Abstract
The latent TGF-β binding proteins (LTBP) −1, −3, and −4 are extracellular proteins that assist in the secretion and localization of latent TGF-β. The null mutation of LTBP-4S in mice causes defects in the differentiation of terminal air-sacs, fragmented elastin, and colon carcinomas. We investigated lung development from embryonic day 14.5 (E14.5) to day 7 after birth (P7) in order to determine when the defects in elastin organization initiate and to further examine the relation of TGF-β signaling levels and air-sac septation in Ltbp4S−/− lungs. We found that defects in elastogenesis are visible as early as E14.5 and are maintained in the alveolar walls, in blood vessel media, and subjacent air–way epithelium. The air-sac septation defect was associated with excessive TGF-β signaling and was reversed by lowering TGF-β2 levels. Thus, the phenotype is not directly reflective of a change in TGF-β1, the only TGF-β isoform known to complex with LTBP-4. Reversal of the air-sac septation defect was not associated with normalization of the elastogenesis indicating two separate functions of LTBP-4 as a regulator of elastic fiber assembly and TGF-β levels in lungs.
Keywords: TGF-β, LTBP-4, elastogenesis, air-sac septation, TGF-β activation
Introduction
The latent transforming growth factor-β (TGF-β) binding proteins (LTBPs) comprise a family of four extracellular matrix proteins, LTBPs −1 to −4 that are structurally similar to the fibrillins (Rifkin, 2005). Both the LTBPs and fibrillins contain multiple calcium-binding epidermal growth factor-like (CB-EGF) domains and signature domains with eight intramolecularly bound cysteine residues (8-Cys domain) (Rifkin, 2005). LTBP-1, −3, and −4, but not LTBP-2 nor the fibrillins, form covalent bonds with latent TGF-β (Rifkin, 2005).
TGF-βs 1, 2, and 3 are all synthesized as homodimeric proproteins with three intermolecular disulfide bonds (Annes et al., 2003). The TGF-β propeptide dimer is cleaved from the mature cytokine in the trans-Golgi, but the propeptide remains tightly bound to the cytokine by non-covalent interactions. This interaction of TGF-β and its propeptide prevents the growth factor from binding to its receptor. Therefore, the propeptide is referred to as the latency associated protein (LAP) and the TGF-β-LAP complex as the small latent complex (SLC). The release of TGF-β from its interaction with LAP, known as latent TGF-β activation, is a crucial step in the regulation of TGF-β activity (Annes et al., 2003). In vivo, the SLC is often bound to a LTBP by disulfide bonds between two cysteines from the LAP dimer and a pair of cysteines from the third 8-Cys LTBP domain (Chen et al., 2005; Gleizes et al., 1996; Munger et al., 1998; Saharinen et al., 1996). This SLC-LTBP complex is known as the large latent complex (LLC).
The LTBPs may direct and facilitate TGF-β action through several complimentary mechanisms. LTBPs enhance TGF-β secretion, as the formation of disulfide bonds with LAP engages what otherwise would be reactive cysteine residues and promotes proper LAP folding (Miyazono et al., 1991). In the extracellular environment, LTBPs interact with several matrix molecules, including fibronectin and fibrillin, thereby targeting latent TGF-β to specific locations for subsequent activation (Isogai et al., 2003; Taipale et al., 1996). LTBP-1 also directly participates in the activation of latent TGF-β by the integrin αvβ6 presumably by anchoring the LLC to the matrix, and allowing the integrin to apply force to the complex (Annes et al., 2004). This force is believed to distort the LAP, promoting release of the TGF-β.
The three TGF-β-binding LTBPs, LTBP-1, −3, and −4, display differences both in their matrix distribution and TGF-β binding. For example, LTBP-1 and −3 effectively bind all three isoforms of TGF-β, whereas LTBP-4 binds only TGF-β1 (Saharinen et al., 1996). LTBP-1 and −4 bind to fibrillin 1, but LTBP-3 does not (Isogai et al., 2003). The kinetics of LTBP assembly into the matrix differ with LTBP-1 incorporated most rapidly and LTBP-3 most slowly (Koli et al., 2005). In addition LTBP-1 and LTBP-4 exist as both long (LTBP-1L and LTBP-4L) and short (LTBP-1S and LTBP-4S) forms initiated from separate promoters ((Koski et al., 1999) and H. von Melchner – unpublished observations). It has been shown that LTBP-1L incorporates into extracellular matrix (ECM) more efficiently than LTBP-1S (Olofsson et al., 1995). However, biological significance of these different forms of LTBP-1 and LTBP-4 is not fully understood.
Phenotypes of mice with null or hypomorphic mutations in LTBP genes (Ltbp) have been interpreted as being consistent with decreased TGF-β activity. Thus, the heart outflow tract abnormalities in Ltbp1L−/− mice (Todorovic et al., 2007), the skeletal abnormalities in Ltbp3−/− (Dabovic et al., 2002) and Ltbp1−/− mice (Drews et al., 2008), and the pulmonary emphysema and the colorectal tumors in Ltbp4S−/− mice ((Sterner-Kock et al., 2002), and H. von Melchner, unpublished observations) are consistent with previously published data on genetically modified mouse models and human tumors with impaired TGF-β signaling (Choudhary et al., 2006; Erlebacher and Derynck, 1996; Filvaroff et al., 1999; Markowitz et al., 1995; Riggins et al., 1997; Yang et al., 2001). However, because of their incorporation into the matrix and their structural resemblance to the fibrillins, certain phenotypes in Ltbp mutant mice may represent the loss of a TGF-β-independent function. Indeed, not all effects of blocking LTBP in cell culture can be replicated by blocking TGF-β (Dallas et al., 1995). Moreover, LTBP-2 does not bind TGF-β, yet the Ltbp2−/− mutation is embryonic lethal, indicating an essential non-TGF-β-related role for this protein (Shipley et al., 2000).
Developmental abnormalities in Ltbp4S−/− mice are twofold: 1) a defective elastic fiber structure and 2) a strong impairment of terminal air-sac septation, first evident at the saccular stage of lung development (Sterner-Kock et al., 2002). Properly organized elastin at the tips of the growing alveolar septae is required for alveolar differentiation (Wendel et al., 2000) and this requirement may account for the defect in terminal lung septation in Ltbp4S−/− animals. However, as TGF-β is a regulator of matrix molecule expression, the elastin anomaly may be TGF-β-dependent. In addition, it has been reported that LTBP-4 binds only TGF-β1 (Saharinen et al., 1996), yet Tgfb1−/− mice have no obvious lung abnormalities (Kulkarni et al., 1993; Shull et al., 1992). This raises an apparent contradiction in interpreting the Ltbp4S−/− lung defects as a consequence of decreased TGF-β.
To clarify the cause of the alveolar septation and elastogenesis defects in Ltbp4S−/− lungs, we examined elastogenesis in lungs from wild type (WT) and Ltbp4S−/− mice at embryonic day (E) 14.5 to postnatal day (P) 7. We found that there was a defect in elastogenesis as early as E14.5–16.5 in the mutant animals in the lung alveolar walls, large airways and blood vessels. Contrary to what we expected, we found increased TGF-β signaling in Ltbp4S−/− lungs. Decreasing TGF-β improved septation of terminal air-sacs but did not reverse the defects in elastogenesis suggesting that alterations in TGF-β signaling and abnormal elastogenesis represent two separable functions of LTBP-4.
Materials and Methods
Drugs and antibodies
TβR1 inhibitor SB431542 was purchased from Sigma-Aldrich, (St. Louis, MO). Antibodies to P-Smad2 and Smad2/3 were purchased from Cell Signaling Technology (Danvers, MA). Anti TGF-β1 was purchased from R&D Systems (Minneapolis, MN).
Mice
The Ltbp4S−/− mice were previously described by Sterner-Kock et al. (Sterner-Kock et al., 2002). Tgfb2+/− mice were purchased from Jackson Labs (Bar Harbor, Maine). All mice were maintained on normal lab diet. For staged embryos, female and male mice were housed together overnight. Noon of the day of vaginal plug appearance was considered 0.5 dpc (days post coitum) or P0.5. Pregnant females were killed by asphyxiation by CO2 and cervical dislocation and the embryos were collected and placed immediately in 10% buffered formalin at room temperature. All procedures were conducted according to the regulations of the NYU Langone Medical Center IACUC.
Genotyping
Mice from Ltbp4S+/− × Ltbp4S+/− crosses were genotyped by PCR using reverse primers 3C7Wt: GGCTCATGCTTGAATGTTCAG and 3C7Tg: ATCATGCAAGCTGGTGGCTG specific for the mutated and the WT allele, respectively, and a common forward primer P3: CCAATCTTGCTTCTTTGCTG AGC. Mice from Tgfb2+/− × Tgfb2+/− crosses were genotyped using forward allele –specific primers for the WT, B2–6F: AATGTGCAGGATAATTGCTGC and the mutant, Neo-1L: CGACCACCAAGCGAAACATCGC, and a common reverse primer B2–6R: AACTCCATAGATATGGGGATGC.
Quantitative real time RT-PCR
RNA was extracted from freshly dissected lungs using Trizol (Invitrogen). Reverse transcription (RT) reactions were performed using 1 µg of RNA and Superscript III Reverse Transcriptase (Invitrogen) at 50° C for 60 minutes. The cDNA produced was used for quantitative real-time RT-PCR (Q-RT-PCR) analysis (Wang et al., 2006). Q-RT-PCR reactions were carried out with specific primers and the Quanti Fast SYBR Green PCR Kit (Qiagen) using an iCycler Thermal Cycler (Bio-Rad). The transcript expression for each target was quantified by comparing the threshold cycle (TC) with that of hypoxanthine guanine phosphoribosyl transferase using the comparative TC method. The primers used are shown in Supplemental Table III.
Histology and Immunohistochemistry
Mouse lungs were inflated with 10% buffered formalin (Sigma-Aldrich) at room temperature through the cannulated trachea under water pressure of 25 cm for day 7 and 15 cm for new-born and E 18.5 lungs. The tissues were fixed in 10% buffered formalin, processed and embedded in paraffin. Five-micrometer sections were used in all studies. For histological and histomorphometric analysis the sections were stained with hematoxilin and eosin (H&E)(Sigma). Elastin was stained using orcinol – new fuchsin technique (Sheehan and Hrapchak, 1980).
Immunohistochemistry with P-Smad2 antibody was performed following the manufacturers protocol. The staining was revealed using ABC Vector Elite Kit (Vector Laboratories, Burlingame, CA).
Western Blot Analysis
Western blot analysis was performed on lung extracts from P7 mice. Tissue was snap-frozen in liquid nitrogen, pulverized using mortar and pestle and resuspended in lysis buffer (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 1% TritonX-100, 1 mM glycerophosphate, 2.5 mM sodium pyrophosphate, 1 mM sodium orthovanadate) containing protease inhibitor cocktail (Roche, Indianapolis, IN). After 10 min incubation on ice, lysates were passed at least 5 times through a 22-gauge needle fitted to a syringe and thereafter centrifuged for 10 minutes at full speed in a micro centrifuge at 4°C. The supernatants were collected and protein concentrations were determined using Pierce BCA kit (Thermo Scientific, Rockford, IL). Equivalent amounts of protein from each sample, corresponding to 50–100 µḷ of lysate, were used for further analysis. Western blotting with P-Smad2 and Smad2/3 antibodies was performed according to manufacturer’s protocol. Immunoreactive bands were revealed using Pierce ECL Western Blotting Substrate (Thermo Scientific). Relative intensity of the bands was evaluated using Kodak 1D 3.5.4 software (Kodak Scientific Imaging System, Rockville, MD). The ratio of the intensity of P-Smad2 versus Smad2/3 bands in Ltbp4S−/− samples was normalized to the ratio calculated for the WT samples.
TβR1 inhibitor treatment
On 16.5 and 18.5 dpc pregnant Ltbp4S+/− females from Ltbp4S+/−× Ltbp4S+/− matings were injected intraperitoneally with 2 mg/kg of SB431542 dissolved in PBS. Control females were injected with PBS. The pups were sacrificed the first day after birth (P0.5), and the lungs were processed as described above.
Histomorphometric analysis
For the assessment of mean terminal sac diameter five lung sections were stained with H&E and 10–12 random fields were photographed under 20X magnification. 2–3 horizontal lines were drawn across each photographed field in areas without large airways or vessels and each intercept of the lines and terminal air-sac walls was counted. The number of lines was multiplied by 580 (which corresponds to the length of the line connecting opposite vertices in a 20x objective microscope field in µm) and divided by the number of intercepts to obtain the mean terminal air-sac diameter.
Transmission electron microscopy
For transmission electron microscopy of lung elastic fibers, lungs were perfused with ice-cold 3% gluteraldehyde in 0.1 M cacodylate buffer (pH 7.4). The left lobe was removed and placed in fresh fixative overnight. Samples were trimmed to 1.5 mm3 pieces and sequentially stained en bloc with 1% osmium tetroxide, 2% tannic acid and 2% uranyl acetate prior to dehydration and Epon embedding as previously described (Davis, 1993). Thin sections (60 nm) were placed on formvar-coated grids and counterstained with 7% methanolic uranyl acetate followed by lead citrate. Sections were viewed using a Tecnai 12 transmission electron microscope at 120 kV and images were digitally captured.
Results
Defective elastogenesis in Ltbp4S−/− lungs
In the initial report on Ltbp4S−/− mice, Sterner-Kock et al. (Sterner-Kock et al., 2002) described abnormal elastic fibrils in the lungs and intestines of 12-week-old mice. As it was not clear whether fragmented elastin was a result of degradation of preformed fibrils or a result of defective elastic fiber formation, we first characterized elastogenesis in the WT and Ltbp4S−/− lungs at P7 and P0.5.
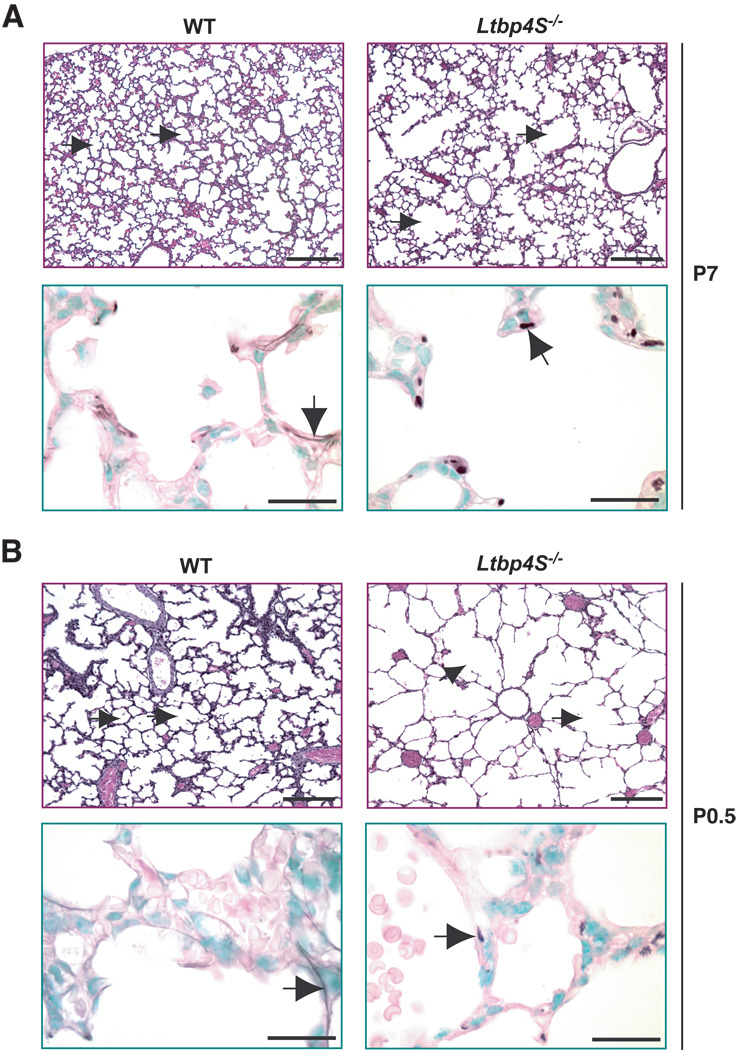
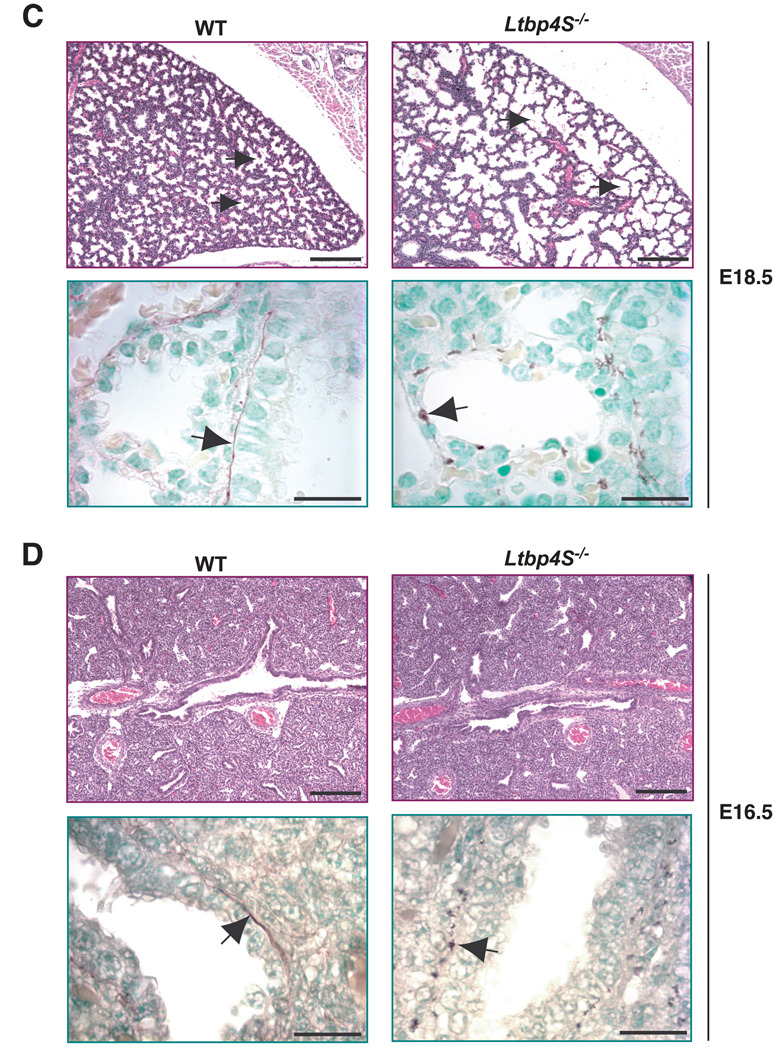
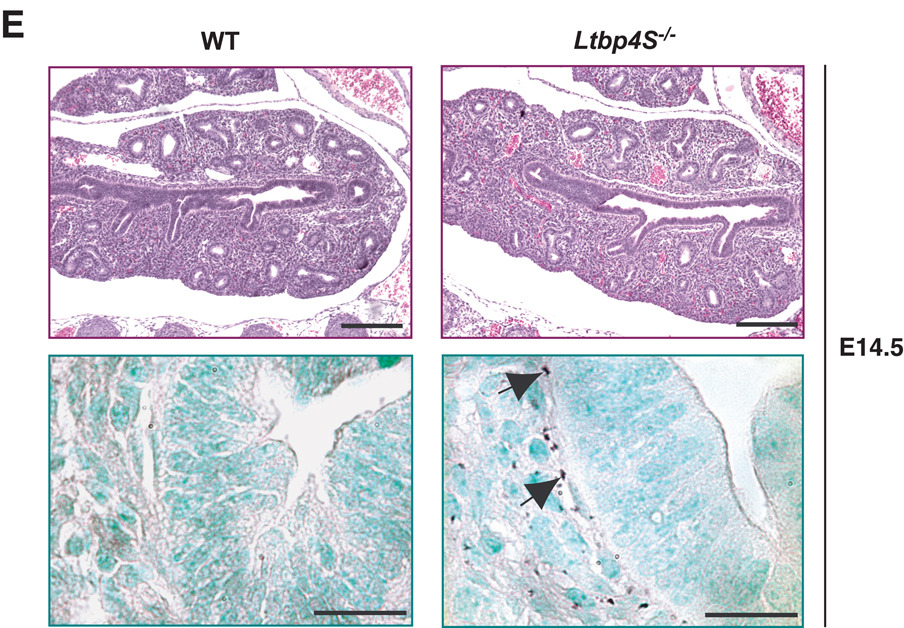
At P7, the impairment of terminal septation in Ltbp4S−/− lungs was obvious with a patch-like distribution of regions undergoing alveolarization interspersed with regions with large terminal air-sacs (Fig. 1A, upper panel). In contrast, alveolarization was uniform throughout WT lungs at this time. In the WT lung at P7, the elastin was assembled into fibrils in the alveolar walls and under the airway epithelium (Fig. 1A, lower panel). In the mutant lung, elastin was not organized into fibrils but rather appeared globular or fragmented (Fig.1A, lower panel). At P0.5, we observed uniform severe enlargement of terminal air-sacs in Ltbp4S−/− compared to WT lungs (Fig. 1B, upper panel). The elastic fibers in WT lungs at P0.5 were thinner, but the overall elastin distribution in the WT and Ltbp4S−/− lungs was similar to that observed at P7 (Fig. 1B, lower panel). Since the defects in lung development and elastin organization were already obvious in newborn Ltbp4S−/− mice, we also analyzed lungs during embryogenesis. At E18.5, air-sac enlargement was already apparent in Ltbp4S−/− compared to WT lungs (Fig. 1C, upper panel). The elastin organization resembled that seen in P0.5 lungs (Fig. 1C, lower panel). At E16.5, Ltbp4S−/− lungs could not be distinguished from WT lungs by histological analysis (Fig. 1D, upper panel). However, in WT lungs, we detected elastic fibers around the airways, including the bronchioles (Fig. 1D, lower panel), whereas at this time the elastin fiber ultrastructure in Ltbp4S−/− lungs was already abnormal with the elastin appearing as globules (Fig. 1D, lower panel). At E14.5, no elastin was detectable in the WT lungs using orcinol-new fucsin, which is probably indicative of fibrils too thin to be visualized by histological staining, since we did detect granules of elastin surrounding the bronchi in the Ltbp4S−/− lungs (Fig. 1E, lower panel). Together, our results indicate that Ltbp4S−/− lung septation is defective starting from E16.5–18.5 and suggest that the abnormality in elastin organization coincides with the beginning of elastogenesis in the lung.
Figure 1.
Defective terminal air-sac septation and elastic fiber formation in Ltbp4S−/− lungs. Upper panels show lung sections stained with H&E, lower panels present orcinol-new fucsin staining of elastin. The arrows in upper panels point to terminal air-sacs; the arrows in the lower panels point to elastin. A. P7 In WT lungs at P7 terminal air-sacs are divided into small units by the process of alveolarization, whereas in Ltbp4S−/− lungs alveolarization is not uniform yielding regions with large terminal air-sacs. Elastin in the WT alveolar walls appears fibrillar, whereas in the Ltbp4S−/− alveolar walls only globules of elastin were observed. B. P0.5 At P0.5, the terminal air-sacs in Ltbp4S−/− lungs are much larger than in WT lungs. The differences in elastin organization between WT and mutant lungs are similar to those illustrated in A. C. E18.5 The difference in terminal air-sac septation and in elastin organization between WT and Ltbp4S−/− lungs was already obvious at E18.5. D and E. E16.5 and E14.5 No differences in WT and Ltbp4S−/− lung histology were observed both at E16.5 and E14.5. However the differences in elastin organization were observed at E16.5 (D) and E14.5 (E). Bars: upper panels − 200 µm, lower panels − 20 µm.
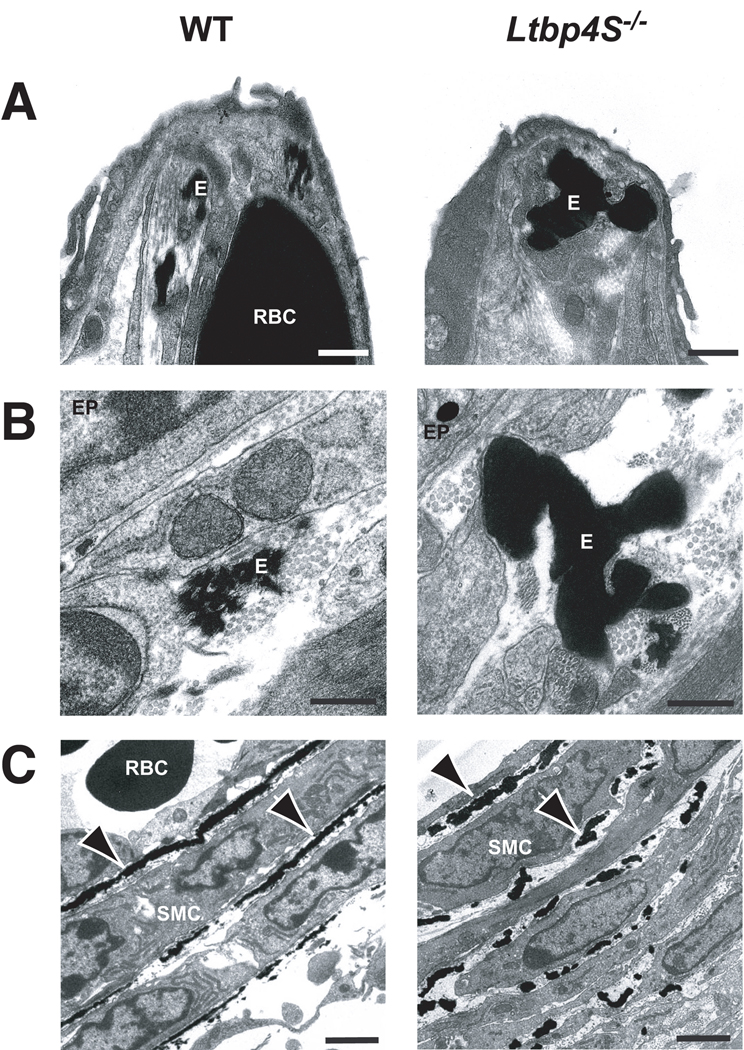
We next examined the elastin fibers in WT and mutant lungs using electron microscopy (EM) at P0.5 (Fig. 2). Differences were apparent in both the alveolae and the airways. In the mutant lung (Fig. 2A), the usual small collection of elastic fibers normally seen in the WT alveolar tips appeared abnormally large and often formed single, fused aggregates of elastin. Similar abnormalities in elastin fiber ultrastructure were observed subjacent to the airway epithelium (EP) (Fig. 2B). Defects in elastin organization were also obvious in the walls of pulmonary blood vessels (Fig. 2C). In WT vessels, elastin was organized into nearly continuous lamellae between the smooth muscle cells (SMCs), whereas in Ltbp4S−/− lungs, the lamellae were not well formed and appeared fragmented.
Figure 2.
Electron micrography of P0.5 WT and Ltbp4S−/− lungs. A Alveolar tips. B Airways. C Blood vessels. The lack of continuous lamellae in the mutant blood vessels is clear in C where there are essentially no ordered lamellae. E – elastin, RBC – red blood cell, SMC - smooth muscle cell, EP – epithelial cell. Arrows point to elastic lamellae in the walls of blood vessels. Bars: A −0.5 m, B - 0.2 µm, C - 2 µm.
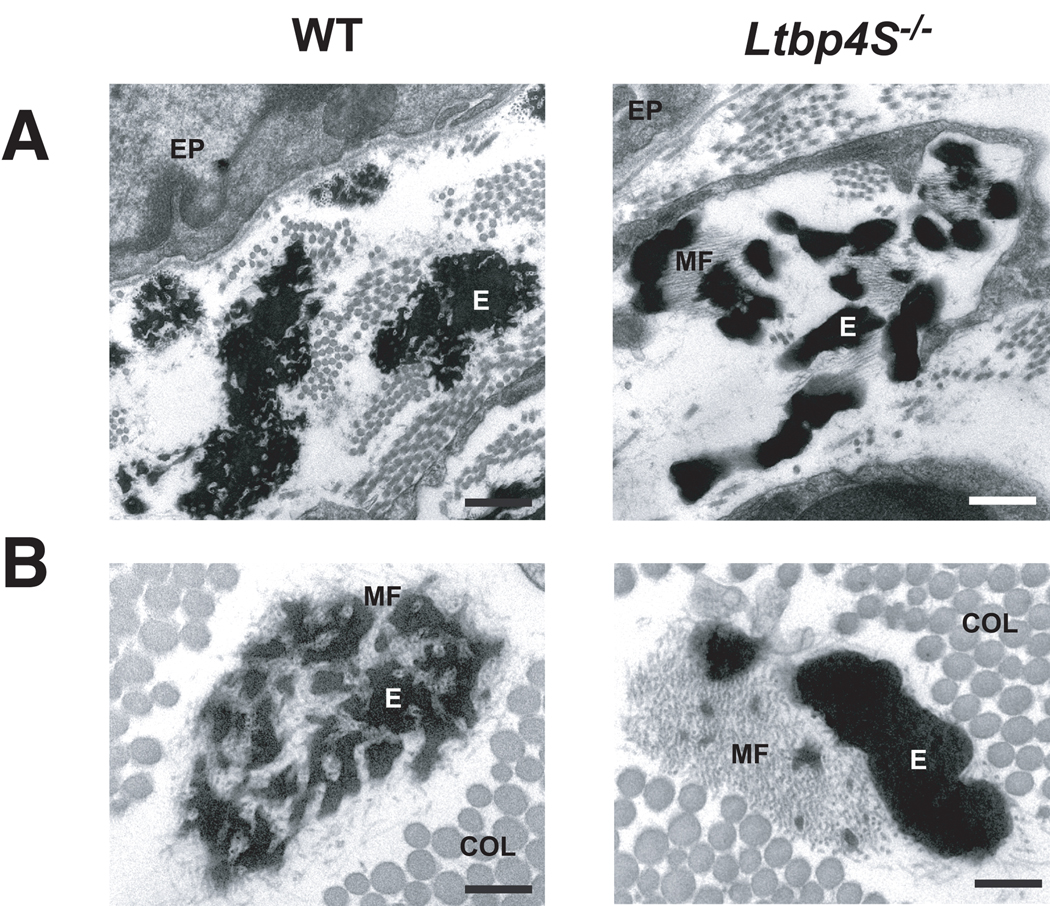
To determine if the defect in elastin organization persists in older animals, we also examined lung and skin from 1-month-old mice by EM. At one month of age, after the elastic fibers are fully developed, the defective assembly of elastin with respect to the microfibrils in Ltbp4S−/− lung was still obvious. In the WT mice, microfibrils are enmeshed in the elastin to form a lace-like structure (Fig. 3A), whereas in Ltbp4S−/− lung, large globules of elastin were localized next to the microfibrils with only a small amount of elastin integrated within the microfibril bundle. A similar abnormality in elastic fiber ultrastructure was observed in the dermis of Ltbp4S−/− mice demonstrating that the elastic fiber defect is not restricted to the lung (Fig. 3B).
Figure 3.
Ultrastructure of elastic fibers in 4-week-old WT and Ltbp4S−/− mice. A - airways, B- dermis. In WT tissues the microfibrils are enmeshed in the elastin, whereas in Ltbp4S−/− tissues large globules of elastin localize next to the microfibrils. COL- Collagen, EP – epithelial cell, E- elastin, MF- microfibrils. Bars: A - 0.5 µm, B – 0.2 µm.
We quantified the expression levels of a number of key proteins known to be involved with elastogenesis using Q-RT-PCR. We examined the mRNA levels for tropoelastin, fibrillin-1 and 2, fibulin-4 and 5, Lox and Loxl-1 (Supp. Table 2). None of the assayed RNA transcripts showed a significant difference between WT and Ltbp4S−/− lung samples at P7. At P0.5, the level of Loxl-1 was considerably higher in the mutant samples compared to WT, whereas other transcripts were expressed at similar levels. As differences in lung maturation are seen at day P7 as well as P0.5, the significance of the Loxl-1 difference is unclear. Therefore, the observed differences in Ltbp4S−/− lung elastin organization cannot be explained by decreased expression of major proteins known to be involved in elastic fiber assembly.
TGF-β levels in Ltbp4S−/− lungs
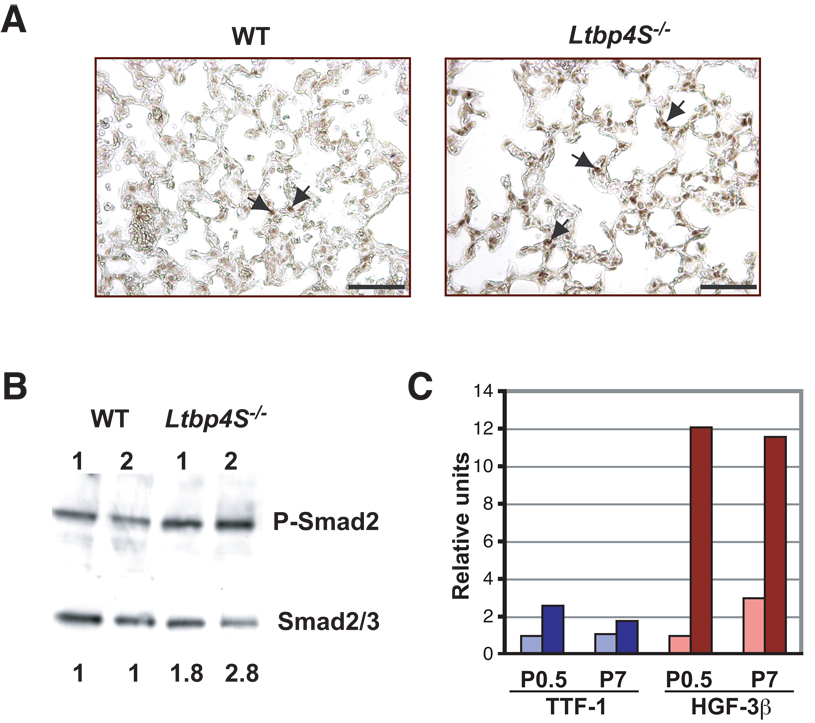
We next examined the level of TGF-β signaling in the WT and Ltbp4S−/− lungs. To assess the levels of TGF-β signaling in Ltbp4S−/− lungs, we characterized the tissue for the TGF-β signaling transducer P-Smad2 by immunohistochemical (IHC) and Western blot analysis. Upon TGF-β binding to its receptors, Smad2 is phosphorylated and is transported to the nucleus (Derynck and Miyazono, 2008). Therefore, both the ratio of P-Smad2 versus total Smad2 and the cellular distribution (cytosolic versus nuclear) of P-Smad2 are indicators of TGF-β signaling. Our IHC analysis using a P-Smad2 antibody on lung sections from 7-day-old mice indicated increased levels of P-Smad2 in Ltbp4S−/− compared to WT lungs, as the intensity of the stain in individual nuclei was greater in the mutant tissue as was the total number of stained nuclei (Fig 4A). To confirm and quantify the IHC data, Western blot analysis of proteins extracted from P7 Ltbp4S−/− and WT lungs was carried out. The blot was first probed with a P-Smad2 antibody and then, after stripping, with a Smad2/3 antibody. The bands were scanned, and after densitometry, we calculated the ratio of P-Smad2 to Smad2/3 (Fig. 4B). Our results showed that the level of TGF-β signaling in Ltbp4S−/− lungs, as indicated by the ratio of P-Smad2/Smad2/3, is higher than in the WT lungs, demonstrating increased levels of active TGF-β.
Figure 4.
Increased TGF-β signaling in Ltbp4S−/− lungs. A Immunohistochemistry with P-Smad2 antibody revealed a higher number of positive cell nuclei in Ltbp4S−/− lungs. The arrows point to the P-Smad2 positive nuclei. B Quantitative Western Blot analysis of P-Smad2 in the lungs from 2 WT and 2 Ltbp4S−/− P7 mice. The numbers at the bottom indicate the ratio of the intensity of P-Smad2 vs. Smad2/3 in the Ltbp4S−/− samples normalized to the P-Smad2 to Smad2/3 ratio in the WT samples. The ratio of the intensity of the P-Smad2 and the Smad2/3 bands was equivalent in both WT samples. The result shown is representative of four experiments using different animals. C Graphic representation of expression of TTF and HNF-3h in WT and mutant lungs. The expression of both genes was enhanced in Ltbp4S−/− mice indicating increased TGF-β levels. Bar: 5 µm.
We also examined the transcript levels of three TGF-β-responsive genes: Pai-1, Ctgf-1 and C-Myc (Supp. Table 3), whose expression levels are often assayed as indicators of TGF-β signaling (Coffey et al., 1988; Duncan et al., 1999; Flaumenhaft and Rifkin, 1991; Wu et al., 2007). At P0.5 a small decrease in expression of C-Myc was observed in Ltbp4S−/− compared to WT lungs, which is consistent with enhanced TGF-β signaling. However, there was a high degree of variability in expression levels of the TGF-β responsive gene Ctgf-1 in different lung samples of the same genotype, and the overall differences between Ltbp4S−/− and WT lungs were not significant. The expression of Pai-1 was decreased in Ltbp4S−/− lungs, which is consistent with decreased TGF-β signaling. Taken together Q-RT-PCR studies did not give a clear indication of a change in TGF-β levels in P0.5 Ltbp4S−/− lungs. At P7, expression levels of Pai-1, C-Myc, and Ctgf-1 were similar in WT and Ltbp4S−/− lungs (Supp. Table 3).
The transcription factors TTF-1 and HNF3β, which are expressed during lung development, are positively regulated by TGF-β1 (Zeng et al., 2001). We observed increased levels of both of these transcription factors in Ltbp4S−/− lungs at P0.5. At P7, TTF-1 expression still remained much higher in mutant lungs compared to WT, whereas the expression of HNF3β was a little less than 2 fold higher in Ltbp4S−/− than in WT lungs (Fig. 4C). These data are consistent with continued enhanced TGF-β signaling in Ltbp4S−/− lungs.
Our results suggested that the LTBP-4 deficit resulted in increased levels of active TGF-β in the lung. We reasoned that if excess TGF-β signaling caused the defect in septation of terminal air-sacs in Ltbp4S−/− lungs, we could normalize the mutant phenotype by lowering TGF-β levels. In order to decrease TGF-β activity in the developing mouse lung, we treated pregnant females from LtbpS4+/− × Ltbp4S+/− crosses with SB431542, a small molecule inhibitor of the TGF-β receptor I (TβRI / Alk 5). We examined the effects of SB431542 on lungs from newborn animals (P0.5), as the Ltbp4S−/− lung phenotype becomes heterogeneous as the animals age, making it difficult to quantify differences at later times. Morphometric studies assessing mean terminal air-sac diameter indicated a small but statistically significant increase (p<0.057) in terminal air-sac septation in the treated Ltbp4S−/− lungs, suggesting that an increase in TGF-β signaling contributed to the Ltbp4S−/− lung developmental defect (Supp. Fig. 1A). There was no difference between SB431542-treated and non-treated WT lungs indicating that decreasing TGF-β signaling with a short treatment of the TβR1 inhibitor did not affect normal lung development. The relatively small decrease in mean diameter of terminal air-sacs in Ltbp4S−/− lungs induced by SB431542 treatment prompted us to investigate whether a further reduction of TGF-β signaling would further improve septation.
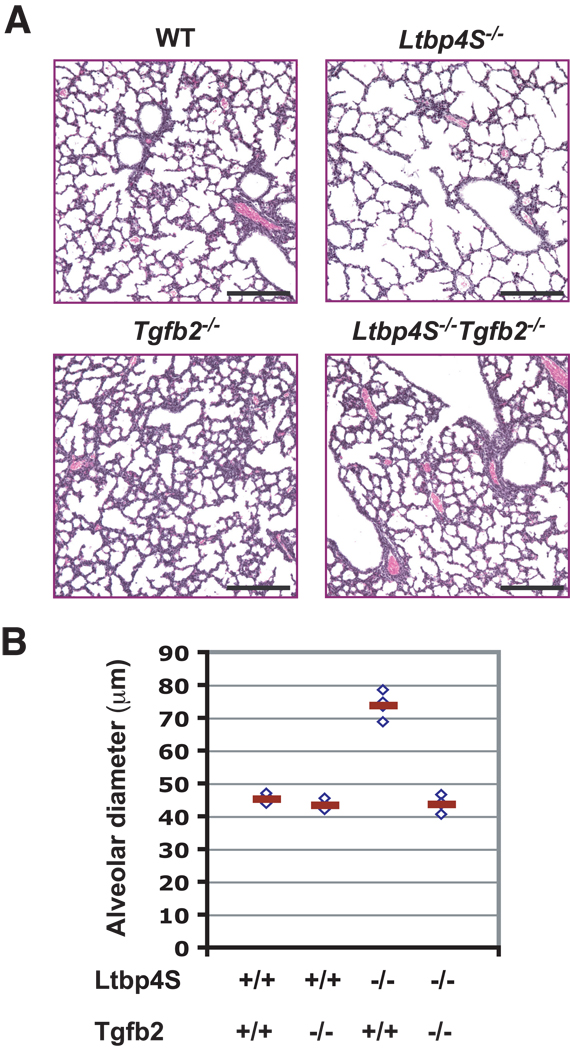
A more effective method to decrease TGF-β levels or signaling earlier during development and to avoid the potential toxicity of the chemical inhibitor is to cross Ltbp4S−/− mice with Tgfb−/− or Tgfr−/− animals. TβR1 is not specific for TGF-β, and both Tgfr1 and Tgfr2 null mutations are lethal early in development. Tgfb1 and Ltbp4 both map to chromosome 7, only 1.1 Mb apart, which makes the generation of Ltbp4S−/−;Tgfb1−/− mice by simple crossing of Ltbp4S+/− and Tgfb1+/− animals difficult. Tgfb3 expression in mouse embryonic lungs decreases in later stages of development and the transcript is not detectable by E 16.5 (Schmid et al., 1991), the stage of development that precedes the lung morphogenesis defect observed in the Ltbp4S−/− embryos. However, Tgfb2 is expressed at high levels in developing mouse lungs and its expression is increased at later stages of development (Schmid et al., 1991). In addition, several studies have indicated an important role of TGF-β2 in lung morphogenesis (Liu et al., 2000; Sanford et al., 1997). Therefore, we reasoned that in order to decrease overall TGF-β levels in lungs at the appropriate time, attenuating TGF-β2 would be the most effective genetic approach. Therefore, we crossed Ltbp4S+/− mice with Tgfb2+/− mice, and we examined the lungs from Ltbp4S−/−;Tgfb2+/− and Ltbp4S−/−;Tgfb2−/− animals. Visual examination of stained lung sections suggested that the loss of a single Tgfb2 allele had a small effect on lung septation (Supp. Fig. 1B). However, when we performed histomorphometric analysis of mean terminal air-sac diameter, we observed no significant differences between Ltbp4S−/−;Tgfb2+/− and Ltbp4S−/−;Tgfb2+/+ lungs. As ablation of one Tgfb2 allele might have been insufficient to produce a biologically significant decrease of TGF-β levels in Ltbp4S−/− lungs, we also examined Ltbp4S−/−;Tgfb2−/− lungs. Tgfb2−/− animals die at birth from multiple organ defects (Sanford et al., 1997). Therefore, we characterized Ltbp4S−/−;Tgfb2−/− lungs before birth, at a late stage of development, E18.5. At E18.5 there was an obvious defect in Ltbp4S−/− lung morphogenesis and elastogenesis (Fig. 5), however histological analysis of E18.5 Ltbp4S−/−;Tgfb2−/− lungs revealed a significant improvement in lung septation compared to Ltbp4S−/− lungs (Fig. 5A). Quantitation by histomorphometric analysis revealed a complete (100%) rescue of terminal air-sac development (Fig. 5B). These results imply that increased, rather than decreased, TGF-β is responsible for the impairment of lung development Ltbp4S−/− mice.
Figure 5.
Rescued lung development in Ltbp4S−/−;Tgfb2−/− lungs.A Histological analysis of WT, Ltbp4S−/−, Tgfb2−/− and Ltbp4S−/−;Tgfb2−/− indicated improved terminal air-sac septation in Ltbp4S−/−;Tgfb−/− compared to Ltbp4S−/− lungs. Bars: 200 µm B Histomorphometric studies showed a complete rescue of lung development in Ltbp4S−/−;Tgfb2−/− lungs, as measured by average terminal air-sac diameter. Four animals of each genotype were analyzed in this study. Bar: 200 µm.
Increased TGF-β signaling may result from either an increase in TGF-β synthesis or an increase in latent TGF-β activation. It has been reported that cultured Ltbp4S−/− lung fibroblasts express elevated levels of TGF-β2 and TGF-β3 (Koli et al., 2004). To assess TGF-β expression in vivo, we analyzed RNA extracted from WT and Ltbp4S−/− lungs by Q-RT-PCR. At P0.5 a small increase in expression of all three TGF-β isoforms was observed in the mutant lungs compared to control, but by P7 the differences were very small (Supp. Table 3).
Elastin Organization in Ltbp4S−/−;Tgfb2−/− Lungs
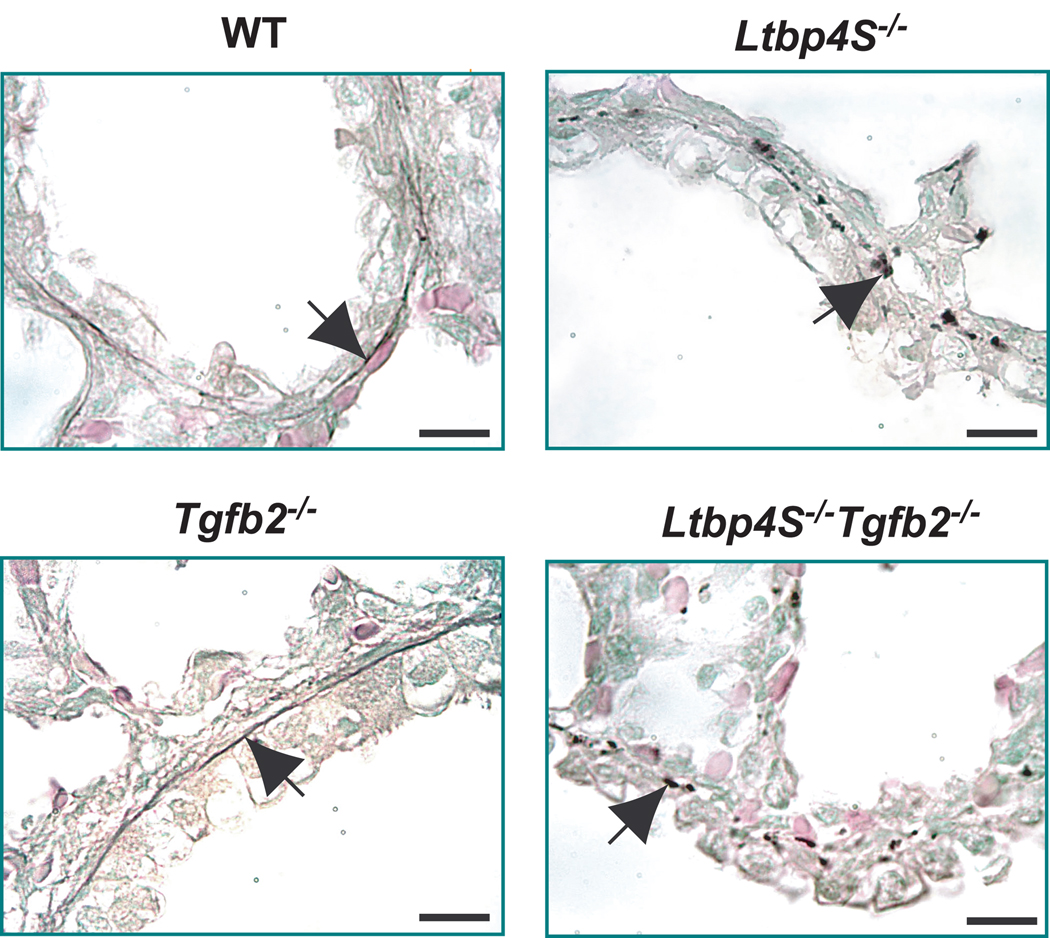
Finally, we examined whether the increased TGF-β levels associated with decreased alveologenesis might also be the cause for defective elastogenesis in Ltbp4S−/− mice and whether the reduction of TGF-β2 levels would improve elastin organization. We found that the elastic fiber organization in E18.5 Ltbp4S−/−;Tgfb2−/− lungs resembled that in Ltbp4S−/−;Tgfb2+/+ lungs (Fig. 6). Thus elastogenesis still appeared to be defective regardless of improved alveolar septation and presumably lower active TGF β levels. Therefore, we suggest that Ltbp-4 plays an important function in elastogenesis distinguishable from its role in the regulation of TGF-β tissue levels.
Figure 6.
Decreasing TGF-β2 levels in Ltbp4S−/− lungs does not rescue elastic fiber formation. Under conditions in which terminal air-sac septation was normalized (Fig. 5A and B), elastogenesis was still defective as indicated by the absence of elastic lamellae. Elastin in sections from P0.5 lungs was stained with orcinol - new fuchsin. The arrows point to the elastic fibers in WT and Tgfb2−/− lungs and to the globular elastin in Ltbp4S−/− and Ltbp4S−/−;Tgfb2−/− lungs. Bar: 10 µm.
Discussion
The experiments presented indicate that the loss of LTBP-4 synthesis in Ltbp4S−/− mouse lungs results in increased TGF-β signaling and an impairment of terminal air-sac development. Decreasing TGF-β expression or signaling in vivo either by genetic or by pharmacological intervention meliorated Ltbp4S−/− lung septation. Ltbp4S−/− mice also display an abnormality in lung elastogenesis apparent as early as E14.5–16.5, which appeared to be independent of TGF-β signaling, as normalization of terminal air-sac septation by decreasing TGF-β levels did not normalize the defects in elastic fiber structure.
Impairment of LTBP function either through null mutations or biochemical inhibition is believed to result in decreased TGF-β action because of faulty secretion, impaired localization, or lack of latent TGF-β activation (Annes et al., 2004; Dabovic et al., 2002; Koli et al., 2005; Miyazono et al., 1991; Sterner-Kock et al., 2002; Taipale et al., 1996; Todorovic et al., 2007). Although blockade of LTBP function may result in decreased signaling in some circumstances (Dabovic et al., 2002; Sterner-Kock et al., 2002; Todorovic et al., 2007), this might not always be the case. It is clear that the SLC can be activated in vivo in the absence of an LTBP, and if sufficient SLC is secreted, it can be activated to provide TGF-β signaling (Mazzieri et al., 2005). Moreover, in several systems, the pathological outcomes of interference with TGF-β localization have been attributed to excessive TGF-β signaling (Cohn et al., 2007; Habashi et al., 2006; Mazzieri et al., 2005; Neptune et al., 2003). The clearest example of this are transgenic mice that produce in the epidermis a truncated LTBP-1, which binds to SLC, but cannot localize to the ECM (Mazzieri et al., 2005). These animals have an early onset of the catagen stage of the hair cycle, concordant with increased TGF-β signaling (Mazzieri et al., 2005). A similar explanation has been proposed for the lung (Neptune et al., 2003), vascular (Habashi et al., 2006) and muscle (Cohn et al., 2007) abnormalities in patients and mice with Marfan syndrome, which is caused by mutations in fibrillin-1 (Dietz et al., 1991; Pereira et al., 1997). In this case, LLCs generate abnormally high levels of active TGF-β, perhaps because of improper targeting of the LLC to defective microfibrils (Mazzieri et al., 2005; Neptune et al., 2003). Decreasing total TGF-β by the administration of either neutralizing antibodies to TGF-β or drugs that decrease TGF-β signaling prevents the development of pathological changes in the affected tissues (Habashi et al., 2006; Mazzieri et al., 2005; Neptune et al., 2003). Therefore, excessive and/or ectopic activation of improperly localized latent TGF-β complexes can result in pathological processes (Habashi et al., 2006; Mazzieri et al., 2005; Neptune et al., 2003).
By decreasing TGF-β with pharmacological intervention using a small molecule inhibitor of TβR1, we improved terminal air-sac septation in Ltbp4S−/− lungs by a small degree, and with complete ablation of TGF-β2, we normalized lung morphology. Thus, for normalization of lung development in Ltbp4S−/− mice, TGF-β may have to be decreased early in embryogenesis and by a significant amount. The initiating events affecting Ltbp4S−/− lung development probably occur before we commenced inhibitor treatment at E16.5, and we may not have maintained sufficiently high levels of inhibitor using our protocols.
Our results indicate complexity in the regulation of TGF-β expression and action. Studies on cultured cells showed that LTBP-4 binds only TGF-β1 (Saharinen et al., 1996). Thus in the absence of LTBP-4, we expected to find a TGF-β1-dependent effect. We were unable to generate mice with compound mutations of TGF-β1 and LTBP-4 in order to specifically decrease TGF-β1 levels in Ltbp4S−/− tissues because Ltbp4 and Tgfb1 are only 1.1 Mb apart on the same chromosome. However, we found that Ltbp4S−/− lung development was rescued by decreasing TGF-β2. The loss of extracellular TGF-β1 and a decrease in the level of active TGF-β1 might stimulate the expression of TGF-β2 in lungs, as has been described for Ltbp4S−/− cells in culture (Koli et al., 2004). Indeed, we did observe small increases in TGF-β2 and TGF-β3 expression in the lungs of P0.5 mice. Many studies have shown that increased synthesis of TGF-β does not necessarily result in increased active TGF-β. As we observed an increase in TGF-β signaling in P7 Ltbp4S−/− lungs, we hypothesize that the apparent increase in TGF-β activity in Ltbp4S−/− lungs is a result of improper latent TGF-β activation, rather than increased TGF-β synthesis. However our results did indicate increased TGF-β signaling in Ltbp4S−/− lung. It is also possible that the TGF-β isoform elevated in Ltbp4S−/− lungs is TGF-β1 and that by eliminating TGF-β2 we normalized the overall levels of TGF-β1, 2, and 3 and consequently restored lung development. Another possibility is that LTBP-4 in vivo or in some cell types can bind not only TGF-β1 but also TGF-β2 and 3. Therefore, additional studies are required to elucidate the complex mechanism of elevated TGF-β signaling in the absence of LTBP-4.
Our results also indicate a critical function of LTBP-4 in elastic fiber assembly. The EM data show that in the absence of LTBP-4, the proper deposition of elastin within the microfibril bundles is impaired. The defect in elastic fiber structure in Ltbp4S−/− lungs resembles that observed in fibulin-5 (Fib-5) null mice and in patients with mutations in the Fib-5 gene (Hu et al., 2006; Yanagisawa et al., 2002). Fib-5 interacts with fibrillin-1 and with tropoelastin and is essential for elastic fiber formation (Wachi et al., 2008; Zheng et al., 2007). In cell culture LTBP-2, which cannot bind TGF-β, can interact with Fib-5 and regulate the deposition of Fib-5 on microfibrils (Hirai et al., 2007). The significance of this interaction in vivo could not be addressed as Ltbp2−/− mice die at an early stage of development, preceding the beginning of elastogenesis (Shipley et al., 2000). LTBP-4 interacts with both fibrillin-1 and −2 and is deposited on microfibrils (Isogai et al., 2003). It is not known whether LTBP-4 interacts with fibulins and elastin, but we propose that this may be an important function of LTBP-4. Further experiments are required to reveal the molecular interactions of LTBP-4 with proteins involved in elastogenesis and to elucidate LTBP-4 function in elastic-fiber formation.
In summary, our results indicate that LTBP-4 has a dual role in lung development by regulating TGF-β activity and elastic fiber formation. Our genetic and EM data strongly imply that LTBP-4 plays a fundamental role in elastogenesis, independent of its function in regulating TGF-β bioavailability. Whether this is influenced by secondary matrix turnover abnormalities induced by TGF-β remains to be established.
Supplementary Material
Acknowledgments
Contract grant sponsor: NIH; Contract grant numbers CA034282 (to DBR) and AR49698 (to DBR, HCD, and FR).
Contract grant sponsor: Canadian Institutes of Health; Contract grant numbers MOP57663 and MOP86713 to ECD. ECD is a Canada Research Chair
References
- Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165(5):723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116(Pt 2):217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ali T, Todorovic V, O’Leary JM, Kristina Downing A, Rifkin DB. Amino acid requirements for formation of the TGF-beta-latent TGF-beta binding protein complexes. J Mol Biol. 2005;345(1):175–186. doi: 10.1016/j.jmb.2004.10.039. [DOI] [PubMed] [Google Scholar]
- Choudhary B, Ito Y, Makita T, Sasaki T, Chai Y, Sucov HM. Cardiovascular malformations with normal smooth muscle differentiation in neural crest-specific type II TGFbeta receptor (Tgfbr2) mutant mice. Dev Biol. 2006;289(2):420–429. doi: 10.1016/j.ydbio.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Coffey RJ, Jr, Bascom CC, Sipes NJ, Graves-Deal R, Weissman BE, Moses HL. Selective inhibition of growth-related gene expression in murine keratinocytes by transforming growth factor beta. Mol Cell Biol. 1988;8(8):3088–3093. doi: 10.1128/mcb.8.8.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn RD, van Erp C, Habashi JP, Soleimani AA, Klein EC, Lisi MT, Gamradt M, apRhys CM, Holm TM, Loeys BL, Ramirez F, Judge DP, Ward CW, Dietz HC. Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nat Med. 2007;13(2):204–210. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabovic B, Chen Y, Colarossi C, Obata H, Zambuto L, Perle MA, Rifkin DB. Bone abnormalities in latent TGF-[beta] binding protein (Ltbp)-3-null mice indicate a role for Ltbp-3 in modulating TGF-[beta] bioavailability. J Cell Biol. 2002;156(2):227–232. doi: 10.1083/jcb.200111080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas SL, Miyazono K, Skerry TM, Mundy GR. Dual role for the latent transforming growth factor-β binding protein in storage of latent TGF-β in the extracellular matrix and as a structural matrix protein. J Cell Biol. 1995;131(2):539–549. doi: 10.1083/jcb.131.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EC. Smooth muscle cell to elastic lamina connections in developing mouse aorta. Role in aortic medial organization. Lab Invest. 1993;68(1):89–99. [PubMed] [Google Scholar]
- Derynck R, Miyazono K. The TGF-β Family. Cold Spring Harbor Laboratory Press; 2008. 520 pp. [Google Scholar]
- Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352(6333):337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- Drews F, Knobel S, Moser M, Muhlack KG, Mohren S, Stoll C, Bosio A, Gressner AM, Weiskirchen R. Disruption of the latent transforming growth factor-beta binding protein-1 gene causes alteration in facial structure and influences TGF-beta bioavailability. Biochim Biophys Acta. 2008;1783(1):34–48. doi: 10.1016/j.bbamcr.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Duncan MR, Frazier KS, Abramson S, Williams S, Klapper H, Huang X, Grotendorst GR. Connective tissue growth factor mediates transforming growth factor beta-induced collagen synthesis: down-regulation by cAMP. Faseb J. 1999;13(13):1774–1786. [PubMed] [Google Scholar]
- Erlebacher A, Derynck R. Increased expression of TGF-beta 2 in osteoblasts results in an osteoporosis-like phenotype. J Cell Biol. 1996;132(1–2):195–210. doi: 10.1083/jcb.132.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filvaroff E, Erlebacher A, Ye J, Gitelman SE, Lotz J, Heillman M, Derynck R. Inhibition of TGF-beta receptor signaling in osteoblasts leads to decreased bone remodeling and increased trabecular bone mass. Development. 1999;126(19):4267–4279. doi: 10.1242/dev.126.19.4267. [DOI] [PubMed] [Google Scholar]
- Flaumenhaft R, Rifkin DB. Extracellular matrix regulation of growth factor and protease activity. Curr Opin Cell Biol. 1991;3(5):817–823. doi: 10.1016/0955-0674(91)90055-4. [DOI] [PubMed] [Google Scholar]
- Gleizes PE, Beavis RC, Mazzieri R, Shen B, Rifkin DB. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-beta binding protein-1 that mediates bonding to the latent transforming growth factor-beta 1. J Biol Chem. 1996;271:29891–29896. doi: 10.1074/jbc.271.47.29891. [DOI] [PubMed] [Google Scholar]
- Habashi JP, Judge DP, Holm TM, Cohn RD, Loeys BL, Cooper TK, Myers L, Klein EC, Liu G, Calvi C, Podowski M, Neptune ER, Halushka MK, Bedja D, Gabrielson K, Rifkin DB, Carta L, Ramirez F, Huso DL, Dietz HC. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312(5770):117–121. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M, Horiguchi M, Ohbayashi T, Kita T, Chien KR, Nakamura T. Latent TGF-beta-binding protein 2 binds to DANCE/fibulin-5 and regulates elastic fiber assembly. Embo J. 2007;26(14):3283–3295. doi: 10.1038/sj.emboj.7601768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Loeys BL, Coucke PJ, De Paepe A, Mecham RP, Choi J, Davis EC, Urban Z. Fibulin-5 mutations: mechanisms of impaired elastic fiber formation in recessive cutis laxa. Hum Mol Genet. 2006;15(23):3379–3386. doi: 10.1093/hmg/ddl414. [DOI] [PubMed] [Google Scholar]
- Isogai Z, Ono RN, Ushiro S, Keene DR, Chen Y, Mazzieri R, Charbonneau NL, Reinhardt DP, Rifkin DB, Sakai LY. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J Biol Chem. 2003;278(4):2750–2757. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- Koli K, Hyytiainen M, Ryynanen MJ, Keski-Oja J. Sequential deposition of latent TGF-beta binding proteins (LTBPs) during formation of the extracellular matrix in human lung fibroblasts. Exp Cell Res. 2005;310(2):370–382. doi: 10.1016/j.yexcr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Koli K, Wempe F, Sterner-Kock A, Kantola A, Komor M, Hofmann WK, von Melchner H, Keski-Oja J. Disruption of LTBP-4 function reduces TGF-{beta} activation and enhances BMP-4 signaling in the lung. J Cell Biol. 2004;167(1):123–133. doi: 10.1083/jcb.200403067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski C, Saharinen J, Keski-Oja J. Independent promoters regulate the expression of two amino terminally distinct forms of latent transforming growth factor-beta binding protein-1 (LTBP-1) in a cell type-specific manner. J Biol Chem. 1999;274(46):32619–32630. doi: 10.1074/jbc.274.46.32619. [DOI] [PubMed] [Google Scholar]
- Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90(2):770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Tseu I, Wang J, Tanswell K, Post M. Transforming growth factor beta2, but not beta1 and beta3, is critical for early rat lung branching. Dev Dyn. 2000;217(4):343–360. doi: 10.1002/(SICI)1097-0177(200004)217:4<343::AID-DVDY2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268(5215):1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Mazzieri R, Jurukovski V, Obata H, Sung J, Platt A, Annes E, Karaman-Jurukovska N, Gleizes PE, Rifkin DB. Expression of truncated latent TGF-{beta}-binding protein modulates TGF-{beta} signaling. J Cell Sci. 2005;118(Pt 10):2177–2187. doi: 10.1242/jcs.02352. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991;10(5):1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Huang XZ, Kawakatsu H, Griffiths MJD, Dalton SL, Wu JF, Pittet JF, Kaminiski N, Garat C, Matthay, Rifkin DB, Sheppard D. The integrin αvβ6 binds and activates latent TGFb1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1998;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY, Dietz HC. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33(3):407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- Olofsson A, Ichijo H, Moren A, ten Dijke P, Miyazono K, Heldin CH. Efficient association of an amino-terminally extended form of human latent transforming growth factor-beta binding protein with the extracellular matrix. J Biol Chem. 1995;270(52):31294–31297. doi: 10.1074/jbc.270.52.31294. [DOI] [PubMed] [Google Scholar]
- Pereira L, Andrikopoulos K, Tian J, Lee SY, Keene DR, Ono R, Reinhardt DP, Sakai LY, Biery NJ, Bunton T, Dietz HC, Ramirez F. Targetting of the gene encoding fibrillin-1 recapitulates the vascular aspect of Marfan syndrome. Nature Genetics. 1997;17(2):218–222. doi: 10.1038/ng1097-218. [DOI] [PubMed] [Google Scholar]
- Rifkin DB. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J Biol Chem. 2005;280(9):7409–7412. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- Riggins GJ, Kinzler KW, Vogelstein B, Thiagalingam S. Frequency of Smad gene mutations in human cancers. Cancer Res. 1997;57(13):2578–2580. [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-beta with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15(2):245–253. [PMC free article] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124(13):2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid P, Cox D, Bilbe G, Maier R, McMaster GK. Differential expression of TGF beta 1, beta 2 and beta 3 genes during mouse embryogenesis. Development. 1991;111(1):117–130. doi: 10.1242/dev.111.1.117. [DOI] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. Theory and practice of histotechnology. St.Louis: Mosby. 1980 [Google Scholar]
- Shipley JM, Mecham RP, Maus E, Bonadio J, Rosenbloom J, McCarthy RT, Baumann ML, Frankfater C, Segade F, Shapiro SD. Developmental expression of latent transforming growth factor beta binding protein 2 and its requirement early in mouse development. Mol Cell Biol. 2000;20(13):4879–4887. doi: 10.1128/mcb.20.13.4879-4887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, Annunziata N, Doetschman T. Targeted disruption of the mouse transforming growth factor-β1 gene results in mulitfocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner-Kock A, Thorey IS, Koli K, Wempe F, Otte J, Bangsow T, Kuhlmeier K, Kirchner T, Jin S, Keski-Oja J, von Melchner H. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev. 2002;16(17):2264–2273. doi: 10.1101/gad.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Saharinen J, Hedman K, Keski-Oja J. Latent transforming growth factor-β1 and its binding protein are components of extracellular matrix microfibrils. J Histochem Cytochem. 1996;44(8):875–889. doi: 10.1177/44.8.8756760. [DOI] [PubMed] [Google Scholar]
- Todorovic V, Frendewey D, Gutstein DE, Chen Y, Freyer L, Finnegan E, Liu F, Murphy A, Valenzuela D, Yancopoulos G, Rifkin DB. Long form of latent TGF-{beta} binding protein 1 (Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development. 2007;134(20):3723–3732. doi: 10.1242/dev.008599. [DOI] [PubMed] [Google Scholar]
- Wachi H, Nonaka R, Sato F, Shibata-Sato K, Ishida M, Iketani S, Maeda I, Okamoto K, Urban Z, Onoue S, Seyama Y. Characterization of the molecular interaction between tropoelastin and DANCE/fibulin-5. J Biochem. 2008;143(5):633–639. doi: 10.1093/jb/mvn014. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39(4):356–362. doi: 10.1016/j.ymeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Wendel DP, Taylor DG, Albertine KH, Keating MT, Li DY. Impaired distal airway development in mice lacking elastin. Am J Respir Cell Mol Biol. 2000;23(3):320–326. doi: 10.1165/ajrcmb.23.3.3906. [DOI] [PubMed] [Google Scholar]
- Wu S, Peng J, Duncan MR, Kasisomayajula K, Grotendorst G, Bancalari E. ALK-5 mediates endogenous and TGF-beta1-induced expression of connective tissue growth factor in embryonic lung. Am J Respir Cell Mol Biol. 2007;36(5):552–561. doi: 10.1165/rcmb.2006-0320OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Davis EC, Starcher BC, Ouchi T, Yanagisawa M, Richardson JA, Olson EN. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature. 2002;415(6868):168–171. doi: 10.1038/415168a. [DOI] [PubMed] [Google Scholar]
- Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153(1):35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Gray M, Stahlman MT, Whitsett JA. TGF-beta1 perturbs vascular development and inhibits epithelial differentiation in fetal lung in vivo. Dev Dyn. 2001;221(3):289–301. doi: 10.1002/dvdy.1140. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Davis EC, Richardson JA, Starcher BC, Li T, Gerard RD, Yanagisawa H. Molecular analysis of fibulin-5 function during de novo synthesis of elastic fibers. Mol Cell Biol. 2007;27(3):1083–1095. doi: 10.1128/MCB.01330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.