Abstract
Krüpple-like transcription factor 5 (KLF5) is a zinc-finger transcription factor promoting cell survival and tumorigenesis in multiple cancers. A high expression level of KLF5 has been shown to be associated with shorter breast cancer patient survival. However, the role of KLF5 and mechanism of KLF5 actions in breast cancer remain unclear. In this study, we found that KLF5 knockdown by small interfering RNA in two breast cell lines, MCF10A and BT20, induces apoptosis. Interestingly, a pro-survival phosphatase, dual specificity mitogen-activated protein kinase phosphatase 1 (MKP-1), is down-regulated by KLF5 ablation. Consistently, KLF5 overexpression increases the MKP-1 protein expression in Hs578T and MCF7. We further found that MKP-1 is essential and sufficient for KLF5 to promote breast cell survival. However, MKP-1 is not a KLF5 direct transcription target because the MKP-1 mRNA level is not regulated by KLF5. By cycloheximide chase assays, we found that KLF5 decreases MKP-1 protein degradation via activating the ERK signaling. Inhibition of pERK by the pharmacological inhibitor U0126 specifically blocks KLF5-induced MKP-1 phosphorylation and stabilization. Additionally, constitutive activation of ERK by constitutively activated MEK1 rescues the KLF5 depletion-induced MKP-1 down-regulation. Consistently, the phosphorylation-deficient MKP-1 mutant cannot be stabilized by KLF5. Finally, the activation of ERK by KLF5 is very likely through the KLF5 direct target gene FGF-BP in breast cells. These findings suggest that KLF5 is a pro-survival factor that promotes breast cell survival partially through pERK-mediated MKP-1 phosphorylation and stabilization. The KLF5-FGF-BP-pERK-MKP-1 signaling axis may provide new therapeutic targets for invasive breast cancer.
The Krüpple-like transcription factor 5 (KLF5/IKLF/BTEB2)2 has been suggested to be an oncogene in multiple carcinomas including the intestinal (1), esophageal (2), bladder (3), and breast (4). A high level of the KLF5 mRNA has been reported to associate with a short survival time in breast cancer patients (4). In addition, KLF5 expression is induced by a number of oncogenes including ERBB2 (5), RAS (6), and WNT (7). Consistently, KLF5 has been shown to promote cell proliferation (3), migration (8), and tumorigenesis (3) in different cell models by regulating gene transcription. KLF5 has been shown to promote cell survival through regulating Survivin (9), Pim1 (10), and PARP1 (11) in different types of cells.
Our previous study showed that KLF5 promotes the TSU-Pr1 bladder cancer cell growth in vitro and in vivo (3). Furthermore, we demonstrated that KLF5 regulates a number of downstream target genes in a microarray study. Following that, we proved that KLF5 promotes breast cell proliferation partially through directly inducing the fibroblast growth factor-binding protein 1 (FGF-BP) transcription in breast cancer.3 FGF-BP was confirmed to be a KLF5-induced gene in the mouse lung in an independent microarray study (13).
Besides FGF-BP, another KLF5 downstream target gene (3), dual specificity mitogen-activated protein kinase phosphatase 1 (MKP-1/DUSP1/CL-100), has been documented to promote cell survival (14). Mitogen-activated protein kinases (MAPKs) are activated via phosphorylation of ERK, p38, and JNK. These MAPKs are inactivated via de-phosphorylation by MKPs including MKP-1 (15). Although pERK usually contributes to cell proliferation and survival, pJNK and pp38 promote cell apoptosis in response to stress (16). The balance between MAPKs and MKPs determines whether cells undergo survival or apoptosis (17). Consistently, MKP-1 has been reported to be overexpressed in many types of cancer including breast cancer (15, 18). It has been shown that MKP-1 is rapidly induced in response to multiple stress stimuli, such as the chemotherapy drugs paclitaxel (14) and cisplatin (19, 20), oxidative stress (21), and UV radiation (22), and contributes to cell survival. The MKP-1 induction by stress is at both transcriptional (23, 24) and post-translational (25, 26) levels and primarily mediated by the activation of ERK signaling. Interestingly, the pERK levels are increased by KLF5 in TSU-Pr1 (3).
Here, we studied the mechanism by which MKP-1 is induced by KLF5 in breast cancer. We showed evidence that KLF5 promotes breast cell survival partially through MKP-1. The induction of MKP-1 by KLF5 in breast cells is at the protein post-translational level but not the transcriptional level. The activation of ERK signaling by KLF5 is essential and sufficient for MKP-1 protein phosphorylation and stabilization in breast cells. We further demonstrated that activation of ERK signaling is likely mediated by the KLF5 direct target gene FGF-BP. Taken together, the KLF5-FGF-BP-pERK-MKP-1 signaling axis may contribute to breast cancer and provide new therapeutic targets for breast cancer.
MATERIALS AND METHODS
Breast Cell Lines and Culture Conditions
The immortalized breast epithelial cell line MCF10A was maintained in Dulbecco's modified Eagle's medium/Ham's F-12 50/50 medium supplemented with 5% horse serum, 0.5 μg/ml hydrocortisone, 10 μg/ml insulin, 20 ng/ml epidermal growth factor, 0.1 μg/ml cholera enterotoxin, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine. The breast cancer cell lines BT-20 and MCF7 were cultured in minimal essential medium containing 5% fetal bovine serum, 0.1 mm non-essential amino acid, 1.5 g/liter sodium bicarbonate, 1 mm sodium pyruvate, 0.01 mg/ml insulin, and 100 units/ml penicillin and 100 μg/ml streptomycin. The breast cancer cell line Hs578T was cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1.5 g/liter sodium bicarbonate, 1 mm sodium pyruvate, 0.01 mg/ml insulin, and 100 units/ml penicillin and 100 μg/ml streptomycin. These cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C.
Immunoblotting and Antibodies
Immunoblotting was performed with 40 μg of proteins. The anti-β-actin and anti-V5 antibodies are from Sigma. The anti-PARP, anti-cleaved caspase 3, anti-pERK, and anti-pMKP-1Ser-359 antibodies are from Cell Signaling (Danvers, MA). The anti-KLF5 rabbit polyclonal antibody has been described previously (27). The anti-MKP-1 antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
siRNA Transfection and Adenovirus Infection
The control luciferase siRNA (Lucsi), KLF5 siRNA (KLF5si) (Dharmacon, Chicago, IL), and MKP-1 siRNA (MKP-1si) (silencer select pre-designed siRNA, Ambion, Austin, TX) were transfected by Lipofectamine 2000 (Invitrogen). The siRNA target sequences were: 5′-AGCTCACCTGAGGACTCACAC-3′, for the human KLF5 gene, 5′-CTTACGCTGAGTACTTCGA-3′ for the luciferase gene, and 5′-GGACTAATCGAGTCAAGCT-3′ for the human MKP-1 gene. The final concentration of Lucsi and KLF5si was 100 nm; and the final concentration of MKP-1si was 10 nm.
The KLF5 and control gfp adenoviruses have been described previously (3). MCF7 and Hs578T cells were infected with adenoviruses in media containing 5% fetal bovine serum. After incubation with the adenoviruses for 4 h, the cells were cultured in normal growth media.
Cycloheximide (CHX) Chase Assays
Hs578T, MCF10A, and HEK293T cells were seeded into a 12-well plate at a density of 1–2.5 × 105 cells per well. After overnight culture, the cells were either transfected with different siRNAs or plasmids or infected with adenoviruses. Two days after transfection or infection, the cells were treated with 50 μg/ml CHX. Total proteins were collected at different time points and subjected to immunoblotting for KLF5, MKP-1, and β-actin.
Reverse Transcriptase-PCR
Total RNAs were isolated using TRIzol® reagent (Invitrogen). Reverse transcriptions were performed using the IscriptTM cDNA synthesis kit (Bio-Rad). Forward primer, 5′-GATCTAGATATGCCCAGTTC-3′, and reverse primer, 5′-CAGCCTTCCCAGGTACACTTG-3′, were used to amplify KLF5 by PCR in a 20-μl volume. Primer sequences for MKP-1 were 5′-CCCGGAGCTGTGCAGCAA-3′ (forward) and 5′-CTGGCCCATGAAGCTGAAGT-3′ (reverse). A total of 32 cycles were used to amplify KLF5 and MKP-1, whereas 28 cycles were used to amplify the β-actin control.
Cell Viability Assay
MCF10A and BT20 cells were transfected with KLF5si, MKP-1si, and Lucsi, respectively, for 5 days before analysis. The SRB assay was used to measure cell viability as described in our previous report (28).
Plasmids and Gene Overexpression by Lentiviruses
The human MKP-1 gene was amplified from IMAGE clone 5296005 with the pfu enzymes by PCR using primers 5′-ttggatccATGGTCATGGAAGTGGGCAC-3′ and 5′-ttctcgagTCAGCAGCTGGGAGAGGTCG-3′. The catalytically inactive MKP-1C258S mutant was generated by PCR using primers 5′-GTTTGTCCACTCCCAGGCAGGCATTTCCCG-3′ and 5′-TGCCTGCCTGGGAGTGGACAAACACCCTTC-3′. The MKP-1S359A/S364A mutant was generated by primers 5′-ttggatccATGGTCATGGAAGTGGGCAC-3′ and 5′-ttctcgagTCAGCAGCTGGGTGCGGTCGTAATGGGTGCCTGAAGGTAGCTCAGCGCAC-3′. The PCR products was digested by BamHI/XhoI and subcloned into the pLenti6/V5-D-TOPO vector and verified by DNA sequencing. The pLenti6/V5-GW/lacZ vector (Invitrogen) was used as a negative control.
A constitutively activated MEK1 was amplified from pMCL-MEK1-ΔED (29) (a gift from Dr. A. E. Aplin, Thomas Jefferson University, Philadelphia, PA) and subcloned into pLenti6/V5-D-TOPO vector. All plasmids were transfected into HEK 293FT packing cells using Lipofectamine 2000. Lentiviruses were collected at 72 h after transfection and used to transduce MCF10A cells in a 6-well plate. Forty-eight h after transduction, the antibiotic blasticidin (10 μg/ml) was added to select drug-resistant populations.
RESULTS
KLF5 Knockdown Induces Apoptosis and Decreases the MKP-1 Expression in Breast Cells
KLF5 has previously been shown to express in estrogen receptor α negative basal-like breast cells.3 To determine whether KLF5 promotes breast cell survival, we knocked down KLF5 in two KLF5 positive breast cell lines, MCF10A and BT20 (30). We examined the levels of apoptosis markers, cleaved PARP, and caspase 3, in the control luciferase siRNA (Lucsi) and well characterized KLF5 siRNA (KLF5si) (3, 31) transfected cells by immunoblotting. We found that KLF5si induces the cleavage of both PARP and caspase 3 compared with Lucsi in MCF10A and BT20 (Fig. 1A). To further confirm that KLF5 knockdown decreases cell survival through inducing apoptosis, we measured cell viability by the SRB assay and Annexin V levels by flow cytometry. Consistent with Western blot results, KLF5si significantly decreases cell viability (Fig. 1B) and increases Annexin V staining (data not shown) in both MCF10A and BT20. Interestingly, the protein expression levels of a potential KLF5 downstream gene, the pro-survival phosphatase MKP-1, are decreased by KLF5si in both cell lines (Fig. 1A).
FIGURE 1.
Knockdown of KLF5 induces apoptosis and down-regulates the MKP-1 protein levels in breast cells. A, knockdown of KLF5 induces the PARP and caspase 3 cleavage and down-regulates the MKP-1 protein levels in MCF10A and BT20. A well characterized KLF5 siRNA was used to knockdown the KLF5 expression in MCF10A and BT20 cells. Luciferase siRNA (Lucsi) was used as the negative control. Protein levels were detected by immunoblotting. B, KLF5 siRNA significantly reduces cell viability in MCF10A and BT20 as determined by the SRB assay. **, p ≤ 0.001 (t test). C, knockdown of MKP-1 induces the PARP and caspase 3 cleavage in both MCF10A and BT20 cell lines compared with the Lucsi negative control and the KLF5si positive control. D, the MKP-1 siRNA significantly reduces cell viability in MCF10A and BT20 compared with the Lucsi negative control and the KLF5si positive control. Data are presented as the mean ± S.D. (error bars) from three independent experiments.
To test if MKP-1 indeed promotes breast cell survival, we knocked down MKP-1 by a pre-designed anti-MKP-1 siRNA in both MCF10A and BT20 and examined apoptosis. As expected, knockdown of MKP-1 also induces the cleavage of both PARP and caspase 3 and the decrease of cell viability like knockdown of KLF5 in both MCF10A and BT20 (Fig. 1, C and D).
KLF5 Promotes Cell Survival Partially through MKP-1
Because silence of KLF5 induces apoptosis and down-regulates the expression of the pro-survival MKP-1 protein in breast cells, we wondered if KLF5 functions partially through MKP-1. We performed a rescue experiment in MCF10A to determine whether MKP-1 overexpression can block the KLF5si-induced apoptosis. The wild-type (WT) MKP-1, the catalytically inactive mutant MKP-1C258S (32), and the lacZ control genes were forced overexpressed in MCF10A populations, respectively, by lentiviruses (Fig. 2A). In line with our previous observation, KLF5si decreases the MKP-1 protein level and induces apoptosis, indicated by cleavage of PARP and caspase 3 and loss of cell viability, in the control LacZ overexpressing cells. As expected, forced overexpression of WT MKP-1 clearly decreases the pERK levels and KLF5si-induced apoptosis (Fig. 2). Similar results were obtained from two stable MKP-1 overexpressing MCF10A clones (data not shown). Unexpectedly, overexpression of the catalytically inactive mutant MKP-1C258S also blocks the KLF5si-induced apoptosis as efficiently as WT MKP-1. As a dominant negative MKP-1 mutant, MKP-1C258S increases the pERK levels (Fig. 2A). Consistently, the expression level of MKP-1C258S is higher than that of WT MKP-1 presumably because a high level of pERK stabilizes the MKP-1 protein (see below in detail). These findings suggest that overexpression of MKP-1 can partially rescue the KLF5si-induced apoptosis in MCF10A.
FIGURE 2.
MKP-1 partially rescues the KLF5 knockdown-induced apoptosis in MCF10A. A, overexpression of either WT MKP-1 or the catalytically inactive MKP-1C258S mutant decreases the KLF5 siRNA-induced PARP and caspase 3 cleavage. MCF10A cell populations stably expressing LacZ, MKP-1, or MKP-1C258S were transfected with Lucsi or KLF5si for 4 days. The apoptosis markers including cleaved PARP and caspase 3 were measured by immunoblotting. B, overexpression of MKP-1 or MKP-1C258S significantly decreases the KLF5 knockdown induced loss of cell viability as shown by the SRB assay. *, p < 0.05 (t test). Data are presented as the mean ± S.D. (error bars) from three independent experiments.
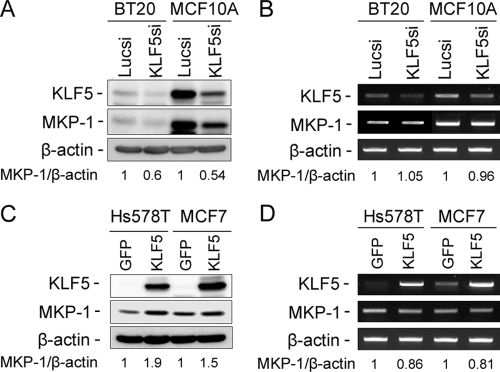
MKP-1 Expression Is Positively Regulated by KLF5 at the Protein Level but Not at the mRNA Level in Breast Cells
KLF5 is a well established transcriptional factor regulating transcription of a number of genes. To test whether MKP-1 is a KLF5 direct transcriptional target, we examined MKP-1 expression at the protein level by Western blot and the mRNA level by semi-quantitative reverse transcriptase-PCR after knocking down and overexpressing KLF5. To our surprise, KLF5si decreases MKP-1 expression at the protein level but not at the mRNA level in both MCF10A and BT20 cell lines (Fig. 3, A and B). Additionally, KLF5 overexpression increases the expression of MKP-1 at the protein level but not at the mRNA level in both Hs578T and MCF7 (Fig. 3, C and D). Finally, we found that KLF5 cannot activate the MKP-1 promoter in MCF7 by dual luciferase reporter assays (data not shown). These results suggest that MKP-1 is not a KLF5 direct transcription target gene in breast cells.
FIGURE 3.
KLF5 up-regulates the MKP-1 expression at the protein level but not at the mRNA level. KLF5 siRNA decreases the MKP-1 protein levels (A) but not mRNA levels (B) in MCF10A and BT20 as measured by immunoblotting and semi-quantitative reverse transcriptase-PCR, respectively. Lucsi was used as a negative control. β-Actin served as the input control. KLF5 overexpression by adenoviruses increases the MKP-1 protein levels (C) but not mRNA levels (D) in Hs578T and MCF7 breast cancer cells. The gfp adenovirus was used as a negative control. The normalized band intensities are shown below each lane (the negative controls are defined as 1). GFP, green fluorescent protein.
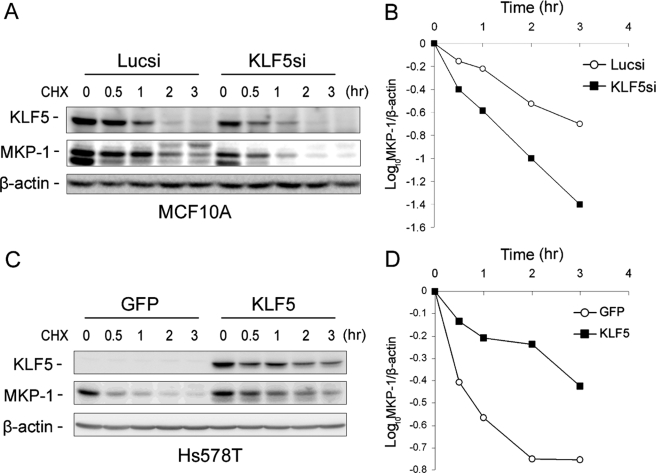
MKP-1 is a short-lived protein (the half-life is about 45 min in fibroblasts) degraded through the ubiquitin proteasome pathway (25). To investigate whether KLF5 regulates MKP-1 protein stability, we performed CHX chase assays and found that KLF5 knockdown in MCF10A cells decreases the endogenous MKP-1 protein half-life (Fig. 4, A and B). Consistently, KLF5 overexpression clearly increases the MKP-1 protein half-life in Hs578T (Fig. 4, C and D). These results suggest that KLF5 decreases MKP-1 protein degradation in breast cells.
FIGURE 4.
KLF5 increases the MKP-1 protein stability in breast cells. A and B, knockdown of KLF5 decreases the protein half-life for MKP-1 in MCF10A as determined by the CHX (50 μg/ml) chase assay. The band intensities were quantified using densitometry. C and D, overexpression of KLF5 increases the half-life of the MKP-1 protein in Hs578T as determined by the CHX chase assay. The cells were infected with Ad-KLF5/gfp and Ad-gfp control adenoviruses, respectively. GFP, green fluorescent protein.
KLF5 Increases MKP-1 Protein Stability through the pERK-mediated MKP-1 Phosphorylation
The proteasomal degradation of MKP-1 has been demonstrated to be inhibited by pERK-mediated MKP-1 phosphorylation at Ser-359 and Ser-364 residues (25). Additionally, the SCFSKP2 E3 ubiquitin ligase has been suggested to promote MKP-1 ubiquitin-mediated degradation (26). We first found that KLF5si does not increase the SKP2 expression levels in MCF10A and BT20 (data not shown). Then, we determined whether KLF5 stabilizes MKP-1 via pERK-mediated MKP-1 phosphorylation because KLF5 up-regulates the pERK levels in TSU-Pr1 (3) and KLF5si decreases the pERK levels in MCF10A (Fig. 2A). Indeed, inhibition of pERK, but not JNK and p38, by pharmacological inhibitors dramatically decreases the MKP-1 protein level in MCF10A (Fig. 5A). Consistently, KLF5si reduces the serum-induced pERK and MKP-1 levels in MCF10A (Fig. 5B). Furthermore, we demonstrated that both the MEK inhibitor U0126 and KLF5si dramatically decrease pERK levels and MKP-1 phosphorylation at Ser-359 by using an antibody that specifically recognizes the pMKP-1Ser-359 (Fig. 5C). Additionally, we found that KLF5 overexpression up-regulates pERK, pMKP-1Ser-359, and total MKP-1 levels in Hs578T. When pERK is inhibited by U0126, KLF5 overexpression fails to increase the levels of pMKP-1Ser-359 and total MKP-1 in Hs578T (Fig. 5D). Finally, the constitutive activation of ERK by overexpression of constitutively activated MEK1 can completely rescue the KLF5si-induced pMKP-1Ser-359 and total MKP-1 down-regulation in MCF10A (Fig. 5E). These results strongly suggest that KLF5 stabilizes MKP-1 through activation of ERK.
FIGURE 5.
KLF5 increases MKP-1 protein stability through activating the ERK signaling in breast cells. A, inhibition of ERK but not JNK and p38 by pharmacological inhibitors reduces the MKP-1 protein level in MCF10A. The cells were serum starved overnight before serum stimulation for 1 h. Dimethyl sulfoxide, U0126 (5 nm, a MEK inhibitor), SP600125 (20 nm, a JNK inhibitor), and SB203580 (20 nm, a p38 inhibitor) were added separately 1 h before serum stimulation. B, knockdown of KLF5 decreases pERK activation and MKP-1 induction by serum. MCF10A cells transfected with KLF5 siRNA and luciferase siRNA were serum starved overnight and cultured in serum-containing media for the indicated time. C, either inhibition of pERK by U0126 or knockdown of KLF5 clearly reduce the pMKP-1Ser-359 and total MKP-1 levels in MCF10A. D, the activation of ERK is essential for KLF5 to up-regulate the pMKP-1Ser-359 and total MKP-1 levels in Hs578T. Hs578T cells were infected with Ad-KLF5/gfp and Ad-gfp control adenoviruses, respectively. The cells were treated with either dimethyl sulfoxide or U0126 (5 nm) as indicated. E, constitutively activated MEK1 (V5 tagged) rescues KLF5 knockdown-induced down-regulation of pMKP-1Ser-359 and total MKP-1. The total ERK level serves as the loading control. GFP, green fluorescent protein.
To further confirm that phosphorylation of MKP-1 at Ser-359/Ser-364 is essential for KLF5-mediated MKP-1 protein stabilization, we generated a MKP-1 mutant in which both Ser-359 and Ser-364 residues were replaced with Ala. As shown in Fig. 6, A and B, WT MKP-1 can be stabilized by KLF5 overexpression in HEK293T cells. However, the MKP-1S359A/S364A mutant cannot be stabilized by KLF5 under the same conditions. Consistently, the mutant MKP-1 shows a shorter half-life than WT MKP-1 presumably due to lack of phosphorylation at Ser-359 and Ser-364 by pERK (Fig. 6, C and D). The degradation of MKP-1S359A/S364A is still through proteasome because the proteasome inhibitor MG132 can dramatically protect it from degradation in HEK293T cells (Fig. 6E).
FIGURE 6.
KLF5 stabilizes WT MKP-1 but not the MKP-1S359A/S364A mutant. A and B, KLF5 increases the WT MKP-1 protein stability as determined by the CHX chase assay. HEK 293T cells (the endogenous KLF5 and MKP-1 expression levels are low) were transfected with MKP-1 and KLF5 expressing constructs. The CHX chase assay was performed 2 days after the transfection. The band intensities were quantified using densitometry. C and D, MKP-1S359A/S364A is less stable than WT MKP-1 and KLF5 cannot stabilize MKP-1S359A/S364A. E, MG132 (20 μm, 4 h treatment) increases the endogenous MKP-1 and the exogenous MKP-1S359A/S364A protein levels in HEK293T cells. GST, glutathione S-transferase.
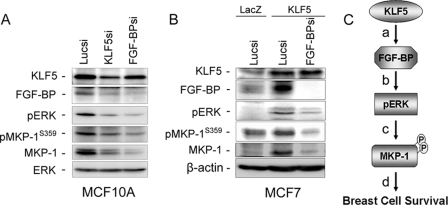
A recent study in our laboratory showed that KLF5 directly regulates transcription of the FGF-BP in breast cells.3 Because FGF-BP has been reported to activate ERK signaling (33), we hypothesized that KLF5 up-regulates the pERK level through FGF-BP. To test this hypothesis, we knocked down both KLF5 and FGF-BP individually in MCF10A. As shown in Fig. 7A, knockdown of either KLF5 or FGF-BP decreases the pERK, pMKP-1Ser-359, and total MKP-1 levels. Additionally, we found that KLF5 cannot increase the MKP-1 protein level in the presence of FGF-BP siRNA in MCF7 (Fig. 7B). Thus, KLF5 may activate ERK-MKP-1 signaling through FGF-BP in breast cells (Fig. 7C).
FIGURE 7.
KLF5 activates ERK-MKP-1 signaling through FGF-BP in breast cells. A, knockdown of either KLF5 or FGF-BP down-regulates pERK, pMKP-1Ser-359, and total MKP-1 protein levels in MCF10A. B, FGF-BP depletion blocks KLF5 overexpression induced up-regulation of the pERK, pMKP-1Ser-359, and total MKP-1 protein levels in MCF7. C, a linear model of KLF5 promoting breast cell survival. a, KLF5 directly induces the FGF-BP transcription. b, FGF-BP activates the ERK signaling. c, the activated ERK phosphorylates MKP-1 at Ser-359 and Ser-364 and prevents its degradation. d, the elevated MKP-1 promotes breast cell survival.
DISCUSSION
Accumulated evidence suggests that KLF5 is a pro-survival factor. First, KLF5 has been shown to promote leukemia cell survival through directly inducing the Survivin gene expression (9). Second, KLF5 was reported to promote cell survival by directly promoting the survival kinase Pim1 expression in the HCT116 colon cancer cell line (10). In addition, KLF5 promotes HeLa and NIH-3T3 cell survival from tumor necrosis factor-α through interacting with PARP1 (11). Finally, the cardiovascular apoptosis is significantly increased in KLF5 heterozygous knock-out mice (11). High expression levels of KLF5 mRNA have been shown to be a prognostic factor for shorter disease-free survival and overall survival in patients with breast cancer (4). Our previous studies suggest that KLF5 is expressed in immortalized breast epithelial cell lines and a subset of estrogen receptor negative breast cancer cell lines (27, 30).3 However, the role of KLF5 and the mechanism by which KLF5 functions in the breast have not been well studied.
In this study, we showed that KLF5 promotes breast cell survival through the FGF-BP-pERK-MKP-1 signaling axis. First, we demonstrated that depletion of endogenous KLF5 or MKP-1 in two breast cell lines, MCF10A and BT20, induces apoptosis (Fig. 1). Next, we found that KLF5 maintains and increases the pro-survival phosphatase MKP-1 protein levels in breast cells (Figs. 1–3). Following that, we demonstrated that MKP-1 can partially rescue KLF5 knockdown-induced apoptosis in MCF10A (Fig. 2). Furthermore, we characterized the mechanism by which KLF5 up-regulates MKP-1 in breast cells and found that KLF5 decreases MKP-1 protein degradation via pERK-mediated MKP-1 phosphorylation (Figs. 4–6). Finally, we showed that KLF5 activates ERK signaling through its direct target gene FGF-BP (Fig. 7).
Multiple lines of evidence suggests that KLF5 up-regulates MKP-1 expression through activating the ERK signaling. First, KLF5 does not directly regulate the MKP-1 gene at the transcriptional level (Fig. 3). Second, KLF5 increases MKP-1 protein stability (Fig. 4). Third, KLF5 maintains and increases pERK levels in multiple breast cell lines, and pERK is sufficient and essential for KLF5-mediated MKP-1 stabilization (Figs. 2 and 5). Additionally, we confirmed that the pMKP-1Ser-359 is regulated by KLF5-FGF-BP-pERK signaling (Figs. 5 and 7). Furthermore, the mutation of two key pERK phosphorylation sites (S359A/S364A) in MKP-1 abrogates KLF5-mediated MKP-1 stabilization (Fig. 6). These findings strongly suggest that KLF5 up-regulates MKP-1 via pERK-mediated MKP-1 protein phosphorylation and stabilization (Fig. 7C).
As described in the Introduction, pERK can increase MKP-1 expression at both transcriptional and post-translational levels (23–26). Indeed, we found that KLF5 increases the MKP-1 mRNA levels in TSU-Pr1 (3). However, MKP-1 mRNA levels are not regulated by KLF5 in tested breast cell lines. It is possible that the signaling transduction from pERK to MKP-1 transcription is inactive in breast cells. The phosphorylation mediated protein stabilization is the primary mechanism by which pERK increases MKP-1 expression in breast cells.
To understand the mechanism by which KLF5 up-regulates ERK signaling, we tested the possibility that KLF5 up-regulates epidermal growth factor receptor, a known KLF5 target gene in squamous epithelial cells (34) and a well known receptor tyrosine kinase activating ERK. However, we did not detect any changes of epidermal growth factor receptor expression when KLF5 is knocked down in MCF10A (data not shown). A recent study in our laboratory indicated that KLF5 directly controls transcription of the FGF-BP gene in breast cells.3 FGF-BP is a secreted protein that can bind to fibroblast growth factors and promote FGF receptor-mediated ERK activation (33). We subsequently tested if FGF-BP mediates ERK activation and MKP-1 stabilization by KLF5. As expected, knockdown of either KLF5 or FGF-BP dramatically decreases the pERK, pMKP-1Ser-359, and total MKP-1 levels in MCF10A (Fig. 7A). Furthermore, knockdown of FGF-BP abrogates the KLF5 overexpression induced up-regulation of pERK, pMKP-1Ser-359, and total MKP-1 in MCF7 (Fig. 7B). These findings support the KLF5-FGF-BP-pERK-MKP-1 linear model proposed in Fig. 7C although it may be oversimplified under physiological conditions.
A surprising finding of this study is that MKP-1 can promote MCF10A cell survival in a phosphatase activity independent manner (Fig. 2). It has been reported that the phosphatase activity is essential for the anti-apoptotic function of MKP-1 after UV treatment in human fibroblasts (35). Because MKP-1 can de-phosphorylate multiple MAPKs, including ERK, JNK, and p38, overexpression of the catalytically inactive MKP-1C258S mutant may increase multiple MAPKs at the same time. Indeed, we found that the pERK levels are dramatically increased in the MKP-1C258S overexpressing MCF10A cells due to the lack of negative feedback control (Fig. 2A). Because the balance between the pro-survival pERK level and the pro-apoptosis pJNK/pp38 levels determines whether cells will live or die (16), it is possible that the elevated pERK levels contribute to the pro-survival function of MKP-1C258S in MCF10A.
Besides MKP-1, KLF5 may promote breast cell survival through other downstream genes and pathways because MKP-1 can only partially rescue KLF5 depletion-induced apoptosis in MCF10A (Fig. 2). Other signal pathways may contribute to the function of KLF5 on cell survival as well. For example, KLF5 has been shown to increase the pAKT levels in TSU-Pr1 (3). AKT has been well documented to promote cell survival through blocking the activation of pro-apoptotic proteins (12). Indeed, we confirmed that both KLF5 and FGF-BP inhibition decreases the pAKT levels in MCF10A (data not shown). Thus, it is most likely that KLF5 promotes breast cell survival through multiple pathways, including FGF-BP-pERK-MKP-1 and FGF-BP-pAKT. Besides FGF-BP, we cannot completely exclude that KLF5 up-regulates the pERK level and breast cell survival through other target genes such as SURVIVIN and PIM1 and other mechanisms, such as interaction with PARP1.
In summary, we showed that KLF5 is a pro-survival transcription factor in breast cells. KLF5 functions partially through pERK-mediated MKP-1 protein phosphorylation and stabilization. Finally, we found that KLF5 may up-regulate the pERK levels through the direct target gene FGF-BP in breast cells. The KLF5-FGF-BP-pERK-MKP-1 signaling axis may provide new therapeutic targets for invasive breast cancer.
Acknowledgments
We are grateful to Dr. A. Aplin for providing the constitutively activated MEK1 plasmid and Dr. P. McKeown-Longo for providing SB203580.
This work was supported by Grant BCTR0503705 from Komen for the Cure, Grant RSG-08-199-01 from the American Cancer Society, and Grant W81XWH-07-1-0191 from the Department of Defense.
3 H. Q. Zheng, Z. Zhou, L. Chaudhury, J. T. Dong, and C. Chen, unpublished data.
- KLF5
- Krüpple-like factor 5
- ERK
- extracellular signal-regulated kinase
- WT
- wild type
- MKP-1
- dual specificity mitogen-activated protein kinase phosphatase 1
- FGF-BP
- fibroblast growth factor-binding protein 1
- PARP1
- poly(ADP-ribose) polymerase 1
- siRNA
- small interfering RNA
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun NH2-terminal kinase
- CHX
- cycloheximide.
REFERENCES
- 1.Chanchevalap S., Nandan M. O., McConnell B. B., Charrier L., Merlin D., Katz J. P., Yang V. W. ( 2006) Nucleic Acids Res. 34, 1216– 1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein B. G., Chao H. H., Yang Y., Yermolina Y. A., Tobias J. W., Katz J. P. ( 2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1784- G1792 [DOI] [PubMed] [Google Scholar]
- 3.Chen C., Benjamin M. S., Sun X., Otto K. B., Guo P., Dong X. Y., Bao Y., Zhou Z., Cheng X., Simons J. W., Dong J. T. ( 2006) Int. J. Cancer 118, 1346– 1355 [DOI] [PubMed] [Google Scholar]
- 4.Tong D., Czerwenka K., Heinze G., Ryffel M., Schuster E., Witt A., Leodolter S., Zeillinger R. ( 2006) Clin. Cancer Res. 12, 2442– 2448 [DOI] [PubMed] [Google Scholar]
- 5.Beckers J., Herrmann F., Rieger S., Drobyshev A. L., Horsch M., Hrabé de Angelis M., Seliger B. ( 2005) Int. J. Cancer 114, 590– 597 [DOI] [PubMed] [Google Scholar]
- 6.Sun R., Chen X., Yang V. W. ( 2001) J. Biol. Chem. 276, 6897– 6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taneyhill L., Pennica D. ( 2004) BMC Dev. Biol. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., Tetreault M. P., Yermolina Y. A., Goldstein B. G., Katz J. P. ( 2008) J. Biol. Chem. 283, 18812– 18820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu N., Gu L., Findley H. W., Chen C., Dong J. T., Yang L., Zhou M. ( 2006) J. Biol. Chem. 281, 14711– 14718 [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y., Hamza M. S., Leong H. S., Lim C. B., Pan Y. F., Cheung E., Soo K. C., Iyer N. G. ( 2008) Oncogene 27, 1– 8 [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T., Nishi T., Nagino T., Sasaki K., Aizawa K., Kada N., Sawaki D., Munemasa Y., Matsumura T., Muto S., Sata M., Miyagawa K., Horikoshi M., Nagai R. ( 2007) J. Biol. Chem. 282, 9895– 9901 [DOI] [PubMed] [Google Scholar]
- 12.Manning B. D., Cantley L. C. ( 2007) Cell 129, 1261– 1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan H., Luo F., Wert S. E., Zhang L., Xu Y., Ikegami M., Maeda Y., Bell S. M., Whitsett J. A. ( 2008) Development 135, 2563– 2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu W., Pew T., Zou M., Pang D., Conzen S. D. ( 2005) J. Biol. Chem. 280, 4117– 4124 [DOI] [PubMed] [Google Scholar]
- 15.Keyse S. M. ( 2008) Cancer Metastasis Rev. 27, 253– 261 [DOI] [PubMed] [Google Scholar]
- 16.Junttila M. R., Li S. P., Westermarck J. ( 2008) FASEB J. 22, 954– 965 [DOI] [PubMed] [Google Scholar]
- 17.Wada T., Penninger J. M. ( 2004) Oncogene 23, 2838– 2849 [DOI] [PubMed] [Google Scholar]
- 18.Loda M., Capodieci P., Mishra R., Yao H., Corless C., Grigioni W., Wang Y., Magi-Galluzzi C., Stork P. J. ( 1996) Am. J. Pathol. 149, 1553– 1564 [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-Pérez I., Martínez-Gomariz M., Williams D., Keyse S. M., Perona R. ( 2000) Oncogene 19, 5142– 5152 [DOI] [PubMed] [Google Scholar]
- 20.Wang J., Zhou J. Y., Wu G. S. ( 2007) Cancer Res. 67, 11933– 11941 [DOI] [PubMed] [Google Scholar]
- 21.Zhou J. Y., Liu Y., Wu G. S. ( 2006) Cancer Res. 66, 4888– 4894 [DOI] [PubMed] [Google Scholar]
- 22.Franklin C. C., Srikanth S., Kraft A. S. ( 1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3014– 3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Gorospe M., Hutter D., Barnes J., Keyse S. M., Liu Y. ( 2001) Mol. Cell. Biol. 21, 8213– 8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Xu J., Zhou J. Y., Liu Y., Wu G. S. ( 2006) Cancer Res. 66, 8870– 8877 [DOI] [PubMed] [Google Scholar]
- 25.Brondello J. M., Pouysségur J., McKenzie F. R. ( 1999) Science 286, 2514– 2517 [DOI] [PubMed] [Google Scholar]
- 26.Lin Y. W., Yang J. L. ( 2006) J. Biol. Chem. 281, 915– 926 [DOI] [PubMed] [Google Scholar]
- 27.Chen C., Sun X., Ran Q., Wilkinson K. D., Murphy T. J., Simons J. W., Dong J. T. ( 2005) Oncogene 24, 3319– 3327 [DOI] [PubMed] [Google Scholar]
- 28.Chen C., Zhou Z., Ross J. S., Zhou W., Dong J. T. ( 2007) Int. J. Cancer 121, 80– 87 [DOI] [PubMed] [Google Scholar]
- 29.Aplin A. E., Stewart S. A., Assoian R. K., Juliano R. L. ( 2001) J. Cell Biol. 153, 273– 282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C., Bhalala H. V., Qiao H., Dong J. T. ( 2002) Oncogene 21, 6567– 6572 [DOI] [PubMed] [Google Scholar]
- 31.Aizawa K., Suzuki T., Kada N., Ishihara A., Kawai-Kowase K., Matsumura T., Sasaki K., Munemasa Y., Manabe I., Kurabayashi M., Collins T., Nagai R. ( 2004) J. Biol. Chem. 279, 70– 76 [DOI] [PubMed] [Google Scholar]
- 32.Slack D. N., Seternes O. M., Gabrielsen M., Keyse S. M. ( 2001) J. Biol. Chem. 276, 16491– 16500 [DOI] [PubMed] [Google Scholar]
- 33.Tassi E., Al-Attar A., Aigner A., Swift M. R., McDonnell K., Karavanov A., Wellstein A. ( 2001) J. Biol. Chem. 276, 40247– 40253 [DOI] [PubMed] [Google Scholar]
- 34.Yang Y., Goldstein B. G., Nakagawa H., Katz J. P. ( 2007) FASEB J. 21, 543– 550 [DOI] [PubMed] [Google Scholar]
- 35.Hamdi M., Kool J., Cornelissen-Steijger P., Carlotti F., Popeijus H. E., van der Burgt C., Janssen J. M., Yasui A., Hoeben R. C., Terleth C., Mullenders L. H., van Dam H. ( 2005) Oncogene 24, 7135– 7144 [DOI] [PubMed] [Google Scholar]