Abstract
AIM: To determine whether SP-TAT-apoptin induces apoptosis and also maintains its tumor cell specificity.
METHODS: In this study, we designed a secretory protein by adding a secretory signal peptide (SP) to the N terminus of Transactivating Transcription (TAT)-apoptin (SP-TAT-apoptin), to test the hypothesis that it gains an additive bystander effect as an anti-cancer therapy. We used an artificial human secretory SP whose amino acid sequence and corresponding cDNA sequence were generated by the SP hidden Markov model.
RESULTS: In human liver carcinoma HepG2 cells, SP-TAT-apoptin expression showed a diffuse pattern in the early phase after transfection. After 48 h, however, it translocated into the nuclear compartment and caused massive apoptotic cell death, as determined by 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and annexin-V binding assay. SP-TAT-apoptin did not, however, cause any cell death in non-malignant human umbilical vein endothelial cells (HUVECs). Most importantly, the conditioned medium from Chinese hamster ovary (CHO) cells transfected with SP-TAT-apoptin also induced significant cell death in HepG2 cells, but not in HUVECs.
CONCLUSION: The data demonstrated that SP-TAT-apoptin induces apoptosis only in malignant cells, and its secretory property might greatly increase its potency once it is delivered in vivo for cancer therapy.
Keywords: Apoptin, Apoptosis, Hepatoma, Human immunodeficiency Virus-Transactivating Transcription protein, Secretory
INTRODUCTION
Apoptin or viral protein 3 (VP3), a protein of 13.6 kDa derived from the chicken anemia virus (CAV), represents a new anti-cancer tool with great potential[1,2]. It appears to have innate tumor-specific, p53-independent[3,4], Bcl-2-enhanced proapoptotic activity[4,5], and hence is of considerable interest for efficient targeting and specific elimination of cancer cells[3,4,6–8]. The antitumor activity of apoptin appears to be linked to its ability to localize in the nuclei of transformed cells, but not in those of primary or non-transformed cells[9]. Therefore, apoptin has been explored to achieve efficient targeting and specific elimination of cancer cells.
To use apoptin in cancer therapy, efficient delivery to or expression of apoptin in cancer cells is required. The Human Immunodeficiency Virus (HIV) Transactivating Transcription (TAT)-derived protein transduction peptide is a small basic peptide that has been successfully shown to deliver a large variety of materials, from small particles to proteins, peptides and nucleic acids, across the cell membrane[10–12]. The region that conveys the cell-penetrating properties appears to be confined to a small (11 amino acids) stretch of basic amino acids (aa 47-57, YGRKKRRQRRR)[13]. This TAT transduction domain has been successfully used to deliver apoptin to cancer cells[14].
In this study, we designed a secretory TAT-apoptin fusion protein by adding a secretory signal to the N-terminal of the recombinant molecule to gain an additive by-stander effect as an anti-cancer therapy. Secreted TAT-apoptin from transformed cells enters un-transformed cancer cells and causes apoptosis. We employed an artificial human secretory signal peptide (SP) whose amino acid sequence and corresponding cDNA sequences were generated by an SP hidden Markov model (SP-HMM)[15]. We demonstrated expression of the secretory fusion protein (SP-TAT-apoptin) and induction of apoptosis by the secreted protein in HepG2 cells.
MATERIALS AND METHODS
Generation and cloning of SP-TAT-apoptin
The human secretory SP was designed and optimized by an HMM that has been used to predict, identify and generate secretory SP sequences[15]. PCR was used to amplify the apoptin gene and to incorporate the TAT transduction domain and SP sequence upstream. The primers were designed based on the published sequences in GenBank (NC_001427), and synthesized by Shanghai Sangon Biological Engineering Technology & Services (Shanghai, China). The first pair of designed primers were: 5′AAGAATGAACGCTCTGCAGGAAGATACTCC-3′ (sense) and 5′-CTGCAGTCTTA TACGCCTTTTTGCGG-3′ (antisense), with a product size of 406 bp. The sense primer contains the TAT transduction domain sequence. The second pair of primers, which incorporated the secretory signal sequence into the TAT-apoptin fusion protein, were: 5′GCTGCTGCTGCTGCTGCTGTGGCCCATGGTG TGGGCCTATGGCAGG-3′ (sense) and the same antisense primer as the first pair, with a product size of 466 bp. The templates used for generating recombinant TAT-apoptin and TAT-GFP in the first round PCR were the apoptin and gfp genes carried on pCDNA3.1-apoptin plasmid[16] and pEGFP plasmid, respectively. The conditions for both rounds of PCR were as follows: 30 cycles of 94°C for 40 s, 56°C for 40 s, and 72°C for 1 min. The PCR products obtained were TOPO® cloned into the pLenti6/V5-D-TOPO® vector (Invitrogen, USA) resulting pLenti6/V5-D-TOPO/SP-TAT-apoptin and pLenti6/V5-D-TOPO/SP-TAT-EGFP. The plasmids were transformed into Stbl3TM Escherichia coli (E.coli) (Invitrogen) by electroporation. The SP-TAT-apoptin cDNA cloned in pLenti6/V5-D- TOPO® vector was confirmed by restriction enzyme digestion and by DNA sequencing.
Cell lines and cell culture
HepG2 human hepatoma cells, human umbilical vein endothelial cells (HUVECs) and Chinese hamster ovary (CHO) cells were purchased from Keygen Company (Nanjing, China). All cells were maintained and grown at 37°C in DMEM (Hyclone, USA), supplemented with 1% penicillin–streptomycin, and 10% fetal bovine serum in an incubator with CO2 controlled at 5%.
Conditioned medium: The conditioned medium from Chinese hamster ovary (CHO) cells transfected with SP-TAT-apoptin. (CHO cells were cultured in a six-well plate. The cells were transfected with the pLenti6/V5-D-TOPO/SP-TAT-apoptin plasmid using the LipofectamineTM 2000 protocol according to manufacturer’s instructions (Invitrogen). After 6 h incubation, the cells were washed with fresh culture medium and cultured for an additional 24 h. The culture supernatants were then collected and added, respectively, to the monolayers of HepG2 cells and HUVECs grown in 24-well plates. )
Stbl3TM Escherichia coli: Stbl3TM E. coli for transfor-mation as this strain is particularly well-suited for use in cloning unstable DNA such as lentiviral DNA containing direct repeats.
Reverse transcriptase-PCR (RT-PCR)
Cells were rinsed twice with PBS and total RNA was isolated from cells using a Simply P Total RNA Extraction Kit (Bioer, Japan) according to the manufacturer’s instructions. One microgram of total RNA was reverse transcribed to first-strand cDNA with Superscript II reverse transcriptase (Invitrogen) at 46°C for 45 min. Synthesized first-strand cDNA was then subjected to PCR analysis using gene-specific primers. The primers used were: CMV forward 5′-CGCAAATGGGCGGTAGGCGTG-3′ and V5(C-term) reverse 5′-ACCGAGGAGAGGGTTAGGGAT-3′, with a product size of 700 bp. The PCR conditions were as follows: 30 cycles at 94°C for 40 s, 56°C for 40 s, and 72°C for 1 min for SP-TAT-apoptin; 25 cycles at 94°C for 30 s, 60°C for 45 s, and 72°C for 1 min for β-actin control. PCR products were run on 1.5% agarose gels containing ethidium bromide and photographed using a Syngene Gene Genius imaging system (Syngene, USA).
Transient transfection and fluorescence microscopy
Cells were cultivated in 24-well culture plates. In each well, the cells were grown at 50%-80% confluency and transfected with 400 ng plasmid DNA pre-incubated with 1.4 μL LipofectamineTM 2000 (Invitrogen), according to the manufacturer’s instructions. Coverslips were placed at the bottom of the wells to allow the cells grow on the slides. Apoptin expression was detected with anti-V5-FITC antibody (Invitrogen) as green fluorescence, and the cell nuclei were stained by propidium iodide (PI) as red fluorescence. The cells were incubated with anti-V5-FITC antibody in the dark for 1 h and washed twice with PBS before staining with PI. Fluorescence images were recorded on a confocal imaging system equipped with krypton–argon laser (Leica SP2 Confocal System, Germany).
Flow cytometry
The loss of cell membrane asymmetry in apoptotic cells was determined by using an Annexin-V FITC Apoptosis Assay Kit (Keygen, China). Apoptin-expressing cells were stained with annexin-V FITC as green fluorescent cells, and the nuclei of the apoptotic cells were stained by PI with red fluorescence. After staining, the cell suspensions were analyzed on a Cytometric FC500 flow cytometer, and 105 events were collected for each sample. Viable cells were defined as annexin-V FITC and PI double-negative events.
Cell viability assay
Cell viability was also determined by the 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye reduction assay which measures mitochondrial respiratory function[17,18]. Exponentially growing cells were plated in 96-well flat bottom plates (Corning, USA) and allowed to adhere for 24 h. At various times post-transfection with the recombinant plasmid, cells were incubated with MTT dye (1 mg/mL) for 2 h and solubilized with 20 μL 10% SDS. A560 was then measured.
DNA laddering assay
DNA fragmentation was detected using an Apoptotic DNA Laddering Kit (Keygen) according to the manufacturer’s instructions. DNA was extracted, separated by 1.5% agarose gel electrophoresis, followed by ethidium bromide staining to visualize the laddered DNA.
Immunocytochemical assay and DAPI staining
At various times post-transfection, cells grown in six-well plates were harvested and treated by trypsinization and resuspended in PBS. The cells were then spread on a slide, fixed by 100% methanol for 5 min at room temperature, stained by the Apoptotic/Necrotic Cell Detection Kit (Keygen), and embedded in resin, after permeabilization, for long-term storage.
Cytotoxicity of SP-TAT was also tested by 2,4-diamidino-2-phenylin-dole (DAPI) staining. Transfected cells grown on coverslips that were placed on the bottom of a 24-well plate were washed with PBS and fixed, and apoptotic cells were differentiated by staining with mounting medium containing DAPI, and visualized using an Olympus AX70 fluorescence microscope.
Protein secretion and activity test
CHO cells were cultured in a six-well plate. The cells were transfected with the pLenti6/V5-D-TOPO/SP-TAT-apoptin plasmid using the LipofectamineTM 2000 protocol according to manufacturer’s instructions (Invitrogen). After 6 h incubation, the cells were washed with fresh culture medium and cultured for an additional 24 h. The culture supernatants were then collected and added respectively to the monolayers of HepG2 cells and HUVECs grown in 24-well plates. These cells had also been washed with PBS before adding the supernatants. At various times post co-culture, these cells were fixed and stained, and apoptin localization and apoptosis were analyzed.
Statistical analysis
ANOVA was performed for multiple group comparison. In conjunction with ANOVA, post hoc pairwise comparisons were performed by Bonferroni’s test, with P < 0.05 regarded as statistically significant.
RESULTS
Generation of SP-TAT-apoptin fusion construct and expression of SP-TAT-apoptin
The human secretory SP was constructed and optimized virtually by the HMM, which has been used to describe, predict, identify, and generate secretory SP sequences[13]. It was inserted at the N terminus of recombinant TAT-apoptin to generate SP-TAT-apoptin fusion protein, and it contained a positively charged N region, a hydrophobic central region, and a C region that contained a cleavage site. Two rounds of PCR were carried out to amplify the apoptin gene and to fuse TAT and the synthetic SP into the construct to create recombinant secretory-TAT-apoptin.
To determine whether the SP-TAT-apoptin cDNA construct generated was expressed in vivo, the HepG2 cell line was transfected with the plenti6/V5-D-TOPO/SP-TAT-apoptin plasmid. Analysis by RT-PCR revealed that SP-TAT-apoptin was expressed 24 h after transfection (data not shown). The expression of SP-TAT-apoptin in HepG2 cells was confirmed by immunofluorescence microscopy (Figure 1).
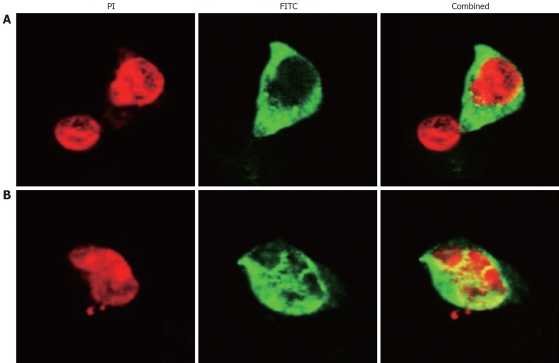
Figure 1.
SP-TAT-apoptin expression in HepG2 cells (× 1000). Cells transfected with plenti6/V5-D-TOPO/SP-TAT-apoptin plasmid and fixed at 24 h (A) and 48 h (B) post-transfection. Recombinant apoptin detected by anti-V5-FITC antibody is shown in green and cell nuclei stained by PI in red. Apoptin protein showed a diffuse pattern in the cytoplasm at 24 h post-transfection, and in the nucleus at 48 h.
Induction of apoptosis by SP-TAT-apoptin
To determine whether SP-TAT-apoptin induces apoptosis and also maintains its tumor cell specificity, HUVECs and HepG2 cells were transfected with the plenti6/V5-D-TOPO/SP-TAT-apoptin plasmid. SP-TAT-apoptin-induced apoptosis was investigated in these two cell lines. Three different assays were used to gauge apoptosis. In the first assay, cell viability was measured by co-staining with annexin-V FITC and PI, followed by flow cytometry. This assay was based on the loss of plasma membrane asymmetry (integrity) as a result of apoptosis. Viable cells were defined as annexin-V FITC and PI double-negative events. As shown in Figure 2A, only HepG2 cells were susceptible to SP-TAT-apoptin-induced apoptosis in a time-dependent manner. These results also confirm previous reports that apoptin has the ability to induce apoptosis specifically in tumor cells[3,4,7,19,20]. In the second assay, cells that were exponentially grown were inoculated in 96-well flat-bottom plates and allowed to adhere for 24 h. After transfection, cell viability was determined at various times by MTT dye reduction assay. Expression of the recombinant protein slightly decreased the viability of HepG2 cells at 24 h post-transfection, and the same was true in HUVECs (Figure 2B). At 48 h post-transfection, the viability of HUVECs was only slightly decreased compared to that at 24 h. However, the viability of HepG2 cells was significantly decreased at 48 h post-transfection. The presence of recombinant apoptin caused a decrease in HepG2 cell viability to < 60% at 72 h and < 50% at 96 h post-transfection (Figure 2B). In the third assay, genomic DNA fragmentation in HepG2 cells and HUVECs was investigated. As shown in Figure 2C, the apoptin fusion protein brought about significant DNA fragmentation in HepG2 cells at 72 h post-transfection. In contrast, detectable apoptotic DNA laddering in HUVECs was not seen during the experiment (data not shown).
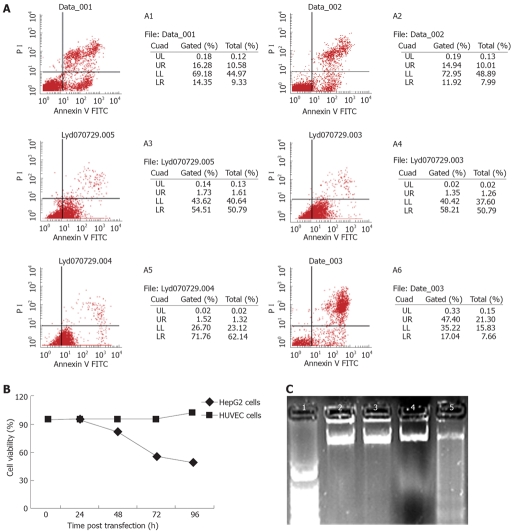
Figure 2.
SP-TAT-apoptin-induced cell death. A: Cell viability measured by flow cytometry. A1 and A2: HUVECs at 48 and 72 h post-transfection; A3-A6: HepG2 cells at 24, 48, 72 and 96 h. HepG2 cells were susceptible to SP-TAT-apoptin-induced apoptosis in a time-dependent manner; B: Cell viability determined by MTT dye reduction assay; C: DNA fragmentation in HepG2 cells demonstrated by agarose gel electrophoresis. Lane 1: 1 kb DNA marker; Lanes 2-5: DNA from cells at 24, 48, 60 and 72 h post-transfection, respectively.
Lack of cytotoxicity of SP-TAT
In order to determine the cytotoxicity of SP-TAT, plenti6/V5-D-TOPO/SP-TAT-apoptin plasmid and plenti6/V5-D-TOPO/SP-TAT-GFP plasmids were transfected into HepG2 cells separately. Robust apoptosis of HepG2 cells was observed, as demonstrated by microscopy at different times after transfection, while in contrast, expression of SP-TAT-GFP did not induce noticeable apoptosis in these cells (Figure 3). Therefore, SP-TAT did not seem to exhibit cytotoxicity in HepG2 cells and apoptosis induced by SP-TAT-apoptin was due to apoptin.
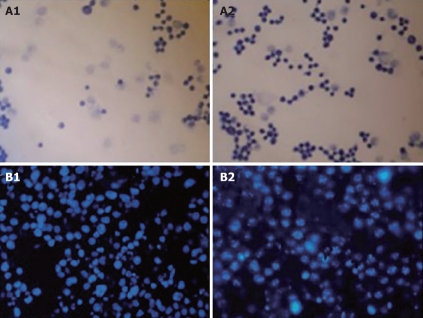
Figure 3.
Cytotoxicity of SP-TAT-apoptin compared to SP-TAT. A: Micrographs of HepG2 cells transfected with SP-TAT-apoptin construct stained by Apoptotic/Necrotic Cell Detection Kit. An inverted microscope (× 400) was used. The nuclei of apoptotic cells were stained deep blue. A1 and A2: HepG2 cells at 24 and 72 h post-transfection; B: HepG2 cells stained with DAPI and observed by fluorescence microscopy (× 400). B1: HepG2 cells 72 h after transfection with plenti6/V5-D-TOPO/SP-TAT-GFP plasmid; B2: HepG2 cells 72 h after transfection with plenti6/V5-D-TOPO/SP-TAT-apoptin. Arrow indicates apoptotic cells.
Secretion of SP-TAT-apoptin and the effect of secreted TAT-apoptin on HepG2 cells
The presence of synthesized SP enables TAT-apoptin to be secreted outside the transfected cells and re-enter adjacent un-transfected HepG2 cells, potentially increasing the efficacy of apoptin when used as cancer therapy. To test its feasibility, the recombinant construct was used to transfect CHO cells and the culture supernatant was collected. HepG2 cells and HUVECs were then co-cultured with this supernatant. At 24 h post co-culture, TAT-apoptin had translocated from the cytoplasm to the nucleus (Figure 4A). At various times post co-culture, cell viability was also determined by MTT dye reduction assay. As shown in Figure 4B, the recombinant protein TAT-apoptin slightly decreased cell viability of HepG2 cells at 24 h post co-culture. In contrast, the viability of HUVECs was actually increased at 24 h, which continued during the course of the experiment. At 48 h post co-culture, viability of HepG2 cells was significantly decreased compared to that of HUVECs. The recombinant apoptin protein decreased HepG2 cell viability to < 50% at 48 h post-transfection.
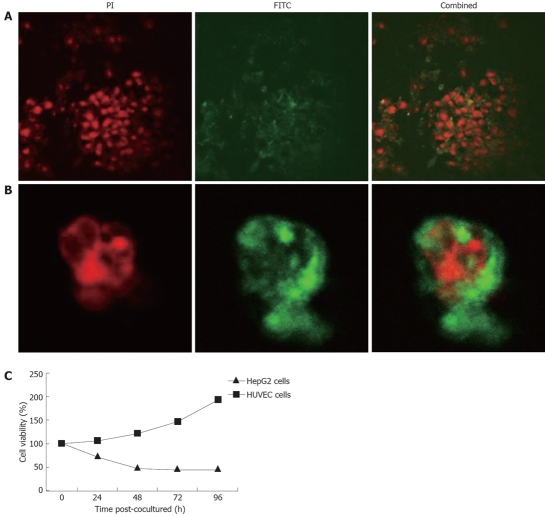
Figure 4.
Cell death induced by secreted TAT-apoptin. A: HepG2 cells immunostained 4 h after co-culture with the supernatant of CHO cells expressing SP-TAT-apoptin. Recombinant TAT-apoptin detected by anti-V5-FITC antibody is shown in green and cell nuclei stained by PI in red (× 1000); B: Translocation of recombinant apoptin in HepG2 cells 24 h after co-culture with the supernatant of CHO cell expressing SP-TAT-apoptin; C: Cell viability determined by MTT dye reduction assay after co-culture with secreted TAT-apoptin.
DISCUSSION
New therapeutic approaches that facilitate selective targeting of cancer cells while sparing normal cells have emerged in recent years. Apoptin represents a new anti-cancer tool in such new approaches with great potential[21–25]. Two routes can be taken using apoptin or its encoding cDNA, i.e. as protein or gene therapy. In any case, efficient systems are required to facilitate the delivery of apoptin to cancer cells or expression of apoptin within these cells[26–30]. The HIV TAT transduction domain has been successfully used to deliver apoptin into cancer cells[14], and no apoptosis of normal cells (HUVECs) was observed with this TAT-apoptin fusion protein. In this study, we generated a cDNA construct of SP-TAT-apoptin. Cancer cells transfected with this construct expressed recombinant apoptin and apoptosis was induced. By incorporating a synthetic SP we also expected apoptin to be secreted from the transfected cells as TAT-apoptin fusion protein and re-enter adjacent untransfected HepG2 cells, which enabled the construct to act as both a protein and gene therapeutic agent, and increased the potency of apoptin in cancer therapy.
SP-TAT-apoptin was expressed in HUVECs and HepG2 cells, and the protein was initially located in the cytoplasm. At 48 h post-transfection, the protein was located in the nucleus of HepG2 cells, which indicated that SP-TAT-apoptin was capable of translocating to the nucleus. SP-TAT-apoptin was also functionally active and efficiently induced HepG2 cell apoptosis, in a time-dependent manner. In HUVECs, SP-TAT-apoptin remained in the cytoplasm and no induction of apoptosis above the background level was observed. Meanwhile, no apoptosis was observed in cells in which SP-TAT-GFP was expressed, which indicates that SP-TAT alone is not cytotoxic for HepG2 cells. Therefore, SP-TAT-apoptin retained the characteristic expression pattern of apoptin and induced apoptosis in cancer cells.
Having a synthetic SP, recombinant apoptin was able to be secreted from transfected cells and re-enter adjacent untransfected HepG2 cells. The recombinant protein was detected in the cytoplasm in HepG2 cells and HUVECs shortly after co-culture of the cells with the cell-free supernatant of the transfected CHO cells. This indicated that the secreted TAT-apoptin fusion protein contained in the CHO cell culture medium was able to enter these cells. The fusion protein was later found in the nucleus of HepG2 cells and induced HepG2 apoptosis. The new secretory characteristic increased the possibility of apoptin being used in cancer gene therapy. However, there are still a large number of unanswered questions regarding the mechanisms and therapeutic usage of apoptin, and further studies are certainly required.
COMMENTS
Background
Apoptin is a protein encoded by Constant Angular Velocity (CAV) and it can cause apoptotic cell death. It has been shown to possess a striking specificity for cancer cells. Apoptin, therefore, has great potential for efficient targeting and specific elimination of cancer cells.
Research frontiers
Human Immunodeficiency Virus (HIV)-Transactiviting Transcription (TAT)-fused apoptin has been shown to possess a striking specificity for cancer cells. However, the cancer killing activity is limited in cells transfected with the apoptin expression construct, which spares the untransfected cancer cells. A secretory TAT-apoptin fusion protein with a secretory signal has an additive by-stander effect as an anti-cancer therapy. Secreted TAT-apoptin from transformed cells enters un-transformed cancer cells and causes apoptosis.
Innovations and breakthroughs
The new secretory characteristic increased the possibility of apoptin being used in cancer gene therapy. However, there are still a large number of unanswered questions regarding the mechanisms and therapeutic usage of apoptin, and further studies are certainly required.
Terminology
Apoptin or VP3 is a protein of 13.6 kDa derived from CAV, represents a new anti-cancer tool with great potentials. It appears to have innate tumor-specific, p53-independent, Bcl-2-enhanced pro-apoptotic activity.
Peer review
The authors investigated the role of secretory TAT-apoptin fusion protein in HepG2 cells. They conclude that such a protein induces apoptosis in HCC cell lines, but not in non-cancer cell line HUVEC. This is a very interesting study, which may be applicable for the treatment of human liver cancer in the future.
Acknowledgments
We thank Dr. Zhi-Xian Sun at the Academy of Military Medical Sciences, China, for providing plasmid pCDNA3.1-apoptin.
Supported by the National Natural Science Foundation of China, No. 30672069 and No. 30470098
Peer reviewer: Gianluigi Giannelli, MD, Dipartimento di Clinica Medica, Immunologia e Malattie Infettive, Sezione di Medicina Interna, Policlinico, Piazza G. Cesare 11, 70124 Bari, Italy
S- Editor Li DL L- Editor Kerr C E- Editor Lin YP
References
- 1.Noteborn MH. Chicken anemia virus induced apoptosis: underlying molecular mechanisms. Vet Microbiol. 2004;98:89–94. doi: 10.1016/j.vetmic.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Olijslagers SJ, Zhang YH, Backendorf C, Noteborn MH. Additive cytotoxic effect of apoptin and chemotherapeutic agents paclitaxel and etoposide on human tumour cells. Basic Clin Pharmacol Toxicol. 2007;100:127–131. doi: 10.1111/j.1742-7843.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 3.Janssen K, Hofmann TG, Jans DA, Hay RT, Schulze-Osthoff K, Fischer U. Apoptin is modified by SUMO conjugation and targeted to promyelocytic leukemia protein nuclear bodies. Oncogene. 2007;26:1557–1566. doi: 10.1038/sj.onc.1209923. [DOI] [PubMed] [Google Scholar]
- 4.Burek M, Maddika S, Burek CJ, Daniel PT, Schulze-Osthoff K, Los M. Apoptin-induced cell death is modulated by Bcl-2 family members and is Apaf-1 dependent. Oncogene. 2006;25:2213–2222. doi: 10.1038/sj.onc.1209258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russo A, Terrasi M, Agnese V, Santini D, Bazan V. Apoptosis: a relevant tool for anticancer therapy. Ann Oncol. 2006;17 Suppl 7:vii115–vii123. doi: 10.1093/annonc/mdl963. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Elojeimy S, El-Zawahry AM, Holman DH, Bielawska A, Bielawski J, Rubinchik S, Guo GW, Dong JY, Keane T, et al. Modulation of ceramide metabolism enhances viral protein apoptin's cytotoxicity in prostate cancer. Mol Ther. 2006;14:637–646. doi: 10.1016/j.ymthe.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Maddika S, Mendoza FJ, Hauff K, Zamzow CR, Paranjothy T, Los M. Cancer-selective therapy of the future: apoptin and its mechanism of action. Cancer Biol Ther. 2006;5:10–19. doi: 10.4161/cbt.5.1.2400. [DOI] [PubMed] [Google Scholar]
- 8.Peng DJ, Sun J, Wang YZ, Tian J, Zhang YH, Noteborn MH, Qu S. Inhibition of hepatocarcinoma by systemic delivery of Apoptin gene via the hepatic asialoglycoprotein receptor. Cancer Gene Ther. 2007;14:66–73. doi: 10.1038/sj.cgt.7700985. [DOI] [PubMed] [Google Scholar]
- 9.Wang QM, Fan GC, Chen JZ, Chen HP, He FC. A putative NES mediates cytoplasmic localization of Apoptin in normal cells. Acta Biochim Biophys Sin (Shanghai) 2004;36:817–823. doi: 10.1093/abbs/36.12.817. [DOI] [PubMed] [Google Scholar]
- 10.Hashida H, Miyamoto M, Cho Y, Hida Y, Kato K, Kurokawa T, Okushiba S, Kondo S, Dosaka-Akita H, Katoh H. Fusion of HIV-1 Tat protein transduction domain to poly-lysine as a new DNA delivery tool. Br J Cancer. 2004;90:1252–1258. doi: 10.1038/sj.bjc.6601680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubo E, Fatma N, Akagi Y, Beier DR, Singh SP, Singh DP. TAT-mediated PRDX6 protein transduction protects against eye lens epithelial cell death and delays lens opacity. Am J Physiol Cell Physiol. 2008;294:C842–C855. doi: 10.1152/ajpcell.00540.2007. [DOI] [PubMed] [Google Scholar]
- 12.Song HY, Lee JA, Ju SM, Yoo KY, Won MH, Kwon HJ, Eum WS, Jang SH, Choi SY, Park J. Topical transduction of superoxide dismutase mediated by HIV-1 Tat protein transduction domain ameliorates 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced inflammation in mice. Biochem Pharmacol. 2008;75:1348–1357. doi: 10.1016/j.bcp.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Ziegler A, Seelig J. Interaction of the protein transduction domain of HIV-1 TAT with heparan sulfate: binding mechanism and thermodynamic parameters. Biophys J. 2004;86:254–263. doi: 10.1016/S0006-3495(04)74101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guelen L, Paterson H, Gaken J, Meyers M, Farzaneh F, Tavassoli M. TAT-apoptin is efficiently delivered and induces apoptosis in cancer cells. Oncogene. 2004;23:1153–1165. doi: 10.1038/sj.onc.1207224. [DOI] [PubMed] [Google Scholar]
- 15.Barash S, Wang W, Shi Y. Human secretory signal peptide description by hidden Markov model and generation of a strong artificial signal peptide for secreted protein expression. Biochem Biophys Res Commun. 2002;294:835–842. doi: 10.1016/S0006-291X(02)00566-1. [DOI] [PubMed] [Google Scholar]
- 16.Sun GJ ,Tong X, Sun ZX, Gene clone and activity assay of apoptin. Junshi Yixue Kexueyuan Yuankan. 2001;2:85–87. [Google Scholar]
- 17.Hayon T, Dvilansky A, Shpilberg O, Nathan I. Appraisal of the MTT-based assay as a useful tool for predicting drug chemosensitivity in leukemia. Leuk Lymphoma. 2003;44:1957–1962. doi: 10.1080/1042819031000116607. [DOI] [PubMed] [Google Scholar]
- 18.Kubota T. (Cancer chemosensitivity test-from laboratory to clinic) Hum Cell. 1995;8:189–194. [PubMed] [Google Scholar]
- 19.Lee YH, Cheng CM, Chang YF, Wang TY, Yuo CY. Apoptin T108 phosphorylation is not required for its tumor-specific nuclear localization but partially affects its apoptotic activity. Biochem Biophys Res Commun. 2007;354:391–395. doi: 10.1016/j.bbrc.2006.12.201. [DOI] [PubMed] [Google Scholar]
- 20.Olijslagers SJ, Zhang YH, Backendorf C, Noteborn MH. Additive cytotoxic effect of apoptin and chemotherapeutic agents paclitaxel and etoposide on human tumour cells. Basic Clin Pharmacol Toxicol. 2007;100:127–131. doi: 10.1111/j.1742-7843.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 21.Danen-Van Oorschot AA, Zhang YH, Leliveld SR, Rohn JL, Seelen MC, Bolk MW, Van Zon A, Erkeland SJ, Abrahams JP, Mumberg D, et al. Importance of nuclear localization of apoptin for tumor-specific induction of apoptosis. J Biol Chem. 2003;278:27729–27736. doi: 10.1074/jbc.M303114200. [DOI] [PubMed] [Google Scholar]
- 22.Alvisi G, Poon IK, Jans DA. Tumor-specific nuclear targeting: promises for anti-cancer therapy? Drug Resist Updat. 2006;9:40–50. doi: 10.1016/j.drup.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Poon IK, Oro C, Dias MM, Zhang J, Jans DA. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 2005;65:7059–7064. doi: 10.1158/0008-5472.CAN-05-1370. [DOI] [PubMed] [Google Scholar]
- 24.He X, Zhang Q, Liu Y, He P. Apoptin Induces Chromatin Condensation in Normal Cells. Virus Genes. 2005;31:49–55. doi: 10.1007/s11262-005-2200-4. [DOI] [PubMed] [Google Scholar]
- 25.Gdynia G, Lehmann-Koch J, Sieber S, Tagscherer KE, Fassl A, Zentgraf H, Matsuzawa S, Reed JC, Roth W. BLOC1S2 interacts with the HIPPI protein and sensitizes NCH89 glioblastoma cells to apoptosis. Apoptosis. 2008;13:437–447. doi: 10.1007/s10495-007-0176-3. [DOI] [PubMed] [Google Scholar]
- 26.Maddika S, Wiechec E, Ande SR, Poon IK, Fischer U, Wesselborg S, Jans DA, Schulze-Osthoff K, Los M. Interaction with PI3-kinase contributes to the cytotoxic activity of apoptin. Oncogene. 2008;27:3060–3065. doi: 10.1038/sj.onc.1210958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddika S, Bay GH, Kroczak TJ, Ande SR, Maddika S, Wiechec E, Gibson SB, Los M. Akt is transferred to the nucleus of cells treated with apoptin, and it participates in apoptin-induced cell death. Cell Prolif. 2007;40:835–848. doi: 10.1111/j.1365-2184.2007.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Backendorf C, Visser AE, de Boer AG, Zimmerman R, Visser M, Voskamp P, Zhang YH, Noteborn M. Apoptin: therapeutic potential of an early sensor of carcinogenic transformation. Annu Rev Pharmacol Toxicol. 2008;48:143–169. doi: 10.1146/annurev.pharmtox.48.121806.154910. [DOI] [PubMed] [Google Scholar]
- 29.Schoop RA, Kooistra K, Baatenburg De Jong RJ, Noteborn MH. Bcl-xL inhibits p53- but not apoptin-induced apoptosis in head and neck squamous cell carcinoma cell line. Int J Cancer. 2004;109:38–42. doi: 10.1002/ijc.11675. [DOI] [PubMed] [Google Scholar]
- 30.Maddika S, Mendoza FJ, Hauff K, Zamzow CR, Paranjothy T, Los M. Cancer-selective therapy of the future: apoptin and its mechanism of action. Cancer Biol Ther. 2006;5:10–19. doi: 10.4161/cbt.5.1.2400. [DOI] [PubMed] [Google Scholar]