Abstract
BACKGROUND
Modern genotyping platforms permit a systematic search for inherited components of complex diseases. We performed a joint analysis of two genomewide association studies of coronary artery disease.
METHODS
We first identified chromosomal loci that were strongly associated with coronary artery disease in the Wellcome Trust Case Control Consortium (WTCCC) study (which involved 1926 case subjects with coronary artery disease and 2938 controls) and looked for replication in the German MI [Myocardial Infarction] Family Study (which involved 875 case subjects with myocardial infarction and 1644 controls). Data on other single-nucleotide polymorphisms (SNPs) that were significantly associated with coronary artery disease in either study (P<0.001) were then combined to identify additional loci with a high probability of true association. Genotyping in both studies was performed with the use of the GeneChip Human Mapping 500K Array Set (Affymetrix).
RESULTS
Of thousands of chromosomal loci studied, the same locus had the strongest association with coronary artery disease in both the WTCCC and the German studies: chromosome 9p21.3 (SNP, rs1333049) (P=1.80×10−14 and P=3.40×10−6, respectively). Overall, the WTCCC study revealed nine loci that were strongly associated with coronary artery disease (P<1.2×10−5 and less than a 50% chance of being falsely positive). In addition to chromosome 9p21.3, two of these loci were successfully replicated (adjusted P<0.05) in the German study: chromosome 6q25.1 (rs6922269) and chromosome 2q36.3 (rs2943634). The combined analysis of the two studies identified four additional loci significantly associated with coronary artery disease (P<1.3×10−6) and a high probability (>80%) of a true association: chromosomes 1p13.3 (rs599839), 1q41 (rs17465637), 10q11.21 (rs501120), and 15q22.33 (rs17228212).
CONCLUSIONS
We identified several genetic loci that, individually and in aggregate, substantially affect the risk of development of coronary artery disease.
Coronary artery disease and its main complication, myocardial infarction, are leading causes of death and disability worldwide.1 Lifestyle and environmental factors play an important role in their development.2 In addition, these complex diseases cluster in families, suggesting a substantial genetic cause.3 Despite extensive exploration of many genes, strong evidence of a molecular genetic association with coronary artery disease or myocardial infarction remains to be obtained.
The recent development of high-density genotyping arrays provides unprecedented resolution for whole-genome assessment of variants associated with common diseases.4 Using the GeneChip Human Mapping 500K Array Set (Affymetrix), which simultaneously types approximately 500,000 genetic variants, the Wellcome Trust Case Control Consortium (WTCCC) recently reported on an analysis of data from approximately 2000 people (and a shared set of 3000 controls) for each of seven complex diseases, including coronary artery disease.5 Several loci showed strong associations with coronary artery disease, but the large number of statistical tests poses the challenge of discriminating between true and false associations. By providing a detailed analysis of the WTCCC data in conjunction with data from another large genomewide association study, the German MI [Myocardial Infarction] Family Study, we sought robust evidence for associations of genetic loci with the risk of coronary artery disease and myocardial infarction.
METHODS
SUBJECTS
All participants in both studies were of white European origin. Detailed descriptions of recruitment and ascertainment in both studies are given in the Supplementary Appendix, available with the full text of this article at www.nejm.org. Local ethics committees approved the study protocols, and all participants gave written informed consent.
WTCCC Study
The 1988 case subjects in the WTCCC study had a history of either myocardial infarction or coronary revascularization before the age of 66 years, as well as a strong family history of coronary artery disease.6,7 Two independent control groups were studied: 1504 controls from the British 1958 Birth Cohort and 1500 controls selected from a sample of blood donors recruited as part of the WTCCC project.5
German MI Family Study
The 875 case subjects in the German MI Family Study were persons who had myocardial infarction before the age of 60 years and at least one first-degree relative with premature coronary artery disease.8,9 The 1644 controls were selected from a well-characterized random sample of German residents, stratified according to sex and age.10
GENOTYPING
All analyses were performed with the use of the GeneChip Human Mapping 500K Array Set, including a StyI and a NspI chip. Details on genotype-calling algorithms, quality criteria (at the level of both the individual and the SNP), and validation steps are described in the Supplementary Appendix. This left 377,857 SNPs in 1926 case subjects and 2938 controls in the WTCCC study and 272,602 SNPs in 870 and 772 case subjects for the StyI chip and the NspI chips, respectively, and from 1644 controls for both chips in the German study.
STATISTICAL ANALYSIS
Before performing the genetic analyses, we examined the data from each cohort individually for population substructure and ascertained that such variation was negligible in both studies (see the Supplementary Appendix). We then undertook three types of genetic analysis. First, after identifying the SNPs with the strongest associations with coronary artery disease in the WTCCC sample, we sought to replicate these loci in the German study (primary analysis). Second, we combined data on other SNPs for which there was evidence of an association in either study, to identify additional loci with a low probability of being falsely positive (combined analysis). Third, we examined the data from both studies for associations with coronary artery disease among SNPs in genes reported to be associated with this disease (candidate-gene analysis).
Primary Analysis
All autosomal SNPs that showed evidence of a significant association with coronary artery disease in the WTCCC study (P<0.001 with use of the two-sided Cochran–Armitage trend test) were assessed for the false-positive-report probability (FPRP).11 (The rationale for the FPRP calculations and the assumptions underlying them are given in the Supplementary Appendix.) A small FPRP suggests that the association of a SNP is unlikely to be a false positive result. A SNP with an FPRP of less than 0.5 (i.e., one for which the chance of a truly positive association was greater than 50%), as well as SNPs within 100 kb in either direction of this SNP, also with P values of less than 0.001, were considered to represent a single locus. Formal replication testing of such loci was then performed in the German study with the use of the lead SNP (defined as the SNP with the lowest FPRP) for each locus and adjustment for multiple testing. A P value of less than 0.05 with use of the two-sided Cochran–Armitage trend test was considered to indicate statistical significance. Haplotype analysis was carried out on replicated loci, as described in the Supplementary Appendix. The power for replication in the German study was estimated for each locus for an odds ratio for myocardial infarction of 1.25 per allele and an adjusted significance level of 0.05.
Combined Analysis
All SNPs that showed evidence of a significant association (P<0.001) with coronary artery disease in the WTCCC study or with myocardial infarction in the German study were combined to assess their combined FPRP. Pooled odds ratios for the risk allele and 95% confidence intervals were calculated within each stratum, according to the study.
Candidate-Gene Analysis
Candidate genes were identified as described in the Supplementary Appendix. On the GeneChip array, we distinguished between SNPs that were identical to those identified in previous studies as showing an association with coronary artery disease and SNPs that were in complete or near-complete linkage disequilibrium with those identified in previous studies as showing an association with coronary artery disease. We used either of two measures of linkage disequilibrium — an r2 value of 0.8 or more or a disequilibrium coefficient (D’) of 0.9 or more — on the basis of data from the International HapMap Project.12
Population Attributable Fractions
Population attributable fractions were estimated in the German study with the use of the lead SNP from each of the three replicated loci. Adjustments were made for age, sex, the interaction between age and sex, the Prospective Cardiovascular Munster (PROCAM) study score, and the Framingham risk score. Further details are given in the Supplementary Appendix.
Data Access
The WTCCC data are accessible at www.wtccc.org.uk/info/summary_stats.shtml. The German MI Family Study data are available to academic researchers in a Web-based database (www.cardiogenics.imbs-luebeck.de).
RESULTS
CHARACTERISTICS OF THE PARTICIPANTS
In both the WTCCC and the German studies, the case subjects were young (mean age at first event, approximately 50 years) and had a strong familial basis for their disease (Table 1).
Table 1. Baseline Characteristics of Case Subjects in the WTCCC Study and the German MI Family Study*.
| Characteristic | WTCCC Study (N = 1926) | German Study (N = 875) |
|---|---|---|
| Sex — no. (%) | ||
| Male | 1527 (79.3) | 591 (67.5) |
| Female | 399 (20.7) | 284 (32.5) |
| Mean age at first event—yr | ||
| Men | 49.4±7.7 | 49.0±7.4 |
| Women | 51.0±7.4 | 52.8±8.2 |
| Mean age at recruitment—yr | 60.1±8.1 | 58.1±8.7 |
| First event—no. (%) | ||
| MI | 1377 (71.5) | 875 (100.0) |
| PCI | 202 (10.5) | |
| CABG | 347 (18.0) | |
| Family history of CAD—no. (%)† | ||
| Father affected | 570 (29.6) | 151 (17.3) |
| Mother affected | 302 (15.7) | 114 (13.0) |
| Both parents affected | 186 (9.7) | 49 (5.6) |
| Sibling affected | 1464 (76.0) | 789 (90.2) |
| Cardiovascular risk factor—no. (%)‡ | ||
| Former or current smoker | 1460 (75.8) | 615 (70.3) |
| Diabetes mellitus | 211 (11.0) | 108 (12.3) |
| Hypertension | 822 (42.7) | 757 (86.5) |
| Hyperlipidemia | 1542 (80.0) | 666 (76.1) |
| Obesity (body-mass index >30) | 448 (23.3) | 197 (22.5) |
| Receipt of lipid-lowering drugs—no. (%) | 1429 (74.2) | 575 (65.7) |
Plus–minus values are means ±SD. Details on the characteristics of controls for each study are given in the Methods section and the Supplementary Appendix. MI denotes myocardial infarction, PCI percutaneous coronary intervention, CABG coronary-artery bypass grafting, and CAD coronary artery disease.
Parental history of CAD was based on reported history; sibling history of CAD was validated by direct examination of records.
Diabetes, hypertension, and hyperlipidemia were identified as risk factors on the basis of the meeting of diagnostic criteria or the receipt of treatment for these conditions. The body-mass index is the weight in kilograms divided by the square of the height in meters.
PRIMARY ANALYSIS
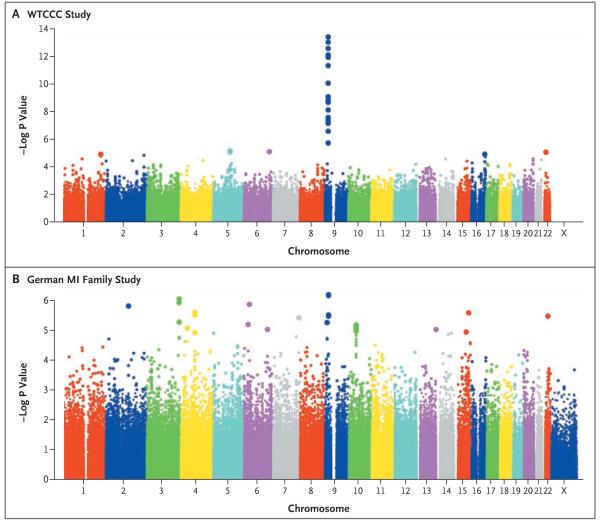
The distribution of P values for the association of SNPs with coronary artery disease in the WTCCC study and with MI in the German Study, according to chromosome, is shown in Figure 1. In the WTCCC study, 396 SNPs were significantly associated with coronary artery disease (P<0.001). Thirty of these SNPs clustering in nine chromosomal regions met the predefined criterion of an FPRP of less than 0.5 (Table 1 in the Supplementary Appendix). We tested these nine loci for a significant association with myocardial infarction in the German study. Three of the loci — chromosomes 9p21.3, 6q25.1, and 2q36.3 — had such an association, even after adjustment for multiple testing for nine loci (Table 2).
Figure 1. Signal-Intensity Plots Showing the Association of Single-Nucleotide Polymorphisms (SNPs) with Coronary Artery Disease or Myocardial Infarction in the Genomewide Association Analysis.
The −log P values are for the association of each SNP with coronary artery disease or myocardial infarction, from two-sided Cochran–Armitage tests for trend. Only SNPs of sufficient quality (see the Supplementary Appendix) are shown. The signal-intensity plots of all SNPs with significant associations (P<0.001) were visually inspected by two independent reviewers in each study to rule out possible artifactual results due to miscalling of genotypes (see Supplementary Appendix). Within each chromosome shown on the x axis, the data are plotted from the p-ter end. The y-axis scale for associations in the WTCCC study (Panel A) differs from the scale for the German MI Family Study (Panel B).
Table 2. Loci from the WTCCC Study with Significant Associations with Coronary Artery Disease That Were Replicated in the German MI Family Study*.
| Chromosome | Lead SNP | Minor Allele in Controls | Risk Allele | Data | Frequency of Minor Allele | Odds Ratio for Risk Allele (95% CI) | Population Attributable Fraction | P Value | |
|---|---|---|---|---|---|---|---|---|---|
| Case Subjects | Controls | ||||||||
| 2 | rs2943634 | A | C | WTCCC | 0.30 | 0.34 | 1.22 (1.11-1.33) | 1.19×10−5 | |
| German | 0.32 | 0.37 | 1.20 (1.06-1.35) | 0.004 | |||||
| Adjusted German | 1.08 (0.90-1.31) | 0.10 | 0.03 | ||||||
| 6 | rs6922269 | A | A | WTCCC | 0.29 | 0.25 | 1.23 (1.13-1.35) | 6.33×10−6 | |
| German | 0.30 | 0.26 | 1.24 (1.09-1.41) | 0.001 | |||||
| Adjusted German | 1.23 (1.01-1.50) | 0.11 | 0.009 | ||||||
| 9 | rs1333049 | C | C | WTCCC | 0.55 | 0.47 | 1.37 (1.26-1.48) | 1.80×10−14 | |
| German | 0.54 | 0.48 | 1.33 (1.18-1.51) | 6.80×10−6 | |||||
| Adjusted German | 1.28 (1.07-1.53) | 0.22 | 6.12×10−5 | ||||||
For each SNP, the odds ratios (with 95% confidence intervals) and P values given for the WTCCC and German data represent the risk of coronary artery disease associated with each copy of the risk allele and are based on the Cochran–Armitage test for trend, without adjustment for covariates. The odds ratios and population attributable fractions for the adjusted German data are based on multivariate analyses with inclusion of all three lead SNPs and with adjustment for age, sex, the interaction between age and sex, the PROCAM study score, and the Framingham risk score. The P values for the adjusted German data were corrected for multiple testing (i.e., the testing of a total of nine loci for association with coronary artery disease) but were not adjusted for covariates. The location of the SNPs and details of their loci are shown in Figure 2 and in Figures 1A and 1B in the Supplementary Appendix. Further details on the SNPs, including genotype frequencies in the two studies, are given in Tables 1 and 3 in the Supplementary Appendix.
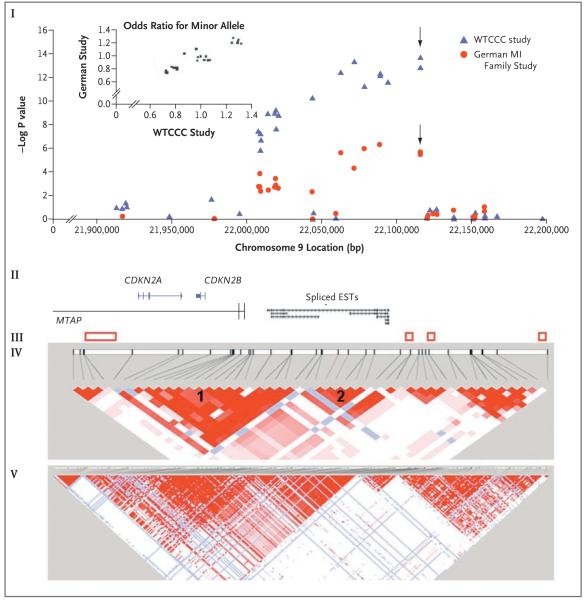
The locus on chromosome 9p21.3 showed the strongest signal in both the WTCCC and the German studies (P=1.80×10−14 and P=3.40×10−6, respectively) (Fig. 1 and Table 2). The combined P value for the association with coronary artery disease of the lead SNP in that locus, rs1333049, was 2.91×10−19, with the risk increased by 36% per copy of the C allele (95% confidence interval [CI], 27 to 46). Approximately 22% of the study participants were homozygous for this allele, with an additional 50% carrying a single copy. In the WTCCC and German studies, a similar pattern of association with respect to direction and magnitude of effect was found across a region of approximately 100 kb on 9p21.3 (Fig. 2). The 19 SNPs showing an association within this region were in strong linkage disequilibrium. However, two blocks could be distinguished, with strong linkage disequilibrium within each block (average D’>0.90) and moderate linkage disequilibrium between blocks (average D’, approximately 0.60) (Fig. 2). Three SNPs in block 1 (rs7044859, rs1292136, and rs7865618) and one SNP in block 2 (rs1333049) were sufficient to tag the region. Haplotype analysis showed that the association was mainly due to two mutually exclusive haplotypes for the four SNPs (TTGG and ACAC) (Supplementary Appendix). In the WTCCC study, the odds ratio for coronary artery disease with the ACAC haplotype (frequency, 0.324 among controls and 0.386 among case subjects), as compared with the TTGG haplotype (frequency, 0.333 among controls and 0.271 among case subjects), was 1.48 (95% CI, 1.34 to 1.64) per copy of the haplotype (P=2.1×10−14). The results of haplotype analysis in the German study were similar (Supplementary Appendix).
Figure 2. Association Signal for Coronary Artery Disease on Chromosome 9.
The −log P values, calculated with the use of the two-sided Cochran–Armitage test for trend, according to the location on chromosome 9 are shown (I). Arrows indicate the lead SNP. The inset graph shows the correlation between the odds ratios for coronary artery disease associated with the minor allele of SNPs from the WTCCC and German studies on chromosome 9. The genomic locations of genes or expressed-sequence tags (ESTs) (according to the Reference Sequence collection of the National Center for Biotechnology Information [NCBI]) are also shown (II). All data are from the Genome Browser of the University of California, Santa Cruz (NCBI build 35). Red boxes represent recombination hot spots, as estimated from HapMap data (Phase II, release 21) (III). Disequilibrium coefficient values for SNPs genotyped in case subjects with coronary artery disease in the WTCCC study, generated with the use of Haploview software, are shown (IV). There were two distinct blocks (1 and 2) of SNPs with significant associations. Disequilibrium coefficients for all SNPs from HapMap CEU data (from persons of Northern and Western European ancestry) across chromosome 9, generated with the use of Haploview software, are shown (V). In IV and V, the strength of the linkage disequilibrium between SNPs increases from white to blue to red (white: disequilibrium coefficient <1 and LOD score <2; blue: disequilibrium coefficient=1 and LOD score <2; pink or light red: disequilibrium coefficient <1 and LOD score ≥2; and bright red: disequilibrium coefficient=1 and LOD score ≥2). References relevant to II, III, and IV are given in the Supplementary Appendix.
The lead SNP on chromosome 6q25.1 (rs6922269, with a combined P value of 2.90×10−8 for the association with coronary artery disease) is in the gene for methylenetetrahydrofolate dehydrogenase (NADP+-dependent) 1–like protein (MTHFD1L). All positive SNPs in the region are located in introns in the middle portion of the gene (Fig. 1A in the Supplementary Appendix). The risk allele (A) for rs6922269 has a prevalence of approximately 25%, with the risk increased by 23% per copy (95% CI, 15 to 33). Haplotype analysis showed that only the two haplotypes carrying the A allele were more frequent in case subjects than in controls, confirming the increased odds ratio for coronary artery disease with the A allele in the single-locus analysis (Supplementary Appendix).
The third replicated locus, on chromosome 2q36.3 (rs2943634, with a combined P value of 1.61×10−7 for the association with coronary artery disease) encompasses a region of 233 kb (Fig. 1B in the Supplementary Appendix). There is only one pseudogene (ENSG00000197218) located within this region. The risk allele (C) for rs2943634 has a prevalence of approximately 65%, with the risk increased by 21% per copy (95% CI, 13 to 30). Haplotype analysis showed that the other associations observed in the linkage-disequilibrium block around rs2943634 are due to linkage disequilibrium with it (Supplementary Appendix).
The association of the loci on chromosomes 9p21.3 and 6q25.1 with myocardial infarction in the German study was not affected by adjustment for cardiovascular risk factors and scores. In contrast, the odds ratio for myocardial infarction at the locus on chromosome 2q36.3 was reduced after such adjustment (Table 2). Further analysis showed that this locus was also significantly related to the body-mass index (P=0.004 in an additive model), the presence or absence of hypertension (P=0.04), and the level of low-density lipoprotein cholesterol (P=0.03). The fully adjusted population attributable fractions for the three loci in the German study are shown in Table 2. The combined fraction for the three loci was 0.38 (95% CI, 0.13 to 0.55). The prediction of myocardial infarction on the basis of the Framingham risk score and the PROCAM study score was substantially improved by adding the predictive information of these three loci to the model (deviance, 191.48; P<1×10−10).
For the six loci with an FPRP of less than 0.5 in the WTCCC study for which the associations were not replicated in the German study, the results of each of the two studies are shown in Table 2 in the Supplementary Appendix. The power to replicate these loci ranged from 43 to 80%.
COMBINED ANALYSIS
The combined analysis of all SNPs identified four additional loci with a high likelihood of association with coronary artery disease (FPRP <0.2) (Table 3, and Table 3 in the Supplementary Appendix). The locus on chromosome 1p13.3 involves the PSRC1 gene, which encodes a proline-rich protein (Fig. 1C in the Supplementary Appendix). The other region of chromosome 1 (1q41) maps to the melanoma inhibitory activity 3 (MIA3) gene (also known as ARNT or TANGO) (Fig. 1D in the Supplementary Appendix). The SNPs associated with chromosome 10q11.21 cluster in a region 100 kb downstream of the CXCL12 gene (also known as the gene for stromal-cell–derived factor 1 precursor) (Fig. 1E in the Supplementary Appendix). Finally, the SNP on chromosome 15q22.33 is an intronic SNP in the SMAD3 gene (Fig. 1F in the Supplementary Appendix). SMAD3 is a transcriptional modulator activated by transforming growth factor β (TGF-β) and activin type 1 receptor kinase.
Table 3. Additional Loci Associated with Coronary Artery Disease from Combined Analysis of Data from the WTCCC and German MI Family Studies*.
| Chromosome | Lead SNP | Minor Allele in Controls | Risk Allele | WTCCC Study | German Study | Combined Analysis | FPRP | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Frequency of Minor Allele among Controls | Odds Ratio for Risk Allele (95% CI) | Frequency of Minor Allele among Controls | Odds Ratio for Risk Allele (95% CI) | Odds Ratio for Risk Allele (95% CI) | P Value | |||||
| 1 | rs599839 | G | A | 0.23 | 1.24 (1.12-1.38) | 0.22 | 1.39 (1.19-1.63) | 1.29 (1.18-1.40) | 4.05×10−9 | 0.0006 |
| 1 | rs17465637 | A | C | 0.29 | 1.23 (1.12-1.34) | 0.26 | 1.15 (1.01-1.32) | 1.20 (1.12-1.30) | 1.27×10−6 | 0.1314 |
| 10 | rs501120 | C | T | 0.13 | 1.24 (1.09-1.41) | 0.16 | 1.54 (1.28-1.86) | 1.33 (1.20-1.48) | 9.46×10−8 | 0.0234 |
| 15 | rs17228212 | C | C | 0.30 | 1.19 (1.09-1.30) | 0.26 | 1.26 (1.11-1.44) | 1.21 (1.13-1.30) | 1.98×10−7 | 0.0178 |
Loci shown are those with a false-positive-report probability (FPRP) of less than 20%, excluding those shown in Table 2. Further details on the single-nucleotide polymorphisms (SNPs), including genotype frequencies, in the two studies are given in Table 3 in the Supplementary Appendix. The odds ratios and 95% confidence intervals for each of the two studies are based on the Cochran–Armitage test for trend and relate to each copy of the risk allele. The combined odds ratios were calculated with the use of stratification according to study, without adjustment for covariates. The calculation of FPRP is described in the Supplementary Appendix.
SUBPHENOTYPE AND SUBGROUP ANALYSIS
In the WTCCC study, approximately 30% of subjects had confirmed evidence of coronary artery disease but had not had a myocardial infarction at the time of recruitment (Table 1). When subjects with coronary artery disease only and those with coronary artery disease and myocardial infarction were analyzed individually, the odds ratios for both phenotypes across all seven chromosomal regions remained significant (Table 4 in the Supplementary Appendix). For the chromosome 15 locus, the effect size was significantly greater (P=0.004) in the subgroup of patients with coronary artery disease only than in the subgroup with coronary artery disease and myocardial infarction. Analysis according to sex in the two studies combined showed that all seven loci affected the risk of coronary artery disease to a similar extent in women and in men (Table 5 in the Supplementary Appendix).
CANDIDATE-GENE ANALYSIS
From a literature search, we identified 142 SNPs, in 91 candidate genes, that had been reported to be associated with coronary artery disease or myocardial infarction. Only 13 of these SNPs are represented on the GeneChip array. For 36 genes, there were no primary or tagging SNPs. For the other genes, we identified 270 SNPs in complete or near-complete linkage disequilibrium with the SNPs that were previously found to be associated with coronary artery disease or myocardial infarction. Although a number of SNPs had a promising association with coronary artery disease in the WTCCC study or with myocardial infarction in the German study, only two linked SNPs (rs17489268 and rs17411031) tagging the Ser447→Ter variant in the lipoprotein lipase gene had a significant association in both studies (Table 6 in the Supplementary Appendix).
DISCUSSION
We jointly analyzed data from two distinct but complementary genomewide association studies of coronary artery disease and myocardial infarction that performed ascertainment in similar ways and that involved the same genotyping platform. Sequential and combined analyses of the two data sets allowed us to identify several new genetic loci, which individually and in aggregate considerably affect the risk of coronary artery disease.
The association of chromosome 9p21.3 with coronary artery disease was the strongest found in the WTCCC study.5 The finding that this locus was also most strongly associated with myocardial infarction in the German study provides compelling proof of its involvement in coronary artery disease. The evidence of association is strong, the risk variant is common, and each copy of the allele substantially increases the probability of the disease. These findings unequivocally demonstrate a major genetic risk variant at this locus.
Indeed, during revision of this manuscript, two other genomewide studies reported a strong association of the same 9p21.3 locus with coronary artery disease and myocardial infarction,13,14 making this the most highly replicated locus for coronary artery disease identified to date. The region contains the coding sequences of genes for two cyclin-dependent kinase inhibitors, CDKN2A (encoding the prototypic INK4 protein p16INK4a) and CDKN2B (encoding p15INK4b), which play an important role in the regulation of the cell cycle and may be implicated, through their role in TGF-β-induced growth inhibition, in the pathogenesis of atherosclerosis.15-17 Although regulation of one or both of the CDKN2 genes may explain the association with coronary artery disease, other explanations also need to be considered, including involvement of the methylthioadenosine phosphorylase (MTAP) gene or of other expressed sequences located in the region. The same region has also recently been associated with increased susceptibility to type 2 diabetes,18-20 raising the possibility of a shared, rather than a single, mechanism causing both coronary artery disease and diabetes.
The association of chromosome 6q25.1 with coronary artery disease maps to the MTHFD1L gene, which encodes the mitochondrial isozyme of C1-tetrahydrofolate (THF) synthase.21,22 The family of C1-THF synthases is used in a variety of cellular processes, particularly the synthesis of purine and methionine.21 Therefore, MTHFD1L activity may also contribute to plasma homocysteine levels,21,23 raising the possibility of a link between MTHFD1L variants and this risk factor for coronary artery disease.24 A preliminary analysis of data from 1070 persons in the AtheroGene study25 has not revealed an association between rs6922269 genotypes and plasma homocysteine levels (Tiret L, Blankenberg S: personal communication). Nevertheless, further studies in a wider range of subjects are needed to investigate this possibility.
Our findings demonstrate the main strength of a genomewide approach — namely, the possibility of identifying hitherto unsuspected loci that increase susceptibility to complex diseases. However, the mechanisms underlying the newly identified associations are often not immediately obvious. Indeed, the mechanisms for the association of signals on chromosomes 9p21.3, 6q25.1, and 2q36.3 with coronary artery disease all require elucidation. Similarly, none of the chromosomal loci identified in the combined analysis have previously been strongly linked to coronary artery disease. However, genes in several of the loci (PSRC1 at 1p13.3, MIA3 at 1q41, and SMAD3 at 15q22.33) play a role in cell growth or inhibition.26-29 These processes are fundamental for the formation and progression of atherosclerotic plaque and also for plaque instability.30 Our results suggest that genetic regulation of these processes plays an important role in the development of coronary artery disease and myocardial infarction.
Some loci from the WTCCC study that we attempted to replicate did not show association in the German study. These negative data underscore the need to view genomewide associations with caution, despite their statistical strength, until they have been replicated in appropriate validation samples. In this context, caution should also be exercised with regard to the four loci identified in our combined analysis.31
Our primary objective was to identify loci with significant associations with coronary artery disease independently of any biologic assumptions. Nonetheless, the genotyping platform also offered an opportunity to examine genetic variants in genes with previously reported associations. Indeed, several showed evidence of an association in one of our studies. However, only SNPs in the lipoprotein lipase gene had evidence of an association in both studies. This finding is in agreement with those in most recent systematic studies that were largely unsuccessful in replicating initial findings in candidate genes.32 However, many of the previously studied gene variants are poorly tagged on the GeneChip array, which clearly fails to cover the full extent of even common variation in these genes.
Whether our findings can be translated into better prevention or treatment for coronary artery disease will become clear only over time and with further research. Although the odds ratios for each locus are modest, as anticipated for a polygenic disorder, the estimates of population attributable fractions for the three validated loci are substantial, both individually and in aggregate. This observation offers the potential for improved overall coronary risk prediction. However, the case subjects in both studies had a strong family history of premature coronary artery disease, which might have enhanced the power to detect an association with coronary artery disease but also might have increased the estimated population attributable risks beyond that of sporadic cases, and further analysis of the loci in a wider range of subjects is necessary. Further studies are also needed to investigate the associations of the loci with other types of atherosclerotic disease, as well as with cardiovascular risk factors and markers. At a genetic level, studies should focus on fine mapping of the associated regions and thorough investigation of candidate genes. Our results provide a framework for all these additional studies.
Our analysis has several important limitations. Although the GeneChip array typed over 500,000 variants, a substantial percentage could not be evaluated, for reasons given in the Supplementary Appendix. Furthermore, to reduce the effect of multiple testing, we used only the rather conservative Cochran–Armitage test for trend (an additive model) to screen the WTCCC data for significant associations. These limitations make it likely that some loci were missed and that further analysis of the data and subsequent validation will reveal other loci.
Nonetheless, by using a sequential strategy of initial replication and subsequent combination of information from the two genomewide association studies, we were able to describe several new genetic loci for coronary artery disease and myocardial infarction that have a considerable effect on the risk of these diseases and that merit indepth follow-up studies. Most important, the finding that a single locus was the strongest signal in two separate studies carries promise for clinically relevant progress in our understanding of the genetics of coronary artery disease. As the current activity in genomewide association studies of complex traits accelerates, our approach may also provide a paradigm for combining the results of such studies to maximize the amount of valuable information that can be extracted from these expensive and laborious experiments.
Supplementary Material
Acknowledgments
Supported by grants from the Wellcome Trust, the National Genome Research Network 2 of the German Federal Ministry of Education and Research, and the Cardiogenics project of the European Union. Recruitment for the WTCCC study was supported by grants from the British Heart Foundation and the U.K. Medical Research Council, and recruitment for the German MI Family Study was supported by grants from the Deutsche Forschungsgemeinschaft and the Deutsche Herzstiftung. We also acknowledge support from the Wellcome Trust Functional Genomics Initiative in Cardiovascular Genetics and the KORA (Cooperative Research in the Region of Augsburg) research platform of the GSF–National Research Centre. Drs. Samani and Ball hold chairs funded by the British Heart Foundation, and Dr. Tobin holds a U.K. Medical Research Council Clinical Scientist Fellowship.
No potential conflict of interest relevant to this article was reported.
We thank Peter Tooze, Andrew Kenniry, Simon Potter, Petra Bruse, Janine Stegmann, Anika Götz, Michaela Vöstner, Klaus Stark, and Viviane Nicaud for assistance.
Footnotes
*Members of the Wellcome Trust Case Control Consortium (WTCCC) and the Cardiogenics Consortium are listed in the Supplementary Appendix, available with the full text of this article at www.nejm.org.
REFERENCES
- 1.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–57. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 2.Yusuf S, Hawken S, Ounpuu S, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–52. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 3.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–6. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 4.Christensen K, Murray JC. What genome-wide association studies can do for medicine. N Engl J Med. 2007;356:1094–7. doi: 10.1056/NEJMp068126. [DOI] [PubMed] [Google Scholar]
- 5.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of diseases seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samani NJ, Burton P, Mangino M, et al. A genomewide linkage study of 1,933 families affected by premature coronary artery disease: The British Heart Foundation (BHF) Family Heart Study. Am J Hum Genet. 2005;77:1011–20. doi: 10.1086/498653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfakih K, Brown B, Lawrance RA, et al. Effect of a common X-linked angiotensin II type 2-receptor gene polymorphism (-1332 G/A) on the occurrence of premature myocardial infarction and stenotic atherosclerosis requiring revascularization. Atherosclerosis. doi: 10.1016/j.atherosclerosis.2007.01.028. in press. [DOI] [PubMed] [Google Scholar]
- 8.Broeckel U, Hengstenberg C, Mayer B, et al. A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet. 2002;30:210–4. doi: 10.1038/ng827. [DOI] [PubMed] [Google Scholar]
- 9.Fischer M, Broeckel U, Holmer S, et al. Distinct heritable patterns of angiographic coronary artery disease in families with myocardial infarction. Circulation. 2005;111:855–62. doi: 10.1161/01.CIR.0000155611.41961.BB. [DOI] [PubMed] [Google Scholar]
- 10.Wichmann HE, Gieger C, Illig T. KORA-gen — resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–S30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 11.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 15.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 16.Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–61. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 17.Kalinina N, Agrotis A, Antropova Y, et al. Smad expression in human atherosclerotic lesions: evidence for impaired TGF-beta/Smad signaling in smooth muscle cells of fibrofatty lesions. Arterioscler Thromb Vasc Biol. 2004;24:1391–6. doi: 10.1161/01.ATV.0000133605.89421.79. [DOI] [PubMed] [Google Scholar]
- 18.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 21.Prasannan P, Pike S, Peng K, Shane B, Appling DR. Human mitochondrial C1-tetrahydrofolate synthase: gene structure, tissue distribution of the mRNA, and immunolocalization in Chinese hamster ovary calls. J Biol Chem. 2003;278:43178–87. doi: 10.1074/jbc.M304319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walkup AS, Appling DR. Enzymatic characterization of human mitochondrial C1-tetrahydrofolate synthase. Arch Biochem Biophys. 2005;442:196–205. doi: 10.1016/j.abb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Randak C, Roschinger W, Rolinski B, Hadorn HB, Applegarth DA, Roscher AA. Three siblings with nonketotic hyperglycinaemia, mildly elevated plasma homocysteine concentrations and moderate methylmalonic aciduria. J Inherit Metab Dis. 2000;23:520–2. doi: 10.1023/a:1005667707355. [DOI] [PubMed] [Google Scholar]
- 24.Fruchart JC, Nierman MC, Stroes ES, Kastelein JJ, Duriez P. New risk factors for atherosclerosis and patient risk assessment. Circulation. 2004;109(Suppl I):III-15–III-9. doi: 10.1161/01.CIR.0000131513.33892.5b. [DOI] [PubMed] [Google Scholar]
- 25.Tiret L, Godefroy T, Lubos E, et al. Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation. 2005;112:643–50. doi: 10.1161/CIRCULATIONAHA.104.519702. [DOI] [PubMed] [Google Scholar]
- 26.Lo PK, Chen JY, Lo WC, et al. Identification of a novel mouse p53 target gene DDA3. Oncogene. 1999;18:7765–74. doi: 10.1038/sj.onc.1203167. [DOI] [PubMed] [Google Scholar]
- 27.Bosserhoff AK, Buettner R. Expression, function and clinical relevance of MIA (melanoma inhibitory activity) Histol Histopathol. 2002;17:289–300. doi: 10.14670/HH-17.289. [DOI] [PubMed] [Google Scholar]
- 28.Qin BY, Lam SS, Correia JJ, Lin K. Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control. Genes Dev. 2002;16:1950–63. doi: 10.1101/gad.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazono K. TGF-beta signaling by Smad proteins. Cytokine Growth Factor Rev. 2000;11:15–22. doi: 10.1016/s1359-6101(99)00025-8. [DOI] [PubMed] [Google Scholar]
- 30.Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation. 2005;111:3481–8. doi: 10.1161/CIRCULATIONAHA.105.537878. [DOI] [PubMed] [Google Scholar]
- 31.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 32.Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007;297:1551–61. doi: 10.1001/jama.297.14.1551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.