Abstract
Neurodegenerative tauopathies, including Alzheimer disease, are characterized by abnormal hyperphosphorylation of the microtubule-associated protein Tau. One group of tauopathies, known as frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17), is directly associated with mutations of the gene tau. However, it is unknown why mutant Tau is highly phosphorylated in the patient brain. In contrast to in vivo high phosphorylation, FTDP-17 Tau is phosphorylated less than wild-type Tau in vitro. Because phosphorylation is a balance between kinase and phosphatase activities, we investigated dephosphorylation of mutant Tau proteins, P301L and R406W. Tau phosphorylated by Cdk5-p25 was dephosphorylated by protein phosphatases in rat brain extracts. Compared with wild-type Tau, R406W was dephosphorylated faster and P301L slower. The two-dimensional phosphopeptide map analysis suggested that faster dephosphorylation of R406W was due to a lack of phosphorylation at Ser-404, which is relatively resistant to dephosphorylation. We studied the effect of the peptidyl-prolyl isomerase Pin1 or microtubule binding on dephosphorylation of wild-type Tau, P301L, and R406W in vitro. Pin1 catalyzes the cis/trans isomerization of phospho-Ser/Thr-Pro sequences in a subset of proteins. Dephosphorylation of wild-type Tau was reduced in brain extracts of Pin1-knockout mice, and this reduction was not observed with P301L and R406W. On the other hand, binding to microtubules almost abolished dephosphorylation of wild-type and mutant Tau proteins. These results demonstrate that mutation of Tau and its association with microtubules may change the conformation of Tau, thereby suppressing dephosphorylation and potentially contributing to the etiology of tauopathies.
One of hallmarks of Alzheimer disease (AD)3 pathology is neurofibrillary tangles, which are composed of paired helical filaments (PHFs), aggregates of the abnormally phosphorylated microtubule-associated protein Tau. Intracellular inclusions comprising Tau are also found in several other neurodegenerative diseases, including Pick disease, progressive supranuclear palsy, corticobasal degeneration, and frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17), collectively called tauopathies (1–3). Identification of Tau as a causative gene of the inherited tauopathy FTDP-17 reveals that Tau mutation is sufficient to cause disease (4–6). However, the impact Tau mutations have on neurodegeneration remains unknown.
Tau proteins in inclusions are hyperphosphorylated, and extensive studies have identified the phosphorylation sites; for example, more than 20 sites have been identified in PHF-Tau obtained from AD brains (7, 8). Tau can be phosphorylated by a variety of protein kinases, including glycogen synthase kinase 3β (GSK3β), cyclin-dependent kinase 5 (Cdk5), mitogen-activated protein kinase, cAMP-dependent protein kinase (PKA), microtubule affinity regulating kinase, and others (9–11). Tau is predominantly phosphorylated on the Ser or Thr residue in Ser/Thr-Pro sequences, suggesting the involvement of proline-directed protein kinases such as GSK3β and Cdk5 in hyperphosphorylation. A critical question is how mutations in Tau induce hyperphosphorylation in brain (12). Early phosphorylation experiments in vitro and in cultured cells have shown that mutant Tau is less phosphorylated than wild-type (WT) Tau (13–18). However, two later studies demonstrated higher phosphorylation of mutant Tau using brain extracts as a source of protein kinases in the presence of protein phosphatase inhibitor okadaic acid (19) or in immortalized cortical cells (20). However, it is not fully understood how mutant Tau becomes highly phosphorylated in vivo.
Tau hyperphosphorylation could also be attributed to reduced dephosphorylation activity. Tau is dephosphorylated in vitro by any of the major four classes of protein phosphatases, PP1, PP2A, PP2B, and PP2C, but PP2A is thought to be the major protein phosphatase that regulates Tau phosphorylation state in brains (21–23). PP2A activity reportedly is decreased in AD brain (24–26), and highly phosphorylated Tau in PHF is relatively resistant to dephosphorylation by PP2A (27). Few studies have been done on dephosphorylation of mutant Tau, however, and thus the mechanism remains unclear. One putative factor involved in mutant Tau dephosphorylation is the peptidyl-prolyl isomerase Pin1. Pin1 catalyzes the cis/trans isomerization of phospho-Ser/Thr-Pro sequences in a subset of proteins (28, 29). Pin1 is involved in AD pathogenesis as shown by the fact that it is found in neurofibrillary tangles and that Tau is hyperphosphorylated in Pin1-deficient mouse brains (30). Pin1 is indicated to facilitate Tau dephosphorylation via PP2A by binding to the phospho-Thr-231-Pro or phospho-Thr-212-Pro site (31–33). The effect of Pin1 on the stability of mutant Tau was recently reported (34), but a detailed analysis of Pin1 action on mutant Tau has not been reported. Another possible factor affecting dephosphorylation of mutant Tau is the binding to microtubules. We previously showed that phosphorylation of Tau is stimulated upon binding to microtubules (35). We thus hypothesized that binding to microtubules may also affect the extent of Tau dephosphorylation.
Here, we examined the effects of Pin1 and binding to microtubules on dephosphorylation of WT and FTDP-17 mutant (P301L and R406W) Tau proteins that had been phosphorylated by Cdk5-p25 or Cdk5-p35. P301L and R406W are two distinct types of FTDP-17 mutants that have been studied well. We show for the first time how the regulation of Tau dephosphorylation can contribute to the observed Tau hyperphosphorylation in tauopathies.
EXPERIMENTAL PROCEDURES
Enzymes, Plasmids, and Chemicals
The catalytic subunit of each of PP1 and PP2A was purchased from Upstate (Lake Placid, NY). Brain extract of Pin1-knockout mice and the preparation of GST-Pin1 have been described (36). Cdk5/p35 and Cdk5/p25 were prepared from Sf9 cells as described (37). GSK3β and the catalytic subunit of PKA were purified from rat brain extract as described (38). GSK3β purchased from Millipore (Billerica, MA) was also used for some experiments.
Expression and Purification of Recombinant Human Tau
cDNAs encoding recombinant human WT and FTDP-17 mutant Tau (P301L and R406W, respectively) with one N-terminal insert and four repeat domains (1N4R, see supplemental Fig. S4A) in the pRK172 bacterial expression vector were used to transform Escherichia coli BL21(DE3)-competent cells (35, 39). ΔMTB, Tau without microtubule-binding repeats, was constructed by PCR with primers 5′-AATAAAAAGATTGAAACCCACAAGCTGAC-3′ (sense) and 5′-ATTCTTCAGGTCTGGCATGG-3′ (antisense). The PCR product was phosphorylated by T4 polynucleotide kinase and ligated into pSG5. The accuracy of the construct was verified by DNA sequencing. Tau proteins were purified from the heat-stable supernatant of the E. coli extract by chromatography on a Mono S column as described (35).
Preparation of Tubulin and Binding of Tau to Microtubules
Tubulin was prepared from porcine brains by three cycles of temperature-dependent polymerization/depolymerization (40). Tubulin was separated from microtubule-associated proteins using a phosphocellulose column (P11, Whatman, Brentford, UK) as described (40). Tau (0.05 mg/ml) was bound to microtubules, which were polymerized from 0.5 mg/ml tubulin with an aid of 20 μm taxol in 20 mm MOPS, pH 6.8, and 1 mm MgCl2 by incubation at 35 °C for 20 min as described (35).
Preparation of Phosphorylated Tau (P-Tau)
Recombinant Tau (0.1 mg/ml) was phosphorylated with Cdk5-p35 or Cdk5-p25 in 20 mm MOPS, pH 6.8, and 1 mm MgCl2, in the presence of 0.1 mm [γ-32P]ATP for 1 h at 35 °C. Excess [γ-32P]ATP was removed by gel-filtration on a Sephadex G-50 column. Fractions containing P-Tau were pooled, stored at −80 °C, and used for dephosphorylation experiments within 10 days.
Dephosphorylation of P-Tau
P-Tau (0.05 mg/ml) was incubated with rat brain extracts at 35 °C for various times in 20 mm HEPES, pH 7.4, 1 mm MgCl2 and 1 mm EDTA. Dephosphorylation was stopped by addition of 4× SDS-PAGE sample buffer. After boiling for 2 min, the samples were subjected to SDS-PAGE. The remaining 32P-Tau was estimated by a FLA-7000 Bioimage Analyzer (Fujifilm, Tokyo, Japan). Dephosphorylation was defined as the amount of phosphate removed from P-Tau during incubation. Dephosphorylation of experimental samples was expressed as a percentage of that of P-WT Tau by WT rat or mouse brain extracts.
Two-dimensional Phosphopeptide Microtubule-associated Protein Analysis
P-Tau separated by SDS-PAGE was digested in gel slices by trypsin (Sigma) in 50 mm NH4HCO3, pH 8.4, at 30 °C overnight. The resulting tryptic peptides were subjected to two-dimensional phosphopeptide mapping using a thin-layer cellulose plate (catalogue number 1.05716.0001, Merck, Darmstadt, Germany) (35). Electrophoresis was performed at pH 1.9, and ascending chromatography was performed in n-butyl alcohol/pyridine/glacial acetic acid/water (150/100/30/120). Phosphopeptide spots were detected, and the intensity was measured by a FLA-7000 Bioimage Analyzer.
RESULTS
Dephosphorylation of FTDP-17 Mutant Tau, P301L, and R406W
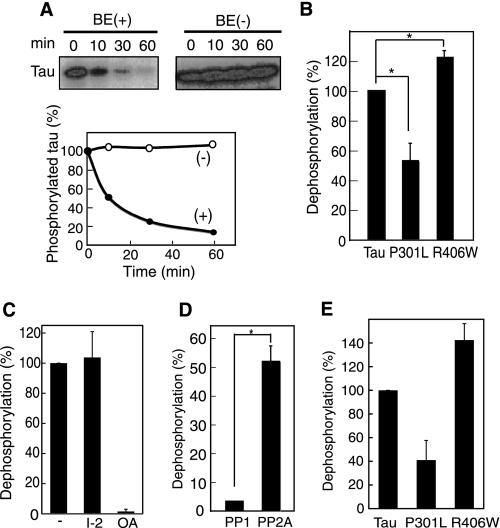
Purified, recombinant Tau, either WT Tau or FDTP-17 mutant Tau P301L or R406W, was phosphorylated in vitro by Cdk5-p25 in the presence of [γ-32P]ATP (supplemental Fig. S1A). About 4 mol of phosphate was incorporated into 1 mol of WT Tau. 32P bound to WT Tau was easily removed by incubation with brain extract (BE) but not without the brain extract (−) (Fig. 1A). The Tau protein was not degraded during incubation. The decrease in the 32P label on Tau was inhibited by the protein phosphatase inhibitor, microcystin LR (supplemental Fig. S2A), indicating that the decrease in the 32P label is dephosphorylation. We measured the dephosphorylation of in vitro phosphorylated P301L and R406W; P-P301L was dephosphorylated slower and P-R406W faster than P-WT Tau (Fig. 1B). These results constitute the evidence of differential rates of dephosphorylation between WT and mutant Taus.
FIGURE 1.
Dephosphorylation of phosphorylated WT and mutant Tau by rat brain extract. A, purified recombinant WT Tau phosphorylated by Cdk5/p25 in the presence of [γ-32P]ATP was incubated without (−) or with rat brain extract (BE) for the indicated times. After the samples were subjected to SDS-PAGE, radioactivity associated with Tau was detected by a FLA-7000 Bioimage Analyzer (upper panels) and quantified (lower panel). B, dephosphorylation of P-WT Tau, P-P301L, and P-R406W by rat brain extract. Dephosphorylation was defined as the signals removed from P-Tau during a 10-min incubation, and the respective dephosphorylation is expressed as the percentage relative to that measured for WT Tau. C, dephosphorylation of P-WT Tau by rat brain extract in the presence of 40 nm inhibitor-2 or 10 nm okadaic acid. D, dephosphorylation of P-WT Tau by the catalytic subunit of PP1 or PP2A (0.1 unit/ml for each). E, dephosphorylation of P-WT Tau, P-P301L, or P-R406W by PP2A.
Tau is mainly dephosphorylated by PP2A (21–23, 25). To determine whether our observed difference in dephosphorylation (Fig. 1B) was due to PP2A activity, we dephosphorylated P-Tau by rat brain extract in the presence of phosphatase inhibitor. Dephosphorylation was inhibited by okadaic acid at 10 nm, the concentration specific to PP2A, but not inhibited by inhibitor-2 (40 nm), a heat-stable inhibitory protein for PP1 (Fig. 1C). We confirmed the results using the purified catalytic subunit of PP1 or PP2A. P-Tau was dephosphorylated by PP2A much faster than by PP1 (Fig. 1D), consistent with previous reports (22, 25). Further, P-P301L was dephosphorylated by PP2A slower and P-R406W faster compared with P-WT Tau (Fig. 1E), as observed in the brain extract experiments (Fig. 1B). These data suggest that PP2A is the major protein phosphatase differentially dephosphorylating these Tau substrates (i.e. FTDP-17 mutants and WT Tau) in brain extract.
Difference in Dephosphorylation Rate among Phosphorylation Sites on Tau
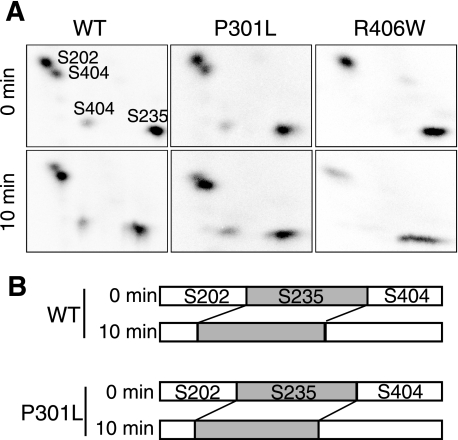
WT and P301L Tau are phosphorylated mainly at Ser-202, Ser-235, and Ser-404 by Cdk5-p25, and R406W is phosphorylated mainly at Ser-202 and Ser-235 (35). We analyzed the dephosphorylation rate at these sites in P301L or R406W by two-dimensional phosphopeptide mapping. Indeed, Ser-202, Ser-235, and Ser-404 were phosphorylated in WT Tau and P301L, and Ser-202 and Ser-235 in R406W (Fig. 2A, 0 min), as shown previously (35). Two-dimensional phosphopeptide patterns after 10 min of dephosphorylation in the brain extract were similar to those before incubation, although there were differences in relative intensity (Fig. 2A). We quantified the phosphorylation signals in each of four spots in WT or P301L and expressed them as a fraction of the sum of these four phosphorylation spots (Fig. 2B). The data clearly show that Ser-202 was dephosphorylated faster than Ser-404. Because R406W lacks Ser-404 phosphorylation, these also results suggest a rationale for why R406W is dephosphorylated faster than WT or P301L.
FIGURE 2.
Phosphorylation site-dependent dephosphorylation of Tau. A, two-dimensional phosphopeptide maps of phosphorylated WT, P301L, and R406W Tau before (0 min) and after a 10-min incubation with the rat brain extract. The radioactivity applied to cellulose plates was adjusted to 1000 cpm. The quantification of the respective spots in P-WT Tau and P-P301L is shown in (B). The relative signal intensity of the Ser-202, Ser-235, and Ser-404 phosphorylation sites is expressed as a fraction of the total signals of these phosphorylation spots in WT and P301L before (0 min) and after (10 min) dephosphorylation. These data are representative of those obtained from three different experiments.
Effect of Pin1 on Dephosphorylation of FTDP-17 Mutant Tau Proteins
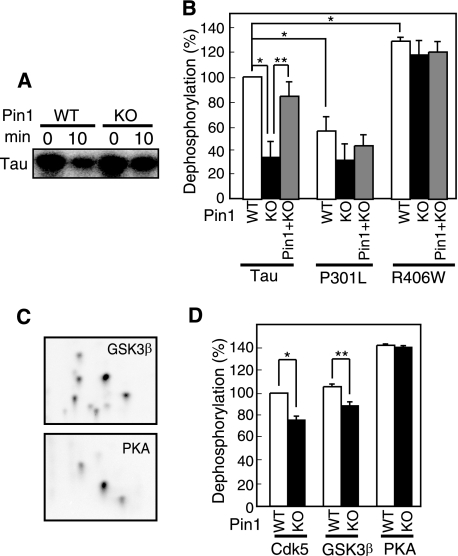
The prolyl-peptidyl isomerase Pin1 facilitates PP2A-dependent dephosphorylation of P-Tau by converting the cis peptide bond of proline at phospho-Ser/Thr-Pro sites to the trans conformation (31), thereby yielding a more kinetically favorable P-Tau substrate. However, its effect on dephosphorylation of mutant Tau has not been examined. We measured dephosphorylation of P-WT Tau and P-mutant Taus in brain extracts from Pin1-KO and WT mice. An incubation time of 10 min was chosen as the optimum condition for detecting the difference (supplemental Fig. S2B). Dephosphorylation of P-Tau was slower in Pin1-KO mouse brain extract compared with WT mouse brain extract (Fig. 3 A and B), and the dephosphorylation rate increased upon addition of purified Pin1 at 2 nm (Fig. 3B, WT), a concentration that is sufficient to restore the dephosphorylation activity in Pin1-KO mice (41). This result is consistent with the report that Pin1 enhances dephosphorylation of P-Tau by PP2A (32). We then compared dephosphorylation of P-P301L and P-R406W in Pin1-KO and WT extracts. Interestingly, there was less of a difference in dephosphorylation between Pin1-KO and WT mouse brain extract in P301L and R406W compared with WT Tau; moreover, addition of purified Pin1 to Pin1-KO mouse brain extract did not stimulate the dephosphorylation rate of P-P301L and P-R406W (Fig. 3B). These results suggest that FTDP-17 mutations confer insensitivity of P-Tau to Pin1-dependent enhancement of dephosphorylation.
FIGURE 3.
Effect of Pin1 on dephosphorylation of P-WT tau and P-mutant Tau proteins. A, autoradiogram of Tau dephosphorylated in Pin1-KO or WT mouse brain extract for 10 min. B, dephosphorylation of P-WT Tau, P-P301L, and P-R406W by brain extract from WT or Pin1-KO mice, or Pin1-KO mouse brain extract supplemented with purified Pin1. C, two-dimensional phosphopeptide maps of Tau phosphorylated by GSK3β or PKA. D, dephosphorylation of WT Tau phosphorylated with Cdk5/p25, GSK3β, or PKA by brain extract from Pin1-KO or WT mice. Dephosphorylation is expressed as the percentage of the dephosphorylation of P-WT Tau by WT mouse brain extract.
We next addressed whether the effect of Pin1 deficiency on dephosphorylation was specific for the phospho-Ser/Thr-Pro sequence. For this experiment, in addition to using Cdk5 we used two other protein kinases, GSK3β, a proline-directed kinase, and PKA, a non-proline-directed kinase, to phosphorylate Tau. About 2–3 mol of phosphate was incorporated into Tau after incubation with these protein kinases (supplemental Fig. S1B), but two-dimensional phosphopeptide maps were different between them as expected based on their different phosphorylation sites (Fig. 3C). Respective phosphopeptide maps are similar to those of GSK3β- and PKA-phosphorylated Tau reported previously (42). P-WT Tau phosphorylated by GSK3β or PKA was dephosphorylated easily by WT brain extract (Fig. 3D). The dephosphorylation of GSK3β-phosphorylated WT Tau as well as Cdk5-phosphorylated WT Tau was decreased in the presence of Pin1-KO mouse brain extract, but this was not the case for PKA-phosphorylated WT Tau, consistent with previous results that the effect of Pin1 on dephosphorylation is specific for certain phosphorylation sequences (31–33).
Dephosphorylation of P-Tau Bound to Microtubules
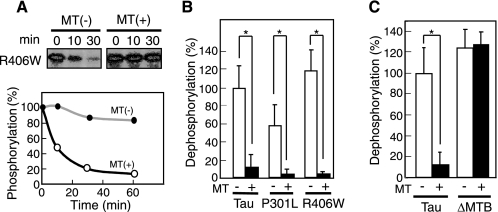
Phosphorylation of Tau is stimulated when Tau is bound on microtubules (40). Further, phosphorylation by Cdk5-p35 or Cdk5-p25 of mutant Tau bound to microtubules is enhanced to the same extent as that of WT Tau (35). We thus hypothesized that binding to microtubules may also affect dephosphorylation of Tau. WT Tau, P301L, or R406W (0.05 mg/ml), each of which had been phosphorylated by Cdk5-p25, was bound to taxol-polymerized microtubules. Under our experimental conditions, essentially 100% of P-Tau was bound to microtubules. When microtubule-bound P-R406W was incubated with rat brain extract, dephosphorylation was almost completely abolished, whereas unbound P-R406W was quickly dephosphorylated (Fig. 4A). Similar results were obtained for P-WT Tau and P-P301L (Fig. 4B).
FIGURE 4.
Dephosphorylation of P-Tau bound to microtubules. A, autoradiogram showing 32P-labeled phosphate remaining on R406W Tau incubated with rat brain extract for 10 or 30 min in the absence (−) or presence (+) of microtubules (MT). The radioactivity was quantified and is presented graphically in the lower panel. B, dephosphorylation of P-WT Tau, P-P301L, or P-R406 by rat brain extract in the absence (−) or presence (+) of microtubules (MT). C, dephosphorylation of P-WT Tau or P-ΔMTB by rat brain extract in the absence (−) or presence (+) of microtubules (MT). Dephosphorylation is expressed as the percentage of the dephosphorylation of WT Tau by rat brain extract. p values were calculated using Student's t test and the bars indicate the means ± S.E. of three experiments; *, p < 0.01.
Several control experiments were performed to substantiate the above results. First, we verified that the tubulin fraction did not contain protein kinases, which could counter any dephosphorylation. In this regard, we checked the phosphorylation state of P-WT Tau after incubation with microtubules; no change was evident, indicating that the tubulin fraction contained neither Tau kinases nor phosphatases (supplemental Fig. S3A). Second, we verified that the GTP used for tubulin polymerization did not act as a competitive substrate for dephosphorylation; we found that dephosphorylation proceeded at a similar rate in the absence or presence of 0.5 mm GTP (supplemental Fig. S3B). Third, we verified that tubulin did not inhibit the protein phosphatases in brain extract. To test this possibility, we used an alternate substrate, neurofilament light chain (NF-L, in this case phosphorylated by Cdk5-p25), which does not interact with microtubules (38). Dephosphorylation of NF-L proceeded in the presence of microtubules, indicating that tubulin does not inhibit brain extract phosphatases (supplemental Fig. S3C). Fourth, it has been suggested that the repeat region of Tau binds to the interior of microtubules when tubulin is co-assembled with Tau (43), and thus we verified that P-WT Tau did not bind inside of microtubules in our system. We added P-WT Tau to pre-assembled microtubules and measured dephosphorylation. Indeed, the order of P-WT Tau addition did not affect its dephosphorylation (supplemental Fig. S3D).
The above results suggested that, upon binding to microtubules, P-Tau changes its conformation such that dephosphorylation is unfavorable. To demonstrate this possibility more clearly, we used a mutant Tau, ΔMTB, in which the microtubule-binding domain (residues 256–367 according to the longest human Tau 441) was deleted (supplemental Fig. S4A). ΔMTB did not bind to microtubules effectively (supplemental Fig. S4B), as shown previously (44), but it was phosphorylated at the same sites as WT Tau, as demonstrated by two-dimensional phosphopeptide mapping (supplemental Fig. S4C). P-ΔMTB Tau and P-WT Tau were dephosphorylated at similar rates when incubated with WT mouse brain extract (Fig. 4C); however, dephosphorylation of P-ΔMTB was not suppressed in the presence of microtubules. Taken together, these results indicate that the binding of P-Tau to microtubules suppresses its dephosphorylation.
Effect of Pin1 on Dephosphorylation of P-Tau Bound to Microtubules
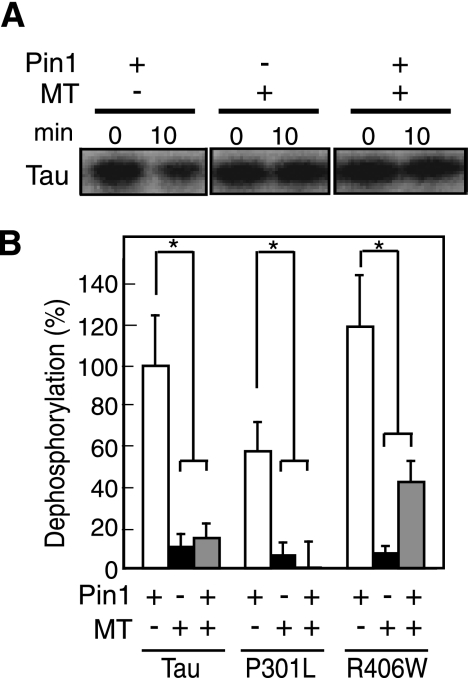
We examined the effect of exogenously added Pin1 on dephosphorylation of P-WT Tau bound to microtubules. Addition of purified Pin1 to Pin1-KO mouse brain extract did not enhance the dephosphorylation of P-WT Tau bound to microtubules (compare lane 6 with lane 4 in Fig. 5A). Dephosphorylated in the absence of microtubules is shown in lane 2 of Fig. 5A. Similar results were obtained with P301L and R406W (Fig. 5B). Although Pin1 appeared to increase the dephosphorylation of R406W, the increase was not statistically significant. Thus, Pin1 could not eliminate the suppression of P-Tau dephosphorylation upon microtubule binding.
FIGURE 5.
Effect of Pin1 on dephosphorylation of P-Tau in the absence or presence microtubules. A, autoradiograms of P-WT Tau on microtubules (MT) incubated with Pin1-KO mouse brain extract supplemented with 2 μm purified Pin1 (+, lanes 5 and 6) or not (−, lanes 3 and 4) for 0 or 10 min. Dephosphorylation of free P-WT Tau is shown in lanes 1 and 2. B, dephosphorylation of P-WT Tau, P-P301L, or P-R406W in the absence (−) or presence (+) of microtubules (MT) by Pin1-KO mouse brain extract supplemented with Pin1 (+) or not (−). Dephosphorylation of free Tau is shown in the left bar of each set of data. Dephosphorylation is expressed as the percentage of the extent of WT Tau dephosphorylation by Pin1-KO mouse brain extract in the absence of microtubules (MT). p values were calculated using Student's t test, and the bars indicate the means ± S.E. of three experiments; *, p < 0.01.
DISCUSSION
We investigated dephosphorylation of P-Taus, namely WT, P301L, and R406W, phosphorylated by Cdk5-p25. Compared with P-WT Tau, P-R406W was dephosphorylated faster and P-P301L slower. The dephosphorylation rate differed between phosphorylation sites; Ser-404 was dephosphorylated slower and Ser-202 was faster than Ser-235. The latter could explain why P-R406W, which lacks Ser-404 phosphorylation, was dephosphorylated faster than P-WT Tau or P-P301L. We confirmed that dephosphorylation of Tau was attenuated in mouse brain extract lacking Pin1. Of import is that the Pin1 deficiency was countered by the FTDP-17 mutation of Tau. Furthermore, dephosphorylation of P-Tau, either WT or mutants, was almost completely abolished upon binding to microtubules. These findings demonstrate that the phosphorylation states of Tau are actively regulated not only by activation of protein kinases but also by dephosphorylation reactions that depend presumably on the conformational state(s) of Tau under normal or pathologic conditions.
Hyperphosphorylation is a characteristic of Tau in tauopathic brains (11, 12, 45). However, it is not known how FTDP-17 mutations induce hyperphosphorylation. There was a discrepancy between in vivo and in vitro phosphorylation status of FTDP-17 Tau (13–16, 45). Two recent reports describe higher phosphorylation of mutant Tau (19, 20), but the molecular mechanism that induces abnormal phosphorylation of mutant Tau remains to be elucidated. Because phosphorylation is a balance between kinase and phosphatase activities, we addressed this issue with respect to dephosphorylation, which for FTDP-17 mutant Taus has not been thoroughly examined. We chose P301L and R406W because they represent two distinct types of FTDP-17 mutation that often have been compared. The early-onset mutation, P301L, is one of the most frequently identified mutations in FTDP-17 and has severe phenotypes (6, 46, 47). By contrast, R406W is late onset and has mild phenotypes (4, 48). These mutations differ in fibrillar formation: P301L can selectively form filaments on its own (45, 49), whereas R406W forms filaments by assembling with WT Tau (45, 50). In vitro studies have demonstrated that both P301L and R406W mutations reduce the ability of Tau to promote microtubule assembly (39, 51), whereas the effect of R406W on microtubules dynamics is comparable to that of WT Tau in cells (52). Here, a difference in dephosphorylation rate was also found between P301L and R406W. P301L was dephosphorylated slower than WT Tau, and R406W was dephosphorylated faster than WT Tau. Very recently, the effect of FTDP-17 mutations on site-specific dephosphorylation was reported using HEK-293 cell extract as a source of protein phosphatases (53). Considering that most FTDP-17 mutants have properties similar to P301L rather than R406W, reduced dephosphorylation of P301L may account, at least in part, for the observed high levels of phosphorylation of mutant Tau in brains afflicted with tauopathies.
Although both P301L and R406W are highly phosphorylated in vivo, their rate of dephosphorylation differed in vitro. Under our in vitro phosphorylation conditions, Cdk5/p25 mainly phosphorylated Ser-202, Ser-235, and Ser-404 in P301L as well as WT Tau. Among these serines, Ser-202 was dephosphorylated faster and Ser-404 was dephosphorylated slower by protein phosphatases in brain extract. The resistance of Ser-404 to dephosphorylation was recently reported (54). Unlike P301L, R406W is not phosphorylated on Ser-404. The lack of Ser-404 phosphorylation in the R406W mutant also has been reported in R406W-overexpressing mouse brain (15, 17, 55). Mutation of Arg-406 in the Ser-Pro-Arg sequence to Trp may disrupt Cdk5 recognition of the phosphorylation site. Clearly, the absence of phosphorylation at a site that is dephosphorylated slower in other Taus results in the apparent faster dephosphorylation of R406W. However, the phosphorylation in the C-terminal tail region involving Ser-396, Ser-400, and Ser-404 is complex. Phosphorylation at Ser-404 affects any subsequent phosphorylation at Ser-400 and then Ser-396 in a hierarchical manner (56). In contrast, it also has been reported that Ser-396 phosphorylation is elevated in R406W when phosphorylated in vitro by brain extract in the presence of okadaic acid (19). It may be interesting to quantify the relative phosphorylation state and dephosphorylation rate of Ser-400 and Ser-396 in R406W, because phosphorylation at Ser-396 and Ser-404 is well known as the PHF1 epitope, one of the abnormal phosphorylation sites in PHF Tau (57).
The site-dependent difference in dephosphorylation rate does not underlie the observed high phosphorylation of mutant Taus, because P301L shares the same phosphorylation pattern as WT Tau. We do not know the answer to explain the issue at present. But the number of microtubule-binding repeats in Tau may be involved in this question. The P301L mutation is located in the C-terminal side of the second microtubule-binding repeat that is found only in four-repeat Tau. This mutation is shown to change the local conformation to increase the propensity of the β-sheet around it, from Lys-298 to Gly-305 (58). This subtle change may increase the interaction of basic amino acids in the repeats, which are the tubulin interacting residues, with phosphorylation sites in the flanking regions. This may result in reduced accessibility by protein phosphatases.
We examined whether Pin1 could distinguish between mutant and WT Taus, because Pin1 facilitates the dephosphorylation of the phospho-Ser/Thr-Pro sequence by PP2A (31). Further, Pin1 dysfunction is indicated in AD pathogenesis, because the protein is detected in neurofibrillary tangles, and Tau is hyperphosphorylated in Pin1-deficient mouse brains (30). We confirmed here that Pin1 enhances dephosphorylation of P-Tau by PP2A, and this enhanced dephosphorylation occurred at proline-directed phosphorylation sites. More important was loss of the Pin1-sensitive enhancement of dephosphorylation in both P301L and R406W. These results predict that P301L and R406W would be phosphorylated more extensively in Pin1 KO mouse brain. We cannot explain why the dephosphorylation of FTDP-17 mutants was insensitive to Pin1, but both P-P301L and P-R406W may have a conformation that cannot be recognized by Pin1. Pin1 binding sites in Tau are reported to be phospho-Thr-231-Pro (30) or phospho-Thr-212-Pro (32, 59). Cdk5 phosphorylation sites at Ser-202, Ser-235 and Ser-404, however, are not included in these Pin1 binding sites. Thus, there may be unidentified Pin1 binding sites among Cdk5-specific phosphorylation sites. It was recently reported that Pin1 differentially affects the in vivo half-life of P301L and R406W; P301L is destabilized by Pin1 deficiency, whereas R406W and WT Tau are stabilized (34). It would be interesting to examine any relationship between Pin1-dependent dephosphorylation of Taus and their stability.
Binding to microtubules remarkably affects dephosphorylation of Tau. In general our results agree with those of Sontag et al. (60) that dephosphorylation of PKA-phosphorylated Tau by purified PP2A is reduced in the presence of taxol-stabilized microtubules; our interpretation of these results, however, differs from theirs. We would like to point out that Sontag et al. (60) collected PKA-phosphorylated Tau, which was still bound to microtubules, by co-sedimentation with the microtubules, and dephosphorylated them after stabilization of microtubules with taxol. They argued that microtubules inhibit PP2A activity using myosin-light chain as a substrate that does not bind to microtubules. In our experiments, however, dephosphorylation of ΔMTB or NF-L, both of which do not bind microtubules, proceeded as fast as soluble Tau even in the presence of microtubules, suggesting that Tau becomes resistant to dephosphorylation by binding to microtubules. The difference may be derived from the protein phosphatase fraction used; we used rat brain extracts that contain various forms of PP2A, including ABαC complex, whereas Sontag et al. (60) used purified ABαC complex. In either case, both datasets suggest that dephosphorylation of Tau is suppressed in the presence of microtubules regardless of whether Tau is phosphorylated by PKA or Cdk5. We think that the suppression is caused by a conformational change in Tau upon microtubule binding, which evades PP2A recognition. Tau has been thought to lack a defined conformation, but some folding of Tau also has been reported (61, 62). It remains unknown, however, whether Tau changes conformation upon binding to microtubules. A recent NMR study revealed two tubulin binding motifs in the proline-rich region flanking the GSK3 phosphorylation site Ser-231 and Cdk5 phosphorylation site Ser-235 (63). As such, it is highly possible that accessibility of kinases or phosphatases to these phosphorylation sites is affected by microtubule binding. Notably, dephosphorylation of Tau bound to microtubules may reflect the situation of PHF Tau, which is resistant to dephosphorylation by protein phosphatases (27). If PHF Tau acquires a conformation similar to that of Tau on microtubules, then investigation of microtubule-dependent suppression of dephosphorylation may help to elucidate the mechanism by which PHF Tau remains hyperphosphorylated.
Taken together, our results suggest a model for how mutant Tau remains highly phosphorylated in the brains of FTDP-17 patients. The phosphorylation state of most FTDP-17 mutants is similar to P301L rather than R406W. Therefore, high phosphorylation state of most mutants may be maintained by a reduced level of dephosphorylation by PP2A. Mutant Tau has a tendency to aggregate irreversibly, resulting in greater resistance to dephosphorylation, leading to hyperphosphorylation. And what of R406W, which appears to bind microtubules with higher affinity than other mutants in cells (52). Therefore, the R406W mutant Tau may undergo a conformational change, similar to other mutants, upon which dephosphorylation becomes insensitive to Pin1. Phosphorylated R406W on microtubules may aggregate with itself or with phosphorylated WT Tau. Interaction between phosphorylated Tau molecules on microtubules may induce oligomerization or PHF-like aggregates that would be relatively resistant to dephosphorylation. In the case of WT Tau, association with microtubules also may initiate hyperphosphorylation. But as it has generally been observed, high phosphorylation of Tau increases its propensity to dissociate from microtubules (9, 11). Compared with mutant Tau proteins, P-WT Tau, upon dissociation from microtubules, is dephosphorylated easily after conversion to the trans form by Pin1, and therefore WT Tau is much less likely to form aggregates i.e. representing the non-pathologic state.
Supplementary Material
This work was supported in part by grants-in-aid for scientific research on Priority Area from MEXT of Japan (to S. H.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
- AD
- Alzheimer disease
- Cdk5
- cyclin-dependent kinase 5
- FTDP-17
- frontotemporal dementia with parkinsonism linked to chromosome 17
- GSK3β
- glycogen synthase kinase 3β
- MT
- microtubule
- PHF
- paired helical filament
- PKA
- cAMP-dependent protein kinase
- PP1
- protein phosphatase 1
- PP2A
- protein phosphatase 2A
- MOPS
- 4-morpholinepropanesulfonic acid
- WT
- wild type.
REFERENCES
- 1.Lee V. M., Goedert M., Trojanowski J. Q. ( 2001) Annu. Rev. Neurosci. 24, 1121– 1159 [DOI] [PubMed] [Google Scholar]
- 2.Goedert M., Spillantini M. G. ( 2006) Science 314, 777– 781 [DOI] [PubMed] [Google Scholar]
- 3.Hasegawa M. ( 2006) Neuropathology 26, 484– 490 [DOI] [PubMed] [Google Scholar]
- 4.Hutton M., Lendon C. L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., Hackett J., Adamson J., Lincoln S., Dickson D., Davies P., Petersen R. C., Stevens M., de Graaff E., Wauters E., van Baren J., Hillebrand M., Joosse M., Kwon J. M., Nowotny P., Che L. K., Norton J., Morris J. C., Reed L. A., Trojanowski J., Basun H., Lannfelt L., Neystat M., Fahn S., Dark F., Tannenberg T., Dodd P. R., Hayward N., Kwok J. B., Schofield P. R., Andreadis A., Snowden J., Craufurd D., Neary D., Owen F., Oostra B. A., Hardy J., Goate A., van Swieten J., Mann D., Lynch T., Heutink P. ( 1998) Nature 393, 702– 705 [DOI] [PubMed] [Google Scholar]
- 5.Poorkaj P., Bird T. D., Wijsman E., Nemens E., Garruto R. M., Anderson L., Andreadis A., Wiederholt W. C., Raskind M., Schellenberg G. D. ( 1998) Ann. Neurol. 43, 815– 825 [DOI] [PubMed] [Google Scholar]
- 6.Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 6469– 6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morishima-Kawashima M., Hasegawa M., Takio K., Suzuki M., Yoshida H., Titani K., Ihara Y. ( 1995) J. Biol. Chem. 270, 823– 829 [DOI] [PubMed] [Google Scholar]
- 8.Hanger D. P., Betts J. C., Loviny T. L., Blackstock W. P., Anderton B. H. ( 1998) J. Neurochem. 71, 2465– 2476 [DOI] [PubMed] [Google Scholar]
- 9.Mandelkow E. M., Biernat J., Drewes G., Gustke N., Trinczek B., Mandelkow E. ( 1995) Neurobiol. Aging 16, 355– 362, discussion 362–363 [DOI] [PubMed] [Google Scholar]
- 10.Imahori K., Uchida T. ( 1997) J. Biochem. 121, 179– 188 [PubMed] [Google Scholar]
- 11.Iqbal K., Alonso Adel C., Chen S., Chohan M. O., El-Akkad E., Gong C. X., Khatoon S., Li B., Liu F., Rahman A., Tanimukai H., Grundke-Iqbal I. ( 2005) Biochim. Biophys. Acta 1739, 198– 210 [DOI] [PubMed] [Google Scholar]
- 12.Avila J. ( 2006) FEBS Lett. 580, 2922– 2927 [DOI] [PubMed] [Google Scholar]
- 13.Matsumura N., Yamazaki T., Ihara Y. ( 1999) Am. J. Pathol. 154, 1649– 1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dayanandan R., Van Slegtenhorst M., Mack T. G., Ko L., Yen S. H., Leroy K., Brion J. P., Anderton B. H., Hutton M., Lovestone S. ( 1999) FEBS Lett. 446, 228– 232 [DOI] [PubMed] [Google Scholar]
- 15.Pérez M., Lim F., Arrasate M., Avila J. ( 2000) J. Neurochem. 74, 2583– 2589 [DOI] [PubMed] [Google Scholar]
- 16.Sahara N., Tomiyama T., Mori H. ( 2000) J. Neurosci. Res. 60, 380– 387 [DOI] [PubMed] [Google Scholar]
- 17.Connel J. W., Gibb G. M., Betts J. C., Blackstock W. P., Gallo J., Lovestone S., Hutton M., Anderton B. H. ( 2000) FEBS Lett. 493, 40– 44 [DOI] [PubMed] [Google Scholar]
- 18.Mack T. G., Dayanandan R., Van Slegtenhorst M., Whone A., Hutton M., Lovestone S., Anderton B. H. ( 2001) Neuroscience 108, 701– 712 [DOI] [PubMed] [Google Scholar]
- 19.Alonso A., del C., Mederlyova A., Novak M., Grundke-Iqbal I., Iqbal K. ( 2004) J. Biol. Chem. 279, 34873– 34881 [DOI] [PubMed] [Google Scholar]
- 20.Krishnamurthy P. K., Johnson G. V. ( 2004) J. Biol. Chem. 279, 7893– 7900 [DOI] [PubMed] [Google Scholar]
- 21.Gong C. X., Grundke-Iqbal I., Iqbal K. ( 1994) Neuroscience 61, 765– 772 [DOI] [PubMed] [Google Scholar]
- 22.Goedert M., Jakes R., Qi Z., Wang J. H., Cohen P. ( 1995) J. Neurochem. 65, 2804– 2807 [DOI] [PubMed] [Google Scholar]
- 23.Kins S., Crameri A., Evans D. R., Hemmings B. A., Nitsch R. M., Gotz J. ( 2001) J. Biol. Chem. 276, 38193– 38200 [DOI] [PubMed] [Google Scholar]
- 24.Gong C. X., Shaikh S., Wang J. Z., Zaidi T., Grundke-Iqbal I., Iqbal. K. ( 1995) J. Neurochem. 65, 732– 738 [DOI] [PubMed] [Google Scholar]
- 25.Gong C. X., Lidsky T., Wegiel J., Zuck L., Grundke-Iqbal I., Iqbal K. ( 2000) J. Biol. Chem. 275, 5535– 5544 [DOI] [PubMed] [Google Scholar]
- 26.Sontag E., Hladik C., Montgomery L., Luangpirom A., Mudrak I., Ogris E., White C. L., 3rd. ( 2004) J. Neuropathol. Exp. Neurol. 63, 1080– 1091 [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto H., Hasegawa M., Ono T., Tashima K., Ihara Y., Miyamoto E. ( 1995) J. Biochem. 118, 1224– 1231 [DOI] [PubMed] [Google Scholar]
- 28.Lu K. P., Zhou X. Z. ( 2007) Nat. Rev. Mol. Cell Biol. 8, 904– 916 [DOI] [PubMed] [Google Scholar]
- 29.Takahashi K., Uchida C., Shin R. W., Shimazaki K., Uchida T. ( 2008) Cell Mol. Life Sci. 65, 359– 375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu P. J., Wulf G., Zhou X. Z., Davies P., Lu K. P. ( 1999) Nature 399, 784– 788 [DOI] [PubMed] [Google Scholar]
- 31.Zhou X. Z., Kops O., Werner A., Lu P.-J., Shen M., Stoller G., Küllertz G., Stark M., Fischer G., Lu K. P. ( 2000) Mol. Cell 6, 873– 883 [DOI] [PubMed] [Google Scholar]
- 32.Smet C., Sambo A. V., Wieruszeski J. M., Leroy A., Landrieu I., Buée L., Lippens G. ( 2004) Biochemistry 43, 2032– 2040 [DOI] [PubMed] [Google Scholar]
- 33.Galas M. C., Dourlen P., Bégard S., Ando K., Blum D., Hamdane M., Buée L. ( 2006) J. Biol. Chem. 281, 19296– 19304 [DOI] [PubMed] [Google Scholar]
- 34.Lim J., Balastik M., Lee T. H., Nakamura K., Liou Y. C., Sun A., Finn G., Pastorino L., Lee V. M., Lu K. P. ( 2008) J. Clin. Invest. 118, 1877– 1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaue F., Saito T., Sato Y., Asada A., Ishiguro K., Hasegawa M., Hisanaga S. ( 2005) J. Biol. Chem. 280, 31522– 31529 [DOI] [PubMed] [Google Scholar]
- 36.Akiyama H., Shin R. W., Uchida C., Kitamoto T., Uchida T. ( 2005) Biochem. Biophys. Res. Commun. 336, 521– 529 [DOI] [PubMed] [Google Scholar]
- 37.Saito T., Onuki R., Fujita Y., Kusakawa G., Ishiguro K., Bibb J. A., Kishimoto T., Hisanaga S. ( 2003) J. Neurosci. 23, 1189– 1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki T., Taoka M., Ishiguro K., Uchida A., Saito T., Isobe T., Hisanaga S. ( 2002) J. Biol. Chem. 277, 36032– 36039 [DOI] [PubMed] [Google Scholar]
- 39.Hasegawa M., Smith M. J., Goedert M. ( 1998) FEBS Lett. 437, 207– 210 [DOI] [PubMed] [Google Scholar]
- 40.Wada Y., Ishiguro K., Itoh T. J., Uchida T., Hotani H., Saito T., Kishimoto T., Hisanaga S. ( 1998) J. Biochem. 124, 738– 746 [DOI] [PubMed] [Google Scholar]
- 41.Fanghänel J., Akiyama H., Uchida C., Uchida T. ( 2006) FEBS Lett. 580, 3237– 3245 [DOI] [PubMed] [Google Scholar]
- 42.Schneider A., Biernat J., von Bergen M., Mandelkow E., Mandelkow E. M. ( 1999) Biochemistry 23, 3549– 3558 [DOI] [PubMed] [Google Scholar]
- 43.Amos L. A., Schlieper D. ( 2005) Adv. Protein Chem. 71, 257– 298 [DOI] [PubMed] [Google Scholar]
- 44.Morfini G., Pigino G., Mizuno N., Kikkawa M., Brady S. T. ( 2007) J. Neurosci. Res. 85, 2620– 2630 [DOI] [PubMed] [Google Scholar]
- 45.Miyasaka T., Morishima-Kawashima M., Ravid R., Heutink P., van Swieten J. C., Nagashima K., Ihara Y. ( 2001) Am. J. Path. 158, 373– 379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bird T. D., Nochlin D., Poorkaj P., Cherrier M., Kaye J., Payami H., Peskind E., Lampe T. H., Nemens E., Boyer P. J., Schellenberg G. D. ( 1999) Brain 122, 741– 756 [DOI] [PubMed] [Google Scholar]
- 47.van Swieten J. C., Stevens M., Rosso S. M., Rizzu P., Joosse M., de Koning I., Kamphorst W., Ravid R., Spillantini M. G., Niermeijer , Heutink P. ( 1999) Ann. Neurol. 46, 617– 626 [DOI] [PubMed] [Google Scholar]
- 48.Reed L. A., Grabowski T. J., Schmidt M. L., Morris J. C., Goate A., Solodkin A., Van Hoesen G. W., Schelper R. L., Talbot C. J., Wragg M. A., Trojanowski J. Q. ( 1997) Ann. Neurol. 42, 564– 572 [DOI] [PubMed] [Google Scholar]
- 49.Rizzu P., Joosse M., Ravid R., Hoogeveen A., Kamphorst W., van Swieten J. C., Willemsen R., Heutink P. ( 2000) Hum. Mol. Genet. 9, 3075– 3082 [DOI] [PubMed] [Google Scholar]
- 50.Aoyagi H., Hasegawa M., Tamaoka A. ( 2007) J. Biol. Chem. 282, 20309– 20318 [DOI] [PubMed] [Google Scholar]
- 51.Hong M., Zhukareva V., Vogelsberg-Ragaglia V., Wszolek Z., Reed L., Miller B. I., Geschwind D. H., Bird T. D., McKeel D., Goate A., Morris J. C., Wilhelmsen K. C., Schellenberg G. D., Trojanowski J. Q., Lee V. M. ( 1998) Science. 282, 1914– 1917 [DOI] [PubMed] [Google Scholar]
- 52.Bunker J. M., Kamath K., Wilson L., Jordan M. A., Feinstein S. C. ( 2006) J. Biol. Chem. 281, 11856– 11863 [DOI] [PubMed] [Google Scholar]
- 53.Han D., Paudel H. K. ( 2009) Neurochem Int. 54, 14– 27 [DOI] [PubMed] [Google Scholar]
- 54.Cole A. R., Soutar M. P., Rembutsu M., van Aalten L., Hastie C. J., McLauchlan H., Peggie M., Balastik M., Lu K. P., Sutherland C. ( 2008) J. Biol. Chem. 283, 18227– 18237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tatebayashi Y., Miyasaka T., Chui D. H., Akagi T., Mishima K., Iwasaki K., Fujiwara M., Tanemura K., Murayama M., Ishiguro K., Planel E., Sato S., Hashikawa T., Takashima A. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 13896– 13901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatebayashi Y., Planel E., Chui D. H., Sato S., Miyasaka T., Sahara N., Murayama M., Kikuchi N., Yoshioka K., Rivka R., Takashima A. ( 2006) FASEB J. 20, 762– 774 [DOI] [PubMed] [Google Scholar]
- 57.Otvos L., Jr., Feiner L., Lang E., Szendrei G. I., Goedert M., Lee V. M. ( 1994) J. Neurosci. Res. 39, 669– 673 [DOI] [PubMed] [Google Scholar]
- 58.Fischer D., Mukrasch M. D., von Bergen M., Klos-Witkowska A., Biernat J., Griesinger C., Mandelkow E., Zweckstetter M. ( 2007) Biochemistry 46, 2574– 2782 [DOI] [PubMed] [Google Scholar]
- 59.Wintjens R., Wieruszeski J. M., Drobecq H., Rousselot-Pailley P., Buée L., Lippens G., Landrieu I. ( 2001) J. Biol. Chem. 276, 25150– 25156 [DOI] [PubMed] [Google Scholar]
- 60.Sontag E., Nunbhakdi-Craig V., Lee G., Brandt R., Kamibayashi C., Kuret J., White C. L., 3rd, Mumby M. C., Bloom G. S. ( 1999) J. Biol. Chem. 274, 25490– 25498 [DOI] [PubMed] [Google Scholar]
- 61.Horowitz P. M., LaPointe N., Guillozet-Bongaarts A. L., Berry R. W., Binder L. I. ( 2006) Biochemistry 45, 12859– 12866 [DOI] [PubMed] [Google Scholar]
- 62.Jeganathan S., von Bergen M., Brutlach H., Steinhoff H. J., Mandelkow E. ( 2006) Biochemistry 45, 2283– 2293 [DOI] [PubMed] [Google Scholar]
- 63.Mukrasch M. D., von Bergen M., Biernat J., Fischer D., Griesinger C., Mandelkow E., Zweckstetter M. ( 2007) J. Biol. Chem. 282, 12230– 12239 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.