Parry et al., 2001).
Recently, a number of automated tools have been described that monitor root curvature and give an output relating to the degree of bending of the root (Basu et al., 2007; Miller et al., 2007). We describe below how the center line description tool we have produced has been extended to recover changes in the curvature of the root and to give an easy-to-interpret output. The core methods have also been extended to determine when the gravitropic response occurred. Automating and objectifying such measures allows more detailed comparison of different phenotypes and raises the possibility of correlating root responses with hormonal and other signals.
Recent improvements in digital camera technology have made it comparatively easy to acquire large, high-quality image sets detailing the dynamics of root growth. However, high-throughput software tools capable of producing objective, quantitative analyses of the resulting images are now required. We have developed such a tool with a view to analyzing sequences of images of Arabidopsis roots growing on agarose plates. The remainder of this paper will explain the tool briefly from a technical perspective and show how to use the tool in practice. This will be followed by two examples of RootTrace in use: first during a straightforward growth experiment, and second to analyze a gravitropic response.
RESULTS AND DISCUSSION
In what follows, we describe the key components of the image-analysis software tool, RootTrace, developed to address the problem of extracting growth parameters from time-series images of Arabidopsis roots growing on agarose plates. This tool takes the form of a Microsoft Windows program with a graphical user interface. The source code is written in Visual C++, and both the executable software and the source code will be released as open source downloads from the Centre for Plant Integrative Biology (www.cpib.ac.uk).
Use is made of the free, open source image-analysis library OpenCV (http://sourceforge.net/projects/opencvlibrary/) to implement some of the basic techniques. It is not necessary for those wishing to use RootTrace to understand the underlying methods in great depth; however, the information in this section may help those interested to understand the situations in which it can be applied and what can cause the software to fail. Further technical details about RootTrace's early development can be found in previous work (French et al., 2008).
Tracing Roots Using Tracking Technologies
RootTrace uses automatic tracking techniques, developed to track moving objects, to trace the line of the root from a user-defined start location to the tip of the root. This process is analogous to tracing the image of the root by placing a pen at the start point and tracing down the root until the tip of the root is detected. At each step along the root, the algorithm builds a probabilistic representation of where the root appears to lie. Image clutter such as reflections and lateral roots may appear in this representation, but the algorithm is designed to be able to represent the locations of both the true target and distractions using a multimodal approach and so increase the chances of it being able to maintain tracking of the root through clutter.
Tracking down the root is accomplished using a well-established particle filter-based method called Condensation (Isard and Blake, 1998). Particle filters represent continuous probability density functions using a set of discrete weighted hypotheses. When tracking a moving target, each hypothesis, or “particle,” contains information about one possible state in which the target might be (one possible position, velocity, etc.) and an associated weight. The weight is assigned by evaluation of an observation model at the target's hypothesized location. Particles more likely to describe the true parameters of the target, therefore, are given a higher weight.
Tracking proceeds via a comparatively simple iterative algorithm. At each time step, a set of particles are randomly selected (with replacement) from those available, taking their weights into account so that high-probability hypotheses will be selected more often than those with low weights. A motion model, capturing the expected movement of the target, is applied to each selected particle to predict where the target would appear in the next image if that particle's hypothesis were correct. As a given particle may be selected more than once, a small random component is included in the motion model to spread the new particles around the predicted target location. The observation model is evaluated at the appropriate location in the next image to calculate the weight of the new hypothesis particle, and the process repeats.
Here, Condensation tracking is employed not to track a moving object through an image sequence but to trace down a root in a single image that is stationary in time. Instead of taking discrete time steps from one image to the next, the RootTrace tracker can be thought of as taking discrete spatial steps, allowing it to “walk down” the root image. At each step, the tracker builds a particle-based representation of a probability density function describing the root's location (Fig. 2B). The Condensation algorithm processes the particles as usual, selecting them according to their associated weight and using a “motion” model to propagate them down the root, predicting its position at each step of the trace. Weighting of particles is achieved by evaluating a suitable observation model that takes as input data at the current particle location in the image and outputs a measure of how well that image area matches a root appearance model. This model can be varied, but using a simple weighted color model that determines how well a circular patch of image matches the expected white color of a root has been found to work well.
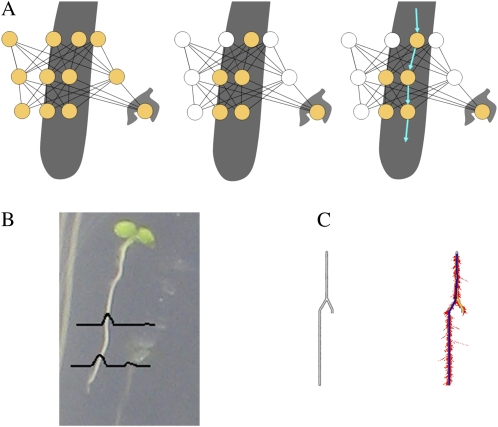
Figure 2.
A, A graph-based approach to root tracing. As the root space is explored, a graph is built of potential root locations at each step (left, yellow circles). This graph is then pruned, removing low-probability locations (middle, white circles). The remaining nodes in the graph can then be traversed to form a path down the primary root (right, arrows). B, Illustration showing a typical probability distribution for a root location at two different steps along the trace. C, Tracing through the particle set by treating it as a connected graph helps the software to avoid becoming trapped at dead ends, such as lateral roots. At left is an unprocessed image of a synthetic root-like structure. At right is a successful tracing (heavy line) through the resulting particle set distribution (dots) after tracking and graph tracing. [See online article for color version of this figure.]
In moving-target tracking, each particle usually contains the target's estimated velocity; in RootTrace, this becomes a vector describing the movement down the root that the particle took at the last step. The motion model describes how this vector is expected to change over time. Like the observation model, this can be varied as required, but a simple model has been found to work well. This model expects the root to generally be growing from the top of the image to the bottom and moves left and right at each step according to a velocity model and some process noise. This process is repeated until the end of the root is reached. Once tracking is complete, the tracker will have explored the area around the length of the root. We can use the locations and weights of all the particles generated by the particle filter to refine the root trace; this will be discussed later.
Automatic Detection of the Root Tip
It is important in a high-throughput, automatic system that the end of the root be located with a high degree of accuracy. Detecting this point allows tracking of the root to stop automatically and a measurement of length and/or other properties to be taken. This is particularly challenging in Arabidopsis images, as the roots can appear partially translucent at times.
While tracking along the root, RootTrace builds and maintains an appearance model of the root by calculating and storing the mean intensity, and the corresponding sd, as the tracking progresses. Prior to tracking, the user is asked to indicate some areas of the image that contain no roots, to allow similar representative background appearance statistics to be calculated. These root and background statistics allow Bayes' rule to be used at each step to calculate a posterior likelihood that the color at the current position belongs to the foreground root appearance model as opposed to the background model. Thresholding with hysteresis (Canny, 1986) is used to “track” this value, allowing it to fall below an upper probability threshold for a number of steps, as long as the likelihood does not fall below a lower threshold. Once the lower threshold is breached, tracking of the root is terminated and the end of the root is set as the last recorded position with a posterior foreground likelihood probability greater than the higher threshold. Therefore, this method is able to adjust to slight changes in root appearance and variations in background appearance across images. It also copes well with isolated noise on the root as the trace proceeds. Although the user must choose the upper and lower threshold values to be used, in practice this is not a difficult task, and often the same general parameters can be used across a variety of data sets. This is especially true because the parameters specify probabilities rather than absolute intensity values.
Finding the Center Line of the Resulting Trace
Once the tracker has reached the tip of the root, there exists an approximation of the probability of the root existing at each point over the area the particle filter has explored. This is a discrete approximation (because it is represented by discrete particles) of a two-dimensional probability density function describing the likelihood of the root existing at points on the image plane. This information can be used to find a path through the particle set and so select a good estimate of the center line of the root.
As the tracker moves down the root, all of the image locations explored at each step (in the form of the particle set considered) are added to a graph. From this probabilistic graph, we can extract an accurate representation of the root center line (Fig. 2A).
The center line is extracted from the graph by first pruning low-probability nodes from the graph (Fig. 2A, middle). This is achieved by calculating the mean and sd values of the particle probability weights at each step down the root and removing those that fall below a fixed cutoff; typically, removing all particles from the graph with a weight lower than 1 sd below the mean works well. Once pruned, Dijkstra's algorithm (Dijkstra, 1959) is used to find the best route through the graph, from the start point until an end node is encountered (Fig. 2C). “Best” is defined here as a combination of shortest distance and highest probability; the weight on each arc is evaluated as the distance of the arc divided by the observation probabilities at each connecting node multiplied together, and the aim of the algorithm is to find the path with the lowest weight.
It is advantageous to use the posttracking graph to trace the root, rather than the maximum a posteriori probability location estimates (e.g. highest weighted particles) from the tracking, as Condensation may follow both the main root and a lateral root or image clutter; the latter, dead-end option may in some cases produce a better measurement model, but if we consider the complete graph we can discount this erroneous path, as it does not allow a complete path from the top of a root to the tip (Fig. 2C). This is analogous to the way people perceive the main root; we can locate it in the context of the complete root image, but shown only segments of the root we are likely to confuse lateral roots with the main root at ambiguous junctions.
In the current implementation, the length measurement is made by calculating the Euclidean distance between the nodes on the chosen path through the graph. However, a number of distance measures could potentially be used (Kimura et al., 1999) and could be added to the system in the future.
Curvature Measures
Once the center line of a root has been identified, further processing is able to measure parameters in addition to the length of the trace. For example, it is possible to quantify curvature parameters for the root. The curvature of the root can be of interest, especially where we are concerned with curvature in response to a stimulus, such as that imposed by altering the direction of gravity stimulus. It is usual to produce this stimulus by rotating the plate on which the roots are growing by 90° and observing the response of the roots. Parameters of interest might include the time the root takes to begin the response, the duration of the response, and the resulting response angle.
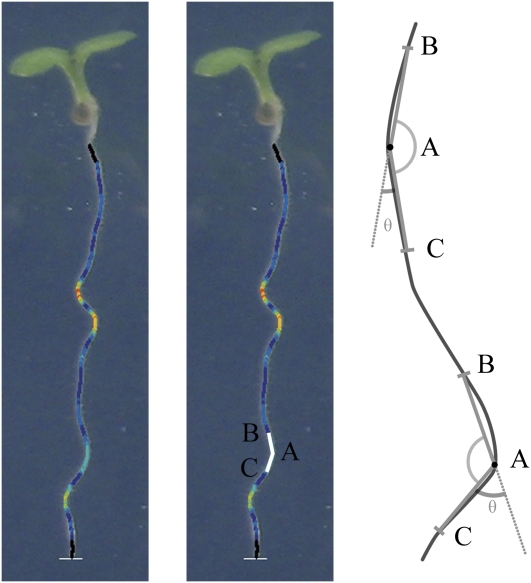
Curvature is measured by calculating local angles at each point along the root trace. For each point along the root, this is achieved by taking points a fixed distance in front of and behind the current point in the trace and calculating the angle between the resulting two lines formed by joining these points to the current point (Fig. 3). The distance over which the angle is measured affects the scale of curvature measured, and this value can be adjusted if required. The recorded angle is 180° minus this value and is marked as θ in Figure 3. Therefore, higher values of θ indicate a more pronounced curvature of the root. Other methods for curvature determination could be used; however, this simple method has been found to work well.
Figure 3.
Illustration of how local root angles are calculated. An angle, BAC, is measured for each point on the trace in turn, by connecting the current point (A) to two points farther up (B) and down (C) the root, as illustrated by the two lines on the center image. The colors overlaid on the root image visualize θ as calculated at each point; dark blue (small angles) through to red (large angles; high curvature). Two examples are also presented on the schematic at right. [See online article for color version of this figure.]
As well as measuring local curvature, the facility to detect the point of one overarching bend in the root allows the system to detect the gravitropic point. We define this as the point along the root that forms a triangle with the maximal area when joined to the start point and end point of the trace. This triangle is formed for all points down the root in the trace, and the point that forms the triangle with the largest area is selected. This method correctly identifies the largest point when the root has one bend sufficiently larger than all others, as is often the case in gravitropic studies. This point is marked on the traced output if the angle θ exceeds a specified threshold (typically 45°; see overlaid circles in Figure 6C below [clarified by arrows]). Additionally, using the length data derived for each root in each image, the time when the root was at the length where this bend occurred can be estimated from this point. This information is used to determine when the gravitational response occurred and is used to produce the response histogram illustrated in Figure 6E below. This point is also marked on the curvature chart (example shown in Fig. 6D below) to show the user where the system believes the strong curvature response occurred in space and time.
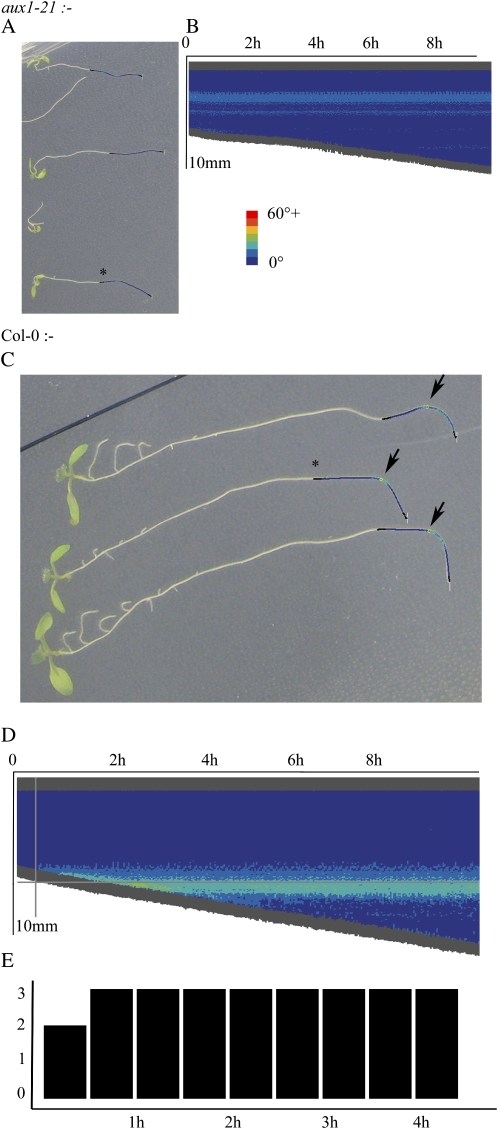
Figure 6.
A, aux1-21 roots show no gravitational response. B, Example of a curvature plot of one of the roots confirms no response. The corresponding root is marked with an asterisk in A. C, Col-0 roots show a clear response to gravity, as indicated by the arrows. D, A Col-0 root shows a gravitational response in the curvature chart; note the emerging curve. The gray cross-hairs indicate the automatically predicted gravitropic point, about 30 min after the response. The corresponding root is marked with an asterisk in C. E, The level of response is confirmed; two of the three roots are marked as having responded after 30 min, and all three roots responded by 1 h. [See online article for color version of this figure.]
Having introduced the fundamentals of how the techniques underlying the software work, the following sections will look at how to take the images and actually use the software, before some example data sets are presented.
Image Requirements
The image-analysis techniques underlying RootTrace have been chosen with robustness to varying image properties and qualities in mind. The root-tracing method builds upon a probabilistic tracking framework. This promotes a flexible and robust method of tracking a target, which in our case is the tracking along the root center line. It is comparatively simple to tune the method to new classes of image by changing the motion and observation models used, and the key practical feature of Condensation is its ability to deal with noise and ambiguity, caused here by branching and crossing roots. However, organizing high-quality images at the point of capture is always advisable, as this ensures that high-quality data are produced. It should be stressed that “high quality” in this context does not necessarily mean high quality in the photographic sense. In image analysis, a high-quality image is one that is well matched to the methods being applied. Avoiding some common pitfalls and understanding some assumptions the program makes here can help RootTrace produce more accurate and robust results.
Figure 4A presents an example of a good-quality image for analysis by RootTrace. It has a number of features that make it suitable for processing by the techniques employed. Although the background is reasonably noisy and produces potentially confusing reflections, these are generally handled well by the RootTrace software. This is because the light roots are a good fit to the observation model used in the tracker. There is also a good level of contrast between the roots and the background, making the root and background appearance models different and so aiding root tip detection. This contrast is achieved by growing the roots on the surface of a transparent medium of solidified agarose (1%, w/v), and photographs are taken with a black background behind the plates (Fig. 1A). In this arrangement, the roots appear light gray in the images over a dark gray background. It has been found that moving the plate slightly away from the black background paper can increase contrast with overhead lighting, and angling the plate slightly away from the lights can help minimize reflections.
Figure 4.
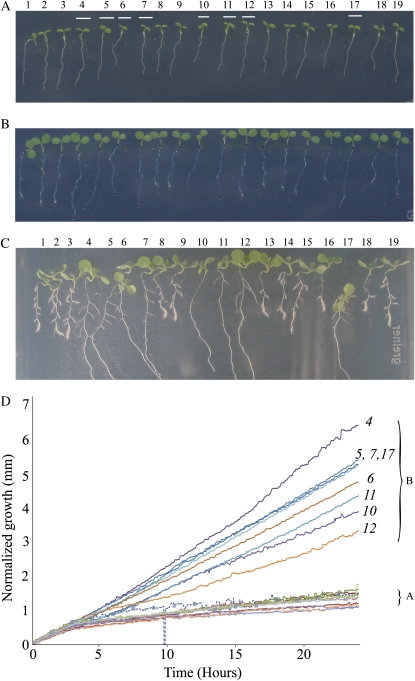
A, Roots at the start of the sequence. Roots that turn out to be the faster growing controls are marked, as referenced from C. These correspond to group B on the graph in D. B, Traced roots 24 h into the time lapse. C, Seven-day-old seedlings. Two groups of roots, differentiated by length, are clearly visible, but several days after the traces presented here. D, Growth profiles for the 19 roots measured over 24 h after DEX treatment. Each line in the graph represents an individual root. The resulting data have been filtered using a median filter over a moving window of five data points. This has the effect of removing occasional extreme peaks caused by root tracing errors, where the end of the roots are missed, for example. A represents the cluster of plots from the transgenic seedlings, and B represents the cluster from the wild-type seedlings. Numbers in italics indicate root numbers. The dotted line indicates a poorly traced root (root 1; Supplemental Video S1). [See online article for color version of this figure.]
While RootTrace can handle roots that cross over each other in a variety of situations, making sure this does not happen at the point of image capture is advantageous, as it removes some potential failure modes from the processing. There is always a danger when roots cross that ambiguous situations will arise that will confuse the tracker. From a biological perspective, avoiding crossovers also means that the roots do not interfere with one another as they grow. Adhering to these guidelines produces the good quality of trace illustrated in Figure 4B. It should be noted that for the data presented here, the plants were grown under continuous light conditions and imaged under the standard growth room lighting (150 μmol m−2 s−1) with no additional lighting required. It may be possible to use the software with images captured in the dark or low light using infrared technology, as long as the images generally have light roots over a darker background.
RootTrace assumes that the position of the plate of roots relative to the camera does not change; the user-determined start points for the tracing procedure are used between frames to locate the roots. This is simple to achieve, as a fixed equipment setup is desirable in order to make use of the ability to automatically capture the images.
Being a time-series sequence, the duration between the captured frames determines the temporal resolution of the resulting data. The optimal frequency of image capture should be tailored to the time scale of the event or parameters to be recorded. If disk space permits, it is better to capture more frequently than will be required. It is always possible to later omit frames during the analysis; recapturing data sets because not enough data were recorded is a time-consuming exercise.
Images saved with JPEG compression can be unsuitable for data processing, as the quality of the image is reduced in an irreversible way. It is this reduction in quality that allows the image file size to be reduced. The JPEG system is designed to reduce the quality of data representation by mostly reducing the amount of data used to represent parts of the image that we as humans perceive least well. One such area that is significantly affected is the spatial resolution of color. However, the proposed tracing technique should be largely tolerant of JPEG artifacts, as the root plate images do not rely heavily on hue information. In these images, the roots are essentially white or light gray and the background is dark gray. This might not be the case for future images, but as in practice the system works well on these images, color information quality, and the consequent effects of JPEG degradation, are less crucial concerns. JPEG images, the most common digital camera image format, can continue to be used; this makes RootTrace more widely applicable. Additionally, the storage space required for a JPEG file is substantially smaller than for its uncompressed equivalent. This is particularly relevant, as higher resolution images tend to produce the best results in RootTrace; early experiments used 7-megapixel camera images, but modern cameras now commonly capture 10 megapixels or more. This degree of fidelity even allows for two plates to be captured within one image. However, JPEG images should be saved with the highest quality settings possible to prevent overdegradation of the image.
Using RootTrace
RootTrace requires minimal interaction from the user in order to process a time series. However, it does still need some information that requires manual entry. The user must select which files to process and must select the areas of the first image that can be used to calculate the background appearance statistics. The points where the tracing of each root must start should then be set. Other parameters may be optimized for use with particularly challenging images. All image-based interactions occur using mouse clicks on the image, and parameters can be set with sliders. The tool offers opportunities to save some settings for future sessions. So, for each root in the image series, interaction is required to specify start points and representative background patches. Calibration information (millimeters per pixel and time between frames) must be entered if required by the user. Beyond this, no interaction is required for the rest of the images in the time series.
Results of length and curvature measures are saved to corresponding data files (.CSV type), which can be opened in common spreadsheet packages. For each input image, a corresponding output image with the traces overlaid is saved. Additionally, preview charts are presented for root lengths, curvature information, and gravitational responses (Figs. 5 and 6).
Figure 5.
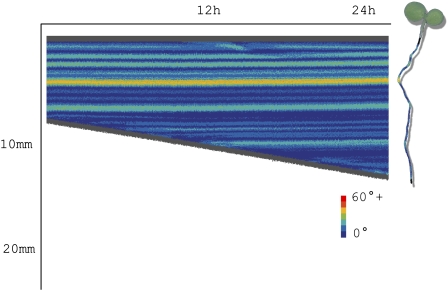
Example of a curvature chart for one root over approximately 1 d. This chart shows the growth over time, with color indicating the local angle formed by the root. Each vertical bar represents a straightened trace of the root at a particular time point. The root on the right illustrates the orientation of the plant relative to the chart. [See online article for color version of this figure.]
As the process, after initialization, is completely automatic, it may be that some of the length measures are erroneous. The most significant cause of errors is a failure to detect the end of the root, although this error rate is still low (3.5% of 400 root images measured did not have their ends correctly identified in one typical sequence). The resulting length measurement errors are often large. Because of the random nature of these errors occurring in the stochastic tracking framework, median filtering of the final results over a small window of measurements (e.g. a window size of five) tends to eliminate these occasional anomalies while not affecting the series of continuous correct measurements. Sometimes it is not possible to remove errors by filtering, and these must be identified by manual inspection of the resulting charts, images, and spreadsheet data.
A quick-start guide to using RootTrace is provided as Supplemental PDF S1.
We will now present two example uses of the software.
Example 1. Quantification of Basal Root Growth
In this experiment, a mixed population of Arabidopsis transgenic seedlings segregating for the SCR:gai-GR-YFP transgene (Ubeda-Tomás et al., 2008) growing on agarose (1%, w/v) were analyzed. Figure 4A corresponds to 3-d-old seedlings passed to 10 μm dexamethasone (DEX)-supplemented agarose. In the presence of DEX, the gibberellin-insensitive repressor gai blocks the gibberellin response in the endodermal cells, resulting in a reduction in root growth with respect to control roots (Ubeda-Tomás et al., 2008). Soon after seedlings were exposed to DEX, the segregating seedlings showed similar root length, as the DEX treatment required a period of time to produce phenotypic alterations in the root (Fig. 4B). It is only after studying the dynamics of root growth over time that the reduction in root growth rate in the transgenic seedlings is detectable with respect to the control seedlings, allowing us to distinguish transgenic from control seedlings.
To quantify the basal root growth over time using RootTrace, a 24-h time series was taken, with images being captured every 3 min using Canon PowerShot A620 cameras. This was controlled using Canon's Remote Capture software, which is supplied with the cameras. The time series was taken in a growth room under constant illumination; no flash was used. The images were seven megapixels in size. The aim of the experiment is to determine the differences in the dynamics of root growth between the segregating transgenic and wild-type seedlings based on their growth rates. Traditional discrimination between transgenic and wild-type seedlings involves allowing the roots to grow for a number of days followed by measurement of the final root length achieved (Fig. 4C). However, simply knowing the final state of the roots does not reveal how one group became longer than the other, for example. Were the shorter roots growing at a constant, slower growth rate, or did their growth rate slow after a period of time? We will demonstrate how providing a high-fidelity growth profile can answer these questions.
Time-lapse images from 1 d of growth were imported into the software, and the start points for the tracing were manually marked. A background region was selected to build a background model. The roots measured were those shown in Figure 4B, which are younger versions of the roots in Figure 4C.
Figure 4B illustrates a typical output from RootTrace. This kind of output is produced for each time point. As well as the processed image, data on growth and curvature information are saved to text (.CSV) files ready for further analysis. The color coding on the roots in Figure 4B relates to the curvature of the roots at that point; the darker blue color indicates low-curvature areas, and brighter yellow and red areas are points of high curvature. This coloring is the same as that defined for the curvature charts in Figure 5.
Figure 4D shows the growth profiles for all of the measured roots. The root profiles clearly cluster together as two distinct groups, termed A and B in Figure 4D. This is clear after only about 10 h. The plots that form cluster A are showing a reduction in root growth with respect to those from group B. This result suggests that roots in group A might correspond to the transgenic seedlings, showing the expected reduction in root growth with respect to wild-type seedlings (group B). This result was validated after checking that roots in group A are the ones showing a reduced root length after 7 d of treatment (roots 1–3, 8, 9, 13–16, 18, and 19 in Fig. 4, A and C), and roots in group B are showing an expected wild-type root length after 7 d (roots 4–7, 10–12, and 17 in Fig. 4, A and C).
The RootTrace data also make it possible to study the dynamics of root growth, giving valuable information about the reduction in root growth in the transgenic seedlings as a response to specific treatment. This information includes temporal data about when the initial response (reduction of root growth) takes place (5–7 h after treatment) and the response over time (transgenic seedlings showed a constantly reduced root growth over time, indicated by the low gradient straight line after 5–7 h in Fig. 4D). The RootTrace data make grouping the roots a simple case of inspecting the graph in Figure 4D, although the changes are not distinguishable to the naked eye at the 24-h point (Fig. 4B).
Example 2. Visualizing and Detecting Curvature Responses
In addition to measuring dynamic changes in root length, many experiments require a quantification of responses to some form of stimulus. Curvature visualization is achieved using heat map charts, as illustrated in Figure 5. To create this chart, at each time point the trace of an individual root is straightened and added to the chart as a vertical line. The chart is an intuitive way of combining growth rate information and curvature information in one place. The color on the chart gives an indication of the local angle as measured on the root at that particular time and location (the local angle is measured as described previously). This visualization enables the researcher to make a quick assessment about the overall conformation of the root and how these curves evolve over time as the root grows. Using this chart, it is easy to see at what time a particular curvature feature evolved; this is useful for seeing at a glance when a gravitropic response began, for example, and the nature of the response (e.g. immediate high curvature or a more gentle, sweeping curve).
Additionally, when studying gravitropism, it is typical to class a root as having responded to gravity when a bend greater than a chosen number of degrees is observed. RootTrace is able to search for these bends automatically at each time step. This allows the number of roots responding to the gravity stimulus to be recorded over time, allowing the study of the dynamics of root gravitropic responses.
To demonstrate the ability of RootTrace to support this kind of study, a plate of two lines of Arabidopsis seedlings, ecotype Columbia (Col-0; wild-type) and aux1-21 (a mutant defective in auxin transport), was rotated 90° after 7 d of vertical growth to provide a gravitropic stimulus. Digital images were captured once every 2 min to monitor root curvature patterns using a Canon G9 12-megapixel consumer digital camera. The time-lapse capture was controlled using third party software (Breeze Systems, 2008) running on a connected laptop in a growth room.
Figure 6 illustrates the analysis possible with RootTrace. Figure 6, A and C, show the traced outputs (only the end of the root is traced, as only the gravitropic point is of interest here). Circles are marked on these traces where a gravitropic point is found (arrows in Fig. 6C). Figure 6, B and D, show the curvature plots for an agravitropic and a gravitropic root, respectively; note the evolving area of high curvature for the wild-type root from approximately 30 min in Figure 6D, consistent with earlier studies (Ishikawa and Evans, 1997), which is absent from Figure 6B. Figure 6E shows a histogram of the number of Col-0 roots responding to gravity with a bend of greater than 45° over time. These analyses are consistent with published results of gravitropic responses of this line (Swarup et al., 2004) and permit a finer granularity of temporal and spatial response analysis.
The gravitropic point is initially measured as occurring at a particular distance down the root length. As RootTrace is also measuring lengths at each time point, this distance can be converted to reveal when a root started to respond. This can then be visualized using a histogram depicting the number of roots responding at successive points in time, or, in combination with the angle of the response, this could be visualized using wheel diagrams at particular time points. In combination with the curvature charts such as in Figure 6D (on which the detected gravitropic point is marked), the user can see the duration and dynamics of the response.
CONCLUSION
We have presented a novel image-analysis-based software tool that is able to quantify root growth rates and offers the possibility of extended analysis of the conformation of the root structure with, for example, curvature measurement and root bend detection.
The software has been designed using robust tracking algorithms as a foundation and can work with standard images from consumer digital cameras. Both the approach of automated image capture and the design of the RootTrace software aim to reduce the work load of the biologist quantifying growth in such sequences while providing an insight into high-resolution growth rate profiles that is challenging to achieve using traditional techniques.
MATERIALS AND METHODS
Plant Growth
Surface-sterilized Arabidopsis (Arabidopsis thaliana) seeds were germinated on half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) and solidified with 1% (w/v) PGP-type agarose (Park Scientific) in environmentally controlled growth conditions (24°C, 150 μmol m−2 s−1, continuous light). Plates were incubated vertically to permit roots to grow on the surface of the medium. For root length analysis, a segregating population of Arabidopsis transgenic lines expressing a SCR:gai-GR-YFP construct was used (Ubeda-Tomás et al., 2008). Seeds were plated as described, then after 3 d they were transferred to fresh medium containing 10 μm DEX. Gravitropism analysis was carried out on Col-0 (wild type) and aux1-21 7-d-old seedlings. Plates were rotated 90° to provide a gravitropic stimulus. All time-lapse imaging was carried out in the growth chamber.
Imaging
All images were taken with a standard consumer digital camera (the particular models used were the Canon Powershot A620 and the Canon Powershot G9). The plate to be imaged was arranged vertically in front of the camera on a shelf in the growth room. It was arranged to minimize reflections from the overhead lights, and if condensation was a problem, the roots were imaged through the agarose side of the plate.
The time lapse was controlled using either Canon's Remote Capture software (shipped with the camera) or PSRemote (www.breezesys.com/PSRemote/index.htm; Breeze Systems, 2008) on a personal computer. The software was set such that the camera parameters, such as focus, aperture, and exposure time, remained constant between exposures. The exposure time was set sufficiently large that flicker from the fluorescent lamps did not affect the images. The equipment was set up so that the plate filled the frame as much as possible while still being in focus. Images were taken by the software and stored directly to the computer hard disk. After the experiment, images were backed up to a networked drive. Postprocessing of the images included cropping around the plate if necessary to save storage space and rotating the images so the roots generally grow from the top of the image to the bottom.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Video S1. A video of the software tracing output for all frames in example 1 compiled into a time-lapse video.
Supplemental PDF S1. Quick-start guide for new users.
Supplementary Material
This work was supported by the Biotechnology and Biological Sciences Research Council and the Engineering and Physical Sciences Research Council as part of their Systems Biology Initiative to establish a number of Centres for Integrative Systems Biology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andrew French (andrew.french@cpib.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wirniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C (2006) Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol 8 249–256 [DOI] [PubMed] [Google Scholar]
- Barron JL, Fleet DJ, Beauchemin SS (1994) Performance of optical flow techniques. Int J Comput Vis 12 43–77 [Google Scholar]
- Barron JL, Liptay A (1994) Optical flow to measure minute increments in plant growth. Bioimaging 2 57–61 [Google Scholar]
- Basu P, Pal A, Lynch JP, Brown KM (2007) A novel image analysis technique for kinematic study of growth and curvature. Plant Physiol 145 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G, Baskin TI (1998) Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol 116 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273 948–950 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Fasano JM, Gilroy S (1998) Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol 116 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze Systems (2008) PSRemote. http://www.breezesys.com/PSRemote/index.htm (June 16, 2009)
- Canny J (1986) A computational approach to edge detection. IEEE Trans Pattern Anal Mach Intell 8 679–698 [PubMed] [Google Scholar]
- Chavarría-Krauser A, Nagel KA, Palme K, Schurr U, Walter A, Scharr H (2007) Spatio-temporal quantification of differential growth processes in root growth zones based on a novel combination of image sequence processing and refined concepts describing curvature production. New Phytol 177 811–821 [DOI] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH (1998) The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA 95 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra EW (1959) A note on two problems in connexion with graphs. Numerische Mathematik 1 269–271 [Google Scholar]
- Draye X (2008) Xavier Draye's profile. http://www.uclouvain.be/en-30461.html (June 16, 2009)
- French AP, Bennett MJ, Howells C, Patel D, Pridmore T (2008) A probabilistic tracking approach to root measurement in images. In P Encarnação, A Veloso, eds, Proceedings of the First International Conference on Biomedical Electronics and Devices, BIOSIGNALS 2008. INSTICC Press, Setubal, Portugal, pp 108–115
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809 [DOI] [PubMed] [Google Scholar]
- International Atomic Energy Agency (2006) Mutational Analysis of Root Characters in Food Plants. International Atomic Energy Agency, Vienna
- Isard M, Blake A (1998) Condensation: conditional density propagation for visual tracking. Int J Comput Vis 29 5–28 [Google Scholar]
- Ishikawa H, Evans ML (1997) Novel software for analysis of root gravitropism: comparative response patterns of Arabidopsis wild-type and axr1 seedlings. Plant Cell Environ 20 919–928 [DOI] [PubMed] [Google Scholar]
- Kimura K, Kikuchi S, Yamasaki S (1999) Accurate root length measurement by image analysis. Plant Soil 216 117–127 [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M (1993) Arabidopsis auxin-resistance gene axr1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364 161–164 [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10 403–413 [DOI] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener H (1938) The action of ethylene on plant growth. Am J Bot 25 711–720 [Google Scholar]
- Miller ND, Parks BM, Spalding EP (2007) Computer-vision analysis of seedling responses to light and gravity. Plant J 52 374–381 [DOI] [PubMed] [Google Scholar]
- Mullen JL, Ishikawa H, Evans ML (1998) Analysis of changes in relative elemental growth rate patterns in the elongation zone of Arabidopsis roots upon gravistimulation. Planta 206 598–603 [DOI] [PubMed] [Google Scholar]
- Muller A, Guan CH, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Parry G, Delbarre A, Marchant A, Swarup R, Napier R, Perrot-Rechenmann C, Bennett MJ (2001) Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J 25 399–406 [DOI] [PubMed] [Google Scholar]
- Qi X, Qi J, Wu Y (2007) RootLM: a simple color image analysis program for length measurement of primary roots in Arabidopsis. Plant Root 1 10–16 [Google Scholar]
- Rashotte AM, DeLong A, Muday GK (2001) Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TJ, McKenna SJ, Hans J, Valentine TA, Bengough AG (2006) Part-based multi-frame registration for estimation of the growth of cellular networks in plant roots. In 18th International Conference on Pattern Recognition. IEEE Computer Society, Los Alamitos, CA, pp 378–381
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O (1998) Changes in auxin response from mutations in an AUX/IAA gene. Science 279 1371–1373 [DOI] [PubMed] [Google Scholar]
- Spalding EP (2009) Phytomorph. http://phytomorph.wisc.edu/ (June 16, 2009)
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al (2004) Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HMO, Haseloff J, Beemster G, Bhalerao R, Bennett MJ (2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7 1057–1065 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Swarup R, Coates J, Swarup K, Beemster G, Bhalerao R, Bennett MJ (2008) Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol 10 625–628 [DOI] [PubMed] [Google Scholar]
- Utsuno K, Shikanai T, Yamada Y, Hashimoto T (1998) AGR, an agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol 39 1111–1118 [DOI] [PubMed] [Google Scholar]
- Van der Laan P (1934) Der Einfluss von Aethylen auf die Wuchsstoffbildung be aena und Vicia. Rec Trav Bot Neerl 31 691–742 [Google Scholar]
- van der Weele CM, Jiang HS, Palaniappan KK, Ivanov VB, Palaniappan K, Baskin TI (2003) A new algorithm for computational image analysis of deformable motion at high spatial and temporal resolution applied to root growth: roughly uniform elongation in the meristem and also, after an abrupt acceleration, in the elongation zone. Plant Physiol 132 1138–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A, Spies H, Terjung S, Küsters R, Kirchgeßner N, Schurr U (2002) Spatio-temporal dynamics of expansion growth in roots: automatic quantification of diurnal course and temperature response by digital image sequence processing. J Exp Biol 53 689–698 [DOI] [PubMed] [Google Scholar]
- Wang L, Uilecan IV, Assadi AH, Kozmik CA, Spalding EP (2009) HYPOTrace: image analysis software for measuring hypocotyl growth and shape demonstrated on Arabidopsis seedlings undergoing photomorphogenesis. Plant Physiol 149 1632–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.