Abstract
Quantitative trait mapping in mice identified a susceptibility locus for gallstones (Lith6) spanning the Apobec-1 locus, the structural gene encoding the RNA-specific cytidine deaminase responsible for production of apolipoprotein B48 in mammalian small intestine and rodent liver. This observation prompted us to compare dietary gallstone susceptibility in Apobec-1−/− mice and congenic C57BL/6 wild type controls. When fed a lithogenic diet (LD) for 2 weeks, 90% Apobec-1−/− mice developed solid gallstones in comparison with 16% wild type controls. LD-fed Apobec-1−/− mice demonstrated increased biliary cholesterol secretion as well as increased cholesterol saturation and bile acid hydrophobicity indices. These changes occurred despite a relative decrease in cholesterol absorption in LD-fed Apobec-1−/− mice. Among the possible mechanisms to account for this phenotype, expression of Cyp7a1 mRNA and protein were significantly decreased in chow-fed Apobec-1−/− mice, decreasing further in LD-fed animals. Cyp7a1 transcription in hepatocyte nuclei, however, was unchanged in Apobec-1−/− mice, excluding transcriptional repression as a potential mechanism for decreased Cyp7a1 expression. We demonstrated that APOBEC-1 binds to AU-rich regions of the 3′-untranslated region of the Cyp7a1 transcript, containing the UUUN(A/U)U consensus motif, using both UV cross-linking to recombinant APOBEC-1 and in vivo RNA co-immunoprecipitation. In vivo Apobec-1-dependent modulation of Cyp7a1 expression was further confirmed following adenovirus-Apobec-1 administration to chow-fed Apobec-1−/− mice, which rescued Cyp7a1 gene expression. Taken together, the findings suggest that the AU-rich RNA binding-protein Apobec-1 mediates post-transcriptional regulation of murine Cyp7a1 expression and influences susceptibility to diet-induced gallstone formation.
Cholesterol gallstone disease is highly prevalent among western societies and represents a substantial and recurrent financial burden to the health care economy (1). Considerable attention has focused on the observations that gallbladder disease and cholelithiasis demonstrate familial clustering and that shared genetic factors account for a significant portion of the risk, although environmental modifiers as well as age, gender, body habitus, and ethnicity are also of major importance (reviewed in Ref. 2). However, despite intensive study over many decades, questions remain regarding the mechanisms by which biliary cholesterol becomes supersaturated, leading to the formation of cholesterol monohydrate crystals that eventually nucleate and result in gallstone formation (reviewed in Ref. 3).
Insight into the genetic factors that underlie cholesterol gallstone disease has emerged from the study of inbred mouse strains where intercrosses between gallstone-susceptible and -resistant strains have generated a comprehensive array of candidate loci through quantitative trait locus mapping, an approach that has yielded at least 23 LITH loci (2, 4). Among the quantitative trait loci identified through this approach is Lith6, a locus on mouse chromosome 6 that mapped to a region that includes Apobec-1 as well as other genes (Pparg and Slco1a1) involved in lipid metabolism (5). Apobec-1, the RNA-specific catalytic deaminase of the mammalian ApoB (apolipoprotein B) mRNA-editing enzyme (6), mediates C to U RNA editing of intestinal ApoB RNA, leading to production of APOB48, the major structural protein on chylomicrons (7). In addition, mouse and rat (but not human) liver exhibit physiologically regulated Apobec-1-dependent ApoB mRNA editing accompanying a range of developmental, nutritional, and hormonal cues, resulting in the ability of murine hepatocytes to synthesize APOB100 and APOB48 in varying proportions with accompanying changes in hepatic VLDL assembly and secretion (reviewed in Ref. 7).
Several lines of evidence suggest a plausible link between the pathways that regulate hepatic VLDL assembly and secretion and those that regulate enterohepatic bile acid metabolism. These include earlier observations in humans that treatment with bile acid sequestrants or interruption of enterohepatic bile acid cycling increases hepatic VLDL production (8, 9). Conversely, treatment of subjects with chenodeoxycholic acid ameliorates hypertriglyceridemia and reduces VLDL secretion through mechanisms that include transcriptional repression of Mttp (microsomal triglyceride transfer protein) gene expression (10–12). In another approach, treatment of wild type mice with an FXR agonist both protected against diet-induced gallstones and decreased plasma triglyceride levels (13). Yet other studies demonstrated that mice with liver-specific inactivation of Mttp are protected against diet-induced gallstones (14), whereas increased expression of hepatic Mttp was found in humans with cholesterol gallstone disease (15). These findings, coupled with earlier observations suggesting that APOB100 is more susceptible (than APOB48) to presecretory degradation in the setting of limiting Mttp expression or function (16, 17), imply the possibility that there may also be an APOB isoform-specific dependence for hepatic lipid mobilization directed toward canalicular secretion.
The current studies were undertaken to examine the importance of Apobec-1 as a genetic modifier of cholesterol gallstone formation in a congenic strain of C57BL/6 mice, in particular its functional significance in relation to the Lith6 locus identified earlier (5). In addition, we were intrigued by the findings that mice genetically engineered to express only APOB100 (but which are Apobec-1-sufficient (18)) exhibited protection from cholesterol gallstone formation compared with mice expressing only APOB48, predominantly through decreased intestinal cholesterol absorption (19). However, our findings illustrate a divergence in the phenotype demonstrated in that report (19). We find that Apobec-1−/− mice demonstrate accelerated gallstone formation and alterations in biliary lipid metabolism that reflect changes in gene expression that are most likely produced by post-transcriptional modifications in Cyp7a1 mRNA expression.
MATERIALS AND METHODS
Animals and Diets
Apobec-1−/− mice (20) were backcrossed >12 generations onto a C57BL/6 background. Male C57BL/6 wild type (WT)2 mice were purchased from Jackson Laboratories (Bar Harbor, ME). Apob100/100 mice were obtained as a generous gift from Dr. Steve Young (UCLA School of Medicine) and are on a mixed background (C57BL/6 and 129/SvJ) (18). All animals were maintained on a 12-h light-dark cycle, in a full barrier facility. 8–10-week-old male Apobec-1−/− and WT mice were fed either a standard rodent chow (PicoLab Rodent Diet 20, fat 4.5%, cholesterol 0.015%) or a lithogenic diet (LD) (Research Diet 960393, fat 18.8%, cholesterol 1.2%, and cholic acid 0.5%) for 2–8 weeks, as indicated.
Gallbladder Bile and Gallstone Analysis
Mice were fasted for 4 h and anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (20 mg/kg). Gallbladders were ligated and punctured at the fundus for collection of gallbladder bile. Bile samples were collected and immediately analyzed (2, 21) by polarizing microscopy using a visual scale with the following criteria: 0 = absence of cholesterol monohydrate crystals (ChMCs), 1 = small number of ChMCs (<3/high power field); 2 = many ChMCs (≥3/high power field); 3 = aggregated ChMCs; 4 = presence of “sandy” light translucent stones or “solid” light opaque stones (21).
Bile Flow, Biliary Lipid Output, and Bile Acid Species Determination
Mice were anesthetized as above after a 4-h fast. An external bile fistula was established surgically via the gallbladder fundus. Hepatic bile was collected for up to 60 min while maintaining body temperature at 37 °C. Hepatic bile volume was determined gravimetrically, assuming a density of 1 g/ml. Bile samples were stored at −80 °C until analyzed. Biliary phospholipid, cholesterol, and total bile acid content were determined enzymatically using a phospholipid B kit (catalog number 990-54009), cholesterol E kit (catalog number 439-17501), and total bile acids kit (catalog number 431-15001), respectively, all from Wako Chemicals (Neuss, Germany). Cholesterol saturation indices in hepatic bile were calculated using published parameters (22). Individual bile salt concentrations were measured via high performance liquid chromatography (HPLC) (23). In brief, 100 μl of diluted hepatic bile (1:20 dilution prepared in methanol) was injected onto a Pursuit C18 reverse phase column (Varian, Lake Forest, CA) using a Rainin HPXL HPLC system (Varian, Lake Forest, CA). Bile acid standards were purchased from Steraloids Inc. (Newport, RI) or from Calbiochem. The hydrophobic index of hepatic bile samples was calculated according to published methods (24).
Hepatic and Intestinal Lipid Determination
Animals were sacrificed as described above after a 4-h fast. Liver and mucosal scrapings were collected from the small intestine and frozen at −80 °C until analyzed. Tissue lipids were extracted into chloroform/methanol (2:1), and triglyceride, cholesterol, free fatty acids, and phospholipids were analyzed enzymatically with an L-type triglyceride H kit (catalog number 993-37592, 993-37492), cholesterol E, HR series NRFA-HR(2) (catalog number 995-34691, 995-34791), and phospholipids B, respectively (Wako Chemicals).
Bile Acid Pool Size and Fecal Bile Acid Output Determination
Bile acid pool size was determined as the sum total bile acid content of the entire small intestine, gallbladder, and liver, which were homogenized and extracted together in ethanol with [24-14C]taurocholic acid (0.025 μCi) added as an internal recovery standard (25). Total bile acid mass was determined enzymatically as above. In addition, fecal bile acid output was determined from stool quantitatively collected from individually housed mice for periods up to 72 h. For each determination, 200-mg triplicate aliquots of dried feces were extracted into ethanol as described (25, 26), and total bile acid mass was again measured enzymatically, normalizing recovery to an internal standard of [14C]taurocholic acid.
Cholesterol Absorption
Cholesterol absorption was measured by a fecal dual isotope ratio method, as described previously (27, 28). Wild type and Apobec-1−/− mice were studied prior to initiating the lithogenic diet and again after 8 weeks on the lithogenic diet. Animals were gavaged with 150 μl of corn oil mixed with 1 μCi of [14C]cholesterol (PerkinElmer Life Sciences) and 2 μCi of [3H]sitostanol (American Radiolabeled Chemicals, Inc.). Feces were collected individually for 48 h after label administration and processed as previously described. The ratio of 14C and 3H in each sample was determined, and cholesterol absorption percentage was calculated as described previously (27, 29).
Gene Expression Analysis
For microarray analysis, total liver RNA was pooled from 4 animals/group. Pooled RNA was hybridized to an Agilent Technologies mouse whole genome microarray (G4122A; Agilent, Palo Alto, CA). Microarray results were confirmed by real time quantitative reverse transcription-PCR (qRT-PCR) using cDNA pooled from 4 or 5 animals/group, and where indicated, the findings were replicated using individual samples. qRT-PCR assays were performed in triplicate on an ABI Prism 7000 sequence detection system using SYBR Green PCR Master Mix (Applied Biosystems) and primer pairs designed by Primer Express software (Applied Biosystems) (listed in supplemental Table 1). Relative mRNA abundance is expressed as fold change compared with mRNA levels in WT mice consuming the same diet, normalized to 18 S ribosomal RNA. Nuclear run-on assays were performed using isolated nuclei freshly prepared from chow-fed WT (n = 5) and Apobec-1−/− mice (n = 5), purified through sucrose gradients, and frozen in liquid nitrogen until used for run-on studies. Nuclear run-on transcription was performed at 30 °C for 30 min in a mixture containing 5 × 107 nuclei; 5 mm Tris, pH 8.0; 2.5 mm MgCl2, 0.15 m KCl, 1 mm each ATP, CTP, and GTP; 150 μCi of [32P]UTP (3000 Ci/mmol, Amersham Biosciences); and 100 units of RNaseOUTTM (Invitrogen). RNAs were denatured and hybridized to immobilized cDNA templates encoding ∼300 bp of Cyp7a1, Gapdh, or plasmid vector control, washed at high stringency (30, 31), and exposed to PhosphorImager screens (Amersham Biosciences).
Microsome Preparation
Liver microsomes were prepared as described (32). In brief, livers from mice of the indicated genotype were quickly removed and homogenized at 4 °C in buffer I containing 40 mm Tris-HCl, 1 mm EDTA, 5 mm dithiothreitol, 50 mm KCl, 50 mm KF, 300 mm sucrose, and 1× protease inhibitor mixture (Roche Applied Science). The homogenate was centrifuged at 20,000 × g for 20 min, and the supernatant was further centrifuged at 108,000 × g for 60 min. The pellet was washed and resuspended in buffer II (buffer I without sucrose), and aliquots representing 25 μg of microsomal protein were separated by 8% SDS-PAGE. Western blot was performed on either whole liver homogenate or microsome preparations with affinity-purified rabbit anti-CYP7A1 IgG (2 μg/ml), a generous gift from Dr. Simon Hui (San Diego State University) and as control either anti-BiP IgG (StressGen) or anti-HSP40 followed by ECL detection reagents (Amersham Biosciences).
Plasmid Construction, in Vitro Transcription, and RNA-Protein UV Cross-linking
cDNA from total mouse liver RNA was used to amplify the Cyp7a1 3′-untranslated region (UTR). The full-length 3′-UTR was cloned behind a green fluorescent protein cDNA as a reporter for mRNA stability studies using actinomycin D (5 μg/ml), as detailed previously (33). Three individual fragments, collectively spanning the entire Cyp7a1 3′-UTR, were used for RNA binding studies. Fragment A spans nucleotides 1642–2314, fragment B spans nucleotides 2287–3274, and fragment C spans nucleotides 3251–4070. Each fragment was cloned into pcDNA3, and the plasmids were linearized and used as templates for in vitro transcription with T7 RNA polymerase. In vitro transcription reactions were conducted for 6 h with [α-32P]UTP, and the resultant RNA product was gel-purified through 5% PAGE, 7 m urea. 32P-Labeled cRNA template (100,000 cpm) was incubated with 50–1000 ng of glutathione S-transferase-APOBEC-1 (34) in a binding buffer containing 10 mm HEPES (pH 7.9), 100 mm KCl, 1 mm EDTA, and 0.25 mm dithiothreitol at room temperature for 15 min. Heparin (3 mg/ml) was then added for another 10 min, and the mixture was digested with RNase T1 (5 units/ml) for another 10 min. The mixture was then subjected to UV cross-linking on ice in a UV Stratalinker 1800 with an energy of 250 mJ/cm2 and resolved by 10% SDS-PAGE.
Adenovirus Infection in Vivo and Co-immunoprecipitation of RNA-Protein Complexes
Recombinant adenoviruses encoding either rat Apobec-1 (Ad-Apobec-1) or β-galactosidase (Ad-β-galactosidase) were prepared as described (35), and 6 × 108 plaque-forming units were injected into Apobec-1−/− mice. 5 days later, animals were anesthetized after a 4-h fast, livers were collected, and Cyp7a1 expression was examined as described above. For co-immunoprecipitation studies, livers from Ad-Apobec-1-infected Apobec-1−/− mice were removed, diced by hand into small (less than 2-mm) pieces, and cross-linked in 1% formaldehyde phosphate-buffered saline complemented with RNaseOUTTM (100 units/ml) and 1× protease inhibitor mixture for 15 min on ice. The cross-link reaction was stopped, and liver pieces were homogenized in 1 ml of polysome lysis buffer containing 10 mm HEPES (pH 7.4), 100 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, 100 units/ml RNaseOUTTM, 1 mmol ribonucleoside vanadyl complex, 0.5% Nonidet P-40. Lysates with 4–5 mg of protein were immunoprecipitated with affinity-purified rabbit anti-APOBEC-1 IgG or control IgG. The RNA-protein complex pellet with protein A beads was treated with proteinase K for 30 min and reverse cross-linked by incubating the mixture at 65 °C for 60 min. RNA from the final pellet was extracted into phenol/choloroform/isoamyalcohol and subjected to reverse transcription-PCR for Cyp7a1 using the primer pairs indicated in supplemental Table 1.
Statistical Analysis
Statistical significance was determined with an unpaired, two-tailed Student's t test. Data are expressed as the mean ± S.E. unless otherwise noted.
RESULTS
Body Weight, Hepatic, and Serum Lipid Profile in LD-fed Apobec-1−/− Mice
Body weight was comparable between the genotypes after 2 weeks of feeding an LD with animals of both genotypes losing weight, but there was a striking decrease in liver weight and liver weight/body weight ratio in Apobec-1−/− mice (Table 1). In addition, serum triglyceride levels were 2-fold higher in both chow- and LD-fed Apobec-1−/− mice compared with WT with comparable serum cholesterol levels, whereas hepatic cholesterol and triglyceride content were reduced in LD-fed Apobec-1−/− mice (Table 1). These findings suggested that LD feeding is accompanied by distinctive alterations in lipid metabolism in Apobec-1−/− mice compared with congenic WT controls.
TABLE 1.
Body weight, liver weight, serum, and hepatic lipid profile of WT and Apobec-1−/− mice on both chow and lithogenic diets
Values represent the mean ± S.E. (n). ALT, alanine aminotransferase. mg/g, mg/g of liver.
| Parameters | Chow |
Lithogenic diet |
p | ||
|---|---|---|---|---|---|
| WT | Apobec-1−/− | WT | Apobec-1−/− | ||
| Body weight (g) | 26.53 ± 0.77 (12) | 26.88 ± 0.88 (13) | 23.64 ± 0.40 (17) | 23.59 ± 0.46 (22) | |
| Liver wt (g) | 1.27 ± 0.04 (12) | 1.19 ± 0.03 (13) | 1.53 ± 0.05 (18) | 1.08 ± 0.03 (22) | p < 0.01a |
| Liver wt (% body wt) | 4.81 ± 0.19 (12) | 4.46 ± 0.11 (13) | 6.76 ± 0.18 (18) | 5.03 ± 0.13 (22) | p < 0.01a |
| Serum cholesterol (mg/dl) | 62.17 ± 2.79 (12) | 65.33 ± 3.04 (13) | 146.37 ± 5.17 (8) | 156.36 ± 7.91 (10) | |
| Serum triglyceride (mg/dl) | 28.43 ± 3.26 (20) | 57.05 ± 5.13 (22) | 24.43 ± 2.74 (12) | 52.84 ± 11.74 (14) | p < 0.05a; p < 0.05b |
| Serum bile acid (μmol/liter) | 6.11 ± 1.21 (4) | 6.88 ± 1.10 (5) | 16.56 ± 3.28 (8) | 22.41 ± 4.46 (10) | |
| Serum ALT (IU/liter) | 10.45 ± 3.97 (7) | 15.86 ± 2.59 (8) | 59.87 ± 11.04 (8) | 49.35 ± 10.30 (14) | |
| Hepatic cholesterol (mg/g) | 1.21 ± 0.06 (5) | 1.31 ± 0.09 (5) | 19.11 ± 0.91 (12) | 15.61 ± 0.64 (15) | p < 0.01a |
| Hepatic triglyceride (mg/g) | 3.71 ± 0.42 (5) | 4.86 ± 0.81 (5) | 15.67 ± 1.28 (12) | 12.73 ± 0.73 (15) | p < 0.05a |
| Hepatic phospholipid (mg/g) | 5.29 ± 0.22 (5) | 5.77 ± 0.31 (5) | 15.24 ± 0.55 (12) | 14.80 ± 0.56 (15) | |
| Hepatic free fatty acid (μmol/g) | 1.05 ± 0.14 (5) | 1.45 ± 0.18 (5) | 3.76 ± 1.15 (12) | 2.91 ± 0.14 (15) | |
a Comparing WT and Apobec-1−/− mice on lithogenic diet.
b Comparing WT and Apobec-1−/− mice on chow diet.
Gallstone Susceptibility Is Increased in LD-fed Apobec-1−/− Mice
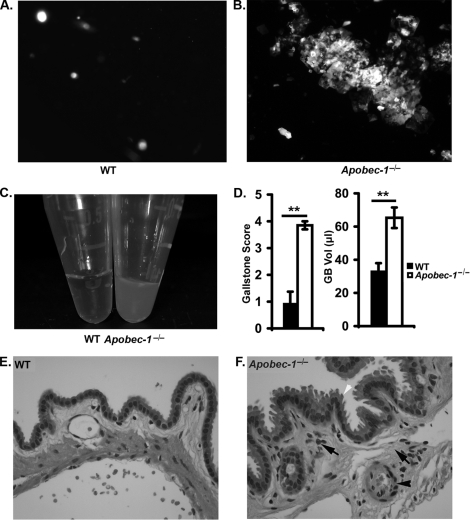
To explore the effects on biliary lipid metabolism, samples of gallbladder bile from LD-fed animals were analyzed by polarizing microscopy, which revealed the presence of abundant aggregated birefringent cholesterol crystals in samples from Apobec-1−/− mice (Fig. 1B) but not in WT samples (Fig. 1A). In addition, bile samples from Apobec-1−/− mice were grossly turbid (Fig. 1C) in comparison with WT controls, and by 2 weeks of LD feeding there were visible stones in 90% of Apobec-1−/− mice (9 of 10 mice) compared with 16% (2 of 12) WT controls. This increased susceptibility to gallstones in Apobec-1−/− mice was evident also in both the higher gallstone score and increased gallbladder volumes (Fig. 1D). In addition, the gallbladder mucosa of LD-fed Apobec-1−/− mice appeared thickened histologically, with increased inflammatory cell infiltration (Fig. 1, E and F).
FIGURE 1.
Biliary cholesterol crystallization, gallstone score, and gallbladder inflammation in LD fed wild type and Apobec-1−/− mice. 11 WT and 10 Apobec-1−/− mice were fed the LD for 2 weeks. Gallbladder bile samples were collected and analyzed under polarizing microscopy. A and B, a few single ChMCs were present in WT gallbladder bile (A), whereas multiple aggregated ChMCs were observed in gallbladder bile from Apobec-1−/− mice (B). C, gallbladder bile appears turbid in Apobec-1−/− compared with WT mice. D, gallstone score (see “Materials and Methods”) and gallbladder volumes reveal a higher gallstone score and larger gallbladder volume in Apobec-1−/− compared with WT mice. E and F, histological examination of gallbladder epithelium following hematoxylin and eosin staining. The black arrows indicate stromal granulocyte infiltration into a thickened gallbladder wall. The black arrowhead indicates submucosal vasodilatation. The white arrowhead indicates mucosal surface indentations noted in Apobec-1−/− but not WT mice (magnification, ×400). **, p < 0.01.
Alterations in Biliary Lipid Secretion in LD-fed Apobec-1−/− Mice
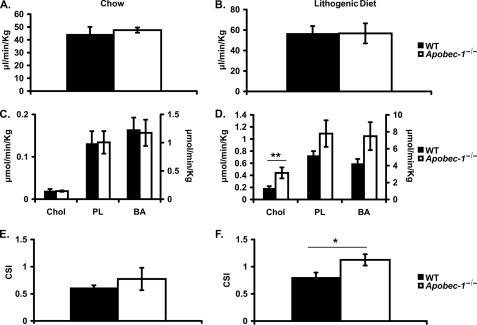
The findings to this point strongly suggest that Apobec-1−/− mice exhibit a striking phenotype of increased gallstone susceptibility upon LD feeding. We next examined individual biliary lipid secretion rates in WT and Apobec-1−/− mice both on chow and LD. The findings reveal comparable bile flow rates in both genotypes on both diets (Fig. 2, A and B) and comparable secretion rates of all biliary lipid classes in chow-fed mice (Fig. 2C). The secretions of phospholipids and bile acids were also comparable in both genotypes on both diets. By contrast, 2 weeks of LD feeding produced an increase in biliary cholesterol secretion in Apobec-1−/− mice (Fig. 2D), which was accompanied by an increase in cholesterol saturation index that was not observed in chow-fed mice (Fig. 2, E and F).
FIGURE 2.
Biliary flow, lipid secretion rates, and hepatic bile cholesterol saturation indices on chow or LD. Hepatic bile was collected as described for 60 min. A and B, biliary flow rates (μl/min/kg of body weight) were similar in Apobec-1−/− and WT mice either on chow (A) or LD diet (B). C and D, biliary cholesterol secretion (μmol/min/kg of body weight) was similar on chow but increased on lithogenic diet in Apobec-1−/− mice compared with WT controls. There were no differences in biliary phospholipids or bile acid secretion on either diet between the two genotypes. E and F, hepatic bile cholesterol saturation indices (CSI) were calculated from Carey's critical tables (see “Materials and Methods”). Apobec-1−/− mice demonstrated increased CSI on LD but not chow, compared with WT mice. Bar graphs indicate the mean ± S.E. Data were from 5 mice/genotype on chow diet and 10 mice/genotype on LD. *, p < 0.05; **, p < 0.01.
Changes in Hepatic Gene Expression in Apobec-1−/− Mice in Response to LD Feeding
In order to begin to understand the complex adaptations that might lead to the increased cholesterol secretion and accelerated formation of gallstones in Apobec-1−/− mice, we turned to microarray analysis of hepatic gene expression in animals consuming either chow or LD. The most significant alterations encountered in chow-fed mice are summarized in Table 2, and those for LD-fed animals are summarized in Table 3. Among the most striking findings is that chow-fed Apobec-1−/− mice demonstrate a 75% decrease in Cyp7a1 mRNA, which encodes the first and rate-limiting enzyme in bile acid synthesis (Table 2). In response to LD feeding, the changes in Cyp7a1 were further magnified (Table 3), suggesting a profound suppression in bile acid synthesis in response to the dietary intervention. There was a nonsignificant trend to increased Apobec-1 mRNA in WT mice following 2 weeks LD feeding (2.7 ± 0.8-fold) and no significant differences in Abcg5 or Abcg8 mRNA abundance or in other canalicular or basolateral cholesterol transporter mRNAs, including Srb1 and Abca1 between WT and Apobec-1−/− mice (Table 3). Thus, the most striking alterations in gene expression in LD-fed Apobec-1−/− mice appear to center around genes regulating bile acid synthesis rather than cholesterol secretion. We will return to this observation below.
TABLE 2.
Liver microarray data for chow
| Gene name | Gene description | Accession number | Ratioa | qRT-PCR | p |
|---|---|---|---|---|---|
| Bile acid synthesis, transport, and metablolism | |||||
| Cyp7a1 | Cholesterol 7α-hydroxylase | NM_007824 | 0.17 | 0.25b | 0.01 |
| Cyp8b1 | Sterol 12α-hydroxylase | NM_010012 | 0.31 | 1.15b | |
| Cyp27a1 | Sterol 27-hydroxylase | NM_024264 | 0.65 | 0.9 | |
| Bsep/Abcb11 | Bile salt export pump | NM_021022 | 0.9 | 0.75 | |
| Ntcp | Na+-taurocholate co-transporting protein | NM_011387 | 0.98 | 1.42 | |
| Oatp4 | Organic anion transporter, 1b2 | NM_178235 | 1.07 | 1.28 | |
| Oatp2 | Organic anion transporter, 1a4 | NM_030687 | 1.5 | 1.69 | |
| Mdr2/Abcb4 | ATP-binding cassette, B4 | NM_008830 | 1.25 | ||
| Mrp2/Abcc2 | ATP-binding cassette, C2 | NM_013806 | 1.1 | ||
| Cyp2b10 | Cytochrome P450, 2b10 | NM_009998 | 0.73 | 2.97 | |
| Cyp3a11 | Cytochrome P450, 3a11 | NM_007818 | 0.7 | 0.51 | |
| Cholesterol synthesis and transport | |||||
| Hmgcr | Hydroxymethylglutaryl-CoA reductase | NM_008255 | 0.7 | 1.64b | |
| Abca1 | ATP-binding cassette, A1 | NM_013454 | 1.4 | 1 | |
| Abcg5 | ATP-binding cassette, G5 | NM_031884 | 1 | 1.3 | |
| Abcg8 | ATP-binding cassette, G8 | NM_026180 | 1 | 1.5 | |
| Srb1 | Scavenger receptor class, B1 | NM_016741 | 1.1 | 1.28 | |
| Scp2 | Sterol carrier protein 2, liver | NM_011327 | 1.3 | 0.66 | |
| Lfabp | Liver fatty acid-binding protein | NM_017399 | 1.2 | 1.06 | |
| Cav1 | Caveolin 1 | NM_007616 | 1.0 | 1.12 | |
| Npc1 | Niemann-Pick type C1 | NM_008720 | 0.9 | ||
| Transcription factors or nuclear receptors | |||||
| Lxrα | Liver X receptor α | NM_013839 | 0.81 | 0.94b | |
| Fxr | Farnesoid X receptor | NM_009108 | 1.3 | 1.16 | |
| Shp | Small heterodimer partner | NM_011850 | 0.66 | 0.37 | |
| Pparα | Peroxisome proliferator-activated receptor α | NM_011144 | 1.5 | 1.6 | |
| Srebp1 | Sterol regulatory element-binding protein 1 | NM_011480 | 0.8 | 0.6b | |
| Srebp2 | Sterol regulatory element-binding protein 2 | NM_033218 | 0.99 | 1.1 | |
| Pxr | Pregnane X receptor | NM_010936 | 1.2 | 1.33 | |
| Lrh-1 | Liver receptor homolog | NM_030676 | 0.98 | ||
| Fatty acids synthesis and oxidation | |||||
| Fasn | Fatty acid synthase | NM_007988 | 1.1 | 0.4 | |
| Ucp2 | Uncoupling protein 2 | NM_011671 | 1.1 | 1.03 | |
| Scd1 | Stearoyl-coenzyme A desaturase 1 | NM_009127 | 0.9 | 0.43 | |
| Scd2 | Stearoyl-coenzyme A desaturase 2 | NM_009128 | 1.1 | 0.58 | |
| Acc | Acetyl-coenzyme A carboxylase α | NM_133360 | 0.9 | ||
| Lipoprotein metabolism | |||||
| ApoB | Apolipoprotein B | XM_137955 | 2.06 | 1.08b | |
| ApoA4 | Apolipoprotein A-IV | NM_007468 | 0.53 | 0.95 | |
| ApoC3 | Apolipoprotein C-III | NM_023114 | 1.2 | 0.7 | |
| ApoC2 | Apolipoprotein C-II | NM_009695 | 1 | 0.92 | |
| Ldlr | Low density lipoprotein receptor | NM_010700 | 0.65 | 0.89 | |
| ApoA1 | Apolipoprotein A | NM_009692 | 0.94 | ||
| ApoE | Apolipoprotein E | NM_009696 | 1.2 | ||
| Others | |||||
| Cyp2C38 | Cytochrome P450, 2C38 | NM_010002 | 0.8 | 0.7 | |
| Cyp4a14 | Cytochrome P450, 4A14 | NM_007822 | 1.6 | 0.4 | |
a Microarray data are expressed as a ratio of Apobec1−/− versus WT. Relative mRNA expression of some genes was confirmed by real time qRT-PCR by using pooled cDNA (5 mice/group). qRT-PCR results are expressed as -fold change normalized to WT control.
b Relative mRNA expression of the indicated genes was confirmed using individual samples (n = 4).
TABLE 3.
Liver microarray data for lithogenic diet for 2 weeks
| Gene name | Gene description | Accession number | Ratioa | qRT-PCR | p |
|---|---|---|---|---|---|
| Bile acid synthesis, transport, and metablolism | |||||
| Cyp7a1 | Cholesterol 7α-hydroxylase | NM_007824 | 0.16 | 0.04b | 0.001 |
| Cyp8b1 | Sterol 12α-hydroxylase | NM_010012 | 0.36 | 0.07 | 0.01 |
| Cyp27a1 | Sterol 27-hydroxylase | NM_024264 | 0.82 | 0.9 | |
| Bsep/Abcb11 | Bile salt export pump | NM_021022 | 2.89 | 1.74 | |
| Ntcp | Na+-taurocholate co-transporting protein | NM_011387 | 1.7 | 0.63 | |
| Oatp4 | Organic anion transporter, 1b2 | NM_178235 | 2.01 | 0.48 | |
| Oatp2 | Organic anion transporter, 1a4 | NM_030687 | 3.49 | 2.55 | |
| Mdr2/Abcb4 | ATP-binding cassette, B4 | NM_008830 | 1.3 | 0.87 | |
| Mrp2/Abcc2 | ATP-binding cassette, C2 | NM_013806 | 1.13 | ||
| Cyp2b10 | Cytochrome P450, 2b10 | NM_009998 | 5 | 19.56 | |
| Cyp3a11 | Cytochrome P450, 3a11 | NM_007818 | 2.5 | 4.29 | |
| Cholesterol synthesis and transport | |||||
| Hmgcr | Hydroxymethylglutaryl-CoA reductase | NM_008255 | 2.09 | 1.84b | 0.04 |
| Abca1 | ATP-binding cassette, A1 | NM_013454 | 2.1 | 1.1 | |
| Abcg5 | ATP-binding cassette, G5 | NM_031884 | 1.6 | 1.03b | |
| Abcg8 | ATP-binding cassette, G8 | NM_026180 | 1.01 | 1.03 | |
| Srb1 | Scavenger receptor class, B1 | NM_016741 | 1.6 | 0.89 | |
| Scp2 | Sterol carrier protein 2, liver | NM_011327 | 2.1 | 1.71 | |
| Lfabp | Liver fatty acid binding protein | NM_017399 | 2.3 | 1.32 | |
| Cav1 | Caveolin 1 | NM_007616 | 1.8 | 0.69 | |
| Npc1 | Niemann-Pick type C1 | NM_008720 | 1.2 | ||
| Transcription factors or nuclear receptors | |||||
| Lxrα | Liver X receptor α | NM_013839 | 0.88 | 0.53b | |
| Fxr | Farnesoid X receptor | NM_009108 | 2.05 | 1.3 | |
| Shp | Small heterodimer partner | NM_011850 | 0.93 | 1.4 | |
| Pparα | Peroxisome proliferator-activated receptor α | NM_011144 | 2.1 | 1.1 | |
| Srebp1 | Sterol regulatory element-binding protein 1 | NM_011480 | 3.64 | 0.91b | |
| Srebp2 | Sterol regulatory element-binding protein 2 | NM_033218 | 0.61 | 0.83 | |
| Pxr | Pregnane X receptor | NM_010936 | 1.7 | 2.25 | |
| Lrh-1 | Liver receptor homolog | NM_030676 | 1.6 | ||
| Fatty acid synthesis and oxidation | |||||
| Fasn | Fatty acid synthase | NM_007988 | 8.1 | 15.1 | |
| Ucp2 | Uncoupling protein 2 | NM_011671 | 0.46 | 0.13 | |
| Scd1 | Stearoyl-coenzyme A desaturase 1 | NM_009127 | 1.6 | 2.07 | |
| Scd2 | Stearoyl-coenzyme A desaturase 2 | NM_009128 | 2.0 | 1.56 | |
| Acc | Acetyl-coenzyme A carboxylase α | NM_133360 | 2.0 | ||
| Lipoprotein metabolism | |||||
| ApoB | Apolipoprotein B | XM_137955 | 1.45 | 0.91b | |
| ApoA4 | Apolipoprotein A-IV | NM_007468 | 2.45 | 2.4 | |
| ApoC3 | Apolipoprotein C-III | NM_023114 | 2.45 | 1.16 | |
| ApoC2 | Apolipoprotein C-II | NM_009695 | 2.43 | 0.84 | |
| Ldlr | Low density lipoprotein receptor | NM_010700 | 0.8 | 1.83 | |
| ApoA1 | Apolipoprotein A | NM_009692 | 1.0 | ||
| ApoE | Apolipoprotein E | NM_009696 | 1.2 | ||
| Others | |||||
| Cyp2C38 | Cytochrome P450, 2C38 | NM_010002 | 4.9 | 1 | |
| Cyp4a14 | Cytochrome P450, 4A14 | NM_007822 | 122.8 | 372.2 | |
a Microarray data are expressed as a ratio of Apobec1−/− versus WT. Relative mRNA expression of some genes was confirmed by real time qRT-PCR by using pooled cDNA (5 mice/group). qRT-PCR results are expressed as -fold change normalized to WT control.
b Relative mRNA expression of indicated genes was confirmed using individual samples (n = 4).
Alterations in Biliary Bile Acid Composition and Hydrophobicity in Apobec-1−/− Mice
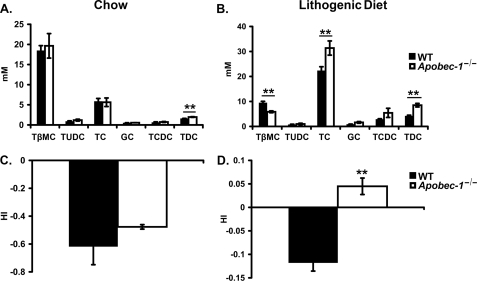
Individual species of biliary bile acid were examined by HPLC, which demonstrated that the primary bile acids β-muricholate and taurocholate together represent the dominant species in chow-fed animals of both genotypes (Fig. 3A), with an increase in taurodeoxycholic acid observed in Apobec-1−/− mice. In response to LD feeding, there was a shift in the bile acid species, with taurocholate representing 58.6 ± 2.2% of the pool in Apobec-1−/− mice compared with 54.7 ± 1.3% in WT controls and a corresponding decrease in β-muricholate to 11.3 ± 0.9% in Apobec-1−/− mice compared with 23.5 ± 1.1% in WT controls (Fig. 3B). In addition, there was a further increase in the relative abundance of taurodeoxycholic acid in Apobec-1−/− mice (Fig. 3B). The changes in bile acid composition upon LD feeding were accompanied by alterations in the hydrophobic index of bile. Although comparable values were found in chow-fed animals (Fig. 3C), there was a significant increase in hydrophobicity noted in Apobec-1−/− mice upon LD feeding (Fig. 3D). These findings suggest that the combination of increased cholesterol secretion and increased bile hydrophobicity might accelerate cholesterol supersaturation in the bile of Apobec-1−/− mice.
FIGURE 3.
Bile acid species distribution and hydrophobicity index. Individual bile acid species were examined in newly secreted bile samples from mice fed chow (A) (n = 5/genotype) or lithogenic diet (B) (n = 7/genotype) by HPLC. Bar graphs represent the mean ± S.E. TβMC, tauro-β-muricholate; TUDC, tauroursodeoxycholate; TC, taurocholate; GC, glycocholate; TCDC, taurochenodeoxycholate; TDC, taurodeoxycholate. Bile salt hydrophobicity indices were comparable in chow-fed animals (C) but increased in Apobec-1−/− mice versus WT controls mice when fed the LD (D). **, p < 0.01.
Bile Acid Pool Size and Synthetic Rates in Apobec-1−/− Mice
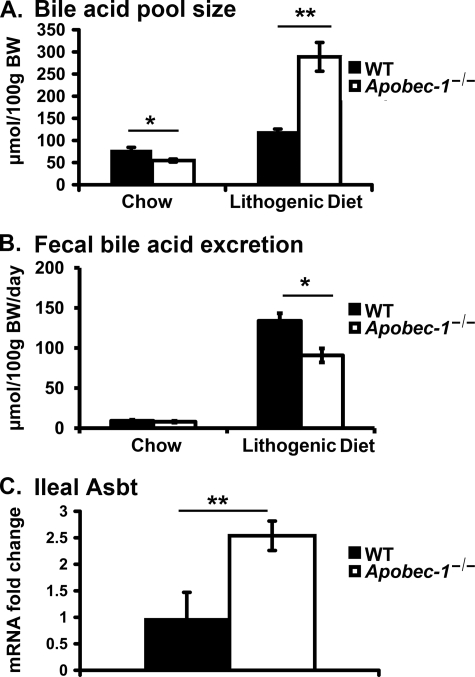
We observed a significant 31% decrease in the total bile acid pool size in chow-fed Apobec-1−/− mice (Fig. 4A), but fecal bile acid excretion rates (at steady state, a surrogate for bile acid synthesis rates) revealed no statistically significant difference between the genotypes (WT 9.2 ± 3.1 μmol/100 g of body weight/day versus 7.9 ± 2.1 μmol/100 g of body weight/day; Fig. 4B, left). The S.D. of these values (25–33%) probably preclude detection of subtle (albeit significant) changes in bile acid excretion rates in chow-fed WT versus Apobec-1−/− mice. By contrast, upon feeding the LD, we observed a 1.4-fold increase in the bile acid pool size in Apobec-1−/− mice, which was accompanied by a significant 32% decrease in fecal bile acid output (Fig. 4, A and B). The combination of an expanded bile acid pool (largely the result of dietary cholate supplementation) in the setting of reduced fecal losses suggests the possibility that enterohepatic bile acid recycling may be increased in Apobec-1−/− mice. As an initial approach to address this possibility, we examined the expression of Asbt, the ileal bile salt transporter mRNA, and found that it was increased 2.5-fold in LD-fed Apobec-1−/− mice (Fig. 4C). These findings collectively suggest that there is adaptive conservation of bile acids (predominantly of dietary origin) resulting from up-regulation of ileal Asbt expression in Apobec-1−/− mice.
FIGURE 4.
Bile acid pool size, fecal bile acid excretion, and ileal Asbt (apical sodium-dependent bile acid transporter) mRNA expression. A, bile acid pool size (μmol/100 g of body weight) was decreased in Apobec-1−/− mice on a chow diet but increased in Apobec-1−/− mice fed an LD for 2 weeks. Liver, gallbladder, and intestine were pooled and subjected to ethanolic bile acid extraction. Total bile acid mass was measured enzymatically. B, fecal bile acid excretion was comparable in chow-fed mice of both genotypes, but LD-fed Apobec-1−/− mice excrete less bile acid than WT controls. Mice were individually housed, and feces were collected up to 72 h. Fecal bile acids were extracted and quantitated enzymatically. C, Asbt mRNA abundance is increased in LD-fed Apobec-1−/− mice. RNA was extracted from ileum of LD-fed WT and Apobec-1−/− mice, and Asbt expression was measured by qRT-PCR. Bar graphs represent the mean ± S.E. from 5 mice/genotype on chow and 4 mice/genotype on LD. *, p < 0.05; **, p < 0.01.
Cholesterol Absorption and Sterol Transporter Expression in Apobec-1−/− Mice
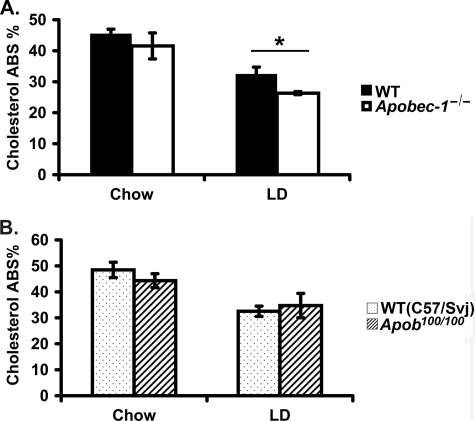
Previous studies examining gallstone susceptibility in the background of hepatic and intestinal production of APOB100-only lipoproteins were undertaken in mice, where the synthesis of APOB48 was eliminated through Apob gene targeting, a mechanism independent of C to U RNA editing (18). In this setting, Apob100/100 mice were found to be protected against diet-induced gallstones in conjunction with a dramatic reduction in intestinal cholesterol absorption (19). Accordingly, we reasoned that cholesterol absorption might be correspondingly altered in the APOB100-only background associated with Apobec-1 deletion. To our surprise, however, this was not the case. Our findings demonstrate that (percentage) cholesterol absorption was indistinguishable in chow-fed wild type and Apobec-1−/− mice, although somewhat lower in LD-fed Apobec-1−/− mice compared with wild type controls (26 ± 0.5% versus 33 ± 2%; Fig. 5A, right). Since these findings were so divergent from those reported in Apob100/100 mice (19) (expanded discussion below), we undertook our own examination of cholesterol absorption in the APOB100-only (Apobec-1 wild type) background, yet again, our findings demonstrated no alteration in cholesterol absorption in either genotype when the mice were fed a chow diet and, equally surprisingly, revealed no differences when these Apob100/100 mice consumed the LD (Fig. 5B). Taken together, these findings suggest that the increased susceptibility to gallstone formation noted in Apobec-1−/− mice is unlikely to be attributed to increased cholesterol absorption. Furthermore, our findings fail to confirm the previously documented decrease in cholesterol absorption in both chow and LD-fed Apob100/100 mice (19).
FIGURE 5.
Cholesterol absorption in Apobec-1−/− mice and in Apob100/100 mice. A, cholesterol absorption in mice of the indicated genotype fed either chow or LD for 8 weeks was measured by fecal dual isotope ratio methodology. There was similar cholesterol absorption in Apobec-1−/− mice and WT controls on the chow diet, but cholesterol absorption was decreased in Apobec-1−/− mice fed the LD. Bar graphs show the mean ± S.E. of data from 8 mice/genotype on chow and 4 mice/genotype on LD. B, cholesterol absorption was similar in Apob100/100 mice and control (C57/129Svj) mice either on chow or LD for 8 weeks. Bar graphs show the mean ± S.E., 5 mice/genotype for each study. *, p < 0.05.
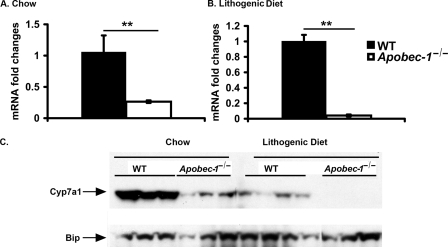
Decreased Cyp7a1 Gene Expression in Apobec-1−/− Mice
Since the most striking effects noted in this study were related to the alterations in bile acid metabolism, we asked whether the decreased expression of Cyp7a1 mRNA in Apobec-1−/− mice (Fig. 6, A and B) was accompanied by a corresponding decrease in protein expression. We found decreased microsomal CYP7A1 protein in chow-fed Apobec-1−/− mice and virtually undetectable levels when Apobec-1−/− mice were fed the LD (Fig. 6C). These findings together point to an unanticipated adaptation in Apobec-1−/− mice in which Cyp7a1 expression is markedly attenuated in chow-fed animals and further suppressed following LD feeding.
FIGURE 6.
Decreased hepatic Cyp7a1 mRNA and protein expression in chow-fed Apobec-1−/− mice and a further decrease in LD-fed mice. A, relative expression of hepatic Cyp7a1 mRNA in chow-fed mice. Bar graphs show the mean ± S.E. of data from four WT and five Apobec-1−/− mice. B, relative expression of hepatic Cyp7a1 mRNA in LD-fed mice. Bar graphs show the mean ± S.E. of data from three WT and five Apobec-1−/− mice. C, Western blot analysis of CYP7A1 protein expression in liver microsomes prepared as described from chow or LD-fed animals. 25 μg of protein was subjected to SDS-PAGE and Western blotted with anti-CYP7A1 IgG and anti-BIP IgG. 3–4 mice from each genotype were studied.
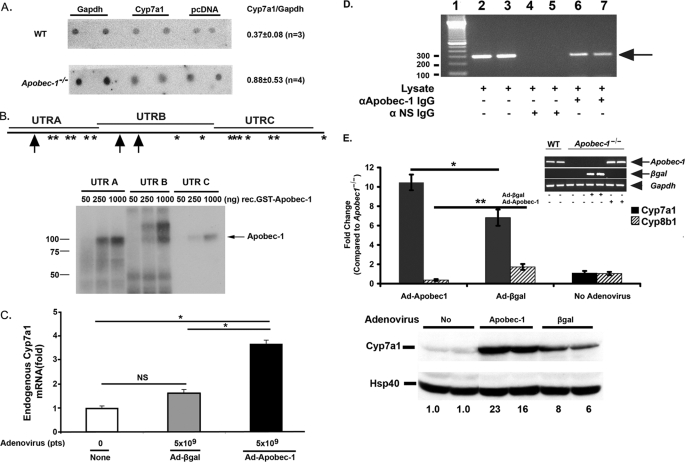
Post-transcriptional Modulation of Cyp7a1 Gene Expression in Apobec-1−/− Mice; APOBEC-1 Binds Cyp7a1 RNA in Vitro and in Vivo
To pursue the mechanisms of the decrease in Cyp7a1 gene expression, we first turned to nuclear run-on analysis to examine whether Cyp7a1 gene transcription was decreased. Our findings revealed no change in Cyp7a1 transcription in nuclei from chow-fed wild type and Apobec-1−/− mice (Fig. 7A). These results suggested the possibility that APOBEC-1 modulates posttranscriptional regulation of Cyp7a1 gene expression. Accordingly, we examined the possibility that APOBEC-1, an RNA-binding protein with affinity for AU-rich targets beyond ApoB mRNA (33, 36), might exhibit RNA binding activity toward the Cyp7a1 3′-UTR, a region of the transcript that contains a large number of AU-rich motifs and the UUUN(A/U)U consensus site for APOBEC-1 binding (36) (Fig. 7B). Recombinant APOBEC-1 was demonstrated to bind to each of the three fragments of murine Cyp7a1 3′-UTR (Fig. 7B), and binding was competed by excess unlabeled competitor (supplemental Fig. 1). Endogenous Cyp7a1 mRNA expression was significantly increased in HepG2 cells following adenovirus-mediated Apobec-1 infection, although a small nonspecific effect (statistically insignificant) was observed following Ad-β-galactosidase transduction (Fig. 7C). We further demonstrated that APOBEC-1 binds Cyp7a1 mRNA in vivo, following immunoprecipitation of Ad-Apobec-1 hepatic lysates with anti-APOBEC-1 IgG, which revealed the presence of Cyp7a1 RNA in the immune pellet (Fig. 7D). This approach was necessary, since APOBEC-1 is a low abundance protein, and co-immunoprecipitation experiments with anti-APOBEC-1 IgG in hepatic lysates from WT animals failed to reveal Cyp7a1 RNA. In addition, control co-immunoprecipitation experiments in nontransduced or Ad-β-galactosidase-transduced Apobec-1−/− mice were negative (data not shown).
FIGURE 7.
Apobec-1-dependent modulation of Cyp7a1 gene expression. A, nuclear run-on assays show similar Cyp7a1 transcription rate in Apobec-1−/− and WT mice. Isolated nuclei were prepared from two separate groups (n = 4 or 5 mice/group for each experiment) of WT and Apobec-1−/− mice and run-on transcripts hybridized to immobilized DNA encoding Gapdh, Cyp7a1, or pcDNA3.1 for background control. Radiolabeled nascent RNA hybridizing to Cyp7a1 versus Gapdh was determined by phosphorimaging and corrected for background control. The data are derived from 3–4 independent experiments from mice of the indicated genotype. The quantitation is indicated on the right as the mean ± S.E. B, top, distribution of Apobec-1 binding sites UUUN(A/U)U in Cyp7a1 3′-UTR is shown. Schematic representation of Apobec-1 binding motif in 3′-UTR of murine Cyp7a1. *, single copy APOBEC-1 binding motif. Arrow, two tandem repeats of APOBEC-1 binding motif. Cyp7a1 3′-UTR was divided into three different fragments (UTRA, UTRB, and UTRC) whose coordinates are detailed under “Materials and Methods.” Bottom, UV-cross-linking assays were performed with radiolabeled cRNA (UTRA, UTRB, or UTRC) and 50–1000 ng of glutathione S-transferase-APOBEC-1. Molecular mass makers are shown on the left. The arrow indicates the cross-linked bands. C, Apobec-1 transfection increases endogenous Cyp7a1 mRNA expression in HepG2 cells. HepG2 cells were infected with 5 × 109 particles (pts) of Ad-β-galactosidase or Ad-Apobec-1 for 48 h. Cyp7a1 mRNA expression was determined by quantitative PCR. Data are normalized to Cyp7a1 mRNA expression in noninfected HepG2 cells. Bar graphs represent the mean ± S.E. from 2–4 independent assays. *, p < 0.05 in the groups of transduced and untransduced HepG2 cells. D, Cyp7a1 mRNA can be coimmunoprecipitated with anti-APOBEC-1 IgG. Liver lysates were freshly prepared from Apobec-1−/− mice following infection with Ad-Apobec-1 and immunoprecipitated with anti-APOBEC-1 IgG or control IgG. RNA was extracted from the immune pellet (or input lysate in lanes 2 and 3) and subjected to reverse transcription-PCR with primers specific to Cyp7a1. The arrow indicates the Cyp7a1 amplified bands. Molecular mass makers are shown on the left. NS, nonspecific IgG. E, hepatic Cyp7a1 mRNA expression (top) and protein expression (bottom) in Apobec-1−/− mice was increased by Ad-Apobec-1 infection. By contrast, Ad-Apobec-1 infection decreased Cyp8b1 mRNA expression in Apobec1−/− mice. Relative expression of hepatic Cyp7a1 and Cyp8b1 mRNA was analyzed by qRT-PCR. Data are normalized to hepatic Cyp7a1 or Cyp8b1 expression of Apobec-1−/− mice without adenovirus infection. Bar graphs show mean ± S.E., 5 mice/group. Filled bar, Cyp7a1; hatched bar, Cyp8b1. The inset images confirm comparable Apobec-1 mRNA expression (reverse transcription-PCR) following Ad-Apobec-1 delivery or of β-galactosidase mRNA expression following Ad-β-galactosidase delivery into Apobec-1−/− liver. The arrows indicate Apobec-1 or β-galactosidase-specific RT-PCR products, and the arrowhead indicates Gapdh-specific RT-PCR product. *, p < 0.05; **, p < 0.01. Hepatic CYP7A1 protein expression was analyzed by Western blot and quantitated by imaging, with HSP40 used as a loading control. The numbers below each lane indicate relative abundance of CYP7A1 after normalizing expression to uninfected Apobec-1−/− mice (no adenovirus) as 1.
Ad-Apobec-1 Rescues Cyp7a1 Gene Expression in Vivo
In order to demonstrate more directly that APOBEC-1 might modify Cyp7a1 gene expression, we examined the consequences of Ad-Apobec-1 rescue in chow-fed Apobec-1−/− mice. This approach revealed a ∼10-fold increase in Cyp7a1 mRNA abundance (Fig. 7E, top), suggesting that Apobec-1 repletion indeed results in increased Cyp7a1 gene expression. We found that injection of control adenovirus also increased Cyp7a1 mRNA expression, but the increase was significantly lower than with Ad-Apobec-1 (Fig. 7E). These differences were even more dramatically illustrated when CYP7A1 protein expression was examined (Fig. 7E, bottom). In addition, Ad-Apobec-1 administration decreased Cyp8b1 mRNA (1.04 (no Ad) versus 1.73 (Ad-β-galactosidase) versus 0.37 (Ad-Apobec-1)) (Fig. 7E), findings consistent with a compensatory down-regulation following rescue of Cyp7a1 expression.
We attempted to pursue the gain-of-function effects of APOBEC-1 on RNA turnover using chimeric green fluorescent protein-reporter constructs cloned upstream of the full-length murine Cyp7a1 3′-UTR, expressed in a variety of cell lines (including HepG2, COS, HeLa, and HEK; data not shown). Steady-state green fluorescent protein mRNA abundance was 2.5-fold greater in the absence of the Cyp7a1 3′-UTR (supplemental Fig. 2A), results similar to those noted in other studies (37). These findings suggest that the 3′-UTR of Cyp7a1 contains destabilizing elements. However, using actinomycin D-treated HepG2 cells, we were unable to show an effect of Ad-Apobec-1 on mRNA half-life of either the endogenous Cyp7a1 or of a chimeric green fluorescent protein-3′-UTR (data not shown), similar to earlier reports suggesting that actinomycin D inhibits a factor(s) required for Cyp7a1 mRNA decay (38). Accordingly, we turned to an alternative approach to examine mRNA decay using transcriptional pulsing in tet-off cells (39). Although this approach revealed that the murine Cyp7a1 3′-UTR conferred instability to a chimeric reporter transcript (supplemental Fig. 2B), there was no effect of Apobec-1 transfection (supplemental Fig. 2C), suggesting that the effects noted in vivo cannot be readily modeled in a cell culture system. We will discuss the implications of these particular findings below.
DISCUSSION
The findings herein demonstrate a dramatic phenotype of increased gallstone susceptibility in Apobec-1−/− mice induced by feeding a lithogenic diet. The suggestion that Apobec-1 expression might be linked to gallstone susceptibility emerged from quantitative trait locus mapping, in which a locus on the middle to distal region of chromosome 6 displayed linkage in D2 × CAST intercrosses, raising the possibility that alterations in hepatic APOB isoforms might in turn lead to alterations in biliary lipid secretion (2, 21). Our a priori objective in initiating these studies was to examine in more detail the role of Apobec-1, in particular to understand the possible pathways by which alterations in the production of APOB isoforms might conceivably lead to changes in canalicular lipid secretion. Our findings demonstrate alterations in bile acid pool size and composition, which, combined with increased biliary cholesterol secretion, increase bile lithogenicity in Apobec-1−/− mice. The current findings, however, are in stark contrast to the phenotype reported in Apob100/100 mice where these animals were protected against diet-induced gallstones (19). Furthermore, and also somewhat unexpectedly, our findings suggest that Cyp7a1 gene expression is reduced in Apobec-1−/− mice, most likely at a post-transcriptional level. Several elements of these findings and our attempts to resolve the underlying mechanisms bear additional discussion.
The first and most striking finding was the increased susceptibility to diet-induced gallstones in Apobec-1−/− mice. In particular, there was a significant 2-fold increase in biliary cholesterol secretion in LD-fed Apobec-1−/− mice, whose basis is not completely explained. We found no change in hepatic mRNA expression for any of several candidate genes implicated in biliary cholesterol transport (including Abcg5/g8, Abca1, and Srb1), although we did not systematically evaluate protein expression or localization of these genes. This caveat notwithstanding, it is possible that the increased cholesterol secretion may reflect alterations in metabolic trafficking rather than changes in the expression of transporter genes. In support of this possibility is the finding that hepatic cholesterol content was decreased in LD-fed Apobec-1−/− mice compared with wild type controls. We also examined the possibility that intestinal absorption of cholesterol might be altered, particularly since studies in Apob100/100 mice demonstrated a dramatic reduction in cholesterol absorption in LD-fed mice, from ∼52% in wild type mice to ∼9% in the Apob100/100 background (19). Those studies also reported that biliary cholesterol secretion was decreased ∼2-fold in LD-fed Apob100/100 mice (19). However, both findings contrast with our results in LD-fed Apobec-1−/− mice. We found no differences in cholesterol absorption in chow-fed wild type and Apobec-1−/− mice, although there was a small, albeit significant, reduction in cholesterol absorption in LD-fed Apobec-1−/− mice. Moreover, we observed no differences in cholesterol absorption in either chow or LD-fed Apob100/100 mice compared with controls. It is worth emphasizing that although the data presented in Fig. 5 suggest that the percentage absorption of cholesterol tended to decrease in animals of all genotypes fed the LD, the net intestinal cholesterol mass absorbed is vastly increased in the cholate-supplemented, LD-fed animals, as evidenced by the dramatic increase in hepatic cholesterol content (Table 1). That said, our findings provide no support for the proposal that ApoB100-only mice, either in the Apobec-1−/− background or Apob100/100 background (Apobec-1-sufficient), exhibit a discernable difference in cholesterol absorption on either diet. We can only speculate on the reasons for the discrepancy in the Apob100/100 background, but subtle differences in strain, bacterial colonization, or other environmental modifiers may be relevant considerations.
We further report heretofore unanticipated alterations in bile acid metabolism in Apobec-1−/− mice and a striking decrease in the expression of Cyp7a1, the enzyme regulating the initial step in the microsomal bile acid synthetic pathway. There was a ∼75% decrease in Cyp7a1 gene expression in chow-fed Apobec-1−/− mice accompanied by a small yet statistically significant decrease (31%) in the bile acid pool size. We found no differences in fecal bile acid excretion despite the decrease in Cyp7a1 mRNA expression in chow-fed Apobec-1−/− mice, but it is worth noting that fecal bile acid output was only decreased by 60% in Cyp7a1−/− mice (25), suggesting that compensatory mechanisms attenuate the net effects on bile acid production, through alternative pathways of bile acid production. The functional implications for bile acid metabolism in chow-fed Apobec-1−/− mice are probably modest, since there was no developmental phenotype in terms of fat malabsorption or skin defects comparable with that observed in Cyp7−/− mice, although it is worth noting that the heterozygous Cyp7+/− mice in those studies were phenotypically normal (40, 41). On the other hand, there was an exaggerated decrease in Cyp7a1 expression in LD-fed Apobec-1−/− mice coupled with an increased bile acid pool (as noted above, probably the result of dietary cholate supplementation), decreased fecal bile acid excretion, and a ∼2.5-fold increase in ileal Asbt mRNA expression. The mechanisms underlying the change in Asbt mRNA expression in Apobec-1−/− mice will require additional study, but the findings suggest that enterohepatic bile acid cycling is increased in LD-fed Apobec-1−/− mice, a possibility consistent with the trend toward increased bile acid secretion noted in LD-fed Apobec-1−/− mice. Taken together, the results demonstrate a range of alterations in bile acid metabolism, including changes in bile acid species and hydrophobicity that probably contribute to the observed increase in gallstone susceptibility in LD-fed Apobec-1−/− mice.
Among the most intriguing questions posed by this study is the mechanism whereby Cyp7a1 gene expression is modulated in the setting of Apobec-1 deletion. The finding that RNA transcription rates were comparable in wild type and Apobec-1−/− mice most plausibly suggests that the decreased Cyp7a1 expression in Apobec-1−/− mice reflects a post-transcriptional mechanism. We have previously demonstrated that APOBEC-1 binds AU-rich RNA templates, particularly those containing the high affinity consensus sequence (UUUN(A/U)U) embedded within its canonical target ApoB RNA as well as others, including c-Myc and Cox-2, and demonstrated that APOBEC-1 binding to these RNA templates also modulates mRNA stability (33, 36). Other work has established a strong precedent for posttranscriptional regulation of Cyp7a1 mRNA, and several authors have pointed out the short half-life (∼30 min to 1 h) of Cyp7a1 mRNA both in cell culture (42) and in vivo in rat (43–45) and hamster liver (46). In particular, the 3′-UTR of murine (as well as human and rat) Cyp7a1 contains several AUUUA motifs, which are characteristic of short lived mRNAs (37, 38, 47). These findings led us to speculate that alteration in Cyp7a1 mRNA stability might account for the changes observed in Apobec-1−/− mice. Nevertheless, we were unable to confirm this prediction experimentally using a gain-of-function approach in HepG2 and other cell lines with (either plasmid- or adenovirus-mediated) Apobec-1 delivery and in models comparing endogenous (i.e. human) Cyp7a1 expression as well as with reporter mRNA constructs cloned in front of the murine Cyp7a1 3′-UTR, using both actinomycin D and transcriptional pulsing to examine mRNA decay.
Previous studies demonstrated that dexamethasone treatment induced a posttranscriptional >20-fold increase in steady-state Cyp7a1 mRNA abundance in L-35 rat hepatoma cells (38). A tetracycline-regulated transcriptional pulsing approach, however, was needed to demonstrate differences in Cyp7a1 mRNA stability, since treatment of L-35 cells with transcriptional inhibitors (such as actinomycin D) obscured differences in mRNA decay, implicating a labile protein(s) for the dexamethasone-dependent increase in Cyp7a1 mRNA stability (38). Although we did not attempt to replicate our findings in this clone of rat hepatoma L-35 cells, one possible explanation for our failure to demonstrate a gain-of-function effect of Apobec-1 on Cyp7a1 mRNA stability in vitro is that the composition, function, or abundance of Apobec-1-interacting proteins (including Acf (Apobec-1 complementation factor)) differs in mouse liver and HepG2 cells. Indirect support for this speculation is that some of these Apobec-1-interacting proteins (such as Apobec-1 complementation factor and HuR) bind Cyp7a1 mRNA,3 although it remains to be determined whether such binding has physiological relevance. In addition, a number of studies have shown that APOBEC-1-mediated C to U ApoB RNA editing requires optimal stoichiometric proportions of trans-acting proteins (34, 48), supporting the prediction that the composition of RNA-binding proteins (some of which may be distinctive to murine hepatocytes) that target murine Cyp7a1 mRNA may in turn be critical determinants of mRNA stability in vivo. Another possibility is that Apobec-1-dependent modulation of Cyp7a1 mRNA expression is mediated indirectly, for example via alterations in micro-RNA expression, which again may differ in regulation or composition between HepG2 cells and murine liver. Further study will clearly be required to resolve these crucial questions.
In conclusion, the current findings raise implications for the regulation of Cyp7a1 gene expression and the integrated role of RNA-binding proteins, including those like Apobec-1 and its partners, which were originally thought to have a restricted function in C to U RNA editing. The range of alternative targets for these gene products continues to evolve.
Supplementary Material
Acknowledgment
We acknowledge with gratitude the contributions, discussion, and input of Dr. Malcolm Lyons during the initiation of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK056260 and R37 HL038180 (to N. O. D.) and P30 DK052574 (to N. O. D. and the Washington University Digestive Diseases Research Core Center, in particular the Morphology and Functional Genomics Cores).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
3 V. Blanc and N. O. Davidson, unpublished observations.
- WT
- wild type
- LD
- lithogenic diet
- HPLC
- high performance liquid chromatography
- qRT-PCR
- quantitative reverse transcription-PCR
- UTR
- untranslated region
- Ad
- adenovirus
- ChMC
- cholesterol monohydrate crystal.
REFERENCES
- 1.Sandler R. S., Everhart J. E., Donowitz M., Adams E., Cronin K., Goodman C., Gemmen E., Shah S., Avdic A., Rubin R. ( 2002) Gastroenterology 122, 1500– 1511 [DOI] [PubMed] [Google Scholar]
- 2.Lyons M. A., Wittenburg H. ( 2006) Gastroenterology 131, 1943– 1970 [DOI] [PubMed] [Google Scholar]
- 3.Zanlungo S., Rigotti A., Nervi F. ( 2004) Curr. Opin. Lipidol. 15, 279– 286 [DOI] [PubMed] [Google Scholar]
- 4.Lammert F., Carey M. C., Paigen B. ( 2001) Gastroenterology 120, 221– 238 [DOI] [PubMed] [Google Scholar]
- 5.Lyons M. A., Wittenburg H., Li R., Walsh K. A., Leonard M. R., Korstanje R., Churchill G. A., Carey M. C., Paigen B. ( 2003) J. Lipid Res. 44, 1763– 1771 [DOI] [PubMed] [Google Scholar]
- 6.Teng B., Burant C. F., Davidson N. O. ( 1993) Science 260, 1816– 1819 [DOI] [PubMed] [Google Scholar]
- 7.Davidson N. O., Shelness G. S. ( 2000) Annu. Rev. Nutr. 20, 169– 193 [DOI] [PubMed] [Google Scholar]
- 8.Angelin B., Einarsson K., Hellström K., Leijd B. ( 1978) J. Lipid Res. 19, 1017– 1024 [PubMed] [Google Scholar]
- 9.Angelin B., Einarsson K., Hellström K., Leijd B. ( 1978) J. Lipid Res. 19, 1004– 1016 [PubMed] [Google Scholar]
- 10.Miller N. E., Nestel P. J. ( 1974) Lancet 2, 929– 931 [DOI] [PubMed] [Google Scholar]
- 11.Watanabe M., Houten S. M., Wang L., Moschetta A., Mangelsdorf D. J., Heyman R. A., Moore D. D., Auwerx J. ( 2004) J. Clin. Invest. 113, 1408– 1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirokane H., Nakahara M., Tachibana S., Shimizu M., Sato R. ( 2004) J. Biol. Chem. 279, 45685– 45692 [DOI] [PubMed] [Google Scholar]
- 13.Moschetta A., Bookout A. L., Mangelsdorf D. J. ( 2004) Nat. Med. 10, 1352– 1358 [DOI] [PubMed] [Google Scholar]
- 14.Amigo L., Castro J., Miquel J. F., Zanlungo S., Young S., Nervi F. ( 2006) Gastroenterology 131, 1870– 1878 [DOI] [PubMed] [Google Scholar]
- 15.Castro J., Amigo L., Miquel J. F., Gälman C., Crovari F., Raddatz A., Zanlungo S., Jalil R., Rudling M., Nervi F. ( 2007) Hepatology 45, 1261– 1266 [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., McLeod R. S., Yao Z. ( 1997) J. Biol. Chem. 272, 12272– 12278 [DOI] [PubMed] [Google Scholar]
- 17.Kulinski A., Rustaeus S., Vance J. E. ( 2002) J. Biol. Chem. 277, 31516– 31525 [DOI] [PubMed] [Google Scholar]
- 18.Farese R. V., Jr., Véniant M. M., Cham C. M., Flynn L. M., Pierotti V., Loring J. F., Traber M., Ruland S., Stokowski R. S., Huszar D., Young S. G. ( 1996) Proc. Natl. Acad. Sci. U. S. A. 93, 6393– 6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H. H., Wang D. Q. ( 2005) Hepatology 42, 894– 904 [DOI] [PubMed] [Google Scholar]
- 20.Hirano K., Young S. G., Farese R. V., Jr., Ng J., Sande E., Warburton C., Powell-Braxton L. M., Davidson N. O. ( 1996) J. Biol. Chem. 271, 9887– 9890 [DOI] [PubMed] [Google Scholar]
- 21.Lyons M. A., Wittenburg H., Li R., Walsh K. A., Churchill G. A., Carey M. C., Paigen B. ( 2003) J. Lipid Res. 44, 953– 967 [DOI] [PubMed] [Google Scholar]
- 22.Carey M. C. ( 1978) J. Lipid Res. 19, 945– 955 [PubMed] [Google Scholar]
- 23.Rossi S. S., Converse J. L., Hofmann A. F. ( 1987) J. Lipid Res. 28, 589– 595 [PubMed] [Google Scholar]
- 24.Heuman D. M. ( 1989) J. Lipid Res. 30, 719– 730 [PubMed] [Google Scholar]
- 25.Schwarz M., Russell D. W., Dietschy J. M., Turley S. D. ( 1998) J. Lipid Res. 39, 1833– 1843 [PubMed] [Google Scholar]
- 26.Setchell K. D., Lawson A. M., Tanida N., Sjövall J. ( 1983) J. Lipid Res. 24, 1085– 1100 [PubMed] [Google Scholar]
- 27.Wang D. Q., Paigen B., Carey M. C. ( 2001) J. Lipid Res. 42, 1820– 1830 [PubMed] [Google Scholar]
- 28.Wang D. Q., Carey M. C. ( 2003) J. Lipid Res. 44, 1042– 1059 [DOI] [PubMed] [Google Scholar]
- 29.Xie Y., Newberry E. P., Young S. G., Robine S., Hamilton R. L., Wong J. S., Luo J., Kennedy S., Davidson N. O. ( 2006) J. Biol. Chem. 281, 4075– 4086 [DOI] [PubMed] [Google Scholar]
- 30.Paulauskis J. D., Sul H. S. ( 1989) J. Biol. Chem. 264, 574– 577 [PubMed] [Google Scholar]
- 31.Shiels B. R., Northemann W., Gehring M. R., Fey G. H. ( 1987) J. Biol. Chem. 262, 12826– 12831 [PubMed] [Google Scholar]
- 32.Straka M. S., Junker L. H., Zacarro L., Zogg D. L., Dueland S., Everson G. T., Davis R. A. ( 1990) J. Biol. Chem. 265, 7145– 7149 [PubMed] [Google Scholar]
- 33.Anant S., Murmu N., Houchen C. W., Mukhopadhyay D., Riehl T. E., Young S. G., Morrison A. R., Stenson W. F., Davidson N. O. ( 2004) Gastroenterology 127, 1139– 1149 [DOI] [PubMed] [Google Scholar]
- 34.Blanc V., Navaratnam N., Henderson J. O., Anant S., Kennedy S., Jarmuz A., Scott J., Davidson N. O. ( 2001) J. Biol. Chem. 276, 10272– 10283 [DOI] [PubMed] [Google Scholar]
- 35.Blanc V., Henderson J. O., Newberry R. D., Xie Y., Cho S. J., Newberry E. P., Kennedy S., Rubin D. C., Wang H. L., Luo J., Davidson N. O. ( 2007) Cancer Res. 67, 8565– 8573 [DOI] [PubMed] [Google Scholar]
- 36.Anant S., Davidson N. O. ( 2000) Mol. Cell. Biol. 20, 1982– 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agellon L. B., Cheema S. K. ( 1997) Biochem. J. 328, 393– 399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker D. M., Wang S. L., Bell D. J., Drevon C. A., Davis R. A. ( 2000) J. Biol. Chem. 275, 19985– 19991 [DOI] [PubMed] [Google Scholar]
- 39.Chen C. Y., Yamashita Y., Chang T. C., Yamashita A., Zhu W., Zhong Z., Shyu A. B. ( 2007) RNA 13, 1775– 1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishibashi S., Schwarz M., Frykman P. K., Herz J., Russell D. W. ( 1996) J. Biol. Chem. 271, 18017– 18023 [DOI] [PubMed] [Google Scholar]
- 41.Schwarz M., Lund E. G., Setchell K. D., Kayden H. J., Zerwekh J. E., Björkhem I., Herz J., Russell D. W. ( 1996) J. Biol. Chem. 271, 18024– 18031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandak W. M., Stravitz R. T., Lucas V., Heuman D. M., Chiang J. Y. ( 1996) Am. J. Physiol. Gastroentest. Liver Physiol. 270, G401– 410 [DOI] [PubMed] [Google Scholar]
- 43.Ness G. C., Pendleton L. C., Li Y. C., Chiang J. Y. ( 1990) Biochem. Biophys. Res. Commun. 172, 1150– 1156 [DOI] [PubMed] [Google Scholar]
- 44.Hylemon P. B., Gurley E. C., Stravitz R. T., Litz J. S., Pandak W. M., Chiang J. Y., Vlahcevic Z. R. ( 1992) J. Biol. Chem. 267, 16866– 16871 [PubMed] [Google Scholar]
- 45.Li Y. C., Wang D. P., Chiang J. Y. ( 1990) J. Biol. Chem. 265, 12012– 12019 [PubMed] [Google Scholar]
- 46.Feingold K. R., Spady D. K., Pollock A. S., Moser A. H., Grunfeld C. ( 1996) J. Lipid Res. 37, 223– 228 [PubMed] [Google Scholar]
- 47.Noshiro M., Nishimoto M., Okuda K. ( 1990) J. Biol. Chem. 265, 10036– 10041 [PubMed] [Google Scholar]
- 48.Sowden M. P., Ballatori N., Jensen K. L., Reed L. H., Smith H. C. ( 2002) J. Cell Sci. 115, 1027– 1039 [DOI] [PubMed] [Google Scholar]
- 49.Yamasaki S., Stoecklin G., Kedersha N., Simarro M., Anderson P. ( 2007) J. Biol. Chem. 282, 30070– 30077 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.