Abstract
Anthocyanins are major pigments in colored grape (Vitis vinifera) berries, and most of them are monomethoxylated or dimethoxylated. We report here the functional characterization of an anthocyanin O-methyltransferase (AOMT) from grapevine. The expression pattern in two cultivars with different anthocyanin methylation profiles (Syrah and Nebbiolo) showed a peak at start ripening (véraison), when the concentrations of all methylated anthocyanins begin to increase. The purified recombinant AOMT protein was active on both anthocyanins and flavonols in vitro, with Km in the micromolar range, and was dependent on divalent cations for activity. AOMT showed a preference for 3′,5′ methylation when a 3′,4′,5′ hydroxylated anthocyanin substrate was tested. In order to assess its in planta activity, we performed transient expression of AOMT in tobacco (Nicotiana benthamiana) leaves expressing the Production of Anthocyanin Pigment1 (PAP1) transcription factor from Arabidopsis (Arabidopsis thaliana). PAP1 expression in leaves induced the accumulation of the nonmethylated anthocyanin delphinidin 3-rutinoside. The coexpression of PAP1 and AOMT resulted in an accumulation of malvidin 3-rutinoside. We also showed that AOMT localized exclusively in the cytoplasm of tobacco leaf cells. These results demonstrate the ability of this enzyme to methylate anthocyanins both in vitro and in vivo, indicating that AOMT plays a major role in anthocyanin biosynthesis in grape berries.
Methylation by S-adenosyl-l-methionine (SAM)-dependent O-methyltransferases (OMTs; EC 2.1.1) is a common modification in plant secondary metabolism, which modulates the physiological properties and the chemical reactivity of phenolic groups (Zhu et al., 1994). Several classifications of plant OMTs have shown that they can be categorized into two major classes (Joshi and Chiang, 1998; Noel et al., 2003; Lam et al., 2007). In the nomenclature proposed by Noel et al. (2003), type 1 OMTs consist of homodimeric OMTs with subunit sizes of about 38 to 43 kD, which do not require divalent cations for activity. This large family includes caffeic acid, flavonoid, coumarin, and alkaloid OMTs (Frick and Kutchan, 1999; Dong et al., 2003). Type 2 OMTs represent a group of lower molecular mass (23–27 kD) and cation-dependent OMTs. Most members of this family have been shown to be specific for CoA esters of phenylpropanoids and have been suggested to be key enzymes in lignin biosynthesis (Ye et al., 1994). Among flavonoids alone, hundreds of O-methylated compounds have been characterized, ranging from monomethylated to polymethylated compounds and belonging to the chalcone, flavone, isoflavone, flavonol, and anthocyanin families (Wollenweber and Dietz, 1981; Markham, 1989; Andersen and Markham, 2006).
Anthocyanins are responsible for the characteristic red, blue, and purple colors of many plant tissues (Harborne and Williams, 2000). They play important roles as pigments of flowers and fruits to attract animals for pollination and seed dispersion and act as protectants against UV irradiation (Winkel-Shirley, 2001). Anthocyanins contribute to the distinctive color and the aesthetic quality of many plant-derived products such as wine. In addition to their direct role in red wine color, anthocyanins also participate in the organoleptic and chemical attributes of wine, because of their interaction with other phenolic and aroma compounds as well as with proteins and polysaccharides (Mazza and Miniati, 1993).
Red grape (Vitis vinifera) varieties usually accumulate 3-O-monoglucoside, 3-O-acetylglucoside, and 3-O-p-coumaroylglucoside derivatives of five anthocyanidins: delphinidin, cyanidin, peonidin, petunidin, and malvidin. The total amount of anthocyanins and relative abundance of single anthocyanins are extremely variable among red- to blue-skinned cultivars; however, monomethylated and dimethylated derivatives are usually largely predominant (Mazza and Miniati, 1993). Methylation is an important aspect of grape anthocyanin biosynthesis, as anthocyanin O-methylation has a slight reddening effect and is known to reduce the chemical reactivity of phenolic hydroxyl groups (Sarni et al., 1995). This modification is proposed to increase the stability of anthocyanins and modify their water solubility, thus significantly contributing to the accumulation of color, a pivotal quality parameter for the red wine industry. Anthocyanin methylation in grapes is affected by genetic factors, malvidin 3-glucoside being the predominant anthocyanin in most cultivars and peonidin 3-glucoside appearing in a more restricted group of genotypes (Mattivi et al., 2006), as well as by environmental and cultural conditions (Downey et al., 2006).
Although many structural and regulatory genes involved in anthocyanin biosynthesis have been cloned from a wide variety of plant species (Mol et al., 1998; Forkmann and Heller, 1999; Winkel-Shirley, 2001), genes responsible for the formation of methylated anthocyanins remain to be characterized. Genetic analyses of anthocyanin biosynthesis in petunia (Petunia hybrida) showed that two pairs of duplicate genes, Mt1/Mt2 and Mf1/Mf2, were responsible for the methylation of the anthocyanin molecule at the 3′ and 5′ positions, respectively (Wiering and de Vlaming, 1977). Genetic relationships among these four genes suggested that if one or both of the Mt genes were dominant, mainly 3′-O-methylated anthocyanins (peonidin or petunidin) accumulated in the flower. The 3′,5′-O-methylated malvidin accumulated as the main pigment only when one or both of the Mf genes were dominant. However, additional investigation of the relationship between the four methylation genes and anthocyanin O-methyltransferase activity in vitro revealed that both the Mt enzymes and the Mf enzymes were capable of methylating the oxygen in the 3′ and 5′ positions of the anthocyanin molecule (Jonsson et al., 1983). The same authors could then separate the four different OMTs in petunia flower extracts using chromatofocusing techniques (Jonsson et al., 1984). A detailed characterization of these enzymes showed that they exhibit different kinetic properties and that their combination reflects the methylation patterns of anthocyanins in flowers in different lines of petunia. However, the genes encoding these OMTs from petunia have not been cloned to date.
Bailly et al. (1997) partially purified an OMT from anthocyanin-producing grape cell suspensions. This enzyme could methylate cyanidin 3-O-glucoside but showed no activity with cyanidin 3-p-coumaroyl-O-glucoside, suggesting that methylation occurred before acylation. Furthermore, the lack of OMT activity when delphinidin was used as a substrate indicated that more than one OMT may be involved in anthocyanin methylation in grape (Bailly et al., 1997). EST and grapevine genome sequencing (Da Silva et al., 2005; Jaillon et al., 2007) have identified a large number of putative OMTs, and expression of some of them is correlated with the accumulation of methylated anthocyanins in berries (Ageorges et al., 2006; Castellarin et al., 2007). In this work, we describe the identification and the biochemical characterization of a novel Mg2+-dependent anthocyanin O-methyltransferase (AOMT) from grapevine. Combining in vitro characterization of recombinant AOMT enzyme with Agrobacterium tumefaciens-mediated transient transformation of tobacco (Nicotiana benthamiana), we show that this grapevine AOMT catalyzes both 3′ and 5′ O-methylation of anthocyanins in vitro and in planta.
RESULTS
Isolation of a Candidate AOMT cDNA from Grapevine
Recently, a high-throughput transcriptomic screening identified a set of genes coexpressed during anthocyanin biosynthesis in grape (Ageorges et al., 2006). One of these genes, corresponding to the partial grape EST BQ796057, was highly similar to the multifunctional phenylpropanoid and flavonoid OMT (PFOMT) from Mesembryanthemum crystallinum (Ibdah et al., 2003), suggesting that this gene could be involved in anthocyanin methylation. A BLAST search of Vitis databases using the grapevine EST BQ796057 as the query sequence identified three overlapping ESTs, named EC932040.1, EC953324.1, and CF603338.1. Their sequences were assembled in a contig, and the full-length cDNA was amplified by reverse transcription-PCR from mature Syrah berries. The predicted amino acid sequence of this candidate AOMT cDNA was the same in the two cultivars and corresponded to a 26.4-kD protein, which contained a conserved domain identified as Methyltransf_3: O-methyltransferase (Pfam 01596). The candidate AOMT belonged to the type 2 family of low-Mr and cation-dependent OMTs. Most members of this family have been shown to be active with CoA esters of phenylpropanoids. However, some type 2 OMTs are also active with flavonoids and/or phenylpropanoid conjugates (Ibdah et al., 2003; Kim et al., 2006; Lee et al., 2008). The candidate AOMT presented 56% and 54% identity with PFOMT from M. crystallinum (Ibdah et al., 2003) and caffeoyl-CoA-OMT (CCoA-OMT) from grapevine (Busam et al., 1997), respectively (Fig. 1A). Phylogenetic analysis of selected members of the plant OMT gene family showed that the grapevine AOMT belongs to a small clade of type 2 OMTs, together with the flavonoid-OMTs PFOMT from M. crystallinum and OMT1 from Arabidopsis (Arabidopsis thaliana; Fig. 1B).
Figure 1.
Comparison of grapevine AOMT with other OMTs. A, The predicted amino acid sequence of AOMT from grapevine (VvAOMT) was aligned with the amino acid sequences of PFOMT from M. crystallinum (McPFOMT) and caffeoyl-CoA-OMT from grapevine (VvCCoAOMT) using ClustalW. Residues shaded in gray indicate identical amino acids. B, Phylogenetic tree of selected OMT cDNA sequences. Type 1 OMTs include caffeic acid-OMT from grapevine (VvCOMT; accession no. AF239740) and Nicotiana tabacum (NtCOMT; AF484252), tricetin-OMT from Triticum aestivum (TaOMT2; DQ223971), flavonoid-OMTs from Catharanthus roseus (CrOMT2; AY127568) and Chrysosplenium americanum (CaOMT2; U16793), and flavonol-OMTs from Arabidopsis (AtFolOMT; U70424) and Oryza sativa (OsFolOMT1;DQ288259). Type 2 OMTs (in boldface) include caffeoyl-CoA-OMT from grapevine (VvCCoAOMT; Z54233), N. tabacum (NtCCoAOMT; U38612), and Populus trichocarpa (PtCCoAOMT; AJ224896), flavonol-OMT from O. sativa (OsFolOMT2; XM_483167), AOMT from grapevine (VvAOMT; FJ460168), PFOMT from M. crystallinum (McPFOMT; AY145521), and flavonoid-OMT from Arabidopsis (AtOMT1; AY087244). The numbers beside the branches represent bootstrap values based on 500 replicates.
Analysis of AOMT Gene Expression during Grape Berry Development
Transcriptomic analyses have shown that AOMT gene expression was induced during anthocyanin accumulation in grape berries, leading to the hypothesis that AOMT could be involved in anthocyanin methylation (Ageorges et al., 2006; Castellarin et al., 2007). In order to gain insight into AOMT gene regulation, Syrah and Nebbiolo grape berries were harvested at different ripening stages and analyzed for both anthocyanin content and AOMT gene expression (Fig. 2). No anthocyanin was detected before véraison, the onset of ripening. From this stage on, anthocyanins started to accumulate in berry skin. Malvidin accumulated in Syrah berries as the major aglycone, reaching 1.12 mg equivalent malvidin per berry (69% of total anthocyanins) in fully mature berries (Fig. 2A). Peonidin content increased significantly during Syrah berry development, reaching 17% of total anthocyanins (Fig. 2A). In Syrah berries, AOMT transcripts were hardly detectable before the onset of ripening (Fig. 2C). After véraison, AOMT expression level increased dramatically during the first weeks and then the expression decreased and remained stable until the end of ripening (Fig. 2C). In Nebbiolo berries, the accumulation of anthocyanins followed the same developmental pattern as in Syrah berries (Fig. 2B). In Nebbiolo, the main anthocyanin was the monomethylated peonidin 3-O-glucoside, which accumulated at concentrations about 2-fold higher than the sum of petunidin 3-O-glucoside and malvidin 3-O-glucoside (Fig. 2B). However, the expression of AOMT in this variety followed the same profile as it did in Syrah (Fig. 2D). Given the fact that AOMT resembled the previously characterized CCoA-OMT (Busam et al., 1997), the expression of the CCoA-OMT gene was investigated during grape berry development as a comparison (Supplemental Fig. S1). Unlike AOMT, the CCoA-OMT gene was strongly expressed in berries a few weeks after flowering, and then its expression decreased rapidly until véraison. During the ripening process, CCoA-OMT transcript accumulation was very weak in grape berries.
Figure 2.
A and B, Accumulation of anthocyanins during Syrah (A) and Nebbiolo (B) berry development. The arrow indicates véraison, the onset of ripening. The content of each anthocyanin is expressed as equivalent (equi.) of malvidin-3-O-glucoside as pure standard. C and D, Quantitative real-time PCR expression profiling of AOMT during Syrah (C) and Nebbiolo (D) berry development. Expression values have been normalized with VvEF1α. Cy, Cyanidin; Dp, delphinidin; Mv, malvidin; Pn, peonidin; Pt, petunidin.
AOMT gene expression was then evaluated in several tissues of Syrah. In vegetative organs, AOMT transcripts could be detected at very low levels in young leaves (Fig. 3). Before véraison, AOMT gene expression was barely detectable in the different berry tissues, although a weak expression could be detected in seeds at the green stage (Fig. 3). During ripening, AOMT transcripts were mainly detected in berry skin, where anthocyanin biosynthesis takes place. As a summary, the expression of AOMT appeared rather fruit specific, with a coincidental timing corresponding with the accumulation of anthocyanin pigments.
Figure 3.
Quantitative real-time PCR expression profiling of AOMT in various Syrah grapevine organs and in different tissues of berry at three stages of development. Expression values have been normalized with VvEF1α and expressed as relative abundance.
Characterization of Recombinant AOMT
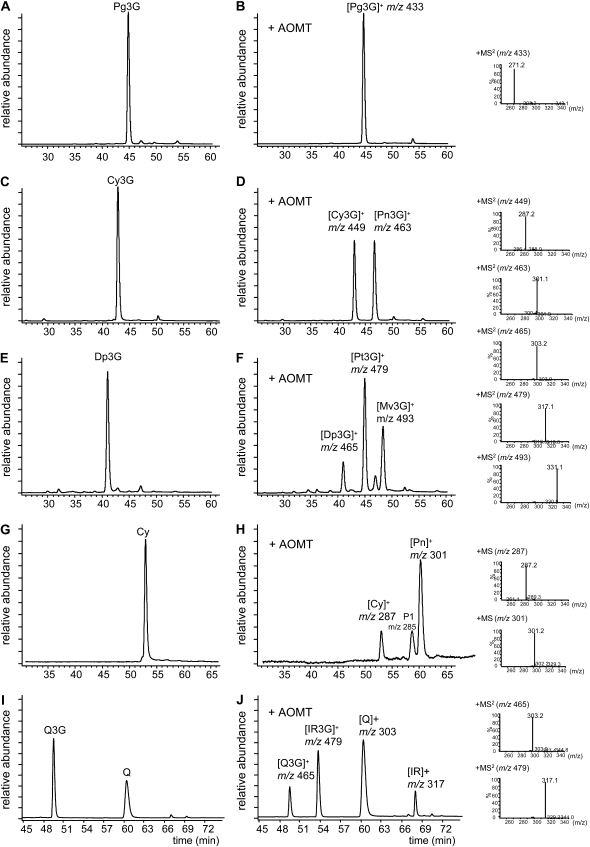
The AOMT coding sequence was cloned into the Gateway-compatible entry vector pDONR207 and then transferred into the destination vector pHNGWA (Busso et al., 2005) to express the corresponding protein in Escherichia coli as a His-tagged NusA fusion protein. The fusion protein was purified by metal affinity chromatography and cleaved on the resin with thrombin to yield purified recombinant AOMT (Supplemental Fig. S2). The activity of AOMT enzyme was then tested in vitro with a number of potential substrates (Table I; Fig. 4), including flavan 3-ols, flavonols, anthocyanidins, and anthocyanins, in the presence of SAM. HPLC-diode array detection (DAD)/electrospray ionization (ESI)-mass spectrometry (MS) was used to analyze reaction products obtained after incubation of the potential substrates without or with AOMT enzyme (Fig. 5). First, AOMT could not utilize flavan 3-ols (catechin and epicatechin) as substrates (Table I). On the other hand, AOMT did not exhibit any methylation activity with the 4′-hydroxylated anthocyanin pelargonidin 3-O-glucoside (Fig. 5, A and B). Indeed, HPLC analysis of the reaction products obtained with pelargonidin 3-O-glucoside detected one major peak, the unmodified substrate (98%), and also 2% of the corresponding aglycone, pelargonidin (Fig. 5, A and B; Supplemental Table S1). On the other hand, over 50% of the cyanidin 3-O-glucoside substrate was converted into peonidin 3-O-glucoside identified by UV-visible and MS spectra (absorbance maximum [λmax] = 516 nm; mass-to-charge ratio [m/z] 463; Fig. 5, C and D). Delphinidin 3-O-glucoside underwent two sequential methylations on the oxygen at the 3′ and 5′ positions to give successively the petunidin 3-O-glucoside (λmax = 525.6 nm; m/z 479) and malvidin 3-O-glucoside (λmax = 527 nm; m/z 493; Fig. 5, E and F). Using anthocyanidin as substrate, AOMT was also able to methylate cyanidin, leading to peonidin (λmax = 523 nm; m/z 301; Fig. 5, G and H). A second product was detected (m/z 285) with a λmax at 487 nm (Fig. 5H, peak P1). This product was not identified but may result from the instability of anthocyanidins under experimental conditions (pH 7.5 in aqueous solution; Awika et al., 2004). In addition to anthocyanin and anthocyanidin substrates, AOMT could methylate the flavonol quercetin 3-O-glucoside to yield its 3′-O-methyl ether isorhamnetin (Fig. 5, I and J). Detailed quantifications of reaction products are presented in Supplemental Table S1. Taken together, these results indicated that recombinant AOMT catalyzed in vitro the 3′ O-methylation of anthocyanins and flavonols with a catechol B-ring (3′,4′ di-OH) and both 3′ and 5′ O-methylation for those showing a pyrogallol (3′,4′,5′ tri-OH) B-ring.
Table I.
Kinetic parameters of AOMT with potential substrates
Data are expressed as means of triplicate assays, and se values are given in parentheses. n.d., Not determined.
| Substrate | Km | Kcat × 10−3 | Kcat/Km | Specific Activity |
|---|---|---|---|---|
| μm | s−1 | m−1 s−1 | pkat mg−1 | |
| Pelargonidin 3-glucoside | n.d. | n.d. | n.d. | 12 (3) |
| Cyanidin 3-glucoside | 43 (3) | 90 (9) | 1,964 (240) | 1,700 (104) |
| Delphinidin 3-glucoside | 44 (6) | 118 (12) | 2,470 (441) | 2,230 (133) |
| Quercetin 3-glucoside | 24 (3) | 100 (5) | 4,250 (120) | 1,880 (67) |
| Cyanidin | 74 (2) | 84 (2) | 1,135 (93) | 1,580 (55) |
| Quercetin | 33 (3) | 112 (10) | 3,420 (80) | 2,120 (144) |
| Myricetin | 19 (2) | 128 (5) | 6,830 (790) | 2,420 (89) |
| Catechin | n.d. | n.d. | n.d. | 8 (3) |
| Epicatechin | n.d. | n.d. | n.d. | 7 (3) |
Figure 4.
Chemical structures of some flavonoid compounds. Listed in boldface are flavonoids assayed as substrates for AOMT. glc, Glucoside.
Figure 5.
Analysis of AOMT in vitro reaction products. HPLC/ESI-MS/DAD analysis of reaction products produced following incubation of potential substrates without (A, C, E, G, and I) or with (B, D, F, H, and J) recombinant AOMT enzyme. Reactions were carried out for 60 min in a total volume of 200 μL, with 200 μm anthocyanin or flavonol substrate, 200 μm SAM, and 5 μg of purified AOMT. Substrates were as follows: A and B, pelargonidin 3-glucoside; C and D, cyanidin 3-glucoside; E and F, delphinidin 3-glucoside; G and H, cyanidin; I and J, quercetin 3-glucoside. Anthocyanins were monitored at 520 nm and flavonols at 360 nm. Reaction products were identified according to their mass fragmentation, UV/visible absorption spectra, and retention times. The data shown are representative of three independent experiments. Pg3G, Pelargonidin 3-glucoside; Cy3G, cyanidin 3-glucoside; Pn3G, peonidin 3-glucoside; Dp3G, delphinidin 3-glucoside; Pt3G, petunidin 3-glucoside; Mv3G, malvidin 3-glucoside; Cy, cyanidin; Pn, peonidin; Q3G, quercetin 3-glucoside; Q, quercetin; IsoR3G, isorhamnetin 3-glucoside; IsoRh, isorhamnetin; P1, unknown product.
Kinetic analyses (Table I) confirmed that AOMT exhibited only background levels of activity with pelargonidin 3-O-glucoside, catechin, and epicatechin. AOMT exhibited equivalent kinetic parameters with cyanidin 3-O-glucoside, delphinidin 3-O-glucoside, quercetin 3-O-glucoside, and the flavonol aglycones quercetin and myricetin (Table I). AOMT activity with cyanidin aglycone was slightly lower than with cyanidin 3-O-glucoside. Km values for these substrates ranged from 20 to 75 μm. Recombinant AOMT was active within a broad range of pH values, with an optimum around 7.5 on both substrates tested (cyanidin 3-O-glucoside and delphinidin 3-O-glucoside; Supplemental Fig. S3). It retained over 25% of its activity at pH 9.4 and at pH 6.25, but activity was dramatically reduced when more acidic conditions were tested (Supplemental Fig. S3).
The influence of different divalent cations on AOMT activity was tested (Tables II and III). AOMT activity was extremely low in the absence of Mg2+ ions, confirming the Mg dependence of AOMT (Table II). The best activity was obtained in the presence of 10 mm MgCl2 (Table II). Incubation of AOMT in the presence of EDTA greatly reduced its activity with both cyanidin 3-O-glucoside and delphinidin 3-O-glucoside substrates. The presence of other divalent cations in addition to Mg2+ strongly decreased activity in the case of Zn2+ and Mn2+, while little effects were observed with Ca2+ (Table III).
Table II.
In vitro AOMT activity in the presence of different MgCl2 concentrations using cyanidin 3-O-glucoside as substrate and expressed as a percentage of the activity with 10 mm MgCl2
| MgCl2 Concentration | Activity |
|---|---|
| mm | % |
| 0 | 5 |
| 2.5 | 24 |
| 5 | 77 |
| 10 | 100 |
Table III.
In vitro AOMT activity in the presence of 10 mm MgCl2 and 10 mm EDTA, ZnCl2, CaCl2, and MnCl2 using cyanidin 3-O-glucoside and delphinidin 3-O-glucoside as substrates
Activity is expressed as a percentage of the activity with 10 mm MgCl2.
| Addition | Cyanidin 3-O-Glucoside | Delphinidin 3-O-Glucoside |
|---|---|---|
| % activity | ||
| EDTA | 16 | 0 |
| ZnCl2 | 30 | 21 |
| CaCl2 | 100 | 83 |
| MnCl2 | 25 | 20 |
Characterization of AOMT Activity in Planta
Recombinant AOMT exhibited flavonol and AOMT activity in vitro. However, in vitro data should be taken with caution for the prediction of the physiological substrates and products of OMTs (Liu and Dixon, 2001). We used Agrobacterium-mediated transient transformation of tobacco to investigate AOMT activity in planta. This approach allowed us to successfully characterize OMT activities in planta (Scalliet et al., 2006; Schmidlin et al., 2008). In order to provide AOMT with anthocyanin substrates, AOMT was coexpressed with Production of Anthocyanin Pigment1 (PAP1), an R2R3 MYB transcription factor from Arabidopsis involved in anthocyanin biosynthesis (Borevitz et al., 2000). Constitutive expression of PAP1 in Arabidopsis and other plant species, including tobacco, petunia, and tomato (Solanum lycopersicum), has been shown to induce anthocyanin accumulation, due to the coordinated up-regulation of genes in the anthocyanin biosynthetic pathway (Borevitz et al., 2000; Xie et al., 2006; Ben Zvi et al., 2008; Zuluaga et al., 2008). No anthocyanin was detected in extracts from leaves expressing the GFP control (Fig. 6A). As expected, Agrobacterium-mediated transient expression of PAP1 in tobacco resulted in significant levels of anthocyanin accumulation in infiltrated leaves (Fig. 6, B and C). The flavonoid composition of transformed tobacco leaves was then determined using both HPLC/ESI-MS/DAD and HPLC/DAD in order to compare the flavonoids produced in PAP1 leaves with those accumulated in leaves coexpressing PAP1 and AOMT. The metabolites were putatively identified from their UV-visible absorption spectra, and comprehensive analysis of mass fragmentation patterns obtained by tandem MS spectroscopy was compared with those of known compounds.
Figure 6.
Characterization of AOMT activity in planta using Agrobacterium-mediated transient transformation. Tobacco leaf sectors (200 mg fresh weight) expressing GFP (A and D) or PAP1 (B and E) or coexpressing PAP1 and AOMT (C and F) were excised 96 h after Agrobacterium-mediated transformation. Anthocyanin and flavonol contents were analyzed using HPLC-DAD. Anthocyanins were monitored at 520 nm and flavonols at 360 nm. The data shown are representative of three independent experiments. Dp3R, Delphinidin 3-O-rutinoside; Pt3R, petunidin 3-O-rutinoside; Mv3G, malvidin 3-O-glucoside; Mv3R, malvidin 3-O-rutinoside; QRH, quercetin 3-O-rutinoside O-hexoside; peak a1, delphinidin 3-O-rutinoside O-glucoside; peak a2, delphinidin 3-O-rutinoside O-hexoside; peak a3, malvidin 3-O-rutinoside O-hexoside; peaks c1, c2, and c3, isomers of caffeoyl quinic acid; peaks f1, f2, and f3, isomers of feruloyl quinic acid. MS/MS spectra of Dp3R, Mv3R, and Mv3G are shown in the middle. ABS, Absorbance; AU, absorbance unit.
Three anthocyanin pigments accumulated in tobacco leaves expressing PAP1 (Fig. 6B). Delphinidin 3-O-rutinoside was the most abundant anthocyanin, amounting to approximately 96% of total anthocyanins in PAP1 leaves. The other two, petunidin 3-O-rutinoside and delphinidin 3-O-rutinoside O-hexoside, were present in trace amounts (Fig. 6B, peaks Pt3R and a1, respectively). Coexpression of PAP1 and AOMT resulted in a marked decrease in the delphinidin 3-O-rutinoside peak (more than 50%) together with an accumulation of malvidin 3-O-rutinoside and malvidin 3-O-glucoside (Fig. 6C). Two other discrete anthocyanins were detected in PAP1- and AOMT-expressing tobacco leaves, at m/z 773 and 801 (Fig. 6C, peaks a2 and a3, respectively). Fragmentation products (Supplemental Table S2) identified delphinidin (m/z 303) and malvidin (m/z 331) as aglycone in the two compounds, respectively, with two sugar moities attached to the anthocyanin, a rutinose (neutral loss m/z 308) and a hexose (neutral loss m/z 162; Supplemental Table S2). In leaves coexpressing PAP1 and AOMT, malvidin conjugates represented about 70% of total anthocyanins and O-methylation of delphinidin conjugates was complete on both 3′ and 5′ positions, as no trace of petunidin conjugates could be detected in PAP1- and AOMT-expressing tobacco leaves (Fig. 6C).
In addition to anthocyanins, two quercetin glycosides and two kampferol glycosides were identified in control tobacco leaves expressing GFP as well as in both PAP1-expressing and PAP1- and AOMT-expressing leaves (Fig. 6, D–F). Quercetin O-rutinoside O-hexoside was the major flavonol detected in all of the transformed tobacco leaves, accounting for approximately 70% of total flavonols in control leaves and 50% in the leaves expressing PAP1. Nevertheless, the same composition in flavonol compounds was observed in leaves expressing PAP1 or coexpressing PAP1 and AOMT. Moreover, two hydroxycinnamic acids, caffeoyl quinic acid and its corresponding methylated form, feruloyl quinic acid, each with three isomers, were identified at 320 nm in 35S-GFP tobacco leaves as well as in both PAP1-expressing and PAP1- and AOMT-expressing leaves (Fig. 6, D–F, peaks c1, c2, c3, f1, f2, and f3). Two isomers of feruloyl quinic acids (Fig. 6, D–F, peaks f2 and f3; Supplemental Table S2) accumulated more in PAP1-transformed and PAP1- and AOMT-transformed leaves than in GFP control leaves. Taken together, these analyses indicated that coexpression of AOMT and PAP1 did not modify significantly the flavonol and phenolic acid profile, in comparison with leaves expressing PAP1 alone.
Subcellular Localization of AOMT
To investigate AOMT subcellular localization, a construct encoding AOMT fused to the N terminus of GFP (AOMT-GFP) was expressed in tobacco using Agrobacterium-mediated transient transformation. Nontargeted GFP control and AOMT-GFP were coexpressed with PAP1 transcription factor in order to investigate both the cellular localization and the enzyme activity of the AOMT-GFP fusion protein. Confocal microscopy imaging of tobacco cells expressing AOMT-GFP showed a cytosolic fluorescence pattern, very similar to that obtained with nontargeted GFP (Fig. 7). HPLC analysis of anthocyanins accumulated in tobacco leaves coexpressing PAP1 and AOMT-GFP (Supplemental Fig. S4) showed that the anthocyanin profile closely resembled that of leaves coexpressing PAP1 and AOMT (Fig. 6C). Taken together, these results indicated that the AOMT-GFP fusion protein retained AOMT activity and was located in the cytosol of transformed cells.
Figure 7.
Subcellular localization of the AOMT-GFP fusion protein. Transient expression of nontargeted GFP (A) and the AOMT-GFP fusion protein (B) following agroinfiltration of tobacco leaves. Abaxial epidermal cell images are projections of 20- × 1-μm optical sections collected by confocal laser scanning microscopy. Bars = 20 μm. [See online article for color version of this figure.]
DISCUSSION
Functional Characterization of a Candidate AOMT from Grapevine
Several recent transcriptomic studies suggested that a putative OMT could be involved in anthocyanin methoxylation in grape berries (Ageorges et al., 2006; Castellarin et al., 2007; Castellarin and Di Gaspero, 2007; Pilati et al., 2007; Cutanda-Perez et al., 2009). In order to characterize this gene, called hereafter AOMT, its cDNA was cloned from Syrah and Nebbiolo grape berries. Phylogenetic analysis of selected members of the plant OMT family showed that AOMT belonged to a subset of type 2 plant OMTs and was distinct from the clade of classical CCoA-OMTs, presumably involved in lignin biosynthesis. In recent years, several OMTs active with flavonoid substrates have been characterized from different plant species, especially OMTs catalyzing the 3′ or 3′,5′ O-methylation of the B-ring of flavonoid substrates. Interestingly, such OMTs have been characterized in both type 1 and type 2 plant OMT families (Muzac et al., 2000; Kim et al., 2006; Lee et al., 2008). Thus, AOMT represents a new member of a small subfamily of type 2 plant OMTs involved in the methylation of flavonoid substrates (Ibdah et al., 2003). Determination of AOMT kinetic parameters showed that Km values and specific activities for all flavonol and anthocyanin substrates tested were similar to values found for other OMTs with their primary substrate (Ibrahim et al., 1998). AOMT exhibited a strict specificity for 3′ and 5′ hydroxyl group methylation in both flavonol and anthocyanin substrates, showing no activity with pelargonidin 3-O-glucoside. Furthermore, AOMT showed roughly equivalent activity with quercetin and quercetin 3-O-glucoside and with cyanidin and cyanidin 3-O-glucoside, suggesting that AOMT could methylate efficiently both glycosylated and aglycone substrates. In grape berries, major anthocyanins are both glycosylated and methylated, raising the question of the order in which these two reactions occur in vivo. Ford et al. (1998) characterized a UDP Glc-flavonoid glucosyl transferase (UFGT) catalyzing the glycosylation of both anthocyanidin and flavonol substrates in vitro. However, UFGT enzyme exhibited a greatly reduced activity with malvidin aglycone compared with petunidin aglycone, even though malvidin 3-O-glucoside is usually the most abundant anthocyanin in grape berry. This suggests that anthocyanin glycosylation may occur prior to methylation in grape berries.
In order to determine the subcellular localization of AOMT, an AOMT-GFP fusion protein was expressed in tobacco. Like the nontargeted GFP control, the AOMT-GFP fusion protein exhibited a clear cytosolic localization, which is very likely to reflect the physiological localization of AOMT, as discussed below. These results suggest that both glycosylation and methylation of anthocyanins take place in the cytosol, although the precise in vivo chronology of these reactions remains to be determined. Several enzymes involved in flavonoid biosynthesis have been shown to be organized into macromolecular complexes optimizing the cooperation between its members (Burbulis and Winkel-Shirley, 1999; Liu and Dixon, 2001; Winkel, 2004). The cytoplasmic localization of the AOMT-GFP fusion protein has led to the suggestion that AOMT may be associated with the endoplasmic reticulum-multienzyme complexes, facilitating rapid methylation and stabilization of its product. Glycosylation and methylation reactions may take place in such complexes, ensuring an efficient biosynthesis of anthocyanins in vivo, although the activity of UFGT with some anthocyanin substrates is low in vitro (Ford et al., 1998).
The AOMT Gene Expression Profile Mirrors Anthocyanin Biosynthesis in Berry
Five major anthocyanins exist in grape berries, which differ in their hydroxylation and methylation patterns at the phenolic B-ring. These anthocyanins include the nonmethylated cyanidin (3′,4′-OH) and delphinidin (3′,4′,5′-OH), the monomethylated peonidin (3′-OCH3 4′-OH) and petunidin (3′-OCH3 4′,5′-OH), and the dimethylated malvidin (3′,5′-OCH3 4′-OH), pelargonidin (4′-OH) being notably absent in all known Vitis genotypes (Macheix et al., 1990). The expression of the AOMT gene was monitored in two genotypes, characterized either by high total anthocyanin content and predominance of the dimethylated malvidin (Syrah; Mattivi et al., 2006) or by low anthocyanin content and predominance of the monomethylated peonidin (Nebbiolo; Guidoni et al., 2008). The AOMT gene was expressed in both varieties, exhibiting similar expression profiles, with a maximum at the onset of ripening, when methylated anthocyanins start to accumulate. Castellarin and Di Gaspero (2007) reported similar expression profiles for this OMT gene during grape berry development in other cultivars presenting different anthocyanin composition. The evolution of the transcriptional level of AOMT through ripening and the relative abundance of methoxylated anthocyanins are compatible with a major role in anthocyanin methylation in grape varieties, irrespective of their different anthocyanin composition.
While the expression pattern and the substrate specificities of AOMT correlated well with the anthocyanin profiles found in grape berries, the situation was less clear for flavonols. Similar to anthocyanins, dihydroxylated or trihydroxylated flavonol compounds on the B-ring (e.g. quercetin and myricetin) can be monomethylated (quercetin to isorhamnetin and myricetin to laricitrin) or dimethylated (myricetin to syringetin). The comparative study of anthocyanin and flavonol methylation patterns over different grape genotypes showed that the ratio of flavonol monomethylation versus total flavonol was positively correlated with the ratio of anthocyanin monomethylation versus total anthocyanin, and the same was true for dimethylation (Mattivi et al., 2006). These results agree with a model where AOMT would also operate on flavonols, with the same methylation patterns as on anthocyanins. The AOMT enzymic activity measured in vitro on both substrates corroborates this hypothesis. However, in grapevine, flavonols and anthocyanins accumulate at different time points in the berry skins: flavonols accumulate at the early stage of development and during ripening, while anthocyanins accumulate only during ripening (Downey et al., 2003). In this context, it is expected that the gene encoding the flavonol methyltransferase would have a different expression pattern from the AOMT gene. Furthermore, Castellarin and Di Gaspero (2007) have shown that AOMT is not expressed in Pinot Gris, although this white genotype was shown to contain methylated flavonols (Mattivi et al., 2006). The possibility that an alternative flavonol-OMT is active in grape berries is thus still open.
Coexpression of PAP1 and AOMT in Tobacco Leaves Induces the Accumulation of Methylated Anthocyanins
In this work, we used tobacco leaves in which anthocyanin biosynthesis was induced by transient expression of the R2R3 MYB transcription factor PAP1 from Arabidopsis to characterize the activity of AOMT in planta. Overexpression of PAP1 in transgenic plants was shown to enhance the accumulation of lignin, hydroxycinnamic acid esters, and flavonoids, including various anthocyanins (Borevitz et al., 2000; Tohge et al., 2005). In the species used in our study (N. benthamiana), the most abundant anthocyanin induced by the overexpression of PAP1 was delphinidin 3-O-rutinoside. The strong accumulation of this nonmethylated anthocyanin in PAP1-expressing tobacco leaves allowed the investigation of AOMT activity in vivo. The malvidin 3-O-rutinoside accumulation following coexpression of PAP1 and AOMT in tobacco leaves confirmed that AOMT alone catalyzed both 3′ and 5′ methylation steps of delphinidin. The predominance of malvidin 3-O-rutinoside indicates a good efficiency for the double meta methylation in vivo, even if residual delphinidin was still detected. These data confirmed the in vitro results that AOMT was able to catalyze two sequential methylations of delphinidin derivatives at the 3′ and 5′ positions to yield malvidin derivatives. However, petunidin 3-O-rutinoside was absent, suggesting a higher efficiency for the second methylation step in planta, which prevented the accumulation of the monomethylated intermediate. In contrast to anthocyanins, coexpression of AOMT and PAP1 did not modify significantly the flavonol and phenolic acid contents in tobacco leaves, in comparison with expression of PAP1 alone. Although AOMT was active with flavonols in vitro, the tobacco leaves, which accumulated only minor amounts of flavonols, did not allow us to confirm this activity in vivo.
Moreover, the coexpression of the AOMT-GFP fusion protein with PAP1 transcription factor in tobacco leaves resulted in the accumulation of methylated anthocyanins, identical to those accumulating after coexpression of AOMT and PAP1. These similar anthocyanin profiles indicated that the AOMT-GFP fusion protein retained AOMT activity and that its cytosolic localization likely reflected the physiological localization of the native AOMT protein.
AOMT Is Likely to Play a Major Role in Anthocyanin Methylation in Grape Berries
Previous works in petunia and grape suggested that different OMTs were responsible for the O-methylation at the 3′ and 5′ positions, respectively, of anthocyanin molecules (Wiering and de Vlaming, 1977; Bailly et al., 1997). In contrast with these predictions, the AOMT described here behaved as a bifunctional 3′,5′-OMT both in vitro and in vivo, suggesting that AOMT alone would be sufficient to perform both methylation steps in grape berries. The absence of cyanidin derivatives in PAP1-expressing tobacco leaves did not allow the investigation of AOMT activity with 3′,4′-dihydroxylated anthocyanins in the tobacco system. However, the AOMT gene was strongly expressed after véraison in Nebbiolo and in Grignolino grapes (Castellarin and Di Gaspero, 2007), which accumulate mainly the monomethylated peonidin 3-O-glucoside as the major anthocyanin. Given the fact that AOMT could efficiently methylate cyanidin 3-O-glucoside in vitro, these results suggest that AOMT is also involved in the methylation of cyanidin derivatives in grape berries. These results can thus be accommodated in a model where AOMT would be the major methyltransferase acting on anthocyanins in grape berries or the main member of a family of enzymes with similar kinetic properties. This model would also explain the fact that among several grape cultivars whose berry anthocyanin composition has been analyzed, accumulation of petunidin in the absence of malvidin was never reported (Wenzel et al., 1987; Mattivi et al., 2006). The predominance and preferential 3′,5′ methylating activity of this AOMT would also explain the common occurrence of malvidin in the genotypes where trihydroxylated anthocyanins are available and the existence of a positive correlation between the 3′ and 3′,5′ anthocyanin methoxylation activity, as estimated by metabolomic data over 64 grape genotypes (elaborated from Mattivi et al., 2006). This model is further supported by several transcriptomic studies, which identified independently the same AOMT as a major candidate for anthocyanin methylation in grape berries (Ageorges et al., 2006; Castellarin et al., 2007; Castellarin and Di Gaspero, 2007; Pilati et al., 2007; Cutanda-Perez et al., 2009). Finally, this model does not rule out the possibility that other OMTs may also participate in anthocyanin methylation in grapevine. Analysis of the grape genome sequence (Jaillon et al., 2007; Velasco et al., 2007) shows that the AOMT gene characterized in this work belongs to a small family of four similar putative OMT genes. Future work will be needed to investigate the roles of the other members of this gene family and their potential role in anthocyanin and/or flavonol methylation in grapevine.
In conclusion, we characterized a grapevine anthocyanin OMT that efficiently catalyzes the methylation of anthocyanins both in vitro and in planta. In addition, we provide evidence that AOMT is likely to play a major role in anthocyanin methylation in grape berries. Methylation tends to shift berry color toward purple, and these changes are transmitted to the wines, where anthocyanins form relatively stable complexes with other grape-derived flavonoids (Heredia et al., 1998; Jensen et al., 2008). Thus, identification of AOMT constitutes an important step in understanding the biosynthesis of major anthocyanins in grape and the determination of one of the key quality parameters of red grapes and wine.
MATERIALS AND METHODS
Chemicals and Radiochemicals
[14C]SAM (55 mCi mmol−1) was from GE Healthcare-Amersham Biosciences. Quercetin 3-O-glucoside, pelargonidin 3-O-glucoside, cyanidin 3-O-glucoside, and delphinidin-3-O-glucoside were from Extrasynthèse. All other chemicals and reagents were from Sigma.
Plant Material, RNA Extraction, and Grape Anthocyanin Analysis
Roots, shoots, leaves, and berries were harvested from grapevine plants (Vitis vinifera ‘Syrah’ [malvidin 3-glucoside as the predominant anthocyanin] and ‘Nebbiolo’ [peonidin 3-glucoside as the predominant anthocyanin]). Syrah plants were grown in the SupAGRO-INRA vineyard (Montpellier, France) on 30 vinestocks of homogeneous vigor. Average daily temperatures in the growing season ranged between 22.4°C (June) and 23.6°C (August), and mean relative humidity values ranged from 55.9% (June) to 67.6% (August). Nebbiolo plants were from a 15-year-old experimental vineyard at Grinzane Cavour (Piedmont, Italy) on east-west oriented rows. Average daily temperatures in the growing season ranged between 17.1°C (May) and 22.8°C (August), and rainfall summed up to 411 mm in the period from May to September. Young leaves were from the third rank, counted from the apex, with mean weight of 0.3 g per leaf. Old leaves were fully expanded leaves with mean weight of 2.8 g per leaf. Berries were collected at nine developmental stages as described previously (Terrier et al., 2005). All collected samples were quickly frozen in liquid nitrogen, ground under liquid nitrogen to a fine powder with a Dangoumau blender, and stored at –80°C until use. Total RNA from Syrah tissues was extracted using the RNeasy Plant Mini kit (Qiagen) following the manufacturer's protocol from 200 mg of starting tissue, and RNA from Nebbiolo tissues was extracted using a modification of the Chang protocol (Carra et al., 2007). Grape anthocyanin analyses of Syrah berries were performed by HPLC on 200 mg of ground frozen tissue as described by Ageorges et al. (2006). In Nebbiolo berries, anthocyanin analyses were done on 250 mg of frozen tissue by HPLC following the protocol of Guidoni et al. (2002).
Cloning of AOMT cDNA and Phylogenetic Analysis
The full-length cDNA of AOMT was amplified from cDNA of mature berries (Syrah) with high-fidelity Taq polymerase (Advantage-HF 2 PCR kit; Clontech) using the forward primer 5′-TTTTCTTGTACGGCAGGCTTA-3′ and the reverse primer 5′-TGAGAATGGATTTAGGCTAATAGAG-3′. The amplified cDNA for AOMT was cloned into the pGEM-T Easy vector (Promega), and the resulting plasmid was sequenced. OMT sequences were obtained from GenBank. Nucleic acid and protein sequences were aligned using ClustalW (Thompson et al., 1994). The Phylo_win program (Galtier et al., 1996) was used to construct phylogenetic trees, using the neighbor-joining method, with 500 bootstrap replicates.
Quantitative PCR Analysis of Gene Expression
In the case of Syrah, RNA was quantified with Ribogreen (Molecular Probes) and reverse transcription was performed in triplicate from each sample from 500 ng of purified RNA using the SuperScript II Reverse Transcription kit (Life Technologies). Triplicates were further pooled for PCR. Gene-specific primers were as follows: AOMT-F (5′-CTCTGCAGGCGCCTCTATTA-3′) and AOMT-R (5′-CCCAAAACAGAGTCTGGACA-3′). Specific annealing of the oligonucleotides was controlled on dissociation kinetics performed at the end of each PCR run. The efficiency of the primer pair was measured on a plasmid serial dilution. PCR was performed in triplicate as described by Fernandez et al. (2007). Gene transcripts were quantified upon normalization to VvEF1α (Terrier et al., 2005) by comparing the cycle threshold of the target gene with that of VvEF1α. Gene expression was expressed as mean and se calculated over all biological and technical replicates. In the case of Nebbiolo, the following modifications to the protocol were applied. Ten micrograms of RNA was reverse transcribed in duplicate using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) and PowerSYBR Green (Applied Biosystems) for quantitation of amplification results. The gene-specific primers for Nebbiolo were AOMT-F′ (5′-GATGAATGTCCCTGTCGATGAG-3′) and AOMT-R′ (5′-GCAAGAGCTGTTGCCAAGAGA-3′).
Characterization of Recombinant AOMT
AOMT cDNA was amplified by PCR using the upstream primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGTTCCGCGTGGATCCATGTCCAGCTCAAGTCATAGG-3′ and the downstream primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCACTAATAGAGGCGCCTGCAGAG-3′ and cloned into pDONR207 Gateway-compatible vector (Invitrogen). AOMT cDNA was sequenced to verify that no mutation had been introduced and transferred into the pHNGWA destination vector (accession no. EU680842; Busso et al., 2005). AOMT enzyme was expressed as His-tagged NusA-fusion protein. Recombinant AOMT was purified using TALON metal affinity resin (Clontech) and characterized after cleavage of the NusA moiety using thrombin (GE Healthcare-Amersham). Purified recombinant AOMT (5 μg) was assayed in a final volume of 200 μL with 200 μm SAM and 200 μm anthocyanin or flavonol substrate in 0.1 m Tris, pH 7.5, containing 10% (v/v) glycerol, 10 mm MgCl2, 14 mm 2-mercaptoethanol, and 1 mg mL−1 bovine serum albumin. Incubations were stopped with 800 μL of methanol containing 0.2 n HCl. For kinetic studies, recombinant AOMT (200 ng) was incubated in a final volume of 25 μL as above, with the exception that 100 μm [14C]SAM (8 mCi mmol−1) was used. Ranges of anthocyanin and flavonol substrate concentrations between 3 and 500 μm were used for Km determination. Reactions were stopped by the addition of 75 μL of 2 n HCl and incubated at 95°C for 30 min for hydrolysis of glucosides. Reaction products were extracted with 100 μL of isoamyl alcohol, and the incorporated radioactivity was measured by liquid scintillation. Km and Vmax values were calculated from Lineweaver-Burk plots.
The pH dependence of AOMT activity was assessed in the pH range 4.5 to 9.4 using MES (4.5–6.5), PIPES (6.6), HEPES (7.0–7.5), and Tris-HCl (7.5–9.4). The effect of divalent cations on enzyme activity was assessed by testing different concentrations of MgCl2 (0, 2.5, 5, and 10 mm). Metal inhibition on enzyme activity was estimated by adding to the reaction mixture containing 10 mm MgCl2 and CaCl2, ZnCl2, MnCl2, or EDTA (final concentration of 2.5, 5.0, or 10 mm).
Transient Expression in Tobacco
For Agrobacterium tumefaciens-mediated transient expression, AOMT cDNA was transferred into the Gateway-compatible binary vector pB2GW7 (Karimi et al., 2002). A PAP1 genomic fragment from Arabidopsis (Arabidopsis thaliana ecotype Columbia) was amplified by PCR using the upstream primer 5′-ACCTTTTACAATTTGTTTATATATTTTACG-3′ and the downstream primer 5′-CAAACCTATACACAAACGCAAACAAATGTT-3′ (Borevitz et al., 2000). The PAP1 gene was then reamplified using the upstream primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATTTGTTTATATATTTTAC-3′ and the downstream primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTACACAAACGCAAACAAATGTT-3′ and cloned into pDONR207 vector. PAP1 was subsequently transferred into the Gateway binary vector pB2GW7 (Karimi et al., 2002). A m-GFP4-HDEL construct was used as a control (Haseloff et al., 1997). All constructs were introduced into Agrobacterium strain C58 (pMP90) by electroporation. Tobacco (Nicotiana benthamiana) leaves were infiltrated with Agrobacterium cultures (optical density at 600 nm = 0.1–0.3) according to Batoko et al. (2000). Discs were punched from tobacco leaves 4 d after Agrobacterium infiltration and analyzed for anthocyanin content.
Subcellular Localization of AOMT
The AOMT coding sequence was amplified by PCR using the upstream primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGTTCCGCGTGGATCCATGTCCAGCTCAAGTCATAGG-3′ and the downstream primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATAGAGGCGCCTGCAGAGCGCGAC-3′. The amplified DNA fragment was first introduced into pDONR 207 (Invitrogen) and then into the Gateway-compatible pK7FWG2 vector (Karimi et al., 2002) in order to obtain a C-terminal GFP fusion protein (AOMT::GFP). The pK7FWG2-AOMT construct was transformed into tobacco using agroinfiltration as described above. A nontargeted GFP construct (mGFP4) was used as a control (Haseloff et al., 1997). Tobacco leaves were examined 48 h after transformation using a Zeiss LSM 510 confocal imaging system attached to a Zeiss Axioplan 2 microscope (Carl Zeiss). GFP was visualized by excitation with the 488 line of a krypton/argon laser and use of a band-pass 505 to 550 emission filter. Projections of optical sections were accomplished using LSM image-processing software (Zeiss).
Analysis of AOMT Reaction Products
The in vitro enzymatic reactions (200 μL) were diluted with 800 μL of methanol containing 0.2 n HCl. The transformed tobacco tissues were resupended in methanol containing 0.2 n HCl (1 mg tissue fresh weight mL−1 solvent). Samples were stirred on a Stuart Tube Rotator SB3 (Bibby Sterilin) in the dark at room temperature for 1 h, centrifuged (13,000g, 15 min, 4°C), and then dried under vacuum at 35°C for 2 h (Genevac). Dried residues were dissolved in 100 μL of 20% methanol containing 1% HCl before HPLC analyses.
Anthocyanins and flavonols were analyzed by HPLC-DAD as described previously (Fournand et al., 2006) and further characterized with HPLC-DAD/ESI-MS. Separations were performed using a Waters Millennium HPLC-DAD system on a 250- × 2-mm (i.d.) Atlantis dC18 column (Waters; 5 μm) with a guard column operated at 30°C. The mobile phase consisted of water:formic acid (98:2, v/v; eluent A) and water:acetonitrile:formic acid (18:80:2, v/v/v; eluent B). Flow rate was 0.25 mL min−1. The elution program was as follows: isocratic for 2 min with 0% B, 0% to 2% B (2–5 min), isocratic with 2% B (5–12 min), 2% to 3% B (12–15 min), 3% to 8% B (15–25 min), 8% to 20% B (25–40 min), 20% to 25% B (40–45 min), isocratic with 25% B (45–55 min), 25% to 65% B (55–70 min), and isocratic with 65% B (70–75 min). ESI-MS/MS analyses were performed with a ThermoFinnigan LCQ advantage mass spectrometer equipped with an electrospray source and an ion trap mass analyzer controlled by the LCQ navigator software. The spectrometer was operated in the positive ion mode (source voltage, 4 kV; capillary voltage, 30 V; capillary temperature, 250°C; sheath gas, 50 arbitrary units; auxiliary gas, 10 arbitrary units). The collision energy for fragmentation used for MS/MS experiments was set at 35%. Anthocyanins, flavonols, and hydroxycinnamic acids were identified according to their mass fragmentation, UV/visible absorption spectra, and retention times, respectively. Anthocyanin quantifications were based on peak areas at 520 nm, whereas quantifications for flavonols and hydroxycinnamic acids were done at 360 and 320 nm, respectively.
Sequence data from this article have been deposited with the GenBank/EMBL data libraries under accession number FJ460168 (AOMT from Syrah). The Nebbiolo sequence differs from Syrah in two single nucleotide polymorphisms (A216C and T441C).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantitative real-time PCR expression profiling of VvCCoAOMT during Nebbiolo berry development.
Supplemental Figure S2. SDS-PAGE of the recombinant AOMT.
Supplemental Figure S3. In vitro pH optimum of AOMT using cyanidin 3-O-glucoside (A) and delphinidin 3-O-glucoside (B).
Supplemental Figure S4. Anthocyanin profiles of transient transformed tobacco coexpressing PAP1 and AOMT (A) or coexpressing PAP1 and AOMT::GFP (B).
Supplemental Table S1. In vitro substrate specificity of the recombinant AOMT.
Supplemental Table S2. Flavonoid profiles in acidic methanol-water extracts of transformed tobacco leaves expressing GFP (control), PAP1, or PAP1 + AOMT.
Supplementary Material
Acknowledgments
We are indebted to Peter Beyer (Center for Applied Biosciences, Freiburg, Germany) and Bilal Camara and Florence Bouvier (Institut de Biologie Moléculaire des Plantes, Strasbourg, France) for the use of their laboratory facilities. We thank Jérôme Mutterer (Institut de Biologie Moléculaire des Plantes, Strasbourg, France) for help with confocal imaging. We thank Pascale Coste, Bernard Delnatte, Denise Hartmann, Charlotte Knichel, Jacky Misbach, and Christian Vivant (INRA, Colmar, France) for assistance with plant material, Mathias de Vismes (INRA, Colmar, France) for help with tobacco transformation, and Francesca Cardinale (University of Torino, Turin, Italy) for advice in the production of the recombinant protein. We also thank Sandrine Vialet (INRA, Montpellier, France) for excellent technical support in quantitative reverse transcription-PCR analysis.
This work was supported by the European Union (grant no. FLAVO 2005–513960) and the Italian Ministry of Research and Education (grant no. PRIN 2005–2005071021_004).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Agnès Ageorges (ageorges@supagro.inra.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ageorges A, Fernandez L, Vialet S, Merdinoglu D, Terrier N, Romieu C (2006) Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci 170 372–383 [Google Scholar]
- Andersen O, Markham KR (2006) Flavonoids: Chemistry, Biochemistry and Applications. CRC Press, Boca Raton, FL
- Awika JM, Rooney LW, Waniska R (2004) Properties of 3-deoxyanthocyanins from sorghum. J Agric Food Chem 52 4388–4394 [DOI] [PubMed] [Google Scholar]
- Bailly C, Cormier F, Do CB (1997) Characterization and activities of S-adenosyl-L-methionine:cyanidin 3-glucoside 3′-O-methyltransferase in relation to anthocyanin accumulation in Vitis vinifera cell suspension cultures. Plant Sci 122 81–89 [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12 2201–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Zvi MM, Negre-Zakharov F, Masci T, Ovadis M, Shklarman E, Ben-Meir H, Tzfira T, Dudareva N, Vainstein A (2008) Interlinking showy traits: co-engineering of scent and colour biosynthesis in flowers. Plant Biotechnol J 6 403–415 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96 12929–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busam G, Junghanns KT, Kneusel RE, Kassemeyer HH, Matern U (1997) Characterization and expression of caffeoyl-coenzyme A 3-O-methyltransferase proposed for the induced resistance response of Vitis vinifera L. Plant Physiol 115 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso D, Delagoutte-Busso B, Moras D (2005) Construction of a set of Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal Biochem 343 313–321 [DOI] [PubMed] [Google Scholar]
- Carra A, Gambino G, Schubert A (2007) A cetyltrimethylammonium bromide-based method to extract low-molecular-weight RNA from polysaccharide-rich plant tissues. Anal Biochem 360 318–320 [DOI] [PubMed] [Google Scholar]
- Castellarin SD, Di Gaspero G (2007) Transcriptional control of anthocyanin biosynthetic genes in extreme phenotypes for berry pigmentation of naturally occurring grapevines. BMC Plant Biol 7 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin SD, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E, Di Gaspero G (2007) Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ 30 1381–1389 [DOI] [PubMed] [Google Scholar]
- Cutanda-Perez MC, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L (2009) Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol Biol 69 633–648 [DOI] [PubMed] [Google Scholar]
- Da Silva FG, Iandolino A, Al-Kayal F, Bohlmann MC, Cushman MA, Lim H, Ergul A, Figueroa R, Kabuloglu EK, Osborne C, et al (2005) Characterizing the grape transcriptome: analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiol 139 574–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N, Anzellotti D, Ibrahim RK, Huner N, Sarhan F (2003) Daphnetin methylation by a novel O-methyltransferase is associated with cold acclimation and photosystem II excitation pressure in rye. J Biol Chem 278 6854–6861 [DOI] [PubMed] [Google Scholar]
- Downey MO, Dokoozlian NK, Krstic MP (2006) Cultural practice and environmental impacts on the flavonoid composition of grapes and wine: a review of recent research. Am J Enol Vitic 57 257–268 [Google Scholar]
- Downey MO, Harvey JS, Robinson SR (2003) Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.). J Grape Wine Res 9 110–121 [Google Scholar]
- Fernandez L, Torregrossa L, Terrier N, Sreekantan L, Grimplet J, Davies C, Thomas MR, Romieu C, Ageorges A (2007) Identification of genes associated with flesh morphogenesis during grapevine fruit development. Plant Mol Biol 63 307–323 [DOI] [PubMed] [Google Scholar]
- Ford CM, Boss PK, Hoj PB (1998) Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J Biol Chem 273 9224–9233 [DOI] [PubMed] [Google Scholar]
- Forkmann G, Heller W (1999) Biosynthesis of flavonoids. In U Sankawa, ed, Polyketides and Other Secondary Metabolites including Fatty Acids and Their Derivatives, Vol 1. Elsevier, Amsterdam, pp 713–748
- Fournand D, Vicens A, Sidhoum L, Souquet JM, Moutounet M, Cheynier V (2006) Accumulation and extractability of grape skin tannins and anthocyanins at different advanced physiological stages. J Agric Food Chem 54 7331–7338 [DOI] [PubMed] [Google Scholar]
- Frick S, Kutchan T (1999) Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J 17 329–339 [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12 543–548 [DOI] [PubMed] [Google Scholar]
- Guidoni S, Allara P, Schubert A (2002) Cluster thinning affects the berry skin anthocyanin composition of Vitis vinifera cv. Nebbiolo. Am J Enol Vitic 53 224–226 [Google Scholar]
- Guidoni S, Ferrandino A, Novello V (2008) Effect of seasonal and agronomical practices on skin anthocyanin profile of Nebbiolo grapes. Am J Enol Vitic 59 22–29 [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55 481–504 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis brightly. Proc Natl Acad Sci USA 94 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia FJ, Francia-Aricha EM, Rivas-Gonzalo JC, Vicario IM, Santos-Buelga C (1998) Chromatic characterization of anthocyanins from red grapes. I. pH effects. Food Chem 63 491–498 [Google Scholar]
- Ibdah M, Zhang XH, Schmidt J, Vogt T (2003) A novel Mg(2+)-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. J Biol Chem 278 43961–43972 [DOI] [PubMed] [Google Scholar]
- Ibrahim RK, Bruneau A, Bantignies B (1998) Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol 36 1–10 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–467 [DOI] [PubMed] [Google Scholar]
- Jensen JS, Demiray S, Egebo M, Meyer AS (2008) Prediction of wine color attributes from the phenolic profiles of red grapes (Vitis vinifera). J Agric Food Chem 56 1105–1115 [DOI] [PubMed] [Google Scholar]
- Jonsson LMV, Aarsman MEG, Poulton JE, Schram AW (1984) Properties and genetic control of four methyltransferases involved in methylation of anthocyanins in flowers of Petunia hybrida. Planta 160 174–179 [DOI] [PubMed] [Google Scholar]
- Jonsson LMV, de Vlaming P, Wiering H, Aarsman MEG, Schram AW (1983) Genetic control of anthocyanin O-methyltransferase activity in flowers of Petunia hybrida. Theor Appl Genet 66 349–355 [DOI] [PubMed] [Google Scholar]
- Joshi CP, Chiang VL (1998) Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol Biol 37 663–674 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY® vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Kim BG, Lee Y, Hur HG, Lim Y, Ahn JH (2006) Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67 387–394 [DOI] [PubMed] [Google Scholar]
- Lam KC, Ibrahim RK, Behdad B, Dayanandan S (2007) Structure, function, and evolution of plant O-methyltransferases. Genome 50 1001–1013 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Kim BG, Chong Y, Lim Y, Ahn JH (2008) Cation dependent O-methyltransferases from rice. Planta 227 641–647 [DOI] [PubMed] [Google Scholar]
- Liu CJ, Dixon RA (2001) Elicitor-induced association of isoflavone O-methyltransferase with endomembranes prevents the formation and 7-O-methylation of daidzein during isoflavonoid phytoalexin biosynthesis. Plant Cell 13 2643–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheix JJ, Fleuriet A, Billot J (1990) Fruit Phenolics. CRC Press, Boca Raton, FL
- Markham KR (1989) Flavones, flavonols and their glycosides. In JB Harborne, ed, Methods in Plant Biochemistry. Academic Press, London, pp 197–235
- Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R (2006) Metabolite profiling of grape: flavonols and anthocyanins. J Agric Food Chem 54 7692–7702 [DOI] [PubMed] [Google Scholar]
- Mazza G, Miniati E (1993) Grapes. In G Mazza, E Miniati, eds, Anthocyanins in Fruits, Vegetables, and Grains. CRC Press, Boca Raton, FL, pp 149–199
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3 212–217 [Google Scholar]
- Muzac I, Wang J, Anzellotti D, Zhang H, Ibrahim RK (2000) Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3′-O-methyltransferase and characterization of the gene product. Arch Biochem Biophys 375 385–388 [DOI] [PubMed] [Google Scholar]
- Noel JP, Dixon RA, Pichersky E, Zubieta C, Ferrer JL (2003) Structural, functional, and evolutionary basis for methylation of plant small molecules. In JT Romeo, ed, Recent Advances in Phytochemistry, Vol 37. Elsevier Science, Oxford, pp 37–58
- Pilati S, Perazzolli M, Malossini A, Cestaro A, Demattè L, Fontana P, Dal Ri A, Viola R, Velasco R, Moser C (2007) Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at véraison. BMC Genomics 8 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarni P, Fulcrand H, Souillol V, Souquet JM, Cheynier V (1995) Mechanisms of anthocyanin degradation in grape must-like model solutions. J Sci Food Agric 69 385–391 [Google Scholar]
- Scalliet G, Lionnet L, Le Bechec M, Dutron D, Magnard JL, Baudino S, Bergougnoux V, Jullien F, Chambrier P, Vergne P, et al (2006) Role of petal-specific orcinol O-methyltransferases in the evolution of rose scent. Plant Physiol 14 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin L, Poutaraud A, Claudel P, Mestre P, Prado E, Santos-Rosa M, Wiedemann-Merdinoglu S, Karst F, Merdinoglu D, Hugueney P (2008) A stress-inducible resveratrol O-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol 148 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N, Glissant D, Grimplet J, Barrieu F, Abbal P, Couture C, Ageorges A, Atanassova R, Léon C, Renaudin JP, et al (2005) Isogene specific oligo arrays reveal multifaceted changes in gene expression during grape berry (Vitis vinifera L.) development. Planta 222 820–831 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima JI, Awazuhara M, Inoue E, Takahashi H, Goodenowe D, Kitayam M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42 218–235 [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J, et al (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2 e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel K, Dittrich HH, Heimfarth M (1987) Die Zusammensetzung der Anthocyane in den Beeren verschiedener Rebsorten. Vitis 26 65–78 [Google Scholar]
- Wiering H, de Vlaming P (1977) Glycosylation and methylation patterns of anthocyanins in Petunia hybrida. II. The genes Mr1 and Mf2. Z Pflanzenzucht 78 113–123 [Google Scholar]
- Winkel BSJ (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55 85–107 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenweber E, Dietz VH (1981) Occurrence and distribution of free flavonoid aglycones in plants. Phytochemistry 20 869–932 [Google Scholar]
- Xie DY, Sharma SB, Wright E, Wang ZY, Dixon RA (2006) Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J 45 895–907 [DOI] [PubMed] [Google Scholar]
- Ye ZH, Kneusel RE, Matern U, Varner JE (1994) An alternative methylation pathway in lignin biosynthesis in zinnia. Plant Cell 6 1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Ezell E, Liehr JG (1994) Catechol-O-methyltransferase-catalyzed rapid O-methylation of mutagenic flavonoids: metabolic inactivation as a possible reason for their lack of carcinogenicity in vivo. J Biol Chem 269 292–299 [PubMed] [Google Scholar]
- Zuluaga DL, Gonzali S, Loreti E, Pucciariello C, Degl'Innocenti E, Guidi L, Alpi A, Perata P (2008) Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct Plant Biol 35 606–618 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.