Abstract
Using a positional cloning approach supported by comparative genomics, we have identified a previously unreported gene, EYS, at the RP25 locus on chromosome 6q12 commonly mutated in autosomal recessive retinitis pigmentosa. Spanning over 2 Mb, this is the largest eye-specific gene identified so far. EYS is independently disrupted in four other mammalian lineages, including that of rodents, but is well conserved from Drosophila to man and is likely to have a role in the modeling of retinal architecture.
Autosomal recessive retinitis pigmentosa is one of the most debilitating hereditary retinal disorders leading to severe visual impairment. To date 26 genetic loci have been implicated in autosomal recessive retinitis pigmentosa; however, with the exception of RP25 (MIM602772; RetNet), the prevalence of each of the reported loci is only 1-5%.
Previous genetic studies mapped the RP25 locus to a ~16-cM region on chromosome 6p12.1-q15 in four Spanish families1. Subsequently, linkage to the same locus was reported in multiple families from various ancestral origins, supporting RP25 as the first major locus for recessive retinitis pigmentosa2-4.
A detailed review of the RP25 interval revealed information on 110 genes, emphasizing the extent of the work required to identify the causative gene. We therefore adopted the approach of (i) exclusion of 15 candidate genes, such as GABRR1, GABRR2, MYO6, EEF1A1, ELOVL4, RIMS1, IMPG1 and LCA5 (C6ORF52)5, on the basis of their known retina-related function or their involvement in other retinal degenerations overlapping with RP25; and (ii) systematic screening of a further 45 genes, leading to the exclusion of 60 of the original 110 genes6. At this stage, mapping of five additional families at the RP25 locus helped refine the disease interval to a 2.67-cM region between D6S257 and D6S1557, thereby focusing our search for the disease-associated gene to a much smaller interval7. In parallel, a high-throughput screening for deletions was undertaken using array comparative genomic hybridization (array CGH). Notably, we found that a ~100-kb clone (chr6tp-19C7) was deleted in all affected members of one of the originally linked families (RP5)6. This suggested that the deleted clone could contain or overlap with the disease-associated gene (provisionally referred to here as the RP25 gene). The genomic sequence spanning this deletion contained six independently predicted genes, Q5T1H1, Q9H557, Q5TEL3, Q5TEL4, Q5VVG4 and Q5T3C8, which became the priority for mutation screening.
We carried out molecular analysis of these six genes in ten previously linked Spanish families, three of which are consanguineous (RP5, RP167 and RP377) and seven nonconsanguineous (RP73, RP214, RP299, RP328, RP349, RP351 and RP355)1,7. In one of these predicted genes, Q9H557, we observed a homozygous 17-bp deletion in family RP214 (Supplementary Fig. 1a online). Additionally, the four predicted exons from the same gene and a single-exon gene, Q5TEL3, failed to amplify in all affected members of family RP5 (Supplementary Fig. 2 online). This confirmed the ~100-kb deletion seen earlier by array CGH and strongly implicated Q9H557 and Q5TEL3 as part of the RP25 gene.
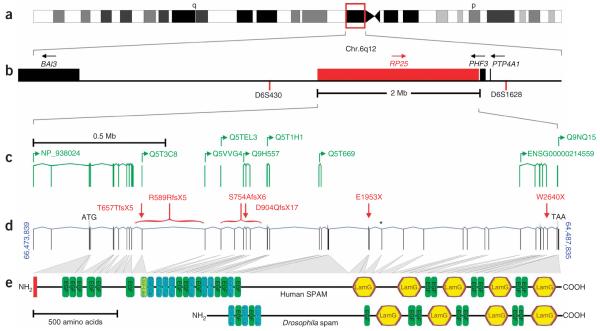
To fully characterize the RP25 gene, we carried out multiple RT-PCRs and RACE using the sequence information of Q9H557 and Q5TEL3 (Supplementary Methods online). This approach, combined with the comparative genomic analysis, allowed us to unravel the structure of this newly identified gene, which we eventually determined to be an unannotated prediction. On assembling all available data we noted that RP25 encompasses 30 exons belonging to nine previously predicted genes and 13 newly reported exons, and spans the interval between 64,487,835 and 66,473,839 on chromosome 6q12 (Fig. 1). Of note, we found that a 1,238-bp segment spanning the 3′ end of exon 29 was entirely absent from the reference human genome assembly but was well represented within trace archive sequences (Fig. 1d). The full coding region of the RP25 gene (~9 kb) was amplified in two overlapping fragments (Supplementary Fig. 3 online). RT-PCR analysis of cDNAs from a variety of normal tissues and cell lines using cDNA specific primers within RP25 (Supplementary Table 1 online) amplified the expected-size product only from the retina and from a photoreceptor-like cell line, Y79 (Fig. 2a).
Figure 1.
RP25 gene structure and domain architecture. (a) Chromosomal region at 6q12. (b) Schematic representation of the genes and microsatellite markers flanking the RP25 gene. (c) Previously predicted genes overlapping RP25. (d) The 43 exons comprising the RP25 gene with the initiation (ATG) and stop codon (TAA) marked within exons 4 and 43, respectively; mutations are indicated in red and the asterisk at exon 29 marks the 1,238-bp segment missing from the human reference assembly. (e) Domain architecture of human SPAM and its Drosophila spam ortholog.
Figure 2.
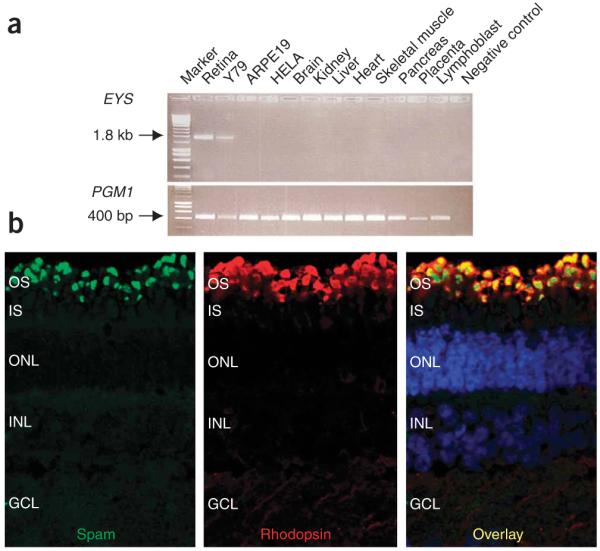
Expression pattern and immunolocalization of spam. (a) EYS (SPAM) expression in different tissues is shown in the upper panel with a specific 1.8-kb product in the retina and in Y79 photoreceptor-like cells. ARPE19 is a retinal pigment epithelial cell line. A 400-bp fragment representing the gene PGM1, which is ubiquitously expressed in all tissues and cell lines, is shown as an amplification control in the lower panel. (b) Subcellular localization of spam to the outer segment of mature porcine retina using antibody to spam (green) and antibody to rhodopsin (red). The overlay shows the localization of spam in the same region as rhodopsin (yellow) in the outer segment of the photoreceptor cell layer.
We then used a combination of methods incorporating direct sequence analysis, array CGH and the multiplex ligation-dependent probe amplification (MLPA) techniques to ensure comprehensive mutation screening of the coding regions and splice sites of the 43 exons comprising RP25 (Supplementary Methods). So far, we have identified six independent mutations, including four deletions and two nonsense substitutions, all leading to premature stop codons in five unrelated families (Table 1 and Supplementary Note online). It is known that mRNA containing premature stop codons undergo nonsense-mediated decay8; therefore, the disease mechanism in these families may be due to complete absence of a functional protein.
Table 1. Mutations identified within the RP25 gene.
| Family ID | Exon | Nucleotide position | Protein alteration | Type of mutation |
|---|---|---|---|---|
| RP214 | 17 | 2710_2726del17 | D904QfsX17 | Homozygous |
| RP5 | 15-19 | 2260-51191_2992+45990 | S754AfsX6 | Homozygous |
| RP328 | 12 | 1971delT | T657TfsX5 | Homozygous |
| RP73 | 12 | [1767-24596_2023+238135] | R589RfsX5 | Heterozygous |
| + | ||||
| 28 | [5857G>T] | E1953X | Heterozygous | |
| RP349 | 41 | 7919G>A | W2640X | Homozygous |
Direct sequence analysis was not appropriate for detecting the large heterozygous deletion in family RP73 or defining the deletion break-points in families RP5 and RP73; hence, we used both array CGH and MLPA. We also used restriction-digest analysis as a second independent method to study the segregation of the nonsense substitutions and the small deletions (Supplementary Fig. 1a-d). None of the identified mutations have been detected in 200 control individuals.
RP25 is predicted to be a multidomain protein containing 3,145 amino acids with at least 21 epidermal growth factor (EGF)-like domains in its N-terminus followed by five C-terminal LamG domains, interspersed by further EGF repeats (Fig. 1e). This unique domain structure9 was also seen in the Drosophila spacemaker (spam) protein (Supplementary Methods), encoded by eys (eyes shut). We have accordingly named the human RP25 gene EYS (encoding the protein SPAM). Drosophila spam is expressed in the eye across diverse insect species with an open rhabdom system, such as fruitflies (Drosophila melanogaster) and houseflies (Musca domestica Linnaeus), in which the rhabdomeres or photoreceptor cells of each ommatidium in the compound eye are separated from each other. In contrast, species with a closed system, such as the mosquito (Anopheles gambiae), rust-red flour beetle (Tribolium castaneum) and the honeybee (Apis mellifera), do not express spam in the eye. The complete loss of eys (spam) converts an open rhabdom system to a closed one, whereas its targeted expression to photoreceptors of a closed system markedly reorganizes the architecture of the compound eyes to resemble an open system10. On the basis of these findings in Drosophila and the RT-PCR data (Fig. 2a), we expected SPAM to localize in the photoreceptor layer, and indeed our immunohistochemical studies confirmed this localization (Fig. 2b).
An apparently intact eys gene is found across the mammalian clade, including monotremes (platypus) and marsupials (opossum) (Supplementary Fig. 4 online). However, despite the mutations and the presumed loss of function associated with human disease, this gene has been dispensed with on at least four separate occasions in the last ~100 million years of mammalian evolution11, including in the armadillo (Dasypus novemcinctus), little brown bat (Myotis lucifugus) and ruminant (cattle and sheep) lineages (Supplementary Methods). Eys has acquired many (≥3) reading-frame disruptions in three rodents (mouse, rat and guinea pig) representing two of the three major rodent clades (Supplementary Fig. 4)11,12. This was also confirmed by failure of PCR amplification of eys from mouse retinal cDNA and further supported by the absence of any immunolocalization signal in the mouse retina (data not shown). EYS is only the fourth mendelian disease-associated human gene whose orthologs are disrupted or absent from rodent genomes13.
In summary, we report the identification and genomic characterization of a previously unreported gene, EYS (encoding SPAM), implicated in autosomal recessive retinitis pigmentosa. The identification of six independent mutations and the presence of linked families from different ancestral origins support EYS as the first major gene reported for autosomal recessive retinitis pigmentosa. With 43 exons, covering 2.0 Mb, this is the largest gene identified to be expressed in the human eye and the fifth largest overall in the human genome. Information about the established function of insect orthologs suggests that EYS may possess similar functions in maintaining the integrity of the photoreceptor cells in human retina.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the families who participated in the study. This study was funded by Fondo de Investigación Sanitaria (PI050857), Spain; Consejería de Salud (PI-0334/2007), Junta de Andalucía, Spain; British Retinitis Pigmentosa Society (grant ref. GR556); Foresight, Dubai; Foundation Fighting Blindness (USA); National Institute of Health Research Biomedical Research Centre for Ophthalmology, The Special Trustees of Moorfields Eye Hospital London; the UK Medical Research Council and EU-Neurotrain (grant ref. MEST-CT-2005-020235); EU-GENORET (grant ref. LSHG-CT-2005-512036). The El Centro de Investigación Biomédica en Red de Enfermedades Raras is an initiative of the Instituto de Salud Carlos III. N.P.C. and E.P. were supported by the Wellcome Trust. We would like also to thank R. Molday (University of British Columbia, Vancouver) for the gift of 1D4 antibody and G. Jeffery for his advice with immunohistochemistry.
Footnotes
Published online at http://www.nature.com/naturegenetics/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.Ruiz A, Borrego S, Marcos I, Antinolo G. Am. J. Hum. Genet. 1998;62:1452–1459. doi: 10.1086/301866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khaliq S, et al. Am. J. Hum. Genet. 1999;65:571–574. doi: 10.1086/302493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barragán I, Marcos I, Borrego S, Antiñolo G. Ophthalmic Res. 2005;37:89–93. doi: 10.1159/000084250. [DOI] [PubMed] [Google Scholar]
- 4.Abd El-Aziz MM, et al. Ann. Hum. Genet. 2007;71:281–299. doi: 10.1111/j.1469-1809.2006.00333.x. [DOI] [PubMed] [Google Scholar]
- 5.den Hollander AI, et al. Nat. Genet. 2007;39:889–895. doi: 10.1038/ng2066. [DOI] [PubMed] [Google Scholar]
- 6.Abd El-Aziz MM, et al. Ann. Hum. Genet. 2008;72:463–477. doi: 10.1111/j.1469-1809.2008.00455.x. published online 29 May 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barragan I, et al. Ann. Hum. Genet. 2008;72:454–462. doi: 10.1111/j.1469-1809.2008.00448.x. published online 29 May 2008. [DOI] [PubMed] [Google Scholar]
- 8.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nat. Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 9.Husain N, et al. Dev.Cell. 2006;11:483–493. doi: 10.1016/j.devcel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Zelhof AC, Hardy RW, Becker A, Zuker CS. Nature. 2006;443:696–699. doi: 10.1038/nature05128. [DOI] [PubMed] [Google Scholar]
- 11.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huchon D, et al. Mol. Biol. Evol. 2002;19:1053–1065. doi: 10.1093/oxfordjournals.molbev.a004164. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, et al. Genome Biol. 2004;5:R47. doi: 10.1186/gb-2004-5-7-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.