Abstract
Hypoxia-inducible factor (HIF) controls an extensive range of adaptive responses to hypoxia. To better understand this transcriptional cascade we performed genome-wide chromatin immunoprecipitation using antibodies to two major HIF-α subunits, and correlated the results with genome-wide transcript profiling. Within a tiled promoter array we identified 546 and 143 sequences that bound, respectively, to HIF-1α or HIF-2α at high stringency. Analysis of these sequences confirmed an identical core binding motif for HIF-1α and HIF-2α (RCGTG) but demonstrated that binding to this motif was highly selective, with binding enriched at distinct regions both upstream and downstream of the transcriptional start. Comparison of HIF-promoter binding data with bidirectional HIF-dependent changes in transcript expression indicated that whereas a substantial proportion of positive responses (>20% across all significantly regulated genes) are direct, HIF-dependent gene suppression is almost entirely indirect. Comparison of HIF-1α- versus HIF-2α-binding sites revealed that whereas some loci bound HIF-1α in isolation, many bound both isoforms with similar affinity. Despite high-affinity binding to multiple promoters, HIF-2α contributed to few, if any, of the transcriptional responses to acute hypoxia at these loci. Given emerging evidence for biologically distinct functions of HIF-1α versus HIF-2α understanding the mechanisms restricting HIF-2α activity will be of interest.
Cells respond to changes in environmental oxygen levels through the coordinated regulation of the expression of a large number of genes with key functions in processes as diverse as proliferation, differentiation, apoptosis, energy metabolism, and growth factor production that are important in physiological and pathophysiological processes spanning embryonic development, adaptation to altitude, wound healing, inflammation, ischemic vascular disease, and cancer (1–3). Central to many of these responses is the transcription factor hypoxia-inducible factor (HIF),2 which is regulated by oxygen through enzymatic post-translational hydroxylation of the α-subunit (4, 5), which in turn regulates its stability and its interaction with coactivators (5–7).
Analyses of HIF-DNA interactions at ∼50 gene loci have defined a core hypoxia response element (HRE), RCGTG that binds HIF (8). However, genome-wide transcript analyses using microarrays have indicated that a much larger number of genes respond to HIF signaling, with significant positive and negative responses extending across several hundreds of transcripts (9–18). Furthermore, recent studies indicate that HIF may affect gene expression profiles indirectly through diverse mechanisms (19–25), raising questions as to the extent of direct versus indirect effects of HIF.
Further complexity is generated by the existence of multiple HIF isoforms, with the best understood being HIF-1α and HIF-2α. These have similar domain architectures and mechanisms of regulation, and both bind to HREs and efficiently activate HRE-linked reporter genes (26, 27). However, they generate different developmental phenotypes upon inactivation (28–31). Although, differential expression may contribute to these differences, recent studies have demonstrated that each isoform appears to have distinct transcriptional targets (11, 32–35). Mechanisms of HIF-α transcriptional selectivity are poorly understood and chromatin immunoprecipitation studies at a limited number of loci have not shown selective binding of the two HIF-α isoforms (12, 36, 37). However, to date, such studies have examined only a small set of gene loci and it remains unclear how HIF-α binding correlates with functional effects on gene expression across the genome.
To address this we have undertaken a genome-wide analysis of HIF-α DNA binding in MCF-7 cells using chromatin immunoprecipitation. Here we report on the distribution of HIF-1α- and HIF-2α-binding sites across more than 25,500 human gene promoters, and on the correlation of HIF-α binding with functional responses to HIF-1α and HIF-2α across the genome (9). The work provides an estimate of the scale of direct versus indirect effects of HIF on early changes in gene expression in response to HIF activation. Despite a large degree of overlap in binding of the two HIF-α isoforms there were striking differences in gene regulation with HIF-2α contributing very little to the overall HIF response.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF7 breast cancer cells were grown in Dulbecco's modified Eagle's medium, 2 mm l-glutamine, and 10% fetal bovine serum (Sigma). Subconfluent cell cultures were exposed to 2 mm dimethyloxalylglycine (DMOG) (Frontier Scientific) for 16 h prior to harvest.
Chromatin Immunoprecipitation
Three independent chromatin immunoprecipitation assays were performed using the Upstate protocol (Millipore). Cells were sonicated in 30-s pulses for a total of 3 min (Sonics & Materials, VCX 500). Chromatin was immunoprecipitated using rabbit polyclonal antisera to HIF-1α (PM14) and HIF-2α (PM9) (36, 38). These antibodies have previously been shown to perform well in chromatin immunoprecipitation assays and to be highly specific for HIF-1α or HIF-2α (36). Preimmune serum was used as a negative control.
Chromatin immunoprecipitation (ChIP)-ChIP Analysis Using Affymetrix Human Promoter 1.0R Microarray
Immunoprecipitated chromatin was amplified using the Sigma Whole Genome Amplification kit according to the manufacturer's instructions. After each stage of amplification, preservation of enrichment of the PHD3 enhancer (36) was demonstrated by quantitative-PCR. 7.5 mg of amplified chromatin was then digested with 150 miliunits of DNase I (Invitrogen) for 35 min at 37 °C, to generate a fragment size of ∼70 bp, labeled, and hybridized to Affymetrix Human promoter 1.0R arrays, and detected according to the Affymetrix user manual.
The ChIP-ChIP peak detection tool CisGenome (39–41) was used to define potential protein-binding regions. Three independent HIF-1α and HIF-2α chromatin immunoprecipitations were compared with three independent control chromatin immunoprecipitations performed using preimmune sera. Quantile normalization was applied prior to analysis. A moving average (MA) statistic was computed for each probe based on a half-window size of 300 bp or 5 probes. Probes with the MA statistic 4 S.D. away from the global mean were used to define protein-binding regions. Peaks were discarded if they contained less than five probes or were less than 100 base pairs in width. Peaks that were separated by less than 300 base pairs or 5 probes were merged. Peaks that had a left-tail false discovery rate >5% were discounted (39–41). The genomic coordinates of all regions were converted into coordinates based on NCBI build 36 (hg18) and mapped to the nearest transcriptional start site.
Independent Confirmation of Chromatin Enrichment by Real-time Quantitative-PCR
Real-time quantitative-PCR for DNA quantification employed SYBR Green gene expression assays on a StepOne thermocycler (Applied Biosystems). Normalization was to β-actin DNA and fold-enrichment at each locus was calculated using the ΔCT method. Primer sequences are given in supplemental materials Table S1.
Protein Abundance Analysis by Immunoblotting
Gal-tagged HIF-1α and HIF-2α were expressed by the rabbit reticulocyte lysate in vitro transcription and translation system (Promega). Relative amounts of each isoform were compared by immunoblot using anti-Gal. These signals were then used to calibrate the abundance of endogenous proteins using the HIF-1α and HIF-2α antibodies. Primary antibodies used were mouse anti-HIF-1α (BD Transduction Laboratories), mouse anti-HIF-2α (26), and mouse anti-Gal (Santa Cruz Biotechnology).
Expression Array
The microarray analysis of gene expression in response to 16 h of 2 mm DMOG or 1% hypoxia in the presence or absence of siRNA-mediated suppression of HIF-1α or HIF-2α has been previously described (9). To correct for multiple testing, we used an arbitrary false discovery rate cut-off of 5% (q value <0.05) to identify probe sets that are significantly up- or down-regulated between two treatments.
RESULTS
ChIP analyses from three independent experiments identified 546 HIF-1α-binding regions (length 128–1738 bp) and 143 HIF-2α-binding regions (length 142–1866 bp) that met stringent criteria of an MA score more than 4 mean ± S.D., a width of 100 bp, or 5 probes or more and a false discovery rate of <5%. Binding regions were annotated using the nearest gene locus, as identified by the shortest distance to a transcriptional start site. The 546 HIF-1α binding sequences mapped to 394 different gene loci and the 143 HIF-2 binding sequences mapped to 134 different gene loci. The results for the top 25 sequences (as ranked by statistical significance) are given in Tables 1 and 2 and a complete list of all binding regions identified in HIF-1α and HIF-2α chromatin immunoprecipitations is provided in supplemental materials Tables S2 and S3. Immunoprecipitation of all of these top 25 sequences for both HIF-1α and HIF-2α was confirmed by quantitative-PCR. Genes at the immunoprecipitated loci included both known and previously unknown HIF-target genes. When stratified by gene ontology using the DAVID Bioinformatics Resources (david.abcc.ncifcrt.gov) (42, 43) a trend toward a higher proportion of binding to HIF-1 versus HIF-2 was observed in genes encoding glycolytic enzymes and oxidoreductase enzymes, as compared with genes encoding molecules involved in angiogenic and hematopoietic pathways. However, the numbers in some functional groups were small, and overall differences were not statistically significant (Fig. 1).
TABLE 1.
Anti-HIF-1α chromatin immunoprecipitation (top 25 gene loci)
The table gives the chromosomal coordinates of each anti-HIF-1α immunoprecipitated locus meeting criteria of an MA Z score of >4, a width of >100 bp or >5 probes, and a false discovery rate of <5%, together with the maximum Z score for anti-HIF-1α and anti-HIF-2α at that gene locus. The presence or absence, and position of any HRE core motif RCGTG within these sequences is given. The table also specifies whether the nearest gene to the immunoprecipitated locus was significantly regulated by exposure of cells to 1% hypoxia or 2 mm DMOG or by transfection of siRNAs directed against HIF-1α, HIF-2α, or both siRNAs in the Affymetrix expression arrays. Note that at a number of loci several sequences were immunoprecipitated that shared proximity to a common transcriptional start site. These loci are listed in order with the most strongly enriched site indicated in bold type and others in normal type.
| HIF-1 | Gene Annotation | Gene name | RefSEQ number | Regulated in expression array | Chromosome | Start | End | Strand | Max Z-score |
RCGTG | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HIF-1 | HIF-2 | ||||||||||
| 1 | ASMT | Acetylserotonin o-methyltransferase | NM_004043 | No | chrX | 1,842,483 | 1,844,214 | + | 17.9 | 21.9 | Yes |
| 2 | PLS3 | Plastin 3 (t isoform) | NM_005032 | No | chrX | 114,898,432 | 114,899,202 | + | 15.8 | 17.4 | Yes |
| 3 | GPI | Glucose-phosphate isomerase | NM_000175 | No | chr19 | 39,541,340 | 3,9542,126 | + | 15.7 | 9.6 | Yes |
| chr19 | 39,540,468 | 39,540,783 | + | 7.4 | No | ||||||
| 4 | DARS | Aspartyl-tRNA synthetase | NM_001349 | Yes | chr2 | 136,458,510 | 136,459,327 | − | 15.5 | 9.8 | Yes |
| 5 | SNAPC1 | Small nuclear RNA activating complex, polypeptide 1, 43 kDa | NM_003082 | No | chr14 | 61,291,513 | 61,291,955 | + | 13.9 | 6.0 | No |
| 6 | SEC61G | Sec61 γ-subunit | NM_001012456 | No | chr7 | 54,794,557 | 54,794,993 | − | 13.4 | 12.2 | Yes |
| 7 | GTF2IRD2B | Gtf2i repeat domain containing 2b | NM_001003795 | No | chr7 | 74,146,820 | 74,147,110 | + | 12.7 | 7.1 | No |
| chr7 | 74,145,407 | 74,145,716 | + | 6.1 | No | ||||||
| chr7 | 74,147,633 | 74,147,933 | + | 5.4 | No | ||||||
| 8 | SAP30 | Sin3a-associated protein, 30 kDa | NM_003864 | Yes | chr4 | 174,527,153 | 174,528,353 | + | 12.6 | 9.3 | Yes |
| 9 | ZMYND8 | Protein kinase C-binding protein 1 | NM_012408 | Yes | chr20 | 45,422,559 | 45,423,749 | − | 12.1 | 15.2 | Yes |
| chr20 | 45,421,122 | 45,421,975 | − | 9.3 | Yes | ||||||
| chr20 | 45,416,738 | 45,416,961 | − | 5.4 | No | ||||||
| chr20 | 45,324,234 | 45,324,808 | − | 5.3 | Yes | ||||||
| chr20 | 45,382,266 | 45,382,561 | − | 5.2 | No | ||||||
| chr20 | 45,381,728 | 45,381,912 | − | 5.0 | No | ||||||
| chr20 | 45,322,893 | 45,323,036 | − | 4.8 | No | ||||||
| chr20 | 45,380596 | 45,380,865 | − | 4.7 | No | ||||||
| 10 | RSBN1 | Round spermatid basic protein 1 | NM_018364 | Yes | chr1 | 114,156,633 | 114,157,374 | − | 11.9 | 6.0 | Yes |
| chr1 | 114,154,609 | 114,155,069 | − | 5.4 | Yes | ||||||
| chr1 | 114,154,093 | 114,154,264 | − | 5.1 | No | ||||||
| 11 | BNIP3L | Bcl2/adenovirus e1b 19-kDa interacting protein 3-like | NM_004331 | Yes | chr8 | 26,297,111 | 26,297,957 | + | 11.9 | 7.9 | Yes |
| 12 | FAM139A | Hypothetical protein flj40722 | NM_173678 | Yes | chr7 | 143,028,228 | 143,029,042 | + | 11.8 | 14.0 | Yes |
| 13 | GTF2IRD2 | Gtf2i repeat domain containing 2 | NM_173537 | No | chr7 | 73,904,992 | 73,905,289 | − | 11.6 | Yes | |
| chr7 | 73,906,359 | 73,906,641 | − | 6.5 | No | ||||||
| 14 | C3ORF28 | Chromsome 3 open reading frame 28 | NM_014367 | Yes | chr3 | 123,584,923 | 123,585,595 | + | 11.6 | 4.9 | No |
| chr3 | 123,586,518 | 123,586,736 | + | 4.7 | No | ||||||
| chr3 | 123,585,892 | 123,586,061 | + | 4.6 | No | ||||||
| 15 | STC2 | Stanniocalcin 2 | NM_003714 | Yes | chr5 | 172,688,209 | 172,689,013 | − | 11.2 | 8.8 | Yes |
| chr5 | 172,689,765 | 172,690,086 | − | 6.0 | Yes | ||||||
| 16 | NARF | Nuclear prelamin a recognition factor | NM_001038618 | Yes | chr17 | 78,008,958 | 78,009,355 | + | 10.9 | No | |
| 17 | TF | Transferrin | NM_001063 | No | chr3 | 134,944,094 | 134,944,856 | + | 10.7 | 13.6 | Yes |
| 18 | HK2 | Hexokinase 2 | NM_000189 | Yes | chr2 | 74,914,233 | 74,914,441 | + | 10.5 | 5.6 | Yes |
| chr2 | 74,912,970 | 74,913,723 | + | 6.9 | No | ||||||
| 19 | INHA | Inhibin, α | NM_002191 | Yes | chr2 | 220,149,206 | 220,150,032 | + | 10.4 | 9.1 | Yes |
| chr2 | 220,150,797 | 220,151,070 | + | 5.2 | No | ||||||
| 20 | PCF11 | Kiaa0824 protein | NM_015885 | No | chr11 | 82,544,764 | 82,545,226 | + | 10.3 | 8.1 | Yes |
| 21 | C9ORF30 | Chromosome 9 open reading frame 30 | NM_080655 | No | chr9 | 102,228,622 | 10,2228,900 | + | 10.3 | 9.7 | Yes |
| chr9 | 102,231,225 | 102,231,536 | + | 5.9 | Yes | ||||||
| 22 | CBWD3 | Cobw domain containing 3 | NM_201453 | No | chr9 | 70,046,581 | 70,047,378 | + | 10.2 | 5.1 | Yes |
| 23 | RAD51L1 | Rad51-like 1 (Saccharomyces cerevisiae) | NM_133510 | Yes | chr14 | 67,356,360 | 67,357,875 | + | 9.8 | 8.2 | Yes |
| 24 | KIAA0195 | Kiaa0195 | NM_014738 | No | chr17 | 70,960,840 | 70,961,202 | + | 9.7 | 11.6 | Yes |
| 25 | S100P | S100 calcium-binding protein p | NM_005980 | Yes | chr4 | 6,721,070 | 6,721,680 | + | 9.7 | 9.4 | Yes |
TABLE 2.
Anti-HIF-2α chromatin immunoprecipitation (top 25 gene loci)
The table gives the chromosomal coordinates of each anti-HIF-2α immunoprecipitated locus meeting criteria of an MA Z-score of >4, a width of >100 bp or >5 probes, and a false discovery rate of <5%, together with the maximum Z-score for anti-HIF-1α and anti-HIF-2α at that gene locus. The presence or absence, and position of any HRE core motif RCGTG within these sequences is given. The table also specifies whether the nearest gene to the immunoprecipitated locus was significantly regulated by exposure of cells to 1% hypoxia or 2 mm DMOG or by transfection of siRNAs directed against HIF-1α, HIF-2α, or both siRNAs in the Affymetrix expression arrays. Note that at a number of loci several sequences were immunoprecipitated that shared proximity to a common transcriptional start site. These loci are listed in order with the most strongly enriched site indicated in bold type and others in normal type.
| HIF-2 | Gene Annotation | Gene name | RefSEQ number | Regulated in array | Chromosome | Start | End | Strand | Max Z-score |
RCGTG | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HIF-2 | HIF-1 | ||||||||||
| 1 | ASMT | Acetylserotonin o-methyltransferase | NM_004043 | No | chrX | 1,842,483 | 1,844,249 | + | 21.9 | 17.9 | Yes |
| 2 | PLS3 | Plastin 3 (t isoform) | NM_005032 | No | chrX | 114,898,432 | 114,899,202 | + | 17.4 | 15.8 | Yes |
| 3 | ZMYND8 | Protein kinase C-binding protein 1 | NM_012408 | Yes | chr20 | 45,422,559 | 45,423,470 | − | 15.2 | 12.1 | Yes |
| chr20 | 45,421,078 | 45,421,975 | − | 12.5 | Yes | ||||||
| 4 | FAM139A | Hypothetical protein flj40722 | NM_173678 | Yes | chr7 | 143,028,228 | 143,029,153 | + | 14.0 | 11.8 | Yes |
| 5 | TF | Transferrin | NM_001063 | No | chr3 | 134944,026 | 134,944,856 | + | 13.6 | 10.7 | Yes |
| 6 | DHX35 | Deah (Asp-Glu-Ala-His) box polypeptide 35 | NM_021931 | No | chr20 | 37022,195 | 37,022,900 | + | 12.7 | 5.6 | No |
| 7 | SEC61G | Sec61 γ-subunit | NM_001012456 | No | chr7 | 54794,671 | 54,794,993 | − | 12.2 | 13.4 | No |
| 8 | HIG2 | Hypoxia-inducible protein 2 | NM_013332 | Yes | chr7 | 127,882,554 | 127,883,114 | + | 12.1 | 9.5 | Yes |
| 9 | CITED2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxyl-terminal domain, 2 | NM_006079 | Yes | chr6 | 139,738,163 | 139,739,480 | − | 12.1 | 8.1 | Yes |
| 10 | RAPGEF1 | Rap guanine nucleotide exchange factor (gef) 1 | NM_005312 | No | chr9 | 133,609,690 | 133,610,065 | − | 12.0 | 6.2 | Yes |
| 11 | CHD1L | Chromodomain helicase DNA-binding protein 1-like | NM_004284 | No | chr1 | 145,181,416 | 145,181,919 | + | 11.9 | 7.5 | Yes |
| 12 | UPK1B | Uroplakin 1b | NM_006952 | No | chr3 | 120,386,306 | 120,387,057 | + | 11.7 | 8.6 | Yes |
| 13 | KIAA0195 | Kiaa0195 | NM_014738 | No | chr17 | 70,960,840 | 70,961,202 | + | 11.6 | 9.7 | Yes |
| 14 | BACE2 | β-Site app-cleaving enzyme 2 | NM_138992 | Yes | chr21 | 41,458,829 | 41,460,026 | + | 11.4 | 8.7 | Yes |
| 15 | C1ORF161 | Chromosome 1 open reading frame 161 | NM_152367 | No | chr1 | 116,455,368 | 116,456,452 | + | 11.2 | 6.9 | Yes |
| 16 | GPRC5A | G protein-coupled receptor, family c, group 5, member a | NM_003979 | Yes | chr12 | 12,916,200 | 12,917,123 | + | 11.2 | 6.1 | Yes |
| 17 | UBC | Ubiquitin c | NM_021009 | No | chr12 | 123,966,902 | 123,967,854 | − | 11.2 | 7.4 | No |
| 18 | S100A4 | S100 calcium-binding protein a4 (calcium protein, calvasculin, metastasin, murine placental homolog) | NM_002961 | Yes | chr1 | 151,784,245 | 151,784,831 | − | 11.1 | 4.7 | Yes |
| 19 | FLJ16641 | Flj16641 protein | NM_001004316 | No | chr3 | 158,017,688 | 158,018,251 | + | 10.8 | 9.3 | Yes |
| chr3 | 158,012,829 | 158,013,289 | + | 8.9 | Yes | ||||||
| chr3 | 158,027,265 | 158,027,560 | + | 7.4 | Yes | ||||||
| 20 | ARRDC3 | Arrestin domain containing 3 | NM_020801 | No | chr5 | 90,612,390 | 90,613,364 | − | 10.6 | 9.4 | Yes |
| 21 | CYP27C1 | Flj16008 protein | NM_001001665 | No | chr2 | 127,700,812 | 127,701,131 | − | 10.5 | 8.3 | Yes |
| 22 | DARS | Aspartyl-tRNA synthetase | NM_001349 | Yes | chr2 | 136,458,799 | 136,459,291 | − | 9.8 | 15.5 | Yes |
| 23 | PLAC8 | Placenta-specific 8 | NM_016619 | Yes | chr4 | 84,253,030 | 84,253,558 | − | 9.8 | 4.8 | Yes |
| 24 | C9ORF30 | Chromosome 9 open reading frame 30 | NM_080655 | No | chr9 | 102,228,622 | 102,228,871 | + | 9.7 | 10.3 | Yes |
| 25 | FLJ39743 | Hypothetical protein flj39743 | NM_182562 | No | chr15 | 96,875,764 | 96,876,378 | − | 9.7 | 5.9 | Yes |
FIGURE 1.
Functional classification of HIF-binding gene loci. The highest stringency (MA score >4) HIF-1α and HIF-2α immunoprecipitating gene loci were stratified by gene ontology using the DAVID Bioinformatics Resource. The number of immunoprecipitating gene loci in each functional category is displayed.
Relationship of Immunoprecipitated DNA Sequences to the HRE Consensus Sequence
Previous studies have identified HIF binding sequences at ∼70 loci, proposed to encode direct HIF transcriptional targets, defining a consensus core HIF-binding motif, RCGTG (8).
As a first step in analyzing these sequences we sought to determine what proportion of the captured DNA sequences contained the RCGTG core motif. Using an MA score of >4 to define the boundaries of immunoprecipitated sequences, we found that 235 (43%) HIF-1-binding sequences and 92 (66%) of the HIF-2-binding sequences contained an RCGTG consensus, many containing multiple motifs; as expected, in each case, this frequency was very much higher than in a control set of sequences taken from the promoter regions of a randomly selected set of genes (p < 10−9). We also observed an increased frequency of the RCGTG motif in the 300 bp (the limit of resolution derived from the DNA fragmentation procedure) flanking each identified sequence (p < 10−3), indicating that many loci are associated with RCGTG motifs just outside the defined sequence, most probably reflecting the use of a very stringent boundary definition. When these flanking sequences were included, the number of sequences that contain an RCGTG motif rose to 382 (70%) for HIF-1α and 121 (85%) for HIF-2α. Thus, whereas the large majority of HIF-binding sequences contain an RCGTG motif, a significant minority apparently do not. Whether these sequences are captured through higher order DNA/protein interactions with HIF bound at HREs, or represent other modes of DNA binding by HIF proteins is unclear.
Overall, however, within the promoter array, only a small proportion (<1%) of DNA sequences containing the core RCGTG motif bound HIF-1α or HIF-2α. We next sought to define features of the DNA sequence that might be associated with immunoprecipitation either by anti-HIF-1α or anti-HIF-2α antibodies. First, we examined the sequences in the immediate vicinity of the RCGTG motifs. All immunoprecipitated RCGTG motifs were extended by 15 base pairs in both directions. However, analysis did not reveal any significant overrepresentation of particular bases beyond the core 5-base pair motif for either HIF-1- or HIF-2-binding sites (supplemental materials Fig. S1).
Second, we tested the predictive value of conservation in determining HIF-binding to the consensus motif. Altogether 82 of the 464 RCGTG consensus motifs that bound HIF-1α and 38 of 188 that bound HIF-2α were within the top 10% most conserved regions of the genome (as defined by PhastCons score). Although this proportion was significantly greater than would be expected by chance alone (p < 0.005), when the same analysis was performed on a set of control sequences from promoters that did not bind HIF, comparable results were obtained, suggesting that, within promoter regions, whether a putative HRE lies within a region of apparent conservation is not a good guide to its ability to bind HIF.
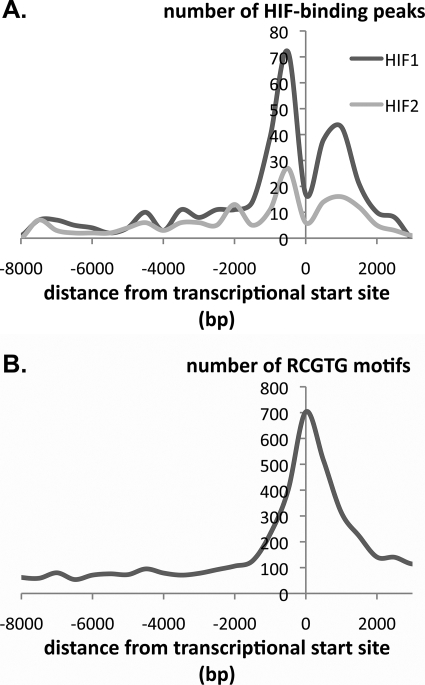
Third, we analyzed the distribution of the captured sequences and associated RCGTG motifs in relation to the transcriptional start site of the closest gene. Clearly the design of the array (intended to represent sequences from −7.5 to +2.5 kb of the transcription start site) places limits on this analysis, although a proportion of the immunoprecipitated sequences lay outside these regions, most likely representing changes in gene annotation between that on which the array was designed and the (latest) annotation that we used for locus assignment. Several interesting features were observed irrespective of whether the precise position of the peak of enrichment profile or the associated HRE was used in the analysis. The distribution of immunoprecipitated sequences was closely similar for both HIF-1α and HIF-2α with the greatest density of HIF-binding sites being observed within 2 kb of the transcriptional start. This region comprised two distinct frequency peaks. One is a sharp peak centered 500 bp upstream of the transcriptional start site, similar to that observed for other transcription factors (44). The other is a broader peak centered 1000 bp downstream of the transcriptional start site. This distribution contrasted markedly with the distribution of all RCGTG motifs at the same set of promoters, which revealed a single peak centered at the transcriptional start site (Fig. 2).
FIGURE 2.
Distribution of HIF-1α- and HIF-2α-binding sites. A, distribution of high stringency (MA score >4) HIF-1α- and HIF-2α-binding sites referred to the nearest transcriptional start site. Number of HIF-binding peaks expressed per 500-bp bin is shown. B, frequency distribution of RCGTG motifs at gene loci that bind HIF-1α or HIF-2α. The number of RCGTG motifs expressed per 500-bp bin is shown.
Finally, to search for overrepresented transcription factor-binding sites that might bind factors cooperating with HIF we identified HIF-binding regions falling within 1 kb of the gene transcriptional start sites (any transcript from the gene). We then extracted the regions 1 kb up- and downstream of the transcriptional start site, omitting known exons (all annotation was taken from the ENSEMBL genome data base (release 51) (45)). We searched these regions using the vertebrate matrices of the TRANSFAC data base (release 2008.4, 621 matrices) (46), using the transcription factor-binding sites perl modules and a 90% scoring threshold cut off, recording, for each factor, and each potential target gene, the number of matches per 1000 non-exonic bases. We compared these numbers to the corresponding numbers for all non-HIF binding gene loci in the genome using a Wilcoxon test implemented in the R software package, using a threshold of p < 8 × 10−5 (p < 0.05 with Bonferroni correction for multiple comparisons).
For all matched HIF regions (combining the HIF-1α and HIF-2α sets), six matrices were identified as overrepresented in the HIF-binding loci (Fig. 5). Three are HIF-binding motifs, further validating the currently held HRE consensus; the others being binding motifs for nuclear respiratory factor-1 (47), Myc intron factor (MIF-1) (48), and E2F transcription factors (49). Given the strict definition of statistical significance, it is also likely that a number of other overrepresented matrices are also valid. Indeed, a number of other E2F matrices, as well as those for an array of previously described HIF-interacting transcriptional cofactors, such as STAT, ETS, and MYC (8), were enriched, but failed to reach the statistical threshold (supplemental materials Table S4). When subgroups of HIF-1α and HIF-2α interacting gene loci were analyzed no significant difference was identified between the two sets of genes, although the numbers in this subgroup analysis were smaller.
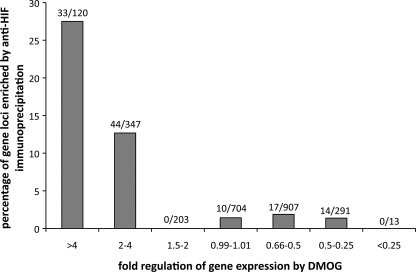
FIGURE 5.
Relationship between amplitude and direction of regulation and chromatin immunoprecipitation by anti-HIF-1α or HIF-2α. Gene loci were assigned to functional groups according to fold up- or down-regulation by DMOG in the Affymetrix expression array. The proportion of gene loci captured by either of the anti-HIF-α immunoprecipitations is given for each of the functional group.
DNA Sequences Immunoprecipitated by Anti-HIF-1α and Anti-HIF-2α Overlap
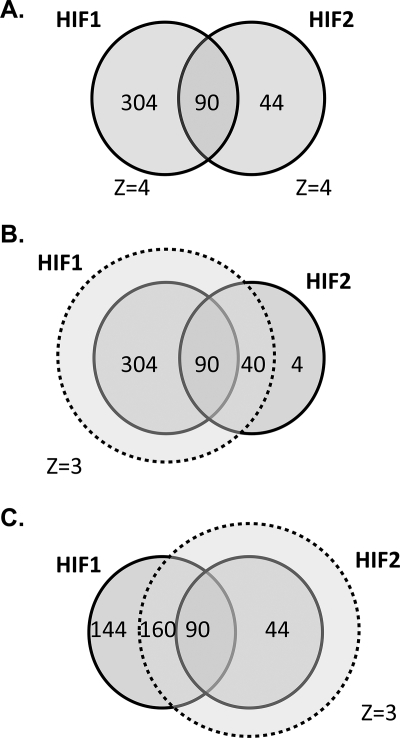
Overall, there was a substantial overlap between the loci immunoprecipitated with the two HIF isoforms. Thus, of the 394 loci that bound HIF-1α and the 134 loci that bound HIF-2α, 90 bound both isoforms. When the stringency of binding for the second isoform was relaxed slightly (MA score >3), the overlap increased, with 250/394 HIF-1α loci binding HIF-2α and 130/134 HIF-2α loci binding HIF-1α. In addition, when both HIF isoforms bind to the same gene locus, they do so at a common site. Thus, of the 90 gene loci that bound both HIF isoforms at the highest level of stringency, all apart from 3 bound HIF-1α and HIF-2α at overlapping sites (Fig. 3).
FIGURE 3.
Intersection between HIF-1α- and HIF-2α-binding gene loci. A, gene loci that bound either HIF-1α or HIF-2α or both isoforms with a Z-score >4. B, defining HIF-1α capture with a Z-score of 3 encompasses almost all HIF-2α binding loci. C, defining HIF-2α capture with a Z-score of 3 excludes almost one-third of HIF-1α binding loci.
Nevertheless, a substantial number of gene loci apparently bound HIF-1α in isolation, despite the similar consensus. We hypothesized that this could be due to higher levels of HIF-1α in the cells, higher affinity of HIF-1α capture by the immunoprecipitating antibody, a generalized higher affinity binding of HIF-1α to the DNA across all loci, or specific differences in affinity for the HIF-α isoforms at particular loci despite a similar consensus.
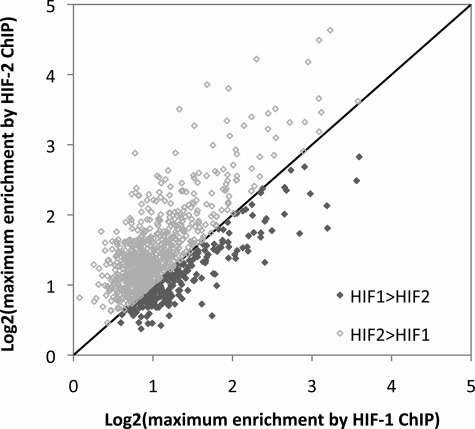
Although we had selected MCF-7 cells for this study on the basis of apparently similar expression of both HIF-α isoforms, we wished to check the relative levels of each isoform using quantitative immunoblotting related to a common standard (supplemental materials Fig. S2). These studies revealed that HIF-2α levels were at least as high as HIF-1α (probably in the region of 3–5-fold higher) indicating that the relative abundance of the two HIF isoforms cannot explain the increased capture by anti-HIF-1α. If the reason for the excess of sites captured by anti-HIF was a greater affinity of the anti-HIF-1α antibody, or a globally higher affinity of HIF-1α for the consensus binding motif, then it would be predicted that enrichment with anti-HIF-1α immunoprecipitation would generally be greater than enrichment by HIF-2α immunoprecipitation across sequences that were immunoprecipitated by both antibodies. We therefore examined all overlapping HIF-1α- and HIF-2α-binding sites for which the MA score was greater than 2. For each of these overlapping binding sites, the maximum fold-enrichment by the two HIF isoforms was compared. In contrast to prediction, across these common sites, HIF-2α showed a trend to greater maximum enrichment than HIF-1α (Fig. 4). Therefore, we conclude that excess HIF-1α-binding sites is not due to better performance of the anti-HIF-1α antibody or higher affinity of HIF-1α for its consensus, and is likely to be due to specific cooperative interactions that regulate (positively or negatively) preferential binding of HIF-α isoforms at certain loci. Interestingly, a weak, but significant positive correlation was also seen between the maximum fold-enrichment by each of the two antibodies suggesting that binding of one isoform to a given sequence is not a result of decreased binding of the other isoform.
FIGURE 4.
Capture by HIF-1α and HIF-2α at common binding sites. Using a Z-score of 2 for each immunoprecipitation identified 992 genomic regions that bound both HIF-1α and HIF-2α. For each of these sequences the maximum enrichment by HIF-2α immunoprecipitation was plotted against that for the HIF-1α immunoprecipitation. More points lie above than below the line of identity indicating greater average enrichment by HIF-2α immunoprecipitation at these common binding loci.
Relationship of Chromatin Immunoprecipitation to the Functional Effects of the HIF-α Isoforms
To better understand the functional consequences of promoter binding by HIF-1α and HIF-2α we next compared chromatin immunoprecipitation data with functional effects on gene expression across the genome, assayed in the same cell line using 2 mm DMOG or 1% hypoxia with or without siRNA to the HIF-α isoforms (9). Overall, 4% of Affymetrix transcripts were significantly up- or down-regulated by one or another of the stimuli with very strong correlations being observed between up-regulation by DMOG and hypoxia and down-regulation by HIF-1α siRNA (9). Genes at loci that were immunoprecipitated by anti-HIF-1α or anti-HIF-2α antibodies were very significantly more likely to be up-regulated by the hypoxia/HIF-response. Thus 20.8% of the loci immunoprecipitated by anti-HIF-1α manifest significant up-regulation by at least one of these stimuli versus 3.6% of those not immunoprecipitated (p < 10−10), whereas for loci immunoprecipitated by anti-HIF-2α the equivalent figures were 32.8 versus 3.7% (p < 10−10). Although, as expected, these correlations are highly significant, the proportion of HIF-binding loci associated with HIF-inducible transcripts was still well below 50%. To explore this further we examined other genes in the vicinity of the immunoprecipitated loci. When all genes within 100 kb of the immunoprecipitated sequences were analyzed, only a few additional regulated transcripts were identified. For instance, at HIF-1α binding loci only 18 further regulated genes were identified in the expression array. Hence, the majority of HIF immunoprecipitating sequences do not regulate expression of a gene within 100 kb of the binding site.
When HIF-1α and HIF-2α binding was separately correlated with functional effects of siRNA suppression a very striking difference was observed. Whereas 15.6% of gene loci that were immunoprecipitated by the HIF-1α antibody were down-regulated by HIF-1α siRNA as compared with 2.3% of gene loci that were not immunoprecipitated (p < 2 × 10−5), only 1.5% of the gene loci immunoprecipitated by the HIF-2α antibody were reduced by HIF-2α siRNA, compared with 0.3% of gene loci that did not immunoprecipitate. Furthermore, when quantitative comparisons were made between maximum fold-enrichment by anti-HIF-α chromatin immunoprecipitation and reduction in gene expression by HIF-α siRNA, a clear correlation was observed for HIF-1α (r = −0.367, p < 0.001) but not HIF-2α (r = −0.183, p = 0.04) where the gradient of any correlation was close to zero (supplemental materials Fig. S3). Similarly no correlation was found between the ratio of combined HIF-1 and -2α siRNA to HIF-1α siRNA (as a measure of HIF-2α activity) and enrichment by anti-HIF-2α ChIP, whereas responses to hypoxia (p = 0.02), DMOG (p = 0.015), combined HIF-1 and -2α siRNA (p < 0.001), and the ratio of combined HIF-1 and -2α to HIF-2α siRNA (as a measure of HIF-1α activity) all correlated with enrichment by HIF-1α ChIP.
Despite the increased binding of HIF-2α compared with HIF-1α at gene loci that bound both isoforms, only one of 90 (TMTC2) was significantly down-regulated by HIF-2α siRNA compared with 25 that were down-regulated by HIF-1α siRNA. Furthermore, TMTC2 was down-regulated by both siRNAs.
In addition, when the top 500 HIF-2α responsive genes (defined as those manifesting the greatest reduction by HIF-2α siRNA) were compared with the 500 least responsive HIF-2α genes no differences were observed either in the numbers reaching an MA score of at least 4 or in the average MA score or fold-enrichment by anti-HIF-2α ChIP. Thus, even the modest effects of HIF-2α on gene regulation that were observed do not appear to relate to HIF-2α binding at these loci, suggesting that these effects of HIF may be indirect. This suggests that although HIF-2α is (on average) binding more strongly than HIF-1α at these loci, HIF-1α is largely or even entirely responsible for transcriptional responses over the time course of the experiments.
Direction of Regulation by Hypoxic Stimuli and HIF Binding
Finally, to pursue the question of the extent to which hypoxia/DMOG inducible and HIF-dependent changes in gene expression are directly or indirectly dependent on HIF binding at the relevant locus, we categorized genes based on the amplitude and direction of changes in expression in response to these stimuli, and determined the proportion of genes in each category that were associated with sequences binding to HIF (Figs. 5 and supplemental S4). This analysis revealed a striking difference between genes that were up-regulated by hypoxic stimuli and those that were down-regulated by these stimuli. Thus for genes that were significantly up-regulated by DMOG, 28% of all loci bound HIF-1α, HIF-2α, or both, whereas for genes that were down-regulated the figure was not significantly different from the 1.4% of genes defined as unregulated (on the basis of fold-regulation by DMOG between 0.99 and 1.01). Grouping of genes by the magnitude of regulation by DMOG revealed that more of the highly up-regulated genes bound HIF (27.5% of those regulated >4-fold by DMOG versus 12.7% of those regulated 2–4-fold and 0% of those regulated 1.5–2 fold; p < 10−12) (Fig. 5). However, no such association was observed among the down-regulated genes where even the most strongly down-regulated genes were no more likely than unregulated genes to bind HIF. Similar results were obtained whether functional responses were stratified on the basis of the response to hypoxia, or suppression of that response by siRNA directed against HIF-α (supplemental materials Fig. S4).
DISCUSSION
Genome-wide analysis of DNA binding to the HIF transcription factors defined more than 600 sites that bound HIF-1α, HIF-2α, or both proteins, although, given the highly stringent criteria we used to define binding sites and the restriction of the tiled array to known or predicted gene promoter regions, the actual number of HIF-binding sites is likely to be substantially greater.
Our results confirmed the core HIF-binding motif, RCGTG (8), and did not define other bases in the immediate vicinity that were significantly overrepresented at bound regions. Nevertheless, HIF binding to this motif was highly selective, indicating that its epigenetic context must in some way define binding. Less than 1% of such motifs within the tiled array bound HIF, and bound sites showed a highly selective distribution in relation to the transcriptional start site. Unexpectedly, binding sites were found to cluster in two distinct regions ∼0.5 kb upstream and 1 kb downstream of the start site, presumably representing the operation of other factors that promote HIF binding within these regions, or restrict accessibility or binding of HIF in the immediate vicinity of the start sites. Indeed, such binding patterns have been previously described for several other transcription factors as well as for histone modifications (50, 51). In silico searches for transcription factor binding motifs that were overrepresented at HIF binding promoters versus equivalent regions at other promoters revealed a number of sites. Interestingly, after the HIF binding consensus itself, the most strongly enriched site was the core binding motif for nuclear respiratory factor-1, a key transcriptional regulator of genes involved in mitochondrial biogenesis and function (47). Whether this reflects specific cooperation in the response to hypoxia, or the operation of physiologically distinct pathways on a group of common genes will require further study.
Pan-genomic comparison of the chromatin immunoprecipitation data with functional responses (9) enabled HIF-binding and transcriptional responses to be compared across the genome. Surprisingly, even though analysis of HIF-binding sequences was restricted to high stringency binding at known or predicted promoter regions, the region of 70% of bound loci were not associated with genes that manifest significant responses to either hypoxia or HIF, within 100 kb of the bound locus. The proportion of HIF-binding loci responding to HIF is comparable with the ratio of Myc binding genes that were differentially modulated in response to Myc (52), where additional transcription factor binding is necessary so that targets are poised to respond (50). Furthermore, although it is also possible that these sites affect gene expression over even longer distances, or are entirely non-functional, the findings also raise the possibility that HIF has as yet unknown effects on genome integrity or function that are mediated by binding at such sites.
Comparison of chromatin immunoprecipitation and functional responses to HIF also allowed the proportion of direct versus indirect HIF-dependent responses to estimated. Across the genome, in the region of 20% of genes reported to demonstrate positive HIF-1α-dependent responses bound HIF at their promoters. Although we cannot be certain that all these HIF-binding sites are functional, the use of highly stringent criteria to define HIF-binding and limited genome coverage by the promoter array would suggest that 20% is more likely to be a lower estimate of the proportion of positive transcriptional responses that are directly dependent on HIF.
These results are in striking contrast with the analysis of genes manifesting down-regulation of expression in response to HIF. Expression array studies have defined similar numbers of genes that are positively and negatively regulated by HIF. In some cases promoter analyses of negatively regulated genes have defined HIF-binding sites and it has been proposed that displacement of more powerful transcriptional activators, or recruitment of corepressors to HIF, accounts for down-regulation of gene expression by HIF (53–55). Our results suggest that these particular mechanisms of HIF-dependent down-regulation of genes are unusual. When promoters of negatively regulated genes were surveyed for HIF binding we did not observe an excess of binding over that in the promoters of genes that were entirely unresponsive to HIF. Thus it appears that for the large majority of genes, HIF-dependent down-regulation of expression is likely to be due to indirect effects “in trans,” rather than direct effects of HIF on the promoter. In keeping with this, a number of genes encoding transcriptional repressors have been identified as positively regulated HIF targets in this and other studies (56, 57). In addition, recent studies have established a role of hypoxia and HIF in the regulation of specific microRNAs, notably miR-210, which act to down-regulate gene expression (58, 59).
Comparison of HIF-1α and HIF-2α DNA binding revealed that whereas many loci bound both HIF-α isoforms, substantially more bound HIF-1α than HIF-2α. Further analysis indicated that this was not due to greater abundance, better antibody capture, or a generally higher affinity of HIF-1α for DNA. Rather, distinct patterns of binding were observed. At common binding sites there was a tendency to greater enrichment of DNA in the anti-HIF2α chromatin immunoprecipitations, suggesting that affinity for HIF-2α was at least as high as for HIF-1α, whereas at other sites, binding was essentially restricted to HIF-1α. How this relates to functional transcriptional selectively is currently unclear.
Recently, studies of selected HIF target genes in mouse embryonic stem cells demonstrated that bound HIF-2α is transcriptionally inactive at the loci analyzed, possibly due to a titratable repressor (12). Although studies in other cells also reported that HIF-2α is transcriptionally inactive on specific HIF target genes (60, 61), we had assumed that this phenomenon would be highly locus specific, because analyses conducted have demonstrated a range of HIF-2α-dependent transcriptional effects under specific circumstances (11, 32, 34, 35). Our genome-wide studies of both chromatin immunoprecipitation and HIF-α-dependent gene expression indicate that this is not the case. Rather, under the conditions of these experiments, HIF-2α appears to exert little or no direct transcriptional activity at HRE-associated targets across the genome despite high affinity binding to these sites. Whether this is due to interaction of HIF-2α with other pathways such as Myc (22, 62) or temporal differences between HIF-1α and HIF-2α activation (34) remains to be determined. Overall, our results provide a framework for a better understanding of the HIF transcriptional cascade that should be of importance in understanding the biology of hypoxia and opportunities for therapeutic manipulation of the response.
Supplementary Material
Acknowledgment
We thank Christopher Pugh for helpful discussions.
This work was supported by The Wellcome Trust and Cancer Research UK.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S4.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S4.
- HIF
- hypoxia-inducible factor
- HRE
- hypoxia response element
- DMOG
- dimethyloxalylglycine
- ChIP
- chromatin immunoprecipitation
- MA
- moving average
- siRNA
- small interfering RNA.
REFERENCES
- 1.Kaelin W. G., Jr. (2008) Nat. Rev. Cancer 8, 865–873 [DOI] [PubMed] [Google Scholar]
- 2.Pouysségur J., Dayan F., Mazure N. M. (2006) Nature 441, 437–443 [DOI] [PubMed] [Google Scholar]
- 3.Bertout J. A., Patel S. A., Simon M. C. (2008) Nat. Rev. Cancer 8, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza G. L. (2007) Science 318, 62–64 [DOI] [PubMed] [Google Scholar]
- 5.Kaelin W. G., Jr., Ratcliffe P. J. (2008) Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 6.Schofield C. J., Ratcliffe P. J. (2004) Nat. Rev. Mol. Cell Biol. 5, 343–354 [DOI] [PubMed] [Google Scholar]
- 7.Lando D., Gorman J. J., Whitelaw M. L., Peet D. J. (2003) Eur. J. Biochem. 270, 781–790 [DOI] [PubMed] [Google Scholar]
- 8.Wenger R. H., Stiehl D. P., Camenisch G. (2005) Sci. STKE 2005, re12. [DOI] [PubMed] [Google Scholar]
- 9.Elvidge G. P., Glenny L., Appelhoff R. J., Ratcliffe P. J., Ragoussis J., Gleadle J. M. (2006) J. Biol. Chem. 281, 15215–15226 [DOI] [PubMed] [Google Scholar]
- 10.Manalo D. J., Rowan A., Lavoie T., Natarajan L., Kelly B. D., Ye S. Q., Garcia J. G., Semenza G. L. (2005) Blood 105, 659–669 [DOI] [PubMed] [Google Scholar]
- 11.Hu C. J., Wang L. Y., Chodosh L. A., Keith B., Simon M. C. (2003) Mol. Cell. Biol. 23, 9361–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu C. J., Iyer S., Sataur A., Covello K. L., Chodosh L. A., Simon M. C. (2006) Mol. Cell. Biol. 26, 3514–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greijer A. E., van der Groep P., Kemming D., Shvarts A., Semenza G. L., Meijer G. A., van de Wiel M. A., Belien J. A., van Diest P. J., van der Wall E. (2005) J. Pathol. 206, 291–304 [DOI] [PubMed] [Google Scholar]
- 14.Imamura T., Kikuchi H., Herraiz M. T., Park D. Y., Mizukami Y., Mino-Kenduson M., Lynch M. P., Rueda B. R., Benita Y., Xavier R. J., Chung D. C. (2009) Int. J. Cancer 124, 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang V., Davis D. A., Haque M., Huang L. E., Yarchoan R. (2005) Cancer Res. 65, 3299–3306 [DOI] [PubMed] [Google Scholar]
- 16.Sung F. L., Hui E. P., Tao Q., Li H., Tsui N. B., Dennis Lo Y. M., Ma B. B., To K. F., Harris A. L., Chan A. T. (2007) Cancer Lett. 253, 74–88 [DOI] [PubMed] [Google Scholar]
- 17.Ragel B. T., Couldwell W. T., Gillespie D. L., Jensen R. L. (2007) Neurosurg. Rev. 30, 181–187 [DOI] [PubMed] [Google Scholar]
- 18.Choi S. M., Oh H., Park H. (2008) FEBS J. 275, 5618–5634 [DOI] [PubMed] [Google Scholar]
- 19.Oikawa M., Abe M., Kurosawa H., Hida W., Shirato K., Sato Y. (2001) Biochem. Biophys. Res. Commun. 289, 39–43 [DOI] [PubMed] [Google Scholar]
- 20.Laderoute K. R., Calaoagan J. M., Gustafson-Brown C., Knapp A. M., Li G. C., Mendonca H. L., Ryan H. E., Wang Z., Johnson R. S. (2002) Mol. Cell. Biol. 22, 2515–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazaki K., Kawamoto T., Tanimoto K., Nishiyama M., Honda H., Kato Y. (2002) J. Biol. Chem. 277, 47014–47021 [DOI] [PubMed] [Google Scholar]
- 22.Koshiji M., Kageyama Y., Pete E. A., Horikawa I., Barrett J. C., Huang L. E. (2004) EMBO J. 23, 1949–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordan J. D., Lal P., Dondeti V. R., Letrero R., Parekh K. N., Oquendo C. E., Greenberg R. A., Flaherty K. T., Rathmell W. K., Keith B., Simon M. C., Nathanson K. L. (2008) Cancer Cell 14, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafsson M. V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J. L., Poellinger L., Lendahl U., Bondesson M. (2005) Dev. Cell 9, 617–628 [DOI] [PubMed] [Google Scholar]
- 25.Kaidi A., Williams A. C., Paraskeva C. (2007) Nat. Cell Biol. 9, 210–217 [DOI] [PubMed] [Google Scholar]
- 26.Wiesener M. S., Turley H., Allen W. E., Willam C., Eckardt K. U., Talks K. L., Wood S. M., Gatter K. C., Harris A. L., Pugh C. W., Ratcliffe P. J., Maxwell P. H. (1998) Blood 92, 2260–2268 [PubMed] [Google Scholar]
- 27.Wenger R. H. (2002) FASEB J. 16, 1151–1162 [DOI] [PubMed] [Google Scholar]
- 28.Scortegagna M., Ding K., Oktay Y., Gaur A., Thurmond F., Yan L. J., Marck B. T., Matsumoto A. M., Shelton J. M., Richardson J. A., Bennett M. J., Garcia J. A. (2003) Nat. Genet. 35, 331–340 [DOI] [PubMed] [Google Scholar]
- 29.Tian H., Hammer R. E., Matsumoto A. M., Russell D. W., McKnight S. L. (1998) Genes & Dev. 12, 3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng J., Zhang L., Drysdale L., Fong G. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8386–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Compernolle V., Brusselmans K., Acker T., Hoet P., Tjwa M., Beck H., Plaisance S., Dor Y., Keshet E., Lupu F., Nemery B., Dewerchin M., Van Veldhoven P., Plate K., Moons L., Collen D., Carmeliet P. (2002) Nat. Med. 8, 702–710 [DOI] [PubMed] [Google Scholar]
- 32.Raval R. R., Lau K. W., Tran M. G., Sowter H. M., Mandriota S. J., Li J. L., Pugh C. W., Maxwell P. H., Harris A. L., Ratcliffe P. J. (2005) Mol. Cell. Biol. 25, 5675–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. (1998) Genes & Dev. 12, 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmquist-Mengelbier L., Fredlund E., Löfstedt T., Noguera R., Navarro S., Nilsson H., Pietras A., Vallon-Christersson J., Borg A., Gradin K., Poellinger L., Påhlman S. (2006) Cancer Cell 10, 413–423 [DOI] [PubMed] [Google Scholar]
- 35.Warnecke C., Zaborowska Z., Kurreck J., Erdmann V. A., Frei U., Wiesener M., Eckardt K. U. (2004) FASEB J. 18, 1462–1464 [DOI] [PubMed] [Google Scholar]
- 36.Lau K. W., Tian Y. M., Raval R. R., Ratcliffe P. J., Pugh C. W. (2007) Br. J. Cancer 96, 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu C. J., Sataur A., Wang L., Chen H., Simon M. C. (2007) Mol. Biol. Cell 18, 4528–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiesener M. S., Jürgensen J. S., Rosenberger C., Scholze C. K., Hörstrup J. H., Warnecke C., Mandriota S., Bechmann I., Frei U. A., Pugh C. W., Ratcliffe P. J., Bachmann S., Maxwell P. H., Eckardt K. U. (2003) FASEB J. 17, 271–273 [DOI] [PubMed] [Google Scholar]
- 39.Ji H., Jiang H., Ma W., Johnson D. S., Myers R. M., Wong W. H. (2008) Nat. Biotechnol. 26, 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji H., Vokes S. A., Wong W. H. (2006) Nucleic Acids Res. 34, e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji H., Wong W. H. (2005) Bioinformatics 21, 3629–3636 [DOI] [PubMed] [Google Scholar]
- 42.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 43.Hosack D. A., Dennis G., Jr., Sherman B. T., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koudritsky M., Domany E. (2008) Nucleic Acids Res. 36, 6795–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hubbard T. J., Aken B. L., Ayling S., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Clarke L., Coates G., Fairley S., Fitzgerald S., Fernandez-Banet J., Gordon L., Graf S., Haider S., Hammond M., Holland R., Howe K., Jenkinson A., Johnson N., Kahari A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Rios D., Schuster M., Slater G., Smedley D., Spooner W., Spudich G., Trevanion S., Vilella A., Vogel J., White S., Wilder S., Zadissa A., Birney E., Cunningham F., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Kasprzyk A., Proctor G., Smith J., Searle S., Flicek P. (2009) Nucleic Acids Res. 37, 690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E. (2006) Nucleic Acids Res. 34, D108–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 48.Reinhold W., Emens L., Itkes A., Blake M., Ichinose I., Zajac-Kaye M. (1995) Mol. Cell. Biol. 15, 3041–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Attwooll C., Lazzerini Denchi E., Helin K. (2004) EMBO J. 23, 4709–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 52.Zeller K. I., Zhao X., Lee C. W., Chiu K. P., Yao F., Yustein J. T., Ooi H. S., Orlov Y. L., Shahab A., Yong H. C., Fu Y., Weng Z., Kuznetsov V. A., Sung W. K., Ruan Y., Dang C. V., Wei C. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peyssonnaux C., Zinkernagel A. S., Schuepbach R. A., Rankin E., Vaulont S., Haase V. H., Nizet V., Johnson R. S. (2007) J. Clin. Invest. 117, 1926–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazure N. M., Chauvet C., Bois-Joyeux B., Bernard M. A., Nacer-Chérif H., Danan J. L. (2002) Cancer Res. 62, 1158–1165 [PubMed] [Google Scholar]
- 55.Chen K. F., Lai Y. Y., Sun H. S., Tsai S. J. (2005) Nucleic Acids Res. 33, 5190–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Löfstedt T., Jögi A., Sigvardsson M., Gradin K., Poellinger L., Påhlman S., Axelson H. (2004) J. Biol. Chem. 279, 39223–39231 [DOI] [PubMed] [Google Scholar]
- 57.Yun Z., Maecker H. L., Johnson R. S., Giaccia A. J. (2002) Dev. Cell 2, 331–341 [DOI] [PubMed] [Google Scholar]
- 58.Camps C., Buffa F. M., Colella S., Moore J., Sotiriou C., Sheldon H., Harris A. L., Gleadle J. M., Ragoussis J. (2008) Clin. Cancer Res. 14, 1340–1348 [DOI] [PubMed] [Google Scholar]
- 59.Kulshreshtha R., Ferracin M., Wojcik S. E., Garzon R., Alder H., Agosto-Perez F. J., Davuluri R., Liu C. G., Croce C. M., Negrini M., Calin G. A., Ivan M. (2007) Mol. Cell. Biol. 27, 1859–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park S. K., Dadak A. M., Haase V. H., Fontana L., Giaccia A. J., Johnson R. S. (2003) Mol. Cell. Biol. 23, 4959–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sowter H. M., Raval R. R., Moore J. W., Ratcliffe P. J., Harris A. L. (2003) Cancer Res. 63, 6130–6134 [PubMed] [Google Scholar]
- 62.Kim J. W., Gao P., Liu Y. C., Semenza G. L., Dang C. V. (2007) Mol. Cell. Biol. 27, 7381–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.