Abstract
Iron stored in phytoferritin plays an important role in the germination and early growth of seedlings. The protein is located in the amyloplast where it stores large amounts of iron as a hydrated ferric oxide mineral core within its shell-like structure. The present work was undertaken to study alternate mechanisms of core formation in pea seed ferritin (PSF). The data reveal a new mechanism for mineral core formation in PSF involving the binding and oxidation of iron at the extension peptide (EP) located on the outer surface of the protein shell. This binding induces aggregation of the protein into large assemblies of ∼400 monomers. The bound iron is gradually translocated to the mineral core during which time the protein dissociates back into its monomeric state. Either the oxidative addition of Fe2+ to the apoprotein to form Fe3+ or the direct addition of Fe3+ to apoPSF causes protein aggregation once the binding capacity of the 24 ferroxidase centers (48 Fe3+/shell) is exceeded. When the EP is enzymatically deleted from PSF, aggregation is not observed, and the rate of iron oxidation is significantly reduced, demonstrating that the EP is a critical structural component for iron binding, oxidation, and protein aggregation. These data point to a functional role for the extension peptide as an iron binding and ferroxidase center that contributes to mineralization of the iron core. As the iron core grows larger, the new pathway becomes less important, and Fe2+ oxidation and deposition occurs directly on the surface of the iron core.
The chemistry of iron and oxygen in a number of non-heme di-iron proteins has been a subject of intense interest because of their varied roles in oxygen activation and catalysis, substrate hydrolysis, oxygen transport, redox reactions, H2O2, and iron detoxification and iron storage. Di-iron centers have similar structural motifs consisting of a combination of carboxylate and histidine ligands that either bind or bridge the two metal ions of the di-nuclear active site; di-iron proteins containing these centers include methane monooxygenase, ribonucleotide reductase, rubrerythrin, stearoyl desaturase, purple acid phosphatase, hemerythrin, the Dps proteins, and ferritins (1–6). Despite their similar di-nuclear centers, each of these proteins fulfills a distinct biological role that seems to be mediated by the nature of the first and second coordination sphere of the di-iron center.
Ferritins are a class of intracellular iron storage and detoxification proteins that facilitate the oxidation of iron by molecular oxygen or hydrogen peroxide to form a hydrous ferric oxide mineral core within their interiors (1–3). Rapid Fe2+ oxidation occurs at the di-iron ferroxidase center located on the H-subunit of the mammalian protein. Unlike the H-chain, the more acidic L-subunit lacks the ferroxidase center but contains a putative nucleation site responsible for slower iron oxidation and mineralization (7). The shapes of both the H- and L-subunit are nearly cylindrical and composed of a four-α-helix bundle containing two antiparallel helix pairs (A, B and C, D) connected by a long non-helical stretch, the BC-loop, between B and C helices. A fifth short helix (E helix) lies at one end of the bundle at 60° to its axis (1, 2).
From an evolutionary point of view, plant and animal ferritins arose from a common ancestor, but plant ferritins exhibit various specific features as compared with animal ferritins. Plant ferritins are observed in plastids (chloroplasts in leaves, amyloplasts in tubers and seeds, etc.) where iron is incorporated into the ferritin shell to form the mineral core, whereas animal ferritins are largely found in the cytoplasm of cell (1, 8). Ferritins from dried soybean and pea seed consist of two subunits of 26.5 and 28.0 kDa, which are designated H-1 and H-2, respectively, share ∼80% amino acid sequence identity (9, 10), and contain ferroxidase centers. The two subunits are synthesized from a 32-kDa precursor with a unique two-domain N-terminal sequence containing a transit peptide (TP)2 and an extension peptide (EP). The TP is responsible for precursor targeting to plastids (11). Upon transport to plastids, the TP is cleaved from the subunit precursor, resulting in the formation of the mature subunit which assembles into a 24-mer apoferritin within the plastids (11, 12). The EP domain is kept at the N-terminal extremity of the mature plant ferritins, but its function remains unknown. It is absent in animal ferritins. For soybean ferritin, further processing of the 26.5-kDa subunit occurs through excising a short C-terminal amino acid sequence (16 amino acid residues) that corresponds to the E helix of the mammalian ferritin subunit (9). Compared with their own precursors, H-1 is devoid of both the N-terminal TP domain and the C-terminal E helix, whereas H-2 lacks the N-terminal TP domain only (13). However, the amino acids constituting the ferroxidase center are strictly conserved in all the plant ferritins except for pea seed ferritin where His-62 replaces Glu-62 (14, 15).
Iron oxidation/mineralization in human ferritin occurs by at least three reaction pathways (16, 17). After Fe2+ binding at the ferroxidase site, the protein-catalyzed oxidation of Fe2+ occurs, H2O2 is the product of O2 reduction, and a mineral core of Fe3+ is produced, written for simplicity as Fe(O)OH(core) (Equation 1) (18, 19). Some of the H2O2 generated in Equation 1 reacts with additional Fe2+ in the Fe2+ + H2O2 detoxification reaction (Equation 2) to produce H2O (16, 20). Once a mineral core of sufficient size has developed, Fe2+ autoxidation becomes significant, and iron oxidation and hydrolysis occurs primarily on the growing surface of the mineral through an autocatalytic process where O2 is reduced completely to H2O (Equation 3) (1, 7, 17).
Based on the above reactions and on identification of intermediates by resonance Raman spectroscopy, Mössbauer spectroscopy, and EXAFS (extended x-ray absorption fine structure), the mechanism of mineral core formation in mammalian ferritins has been reasonably well established (18, 21–23). Initially two Fe2+ are oxidized by O2 at the ferroxidase center to form a transient μ-1,2-peroxodi-Fe3+ intermediate, which rapidly decays to form a μ-oxo di-Fe3+ complex(es), releasing H2O2 to the solution. The μ-oxo(hydroxo) bridged di-Fe3+ dimer(s) then translocates from the ferroxidase center to the inner cavity to form an incipient core, which ultimately leads to the formation of the mineral core itself. However, this mechanism of oxidative deposition of iron is only applicable to ferritins such as horse spleen ferritin and human H-chain ferritin, which regenerate activity at their ferroxidase centers. In contrast, the ferroxidase centers of human mitochondrial ferritin (24), Escherichia coli bacterioferritin (25), E. coli bacterial ferritin A (26), and pea seed ferritin (PSF) (27) lack significant regeneration activity, and Fe3+ produced at their centers migrates with difficulty from the center to the cavity to form the initial core. These observations raise the question as to whether alternate pathways exist for core formation in these proteins, in particular in phytoferritins, the focus of this work.
In the present study we have looked for alternate mechanisms of iron deposition in PSF and demonstrate that ferritin-ferritin aggregation occurs during the oxidative deposition of iron in PSF when more than 48 Fe2+/shell are added to the apoprotein (apoPSF), an amount exceeding the 24 ferroxidase center binding capacity for Fe2+/3+. Such aggregation was also induced by the addition of Fe3+ directly to PSF. The data indicate that binding occurs on the outer surface of the ferritin shell at sites involving the extension peptide. Upon standing, the iron induced aggregate reversibly dissociates to its original undissociated form, whereas the iron migrates to the mineral core. This finding represents an alternate pathway for iron deposition in phytoferritin.
EXPERIMENTAL PROCEDURES
Preparation of Holo and Apo Pea Seed Ferritin and the EP
Pea seed ferritin was purified as recently described (27). Apoferritin was prepared using sodium dithionite in 0.1 m Mops buffer (pH 7.9) under anaerobic conditions (26, 28) followed by dialysis against the working buffers indicated in the figure captions. Both holoferritin and apoferritin concentrations were determined according to the Lowry method using bovine serum albumin as the standard. Holo pea seed ferritin with different sizes of iron core was prepared as previously described (29). The EP of PSF with sequence TTAPLTGVIFEPFEEVKKDYLAVP was purchased from Scilight Biotechnology (Beijing, China). Its purity was more than 95% as indicated by high performance liquid chromatography and mass spectroscopy.
Preparation of Pea Seed Ferritin with the EP Deleted
Typically, purified holo pea seed ferritin (3.0 mg) was added to 0.24 ml of Alcalase 2.4L (Novozymes, Denmark) and incubated at 60 °C for 5 min followed by phenylmethanesulfonyl fluoride at a final concentration of 2 mm to stop the proteolysis reaction. To separate the pea seed ferritin subunit with the EP deleted, the above solution was diluted 500-fold with distilled water and then ultrafiltrated to its former concentration using an YM-100 membrane (Millipore Corp., Bedford, MA). Finally, SDS-PAGE was performed to examine the purity of the protein under reducing conditions using 15% mini-slab gels as shown in supplemental Fig. S1. The N-terminal amino acid sequences of the resultant two new subunits treated with the enzyme are listed in supplemental Fig. S2, confirming that EP had been removed from both.
Polyacrylamide Gel Electrophoresis
The molecular weight of the native pea seed ferritin was estimated by PAGE using a 4–20% polyacrylamide gradient gel run at 40 V for 10 h at 4 °C and employing Tris-HCl (25 mm, pH 8.3) as running buffer. Gels were stained with Coomassie Blue R-250. Gel electrophoresis under denaturing conditions was carried out with 12.5% polyacrylamide-SDS gel as reported by Laemmli (30). Protein samples (∼20 μg) were suspended in 50 μl water to which was added 100 μl of sample buffer containing 25% glycerol, 12.5% 0.5 m Tris-HCl, pH 6.8, 2% SDS, 1% bromphenol blue, and 5% β-mercaptoethanol. After the solution was boiled for 10 min, the supernatant was isolated by centrifugation at 10,000 × g for 20 min.
Sequence of N-terminal Amino Acid
The amino acid sequence of the N terminus was determined on a protein sequencer (Applied Biosystems Procise-PROCISE 491) using automated Edman degradation. After SDS-PAGE, protein was transferred to a polyvinylidene difluoride membrane (Millipore) and stained with Coomassie Brilliant Blue R250. The two subunits were eluted from the membrane, and the sequence of the 10 amino acids at the N terminus was determined for each subunit.
Stopped-flow Light Scattering
Stopped-flow light scattering measurements experiments were performed with a pneumatic drive Hi-Tech SFA-20 M apparatus in conjunction with a Cary Eclipse spectrofluorimeter (Varian). To observe protein aggregation induced by iron, equal 140-μl volumes of a weakly acidic FeSO4 (pH 2.0) solution or FeCl3 (pH 2.0) and buffered apoferritin/holoferritin with different sizes of iron core were mixed at 25 °C in the thermostatted sample compartment containing an 80-μl quartz stopped-flow cuvette. All quoted concentrations are final concentrations after mixing the two reagents. The mixing dead time was determined to be 6.8 ± 0.5 ms using the dichloroindophenol and ascorbic acid test reaction (31). Both excitation and emission wavelengths were set at 680 nm, and the time-dependent change in scattering light at a 90° angle, perpendicular to the beam, was recorded (32). The kinetic data were curve-fitted with Origin 7.5 software (Micro Cal Inc.). The initial rates of PSF aggregation were obtained from the linear A1 term of a third-order polynomial curve fitted to the experimental data, namely Y = A0 + A1t + A2t2 + A3t3 and dY/dt = A1 + 2A2t + 3A3t2 (at t = 0, (dY/dt)0 = A1). Here t is the time in seconds, and Y is the change in scattered light.
Light Scattering
Effects of EDTA and NaCl on ferritin aggregation induced by ferric ion were observed with the Cary Eclipse spectrofluorimeter at room temperature with both excitation and emission wavelengths set at 680 nm (32). The time-dependent change in scattering intensity was recorded upon mixing 96 Fe2+ or Fe3+/shell with 2 ml of apoferritin in 100 mm Mops (pH 7.0). When the scattering intensity of above solution remained unchanged, 10 μl of EDTA or NaCl was added to make final concentrations of 1 and 10 mm, respectively.
Dynamic Light Scattering Experiments
The dynamic light-scattering measurements were performed at 25 °C using a Viscotek model 802 dynamic light-scattering instrument (Viscotek Europe Ltd.). The instrument calculates the translational diffusion coefficient DT of the molecules in the sample cell from the autocorrelation function of scattered light intensity data. The hydrodynamic radius RH of the scattering particle is derived from DT, using the Stokes-Einstein relationship, DT = kBT/6πηRH, where kB is the Boltzmann constant, T the absolute temperature in kelvin, and η is the viscosity of the solvent. The protein concentration was 0.5 μm in mixing buffer solution containing Mops (100 mm) at pH 7.0. Protein solutions were filtered through 100-nm pore-size filters (Whatman). To reduce interference with bubbles or dust, each measurement was averaged over 20 runs, each for 5 s using the software OmniSIZE 2.0. The hydrodynamic radius RH was calculated from the regularization histogram method using the spheres model, from which an apparent molecular mass was estimated according to a standard curve calibrated from known globular proteins. The OmniSIZE 2.0 software was used to calculate the size distribution of aggregated protein from the reduction in pH or the addition of ferric ion. Unless otherwise stated, all samples were allowed to stand for 1 h before DLS measurements to ensure that the association reactions were complete.
Fluorescence Titration and Oxidative Deposition of Iron in Ferritin
Fluorescence titration experiments were performed aerobically using the Cary Eclipse spectrophotometer (3, 24). The concentration of the EP was 75 μm in 0.10 m NaCl and 0.10 m Mops, pH 7.0, at 25 °C. The titrations were conducted by adding 2.0 μl increments of Fe3+ (7.5 mm) to 1.00 ml of the EP. After each Fe3+ addition, fluorescence spectra were scanned. Iron oxidation experiments were performed by measuring the absorbance at 300 nm using the Hi-Tech SFA-20 M stopped-flow accessory on a Varian Cary 50 spectrophotometer (Varian) at 25 °C (27). Final concentrations of iron and ferritin solution were 48 and 0.5 μm, respectively. Data were acquired every 12.5 ms (the shortest acquisition time possible with the Cary 50). The spectrophotometer was operated in single beam mode and zeroed before each kinetic run with a cuvette containing apoferritin in 100 mm Mops (pH 7.0).
RESULTS
During our studies of alternate mechanisms of iron deposition in PSF, we obtained evidence for protein association during Fe2+ oxidation. To study this phenomenon in detail we undertook dynamic and stopped-flow light scattering experiments. The stopped-flow experiments were performed in which Fe2+ was rapidly mixed with apoPSF under aerobic conditions (Fig. 1). The light scattering changed little within the 350-s time frame of the experiment when ≤48 Fe/shell was mixed with the apoprotein (as also found using H2O in place of the iron solution). In contrast, the light scattering intensity increased markedly due to protein aggregation when apoferritin was rapidly mixed aerobically with a series of Fe2+ solutions to give ratios from 72 to 168 Fe2+/shell, the rate and magnitude of the increase being a strong function of the ratio beyond 48 Fe2+/shell (Fig. 1A). Because the 24 di-nuclear ferroxidase centers of PSF have a maximal binding capacity of 48 Fe3+ and are located in the interior of the protein (27), these results suggest that protein-protein association is facilitated by binding of the additional iron on the exterior surface of protein shell (more later). All of the kinetic curves are sigmoidal and contain an initial lag phase (Fig. 1B) attributed to the slow oxidation of Fe2+ at exterior sites (see below).
FIGURE 1.
Time course (350 s (A) and 50 s (B)) of the PSF aggregation during Fe2+ oxidation. Aggregation was initiated by mixing apoPSF and Fe2+ aerobically, and association was followed by the intensity of scattered light at a 90° to the incident beam. The curve represents an average of six experimental measurements. Conditions: [apoPSF] = 0.5 μm in 100 mm Mops (pH 7.0), 24–84 μm FeSO4, 25 °C.
In another series of experiments 96 Fe2+/shell were added to apoPSF and holo pea seed ferritin samples containing pre-existing cores ranging from 100 to 1600 Fe3+. The data show that the intensity of scattered light and, thus, the amount of aggregation decreases as the core increases in size (Fig. 2), an observation consistent with increased iron binding and oxidation on the surface of the mineral in preference over sites on the exterior of the protein shell.
FIGURE 2.
Scatter light intensity (protein aggregation) versus the size of iron core of holo pea seed ferritin. Inset, time course of aggregation of PSF with different size iron cores during Fe2+ oxidation. The PSF aggregation was initiated by mixing Fe2+ anaerobically with PSF having different sizes of mineral core (a, 0; b, 100; c, 200; d, 300; e, 400; f, 800; g, 1600 Fe3+/shell). Association was followed by the intensity of scattered light at a 90° to the incident beam. The curve represents an average of six experimental measurements. Conditions: [PSF] = 0.5 μm in 100 mm Mops (pH 7.0), 48 μm FeSO4, 25 °C.
Stopped-flow light scattering experiments were carried out to determine whether Fe2+ or Fe3+ is responsible for the observed protein aggregation. Light scattering intensity increases slowly when Fe2+ is added aerobically to apoPSF (96 Fe2+/shell) to produce Fe3+ (Fig. 3A), whereas the addition of Fe2+ anaerobically causes no change (Fig. 3B). A rapid increase in intensity was observed when Fe3+ was directly added under the same conditions (Fig. 3C). The kinetic curve for aerobic addition of Fe2+ to apoPSF is sigmoidal and requires ∼100 s for the reaction to go to completion. We assign the initial slow induction period to Fe2+ oxidation.
FIGURE 3.
Comparison of three kinetic curves for PSF association under different experimental conditions. A, apoPSF mixed with Fe2+ aerobically. B, apoPSF mixed with Fe2+ anaerobically. C, apoPSF mixed with Fe3+ aerobically. The curve represents an average of six experimental measurements. Condition: [apoPSF] = 0.5 μm in 100 mm Mops (pH 7.0), 48 μm FeSO4 or FeCl3, 25 °C.
When Fe3+ is added directly to the protein, the kinetic curve is hyperbolic, and the reaction is complete within 10 s (Fig. 3C). Thus, Fe3+ facilitates protein-protein association, not Fe2+. Consistent with this idea, a progressive increase in scattering intensity was observed upon the addition of increasing amounts of Fe3+ to apoPSF (Fe3+/shell ratios from 48/1 to 120/1) (Fig. 4A). The initial rate, ν0, increased in a biphasic manner with increasing Fe3+/shell ratio (Fig. 4B). Above a Fe3+/shell ratio of ∼84, aggregates are formed more rapidly and by a process that appears to be first order in iron concentration.
FIGURE 4.
A, scatter light intensity (PSF aggregation) induced by Fe3+ as a function of time. The PSF aggregation was initiated by mixing apoPSF and Fe3+ aerobically, and association was followed by the intensity of scattered light at a 90° to the incident beam. The curve represents an average of six experimental measurements. Conditions: [apoPSF] = 0.5 μm in 100 mm Mops (pH 7.0), 24–60 μm FeCl3, 25 °C. B, a plot of initial rate (ν0) as the change in scattered light intensity per second against the Fe3+/protein ratio. C, the kinetic data of 96 Fe3+/shell sample were best fitted to the model of three sequential reactions with the residual to the fit shown at the bottom. The determined kinetic parameters are k1 = 9.38 ± 0.79, k2 = 0.678 ± 0.034, and k3 = 0.0739 ± 0.0033 s−1; IB = 101.0 ± 3.1, IC = 219.7 ± 2.5, and ID = 331.6 ± 1.6.
The kinetics for the 96 Fe3+/shell sample was best fitted to the sequential reaction scheme shown in Equation 4 (Fig. 4C),
 |
In Equation 4, A represents a monomeric “Fe3+-protein” complex in which Fe3+ is bound to the outer surface of the ferritin 24mer shell that reacts to form aggregates B, C, and D of increasing size. The light scattering intensity, Y(λ,t), as a function of time was fitted to the equation Y(λ,t) = IB(λ)[B(t)] + IC (λ)[C(t)] + ID(λ)[D(t)], where the Ii are molar intensity constants for the light scattering of species B, C, and D at the specified wavelength and B(t), C(t), and D(t) are their concentrations as a function of time as given elsewhere (22). Best fit values were k1 = 9.38 ± 0.79, k2 = 0.678 ± 0.034, and k3 = 0.0739 ± 0.0033 s−1 and IB = 101.0 ± 3.1, IC = 219.7 ± 2.5, and ID = 331.6 ± 1.6. The scattering intensity follows the order ID > IC > IB as expected for aggregates of increasing size. Very similar curve fitting results were obtained with protein samples from three different protein preparations/purifications, indicating that the observed kinetics are not sample-dependent. In contrast, a significantly poorer fit to the data and corresponding large residual were obtained with the simpler scheme A → B → C (not shown). Similarly, poorer fits were obtained with the second-order reaction scheme 2A → B (not shown), indicating a strong correlation between the fitting parameters and inappropriateness of these models. The above model also describes the kinetic behavior at iron/shell ratios of 108/1 and 120/1. The same trends of k1 > k2 > k3 and ID > IC > IB were observed with these samples as for the 96 iron/shell sample.
Protein association facilitated by Fe3+ was also investigated with DLS. Because light scattering reaches its maximal value at 60 s after introduction of Fe3+ to apoPSF (Fig. 4A) and remains stable for more than 1 h (data not shown), samples were allowed to stand for 1 h at room temperature before DLS measurements. All DLS measurements were performed at pH 7.0. Two populations with RH values of 9.6 and 61.2 nm are evident in the scattered light intensity distribution curve of the apoPSF sample (Fig. 5A). The corresponding mass distribution curve (supplemental Fig. S3A) shows that the population centered at 9.6 nm is rich in monomers, comprising about 97% of the total (Table 1), whereas the second population having RH = 61.2 nm represents a small amount (∼3%) of aggregates composed of ∼110 monomers (Table 1). This result indicates that at pH 7.0, essentially all of apoPSF molecules exist in a dissociated state. In contrast, the size distribution is markedly altered toward large aggregates upon introduction of 96 Fe3+/shell to apoPSF (Fig. 5B and supplemental Fig. S3B). The population with RH = 105 nm represents ∼55% of the total and corresponds to a large aggregate of ∼400 monomers (Table 1). Thus, around half of PSF molecules participate in protein association upon treatment with Fe3+, indicating that Fe3+ triggers the formation of large protein assemblies, a finding in accord with the stopped-flow results of Figs. 1, 3, and 4.
FIGURE 5.
Relative scattered light intensity distribution curves for apoPSF (A) and apoPSF plus Fe3+ ion (B). Conditions: [apoPSF] = 0.5 μm in buffer containing 100 mm Mops (pH 7.0), 0 or 48 μm FeCl3.
TABLE 1.
Dynamic light scattering results from supplemental Fig. S3 showing mass distributions of different particle sizes of apoPSF and apoPSF plus 96 Fe3+/shell under the same experimental conditions as used in Fig. 5
| Sample (pH 7.0) | Rh | % Areaa | Apparent Mrb | Approx no. of monomersc |
|---|---|---|---|---|
| nm | ||||
| ApoPSF | 9.6 ± 1.1 | 97.2 ± 1.1 | 690 ± 110 | 1 ± 0.2 |
| 61.2 ± 2.5 | 2.8 ± 1.1 | 55,800 ± 9,200 | 110 ± 19 | |
| ApoPSF + 96 Fe3+ | 8.9 ± 1.5 | 45.1 ± 2.8 | 580 ± 120 | 1 ± 0.3 |
| 105.0 ± 8.0 | 54.9 ± 2.8 | 197,000 ± 30,000 | 395 ± 60 |
a Percent area under each of the peaks in the mass distribution curve of supplemental Fig. S3 corresponding to the percentage of species present.
b Spherical geometry is assumed.
c Estimated number of monomeric PSF molecules that constitute the aggregate. A monomer molecular weight of ∼500 is assumed.
We tested the hypothesis that iron added to the apoprotein in excess of the 48 Fe3+/shell required to saturate the 24 ferroxidase centers (27) binds on the outer surface of the protein where it facilitates protein aggregation. To apoPSF were added 96 Fe2+/shell aerobically, and after the light scattering intensity reached a plateau at 10 s, 1 mm EDTA was added to the solution at a concentration ∼20-fold higher than iron concentration of 48 μm (Fig. 6, curve b). The EDTA causes a gradual decrease in the light scattering intensity to ∼50% within 45 min. In a parallel experiment 10 mm NaCl was used instead of 1 mm EDTA, and no decrease in the light scattering intensity was observed (Fig. 6, curve a), indicating that the dissociation caused by EDTA is not derived from the change of ionic strength. In control experiments with holo pea seed ferritin, no change in scattered light intensity or decrease in 300-nm absorbance from iron in the mineral core was observed upon the addition of EDTA (not shown), indicating that iron located in the interior of the protein is not readily removed by EDTA. Thus, the iron responsible for aggregation is most likely bound on the surface of the protein where EDTA can access and remove it.
FIGURE 6.
Effects of EDTA and NaCl addition and EP deletion on PSF aggregation induced by Fe2+ oxidation. a, 48 μm Fe2+ added aerobically to 0.5 μm apoPSF followed by the addition of 10 mm NaCl. b, 48 μm Fe2+ added aerobically to 0.5 μm apoPSF followed by addition of 1 mm EDTA. c, 48 μm Fe2+ or Fe3+added aerobically to 0.5 μm EP-deleted apoPSF. Conditions: 100 mm Mops (pH 7.0), 25 °C.
To further establish the involvement of protein association in Fe2+ oxidation, a plot of initial rate of Fe2+ oxidation by oxygen as a function of protein concentration was made at a fixed Fe2+ concentration of 96 μm or at a fixed ratio of 96 Fe3+/shell (supplemental Fig. S4). In both instances the initial rates of iron oxidation show a hyperbolic dependence on protein concentration indicative of saturation kinetics, a result confirming a causal relationship between iron oxidation and protein association.
The three-dimensional model predicted for PSF shows that the EP is located on the outer surface of protein (15), raising the question of whether the peptide contains the binding sites for Fe2+ and Fe3+. Accordingly, apoPSF with the EP deleted was rapidly mixed with 96 Fe2+ or Fe3+/shell aerobically in the stopped-flow apparatus. In contrast to wild-type PSF, no change in the light scattering intensity was observed with the EP0deleted protein (Fig. 6, curve c), a result indicating that the extension peptide is an essential structural component for iron binding and protein aggregation.
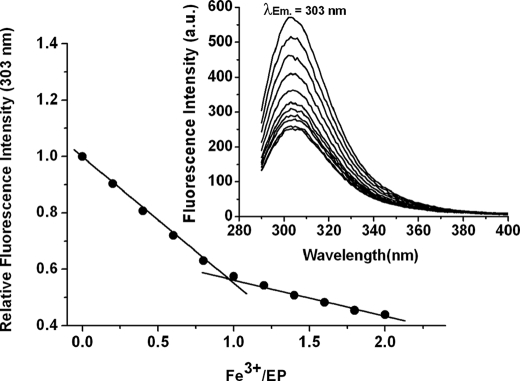
To confirm the above idea, the EP (sequence TTAPLTGVIFEPFEEVKKDYLAVP) was synthesized, and a binding study with Fe3+ carried out. The fluorescence spectra of the EP containing different amounts of Fe3+ are shown in Fig. 7 (inset). Maximal emission occurs at 303 nm for the EP, indicating that most of the observed fluorescence is contributed by the sole tyrosine residue at position 20. The fluorescence quenching titration of Fig. 7 demonstrates that Fe3+ binds to the EP with a 1:1 stoichiometry.
FIGURE 7.
Fluorescence quenching upon Fe3+ addition to the EP. Conditions: λEx = 280 nm, λEm = 303 nm, slits for excitation and emission of 5 and 10 nm, respectively, 75 μm EP, 0–150 μm FeCl3 in 100 mm NaCl and 100 mm Mops, pH 7.0, 25 °C.
In a parallel experiment, under the same conditions as the stopped-flow light scattering experiments shown in Fig. 1, Fe2+ oxidation in both wild-type apoPSF and EP-deleted apoPSF was monitored by absorbance at 300 nm as recently described (Fig. 8) (27). Biphasic absorption kinetics is observed during the same 10–20-s time frame of the lag phase in the light scattering kinetics of Fig. 3A. The initial rapid rise in absorbance is attributed to fast Fe2+ oxidation at the ferroxidase site (27, 33, 34) and, as expected, is nearly identical with both wild-type PSF and EP-deleted PSF because they both have intact ferroxidase centers. However, iron oxidation in the second phase catalyzed by the wild-type protein is about 2-fold faster than that of EP deleted PSF, indicating that the EP catalyzes Fe2+ oxidation in phase 2. As the iron flux into ferritin increases from 96 to 400 Fe2+/protein shell, the disparity in iron oxidation rate between the two proteins becomes much larger (data not shown). These results are consistent with the lag phase in Fig. 3A corresponding to Fe2+ oxidation catalyzed by the EP portion of the protein structure.
FIGURE 8.
Kinetic curves of Fe2+ oxidation by O2 in wild-type PSF and PSF with the EP deleted. Conditions: final (wild type apoPSF or apoPSF without the EP) = 0.5 μm in 100 mm Mops (pH 7.0), 48 μm FeSO4, 25 °C.
To determine whether ferritin association during the oxidative deposition of iron in the protein is reversible, a stopped-flow experiment was carried out in which 96 Fe2+/protein shell were rapidly mixed with apoPSF. The rapid initial rise in intensity was followed by a slow decrease over a period of 14 h to its original value (Fig. 9). Associated with this decay in scattered light intensity was an increase in absorbance at 300 nm because of growth in the mineral core (supplemental Fig. S5). DLS measurements confirmed the reversibility of protein association/dissociation induced by iron (Fig. 10). The mass distribution curve and corresponding parameters of this reversible process are shown in supplemental Fig. S6 and Table S1, respectively. Nearly identical dissociation kinetics was observed with samples prepared by adding Fe3+ directly.
FIGURE 9.
Time-course of the formation and dissociation of PSF aggregates during the oxidative deposition of iron in the protein shell. The curve represents an average of six experimental measurements. Conditions: 0.5 μm apoPSF in 100 mm Mops (pH 7.0), 48 μm Fe2+, 25 °C.
FIGURE 10.
Relative scattered light intensity showing different particle size distributions induced by aerobic addition of Fe2+ to apoPSF at different times. Conditions: 0.5 μm apoPSF in 100 mm Mops (pH 7.0), 48 μm Fe2+, 25 °C.
DISCUSSION
The present work demonstrates that PSF has assembly properties not previously recognized. Significantly, the aggregation of PSF is induced by iron, the natural substrate for ferritin. The sigmoidal association curve of Fig. 1 is only observed upon the addition of more than 48 Fe2+/shell plus O2 to apoPSF, a consequence of the low regeneration activity of the ferroxidase center of PSF (27), causing the excess Fe2+ to bind at exterior sites where it is slowly oxidized, producing the initial lag phase in the light scattering kinetics (Fig. 1B). This initial oxidation takes about 10 s for 96 Fe2+/shell (Fig. 3A) and is much faster than the ∼10 min required for Fe2+ autoxidation under these conditions (data not shown). Thus, PSF contains at least two kinds of centers for Fe2+ oxidation; one is located on the outer surface, and the other is located at the ferroxidase centers. Consistent with this view, the kinetic curves corresponding to PSF aggregation induced by ferric ion are hyperbolic and much faster than the sigmoidal curves produced by Fe2+ plus O2 (Figs. 1, 3C, and 4A). The appreciably different kinetic behavior observed by Fe2+ versus Fe3+ indicates that PSF aggregation is triggered only by the latter. As expected, aggregation does not occur with PSF when 96 Fe2+/protein shell are added to apoPSF under anaerobic conditions (Fig. 3B).
Likewise, protein aggregation is initiated by Fe3+ only when more than 48 Fe3+/protein shell are added to apoPSF (Figs. 4 and 5), further emphasizing that it is Fe3+ bound at exterior sites that facilitates protein-protein intermolecular interactions. The addition of EDTA results in significant dissociation (Fig. 6, curve b), albeit at a relatively slow rate compared with association, perhaps because of reduced accessibility of the Fe3+ to EDTA in the aggregate.
Among several tested models, protein association induced by Fe3+ was best described by a sequential aggregation model given by equation 4 (Fig. 4C) in which intermediates B and C of increasing size are involved, ultimately leading to the final aggregate D. Support for the model comes from DLS results (Fig. 5) showing that upon aerobic treatment with 96 Fe2+/shell for 1 h, the size (RH = 61.2 nm) of the second population in apoPSF increased to 105.0 nm, representing ∼ 55% of the total, accompanied by a decrease in the first population at 9.9 nm corresponding to species A. The population centered at RH = 61.2 nm most likely contains the intermediates B and C, whereas the final aggregate D corresponds to the species with RH = 105 nm (Fig. 5) and is composed of an assembly of ∼400 monomers (Table 1).
Removal of the EP from PSF eliminates protein aggregation in the presence of Fe3+ (Fig. 6, curve c), demonstrating that the EP is an essential structural component of the exterior iron binding site. Fluorescence titration shows that synthesized EP complexes Fe3+ with a 1:1 stoichiometry (Fig. 7). Moreover, EP-deleted PSF exhibits a significantly lower rate of iron oxidation in the second phase as compared with the intact wild-type ferritin (Fig. 8), indicating that the EP domain not only participates in the binding of iron but also its oxidation and, thus, serves as a second center of iron oxidation in addition to the ferroxidase center.
The P-helix of the EP of the 28.0-kDa subunit is flanked by proline residues (X and L), accounts for nearly half of the 24 EP residues, and is folded back on the outer surface of the subunit (15). It contains two Glu residues (O and N) and one Asp residue (S) in reasonable proximity to one another but no His residue (15). These three acidic residues represent possible ligands for Fe3+ binding in which individual ferritin molecules are bridged through coordination bonds to iron to form the aggregate. Ferric ion, because of its hard character and greater positive charge density, is better suited than Fe2+ for binding at these residues and inducing aggregation. At pH 7.0, PSF is anionic (pI = 5.86), and therefore, the more effective charge neutralization afforded by the binding of Fe3+ relative to Fe2+ undoubtedly plays some role in facilitating protein association.
Protein aggregation initiated by the aerobic addition Fe2+ is a strong function of the age of the aggregate and is fully reversible upon standing (Figs. 9 and 10). Complete dissociation was likewise observed with protein aggregation from the direct addition of Fe3+ (data not shown). Disassociation of the aggregate occurs simultaneously with an increase in 300-nm absorbance from growth of the mineral core (supplemental Fig. S5). Based on these findings, a new four-step pathway corresponding to iron oxidative deposition in ferritin through protein-protein association induced by Fe3+ is proposed as elaborated in Scheme 1. Step 1 is the binding reaction of excess Fe2+ with the EP domain located on the exterior surface of PSF at iron/protein ratios exceeding 48 Fe2+/shell. Fe2+ oxidation by O2 under the catalytic action of the EP represents the second step (Figs. 7 and 8) to produce species A. The resulting bound Fe3+ triggers protein association through intermediates B and C, thereby forming PSF aggregate D as described by Equation 4 (Fig. 4C), which corresponds to step 3. After Fe3+ is transferred from the outer surface of protein to the inner cavity, dissociation of the aggregate into monomer is the fourth step (Figs. 9 and 10).
SCHEME 1.
A proposed pathway of Fe2+ oxidative deposition in PSF through protein association at high Fe2+ flux (more than 48 Fe2+/shell) into ferritin.
It is noteworthy that this pathway is operable with both apo- and holoferritin (Figs. 1, 2, and 4) only when the amount of added iron is in excess of that required to saturate the 24 canonical ferroxidase centers. However, this pathway is distinct from the previous mineralization reaction corresponding to Equation 3 where Fe2+ autoxidation directly occurs on the surface of the core. As the size of iron core increases, the mineral surface autoxidation reaction becomes increasingly important as previously described (17, 19, 26), and as expected, there is less iron induced aggregation (Fig. 2). Once the protein attains a core of 1600 Fe3+/shell, little aggregation was observed (Fig. 2).
The present findings are in accord with those of Harrison and co-workers (35) showing that, upon mixing iron-poor and iron-rich ferritin molecules, Fe3+ migrates from the poor to the rich. Increased stability and crystallinity and decreased surface-free energy favors large mineral cores at the expense of small ones. Such transfer of iron may occur through protein-protein association as found in the present study. The reversible aggregation observed for PSF does not derive from permanent disulfide or di-tyrosine cross-linkages as reported for some ferritins damaged during the oxidative deposition of iron in these proteins (36–38). No disulfide and di-tyrosine moieties were observed in our samples nor was there evidence for protein damage in SDS or native PAGE gels of apoPSF treated aerobically with 96 Fe2+/shell compared with untreated apoPSF (data not shown).
In closing, the present study demonstrates that the aerobic addition of Fe2+ or Fe3+ to PSF with its ferroxidase centers saturated with iron (48 iron/shell) results in protein association caused by the binding of Fe3+ to the outer surface of the protein at the extension peptide. This iron ultimately is deposited in the mineral core by a process that represents a new mechanism for the oxidative deposition of iron in ferritin.
Supplementary Material
This work was supported by China High-Tech (863) Project 2007AA10Z333 (to G. Z.) and National Key Technology R&D Program 2006BAD27B04 (to G. Z.). This project was also supported by National Institutes of Health Grant R01-GM20294 (NIGMS, to N. D. C.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
Footnotes
- TP
- transient peptide
- EP
- extension peptide
- Mops
- 3-(N-morpholino)propanesulfonic acid
- DLS
- dynamic light scattering
- PSF
- pea seed ferritin
- apoPSF
- apo pea seed ferritin.
REFERENCES
- 1.Harrison P. M., Arosio P. ( 1996) Biochim. Biophys. Acta 1275, 161– 203 [DOI] [PubMed] [Google Scholar]
- 2.Chasteen N. D., Harrison P. M. ( 1999) J. Struct. Biol. 126, 182– 194 [DOI] [PubMed] [Google Scholar]
- 3.Zhao G., Ceci P., Ilari A., Giangiacomo L., Laue T. M., Chiancone E., Chasteen N. D. ( 2002) J. Biol. Chem. 277, 27689– 27696 [DOI] [PubMed] [Google Scholar]
- 4.Solomon E. I., Brunold T. C., Davis M. I., Kemsley J. N., Lee S. K., Lehnert N., Neese F., Skulan A. J., Yang Y. S., Zhou J. ( 2000) Chem. Rev. 100, 235– 350 [DOI] [PubMed] [Google Scholar]
- 5.Kolberg M., Strand K. R., Graff P., Andersson K. K. ( 2004) Biochim. Biophys. Acta 1699, 1– 34 [DOI] [PubMed] [Google Scholar]
- 6.Kopp D. A., Lippard S. J. ( 2002) Curr. Opin. Chem. Biol. 6, 568– 576 [DOI] [PubMed] [Google Scholar]
- 7.Crichton R. R., Herbas A., Chavez-Alba O., Roland F. ( 1996) J. Biol. Inorg. Chem. 1, 567– 574 [Google Scholar]
- 8.Waldo G. S., Wright E., Whang Z. H., Briat J. F., Theil E. C., Sayers D. E. ( 1995) Plant Physiol. 109, 797– 802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda T., Goto F., Yoshihara T. ( 2001) J. Biol. Chem. 276, 19575– 19579 [DOI] [PubMed] [Google Scholar]
- 10.Laulhere J. P., Laboure A. M., Briat J. F. ( 1989) J. Biol. Chem. 264, 3629– 3635 [PubMed] [Google Scholar]
- 11.Ragland M., Briat J. F., Gagnon J., Laulhere J. P., Massenet O., Theil E. C. ( 1990) J. Biol. Chem. 265, 18339– 18344 [PubMed] [Google Scholar]
- 12.Lescure A. M., Proudhon D., Pesey H., Ragland M., Theil E. C., Briat J. F. ( 1991) Proc. Natl. Acad. Sci. U. S. A. 88, 8222– 8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda T., Goto F., Yoshihara T., Ezure T., Suzuki T., Kobayashi S., Shikata M., Utsumi S. ( 2007) Protein Expr. Purif. 56, 237– 246 [DOI] [PubMed] [Google Scholar]
- 14.van Wuytswinkel O., Savino G., Briat J. F. ( 1995) Biochem. J. 305, 253– 261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobreaux S., Yewdall S. J., Briat J. F., Harrison P. M. ( 1992) Biochem. J. 288, 931– 939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G., Bou-Abdallah F., Arosio P., Levi S., Janus-Chandler C., Chasteen N. D. ( 2003) Biochemistry 42, 3142– 3150 [DOI] [PubMed] [Google Scholar]
- 17.Yang X., Chen-Barrett Y., Arosio P., Chasteen N. D. ( 1998) Biochemistry 37, 9743– 9750 [DOI] [PubMed] [Google Scholar]
- 18.Treffry A., Zhao Z., Quail M. A., Guest J. R., Harrison P. M. ( 1997) Biochemistry 36, 432– 441 [DOI] [PubMed] [Google Scholar]
- 19.Xu B., Chasteen N. D. ( 1991) J. Biol. Chem. 266, 19965– 19970 [PubMed] [Google Scholar]
- 20.Zhao G., Arosio P., Chasteen N. D. ( 2006) Biochemistry 45, 3429– 3436 [DOI] [PubMed] [Google Scholar]
- 21.Hwang J., Krebs C., Huynh B. H., Edmondson D. E., Theil E. C., Penner-Hahn J. E. ( 2000) Science 287, 122– 125 [DOI] [PubMed] [Google Scholar]
- 22.Bou-Abdallah F., Zhao G., Mayne H. R., Arosio P., Chasteen N. D. ( 2005) J. Am. Chem. Soc. 127, 3885– 3893 [DOI] [PubMed] [Google Scholar]
- 23.Pereira A. S., Small W., Krebs C., Tavares P., Edmondson D. E., Theil E. C., Huynh B. H. ( 1998) Biochemistry 37, 9871– 9876 [DOI] [PubMed] [Google Scholar]
- 24.Bou-Abdallah F., Santambrogio P., Levi S., Arosio P., Chasteen N. D. ( 2005) J. Mol. Biol. 347, 543– 554 [DOI] [PubMed] [Google Scholar]
- 25.Yang X., Le Brun N. E., Thomson A. J., Moore G. R., Chasteen N. D. ( 2000) Biochemistry 39, 4915– 4923 [DOI] [PubMed] [Google Scholar]
- 26.Treffry A., Zhao Z., Quail M. A., Guest J. R., Harrison P. M. ( 1998) FEBS Lett. 432, 213– 218 [DOI] [PubMed] [Google Scholar]
- 27.Li C., Hu X., Zhao G. ( 2009) Biochimie 91, 230– 239 [DOI] [PubMed] [Google Scholar]
- 28.Bauminger E. R., Harrison P. M., Hechel D., Nowik I., Treffry A. ( 1991) Biochim. Biophys. Acta 1118, 48– 58 [DOI] [PubMed] [Google Scholar]
- 29.Lee J., Chasteen N. D., Zhao G., Papaefthymiou G. C., Gorun S. M. ( 2002) J. Am. Chem. Soc. 124, 3042– 3049 [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U. K. ( 1970) Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 31.Tonomura B., Nakatani H., Ohnishi M., Yamaguchi-Ito J., Hiromi K. ( 1978) Anal. Biochem. 84, 370– 383 [DOI] [PubMed] [Google Scholar]
- 32.Ivanova E., Jowitt T. A., Lu H. ( 2008) J. Mol. Biol. 375, 229– 239 [DOI] [PubMed] [Google Scholar]
- 33.Zhao G., Su M., Chasteen N. D. ( 2005) J. Mol. Biol. 352, 467– 477 [DOI] [PubMed] [Google Scholar]
- 34.Aitken-Rogers H., Singleton C., Lewin A., Taylor-Gee A., Moore G. R., Le Brun N. E. ( 2004) J. Biol. Inorg. Chem. 9, 161– 170 [DOI] [PubMed] [Google Scholar]
- 35.Bauminger E. R., Harrison P. M., Hechel D., Nowik I., Treffry A. ( 1991) Proc. Biol. Sci. 244, 211– 217 [DOI] [PubMed] [Google Scholar]
- 36.Petsev D. N., Thomas B. R., Yau S., Vekilov P. G. ( 2000) Biophys. J. 78, 2060– 2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niitsu Y., Listowsky I. ( 1973) Biochemistry 12, 4690– 4695 [DOI] [PubMed] [Google Scholar]
- 38.Welch K. D., Van Eden M. E., Aust S. D. ( 2001) Free Radic. Biol. Med. 31, 999– 1006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.