Abstract
Latent transforming growth factor (TGF) β-binding proteins (LTBPs) interact with fibrillin-1. This interaction is important for proper sequestration and extracellular control of TGFβ. Surface plasmon resonance interaction studies show that residues within the first hybrid domain (Hyb1) of fibrillin-1 contribute to interactions with LTBP-1 and LTBP-4. Modulation of binding affinities by fibrillin-1 polypeptides in which residues in the third epidermal growth factor-like domain (EGF3) are mutated demonstrates that the binding sites for LTBP-1 and LTBP-4 are different and suggests that EGF3 may also contribute residues to the binding site for LTBP-4. In addition, fibulin-2, fibulin-4, and fibulin-5 bind to residues contained within EGF3/Hyb1, but mutated polypeptides again indicate differences in their binding sites in fibrillin-1. Results demonstrate that these protein-protein interactions exhibit “exquisite specificities,” a phrase commonly used to describe monoclonal antibody interactions. Despite these differences, interactions between LTBP-1 and fibrillin-1 compete for interactions between fibrillin-1 and these fibulins. All of these proteins have been immunolocalized to microfibrils. However, in fibrillin-1 (Fbn1) null fibroblast cultures, LTBP-1 and LTBP-4 are not incorporated into microfibrils. In contrast, in fibulin-2 (Fbln2) null or fibulin-4 (Fbln4) null cultures, fibrillin-1, LTBP-1, and LTBP-4 are incorporated into microfibrils. These data show for the first time that fibrillin-1, but not fibulin-2 or fibulin-4, is required for appropriate matrix assembly of LTBPs. These studies also suggest that the fibulins may affect matrix sequestration of LTBPs, because in vitro interactions between these proteins are competitive.
Fibrillin microfibrils are ubiquitous structural elements in the connective tissue. Fibrillin microfibrils provide organs with tissue-specific architectural frameworks designed to support the mature functional integrity of the particular organ. In addition, fibrillin microfibrils contribute to proper developmental patterning of organs by targeting growth factors to the right location in the extracellular matrix (1, 2).
Molecules of fibrillin-1 (3), fibrillin-2 (4, 5), and fibrillin-3 (6) polymerize to form the backbone structure of microfibrils. Latent TGFβ-binding protein (LTBP)3-1 associates with fibrillin microfibrils in the perichondrium and in osteoblast cultures (7, 8), and LTBP-1 and LTBP-4 interact with fibrillin (9). Other proteins associated with fibrillin microfibrils include the fibulins (10, 11), microfibril-associated glycoprotein-1 and -2 (12, 13), decorin (14), biglycan (15), versican (16), and perlecan (17). It is likely that one function of these associated extracellular matrix molecules is to connect the fibrillin microfibril scaffold to other architectural elements in tissue- and organ-specific patterns.
In addition to performing architectural functions, fibrillins bind directly to prodomains of bone morphogenetic proteins and growth and differentiation factors (18, 19) and LTBPs bring with them the small latent TGFβ complex (20), suggesting that the microfibril scaffold may position, concentrate, and control growth factor signaling. Studies of fibrillin-1 (Fbn1) and fibrillin-2 (Fbn2) mutant mice demonstrate that loss of fibrillins results in phenotypes associated with dysregulated TGFβ (21–23) or bone morphogenetic protein (24) signaling. Microfibril-associated glycoprotein-1 (Magp-1) null mice reveal phenotypes that may also be related to abnormal TGFβ signaling (25).
In a previous study (9), we determined that the binding site for LTBP-1 and -4 is contained within a specific four-domain region of fibrillin-1. In this study, we performed additional experiments to more precisely define the LTBP binding site. At the same time, we compared binding of fibulins to fibrillin, because the region in fibrillin-1 that was suggested to contain the fibulin binding site (11) was very close to our region of interest for LTBP binding. Our results demonstrate that LTBPs and fibulins compete for binding to fibrillin-1. However, the proteins tested (LTBP-1, LTBP-4, fibulin-2, fibulin-4, and fibulin-5) displayed “exquisite specificities” in their interactions with fibrillin-1.
To test the potential significance of these interactions with fibrillin-1, we investigated matrix incorporation of LTBPs in cell cultures obtained from wild type, Fbn1 null, Fbn2 null, fibulin-2 (Fbln-2) null, and fibulin-4 (Fbln-4) null mice. In addition, we examined the distribution of LTBPs in Fbn1 null and Fbn2 null mice.
EXPERIMENTAL PROCEDURES
Recombinant Proteins
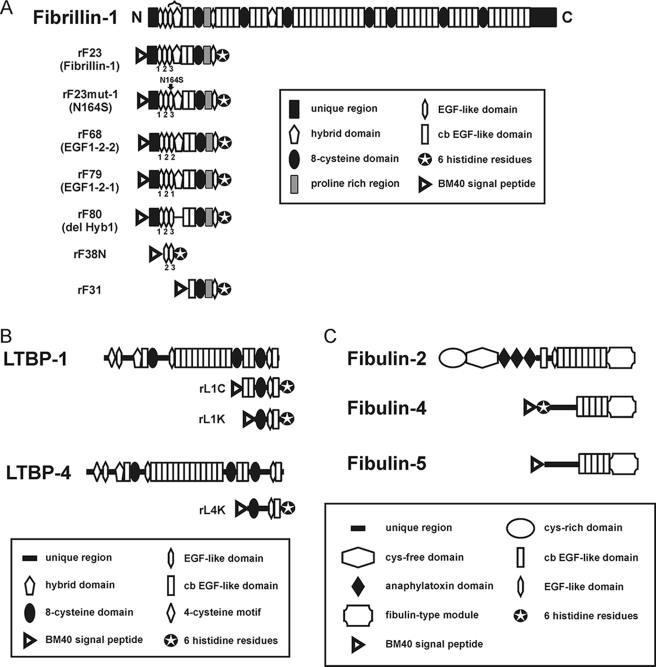
Construction and purification of recombinant fibrillin-1 polypeptides rF23 (10) and rF31 (26) were previously described. New recombinant fibrillin-1 polypeptides were generated with specific mutations designed within the context of rF23. Generation of these new expression constructs is described in the following paragraphs. Primers used for these constructs are listed in supplemental Table S1. Relevant amino acid sequences of these recombinant peptides are listed in supplemental Table S2. Domain structures of fibrillin-1 recombinant polypeptides are shown schematically in Fig. 1A. All fibrillin-1 recombinant polypeptides were harvested from the media of stably transfected 293/EBNA cells and were purified using chelating chromatography, followed by molecular sieve chromatography, as we have described previously (9). Protein solutions were judged for purity by SDS-PAGE (data not shown) and were quantitated by amino acid composition and/or by BCA assay with bovine serum albumin as the standard (Pierce).
FIGURE 1.
Schematic diagrams of the fibrillin-1 recombinant polypeptides (A), LTBP recombinant polypeptides (B), and fibulin recombinant proteins (C) used in this study. Domain modules contained within each peptide are depicted. In A, EGF3 and Hyb1 are bracketed at the top of the diagram. Exact amino acid sequences of the mutated peptides (rF23mut1, rF68, rF79, and rF80) are shown in supplemental Table S2.
For construction of rF68 (EGF1-2-2: substitution of EGF3 with EGF2 within the context of rF23), both rF23 and an intermediate construct, rF67, were used as PCR templates. To construct rF67 (deletion of EGF3 within the context of rF23), the rF23 expression construct was used as a template for PCR, using primers pCEP-5′ and JE2-H1-AS, which together generated a NheI-BbsI fragment. Additionally, primers JH1-S and pCEP-3′ were used to generate a BbsI-XhoI fragment. Finally, a NheI-XhoI-restricted pCEPSP expression vector was ligated with the NheI-BbsI-restricted PCR product and the BbsI-XhoI PCR product to generate pCEPSP-rF67. To continue with construction of rF68, pCEPSP-rF23 was used as a template to generate a NheI-BsmBI PCR product using primers pCEP-5′ and JE2-E2-AS. Primers JE2-S and pCEP-3′ were used to make a BsmBI-XhoI PCR product. Finally, a NheI-XhoI restricted pCEPSP expression vector was ligated with the NheI-BmsBI-restricted PCR product and the BsmBI-XhoI-restricted PCR product to yield pCEPSP-rF68.
For construction of rF79 (EGF1-2-1: substitution of EGF3 with EGF1 within the context of rF23), an intermediate construct, rF69, was used as a PCR template. To generate rF69, primers JE1S and JE1-H1-AS were used along with the rF23-pCEPSP expression construct as template to generate an XbaI/BsmBI-BbsI PCR product. Similarly, primers JH1-S and pCEP3′ were used along with the pCEPSP-rF23 expression construct as template to generate a BbsI-XhoI PCR product. Finally, a XbaI-XhoI-restricted pBluescript II SK(+) cloning vector was ligated with the XbaI/BsmBI-BbsI PCR product and the BbsI-XhoI PCR product to yield pBluescript II-rF69. Next, a third PCR product was generated with primer pCEP-5′ and JE2-E1-AS with pCEPSP-rF23 as template to generate a NheI-BsmBI product. Then the NheI-BsmBI-restricted fragment was ligated with the BsmBI-XhoI-restricted pBluescript-rF69 fragment and a NheI-XhoI restricted pCEPSP vector to yield pCEPSP-rF69. Then to create rF79 (revision of rF69 to juxtapose exon boundaries exactly), two initial PCR products were made using pCEPSP-rF69 as template. Primers pCEP-5′ and rF79AS generated one product, and primers rF79S and pCEP-3′ generated the other product. Equimolar amounts of the two products were mixed to generate a third product by overlap extension PCR, which was then restricted with NheI-XhoI and cloned into the pCEPSP expression vector to generate pCEPSP-rF79.
For construction of rF80 (deletion of Hybrid1), pCEPSP-rF23 was used as a template along with primers pCEP-5′ and JE3-cb1-AS to generate a NheI-BbsI PCR product. Another PCR product used primers Jcb1-S and pCEP-3′ with template pCEPSP-rF23 to yield a BbsI-XhoI product. The two PCR-restricted products were ligated with the NheI-XhoI-restricted pCEPSP expression vector to yield pCEPSP-rF80.
For construction of rF23mut1 (EGF3 N164S within the context of rF23), primers pCEP-5′ and N164SmutAS were used with template pCEPSP-rF23 to generate one PCR product. Primers N164SmutS and pCEP-3′ were used with template pCEPSP-rF23 to generate another PCR product. The two PCR products were then mixed in equimolar amounts to generate a product by overlap extension PCR, which was then restricted with NheI and XhoI. The digested product was then cloned into the pCEPSP expression vector to yield pCEPSP-rF23mut1.
Recombinant LTBP-1K and LTBP-4K were previously described (9). Schematic representations of these recombinant polypeptides are shown in Fig. 1B.
Recombinant human fibulin-2 (27) and human fibulin-5 (28) were prepared and purified as described previously. The cDNA encoding human fibulin-4 (29) was cloned into an N-terminal His6-myc-enterokinase-tagged expression vector based on pCEP-Pu (30) and used to transfect HEK293 EBNA cells. His-tagged human fibulin-4 was purified using nickel-nitrilotriacetic acid-agarose (Qiagen, Valencia, CA) followed by molecular sieve chromatography on a Superose 12 column (HR16/50) equilibrated in 2 m urea, 50 mm Tris-HCl, pH 8.0. Schematic representations of these recombinant proteins are shown in Fig. 1C.
Antibodies
Antibodies specific for fibrillin-1 (pAb 9543), fibrillin-2 (pAb 0868), fibulin-4 (pAb 1147), and fibulin-5 (pAb 1151) were previously characterized (5, 28). For this study, we generated polyclonal antibodies in rabbits using recombinant LTBP-1C (pAb 8579) and recombinant LTBP-4K (pAb 2101). New Zealand White rabbits were immunized several times with 100 μg of immunogen emulsified in an equal volume of TiterMax Gold adjuvant (Sigma) and injected subcutaneously in several sites. These LTBP antisera demonstrated minimal cross-reactivity with the other recombinant polypeptide in enzyme-linked immunosorbent assay (supplemental Fig. S1) or in Western blotting (data not shown). Lack of cross-reactivity was also indicated by immunofluorescence of tissues (Fig. 4). Monoclonal anti-fibronectin (monoclonal antibody 84) was produced and characterized in our laboratory (data not shown).
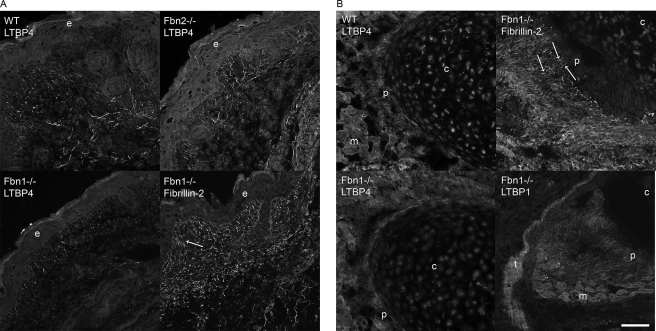
FIGURE 4.
Immunofluorescence of tissues from wild type, Fbn1 null, and Fbn2 null mice. A, P0 skin showed equivalent patterns of pAb 2101 LTBP-4 positive fibrils in both wild type and Fbn2 null sections. Fibrillin-2 staining was used to identify positive microfibrillar staining in Fbn1 null skin; characteristic fibrillin fibrils perpendicular to the dermal-epidermal junction are indicated (arrow), and typical thicker and longer fibrils can be seen in the deeper dermis. LTBP-4 staining patterns in skin were very similar to these fibrillin patterns at the dermal-epidermal junction and in the deeper dermis. In contrast to wild type and Fbn2 null skin sections, many fewer LTBP-4 positive fibrils were seen in Fbn1 null sections. B, LTBP-4 staining was also apparently reduced in sections of perichondrium from Fbn1 null compared with wild type mice. Fibrillin-2 staining showed positive fibrils (arrows) in the perichondrium of Fbn1 null mice. pAb 8579 LTBP-1 staining of perichondrium and tendon in Fbn1 null sections were fibrillar and similar to fibrillin staining patterns. e, epidermis; c, cartilage; p, perichondrium; m, muscle; t, tendon. Bar, 50 μm.
Surface Plasmon Resonance
Binding analyses were performed using a BIAcoreX instrument (BIAcore AB, Uppsala, Sweden). Recombinant LTBP-1 (rL1K), LTBP-4 (rL4K), full-length fibulin-2, -4, or -5 peptides were covalently coupled to CM5 sensor chips (research grade) using the amine coupling kit following the manufacturer's instructions (BIAcore AB). Resonance units coupled to the chips were 638 and 1455 resonance units for rL1K; 630 for rL4K, 3400 for fibulin-2; 2700 for fibulin-4; and 1225 for fibulin-5. Binding responses due to analyte interaction with the surface coupled ligand were normalized by subtraction of background binding to uncoupled control flow cells.
Binding assays were performed at 25 °C in 10 mm Hepes buffer, pH 7.4, containing 0.15 m NaCl, 3 mm EDTA, and 0.005% (v/v) P20 surfactant (HBS-EP buffer, BIAcore AB). Fibrillin peptides were diluted in HBS-EP buffer and then injected at different concentrations and at constant flow rates over immobilized LTBP peptides. For competition assays, the fibrillin peptide, rF23 (at a constant concentration of 80 nm), was preincubated with the competitor LTBP peptide, rL1K, at concentrations of 50–800 nm for ∼1 h prior to injection. To account for variations of the fibrillin signal due to buffer changes caused by the addition of different amounts of competitor, we generated a buffer-matched control without competitor in which the maximum response was set in each case as the 100% reference signal. The surface was regenerated with a pulse of 10 mm glycine, pH 1.7. Kinetic constants were calculated by nonlinear fitting (1:1 interaction model with mass transfer) to the association and dissociation curves according to the manufacturer's instructions (BIAevaluation 3.0 software). Apparent equilibrium dissociation constants (KD) were then calculated as the ratio of kd/ka.
Cell Cultures
Fibroblasts were derived from explant cultures of neonatal skin obtained from Fbn1 (31) or Fbn2 (24) null mice. Primary embryonic fibroblasts were established from embryonic day 18.5 fibulin-2 null (32) and fibulin-4 null4 embryos after digestion of the minced carcasses with trypsin-EDTA and DNase. Cells were cultured in Dulbecco's modified Eagle's medium (MediaTech, Hendon, VA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and penicillin/streptomycin (MediaTech).
Immunofluorescence
1 × 105 cells in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin-streptomycin were plated onto 4-well permanox Lab-Tek chamber slides (Nalge Nunc, Rochester, NY) and were grown for 4, 9, or 13 days. Slides were fixed in cold methanol for 10 min and incubated in primary antibody for 3 h at room temperature. All of the primary antibodies were diluted 1:200 in PBS. Secondary antibodies, Alexa Fluor 488 goat anti-rabbit IgG, or Alexa Fluor 488 goat anti-mouse IgG (Invitrogen), were diluted 1:1000 in PBS and incubated on the sections for 30 min at room temperature. Slides were coverslipped with ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen).
Tissues were snap frozen in isopentanes and were then embedded in Tissue-Tek O.C.T. compound (Fisher Scientific, Pittsburgh, PA). Four-μm cryosections were cut using a Leica CM1850 cryostat. Sections were air dried overnight at room temperature and were then fixed for 10 min in cold acetone and incubated in primary antibodies diluted 1:200 in PBS for 3 h at room temperature in a humidified chamber. After washing in PBS, sections were incubated for 30 min at room temperature in Alexa 488 goat anti-rabbit secondary antibody diluted 1:1000 in PBS. The sections were washed thoroughly and coverslipped using ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole. Slides were viewed on a Zeiss AxioVert 200 microscope, and micrographs were recorded digitally using AxioVision software (version 4.5).
Enzyme-linked Immunosorbent Assay and Western Blotting
Enzyme-linked immunosorbent assays and Western blotting were performed as previously described (16, 18).
Real-time PCR
Cells were seeded on 6-well plates at 1 × 106 cells/well and grown for 4 days. Total RNA preparations from mutant and wild type cells were generated by pipetting 1 ml of TRIzolTM reagent (Invitrogen) onto each well following the manufacturer's protocol and subsequent sample purification using the RNeasyTM kit (Qiagen). RNA samples were quantified by photospectrometry and 0.5 μg of RNA per sample was reverse transcribed using the Bio-Rad iScriptTM cDNA synthesis kit. Samples in triplicate were amplified using the iTaqTM SYBR Green Supermix (Bio-Rad) in an iQTM5 Multicolor Real-time PCR Detection System (Bio-Rad). Analysis of data were performed using the 2−ΔΔCt method (33) and quantitated relative to the glyceraldehyde-3-phosphate dehydrogenase gene. Gene expression was normalized to wild type cell lines, which provided an arbitrary constant for comparative fold-expression.
Circular Dichroism Spectroscopy
Recombinant fibrillin-1 polypeptides rF23, rF23mut1, rF68, and rF79 were dialyzed into 10 mm NaPO4, 100 mm sodium fluoride, pH 7.4. Samples were analyzed in a 0.1-mm cell at 25 °C, with CD measurements taken from 260 to 190 nm using a Jasco J-500A instrument. Concentrations were determined by amino acid analysis.
RESULTS
Extracellular Matrix Incorporation of LTBPs Requires Fibrillin-1
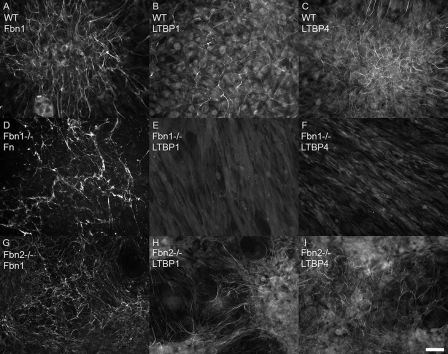
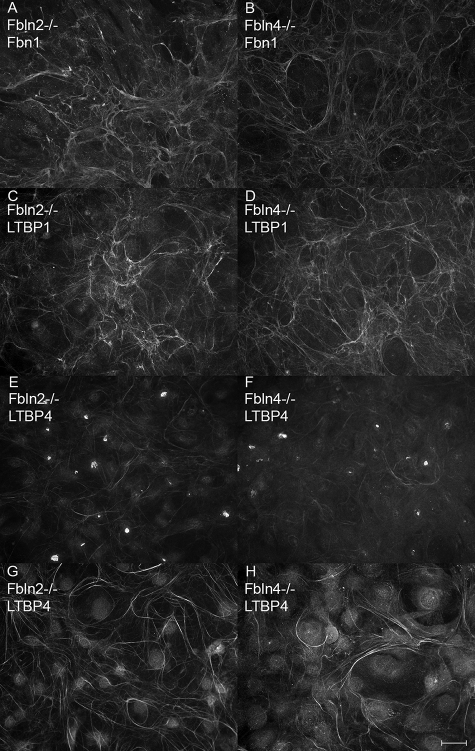
Dermal fibroblasts established from neonatal wild type, fibrillin-1 (Fbn1) null, and fibrillin-2 (Fbn2) null mice were examined by immunofluorescence, Western blotting, and quantitative real-time PCR, to determine the status of LTBPs in the absence of fibrillin-1. Fibroblasts were tested by immunofluorescence after 4, 9, and 13 days in culture. Wild type extracellular matrix contained fibrillin-1 positive microfibrils (Fig. 2A). Immunofluorescence staining with antibodies specific for LTBP-1 or LTBP-4 demonstrated matrix incorporation of both LTBPs in wild type cultures grown for 9 days (Fig. 2, B and C). Fbn1 null fibroblasts demonstrated no fibrillin-1 staining and were also negative for fibrillin-2 staining, consistent with low expression levels of Fbn2 (data not shown). No matrix incorporation of LTBP-1 or LTBP-4 was found in Fbn1 null cultures, even after 13 days in culture (Fig. 2, E and F). Fbn1 null fibroblasts did, however, assemble a matrix containing fibronectin (Fig. 2D). In contrast, Fbn2 null cultures grown for 9 days elaborated an extensive fibrillin-1 containing matrix (Fig. 2G), in which both LTBPs were assembled (Fig. 2, H and I), just like the wild type cultures. 4-Day cultures yielded similar results, except that matrix incorporation of LTBP-4 appeared less abundant (data not shown).
FIGURE 2.
Matrix deposition of fibrillin-1, LTBP-1, and LTBP-4 by fibroblast cultures. Immunofluorescence, using antibodies specific for fibrillin-1 (pAb 9543) (A and G), LTBP-1 (pAb 8579) (B, E, and H), and LTBP-4 (pAb 2101) (C, F, and I), demonstrated incorporation of all three proteins into fibrillar matrices in wild type fibroblasts (A–C) grown for 9 days. In Fbn1 null fibroblasts (D–F) grown for 13 days, no matrix incorporation of LTBPs was visible (E and F), even though fibronectin fibrils were present in the Fbn1 null matrix (D). Fbn2 null fibroblasts (G–I), grown for 9 days, assemble fibrillin-1 positive fibrils (G) as well as LTBP-1 (H) and LTBP-4 (I) positive fibrils. Bar = 20 μm.
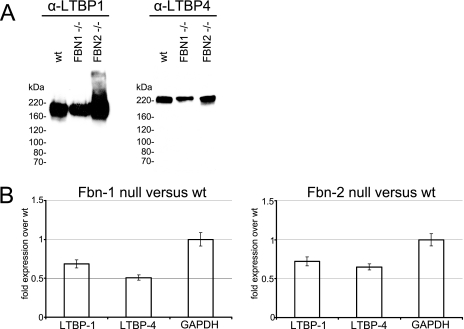
Western blots of Fbn1 null cultures demonstrated that LTBPs were secreted into the medium (Fig. 3A), even though LTBP proteins were not incorporated into the Fbn1 null extracellular matrix (Fig. 2, E and F). To quantitate the relative levels of LTBPs in wild type, Fbn1 null, and Fbn2 null cultures, quantitative reverse transcriptase-PCR was performed. Results showed similar expression levels of LTBP-1 and LTBP-4 in Fbn1 and Fbn2 null cultures (Fig. 3B). Because LTBPs were easily detected in the Fbn2 null matrix by immunofluorescence, lack of matrix incorporation of LTBPs in the Fbn1 null fibroblasts is likely not due to reduced expression levels of LTBPs in the Fbn1 null cells.
FIGURE 3.
A, Western blot analyses of medium proteins from wild type, Fbn1 null, and Fbn2 null fibroblasts. Equal numbers of cells from the different cell lines were aliquoted into wells in serum-containing medium. The next day serum-containing medium was replaced with serum-free medium. Serum-free medium was collected after 2 more days in culture. Total medium proteins were precipitated, applied to 5% SDS-PAGE, transferred to nitrocellulose, and blotted with antibodies specific for LTBP-1 and LTBP-4. Results show that wild type (wt), Fbn1 null, and Fbn2 null fibroblasts secrete LTBP-1 and LTBP-4 into the medium. B, quantitative real-time PCR from Fbn1 null fibroblasts and Fbn2 null fibroblasts compared with wild type littermate fibroblasts. Expression levels of LTBP-1 and LTBP-4 were comparable in Fbn1 null and Fbn2 null fibroblasts. The experiments were conducted in triplicate. Average values from these experiments are shown, with error bars. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was quantitated as a control.
Early postnatal tissues were also examined to determine whether LTBPs are present in matrices devoid of fibrillin-1. In tissues containing fibrillin-1 (that is, in wild type as well as in Fbn2 null tissues), LTBP-4 antibodies stained the matrix in tissue-specific fibrillar patterns, similar to fibrillin staining patterns. LTBP-4 was detected in fibrillar patterns in the dermis of wild type and Fbn2 null (Fig. 4A) mice and in wild type perichondrium (Fig. 4B). In Fbn1 null skin (Fig. 4A) and Fbn1 null perichondrium (Fig. 4B), fibrillin-2 antibody staining patterns were abundant and fibrillar, similar to fibrillin-1 staining patterns in wild type tissues (data not shown). Fibrillin-2 staining was used to identify fibrillar bundles of microfibrils in Fbn1 null tissues. Although LTBP-4 antibodies stained a subset of fibrillin microfibrils, the fibrillar staining patterns were similar to fibrillin. In contrast, LTBP-1 was apparent at the dermal-epidermal junction in skin, but was not present in dermal elastic fibers (data not shown), indicating that LTBP-1 is also associated with a subset of fibrillin microfibrils. LTBP-1 was detected in fibrillar patterns, similar to fibrillin, in the perichondrium and tendon (Fig. 4B). We previously used double immunostaining as well as immunoelectron microscopy to demonstrate colocalization of LTBP-1 and fibrillin-1 in human perichondrium/tendon (9).
In Fbn1 null skin and perichondrium, numbers of LTBP-4 positive fibrils appeared to be significantly reduced, compared with wild type tissues (Fig. 4, A and B). Numbers of LTBP-1 positive fibrils in the perichondrium and tendon seemed equivalent in both wild type and Fbn1 null mice (Fig. 4B and data not shown).
Interrogation of the LTBP-1 Binding Site in Fibrillin-1
Previous data indicated that the LTBP binding site is contained within a region of fibrillin-1 that is composed of four domains: EGF2, EGF3, Hyb1, and cbEGF1 (9). To more precisely define the LTBP binding site, additional recombinant fibrillin-1 polypeptides (Fig. 1) were stably expressed and purified. These new mutated recombinant polypeptides were constructed within the domain context of rF23. The contribution of EGF3 to the LTBP binding site was tested using rF68 (EGF1-2-2) and rF79 (EGF1-2-1), which substituted either EGF1 or EGF2 for EGF3. In addition, rF23mut1 was generated to test whether a naturally occurring mutation in FBN1 (N164S), associated with dominant ectopia lentis (34), affects LTBP binding. Deletion of the first hybrid domain was tested using rF80.
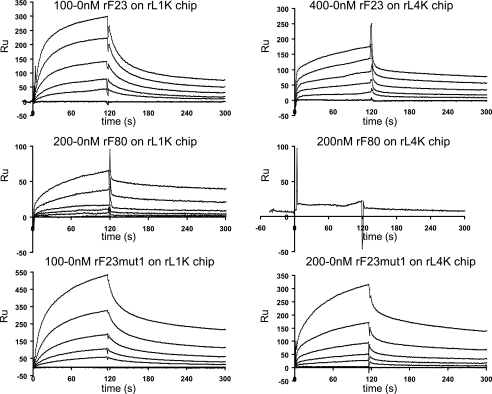
KD values for the association of rF23 and of the mutated rF23 polypeptides with LTBP-1 (rL1K bound to the solid substrate) and with LTBP-4 (rL4K bound to the solid substrate) were determined using surface plasmon resonance. Equivalent high affinity binding of rF23 to both LTBP-1 (21 nm) and LTBP-4 (24 nm) was found. For rF68, averaged KD values were 26 nm for LTBP-1 and 44 nm for LTBP-4, indicating that substitution of EGF3 with EGF2 had no effect on binding to LTBP-1 and reduced binding somewhat to LTBP-4. Similarly, for rF79, KD values were determined to be unaffected for LTBP-1 (23 nm), whereas the KD for LTBP-4 (70 nm) indicated reduced affinity. N164S in rF23mut1 (the mutation associated with dominant ectopia lentis (34)) did not perturb binding to LTBP-1, but this mutation substantially reduced binding to LTBP-4. Deletion of Hyb1 in rF80 abolished binding to LTBP-4 and reduced binding 5-fold to LTBP-1. In addition, a small peptide (rF38N) consisting only of EGF2 and EGF3 demonstrated binding, albeit somewhat reduced, to LTBP-1. These data are summarized in Table 1, and representative binding curves are shown in Fig. 5.
TABLE 1.
KD values (in nanomolar) of interactions determined by surface plasmon resonance
Analytes, listed in the column on the left, were titrated and flowed over chips coupled with the ligands listed in the top row. One experiment consisted of a set of titrations. Interactions that were not tested are listed as not determined. “No binding” indicates that all titrations yielded flat responses.
| rL1K | rL4K | Fibulin-2 | Fibulin-4 | Fibulin-5 | |
|---|---|---|---|---|---|
| rF23 | 21 ± 2 (3)a | 24 ± 5 (4) | 160 | 54 ± 6 (5) | 23 |
| rF23 BNPS cleaved | 213 ± 10 (3) | 72 | NDb | ND | ND |
| rF38N | 65 | ND | ND | ND | ND |
| rF68 | 26 ± 16 (5) | 44 ± 14 (2) | 1260 | 475 | 258 |
| rF79 | 23 ± 8 (2) | 70 | No binding | 67 | 62 |
| rF80 | 104 ± 40 (7) | No binding | No binding | 150 ± 50 (2) | 90 |
| rF23mut1 | 25 ± 5 (3) | 348 ± 221 (2) | 1530 | 430 | 298 |
| rF31 | No binding | ND | ND | No binding | No binding |
| rL1K | ND | ND | No binding | No binding | No binding |
| rL4K | ND | ND | ND | ND | No binding |
a Numbers of experiments that were performed are listed in parentheses.
b ND, not determined.
FIGURE 5.
BiaCore sensorgrams of titrations of rF23, rF80, and rF23mut1 on LTBP-1- and LTBP-4-coated chips. Each experiment was performed as a series of titrated analytes in solution, flowed over a chip coupled with ligand. Titrations are indicated above each set of representative binding curves. Values listed in Table 1 were obtained from experiments similar to the ones shown in this figure. In some cases, titrations were performed multiple times. RU, resonance units.
To confirm the important contribution of the first hybrid domain in fibrillin-1 to interactions with LTBPs, rF23 was cleaved with BNPS (Skatole), which cleaves at tryptophan residues. Binding of rF23 to LTBP-1 was diminished 10-fold, whereas binding to LTBP-4 was diminished 3-fold. The only tryptophan residue present in rF23 is in Hyb1 (see the sequence in supplemental Table S2). Chemical cleavage of rF23 with this reagent was not complete (data not shown), so the residual binding that was detected may be attributed to the uncleaved rF23 present in the sample used for these studies.
LTBPs and Fibulins Compete for Binding to Fibrillin-1
The interaction between fibulin-2 and fibrillin-1 was suggested to be mediated by a region close to Hyb1 (10). Additional studies implicated Hyb1 and the adjacent cbEGF domains in the binding of fibulin-2, fibulin-4, and fibulin-5 to fibrillin-1 (11). Therefore, we tested whether binding to fibulins was affected by the mutations generated in rF68, rF79, rF80, and rF23mut1. KD values for these interactions are shown in Table 1.
In contrast to the binding results obtained with the mutated peptides and LTBP-1, binding of fibrillin-1 to fibulin-2, fibulin-4, and fibulin-5 was more sensitive to mutations affecting EGF3. Substitution of EGF3 with EGF2 (rF68) led to 8–11-fold reduced binding of fibrillin-1 to all of these fibulins. Substitution of EGF3 with EGF1 (rF79) resulted in no binding of fibrillin-1 to fibulin-2, no change in binding to fibulin-4, and a 3-fold reduction in binding to fibulin-5. N164S in rF23mut1 revealed an 8–13-fold reduction in binding to these three fibulins. Deletion of Hyb1 in rF80 resulted in the abolition of fibrillin-1 binding to fibulin-2, a 3-fold reduction in binding to fibulin-4, and a 4-fold reduction in binding to fibulin-5.
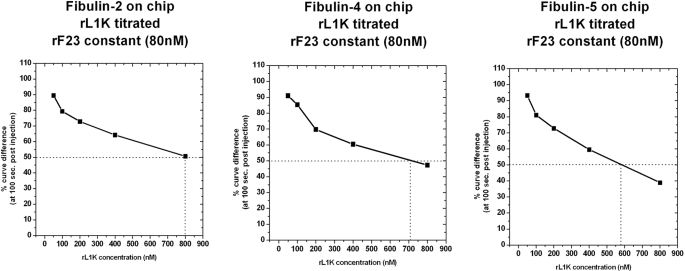
Although the binding sites in fibrillin-1 exhibited exquisite specificities for the LTBPs and fibulins, the interactions were competitive. Binding of rF23 (80 nm) to fibulins -2, -4, and -5 could be inhibited by LTBP-1 (Fig. 6). 50% inhibition of the fibulin-2/fibrillin-1 interactions was achieved by 800 nm LTBP-1, of the fibulin-4/fibrillin-1 interaction by 710 nm LTBP-1, and of the fibulin-5/fibrillin-1 interaction by 580 nm LTBP-1. LTBP-4 could inhibit the fibulin-4/fibrillin-1 and fibulin-5/fibrillin-1 interaction, although it was around 2-fold less effective than LTBP-1 (data not shown). Inhibition of the fibulin-2/fibrillin-1 interaction by LTBP-4 was not tested.
FIGURE 6.
Competition between LTBP-1 and fibrillin-1 for fibulins. Fibulin-2, fibulin-4, and fibulin-5 were each used to coat a BiaCore chip. A constant amount (80 nm) of recombinant fibrillin-1 (rF23) was preincubated with increasing amounts (50–800 nm) of LTBP-1 competitor (rL1K). Binding of rF23 without competitor is set as 100%, and results with competitor LTBP-1 are shown as a percentage of this maximal binding. Titrations of LTBP-1 show that LTBP-1 binding to fibrillin-1 competes with fibrillin-1 binding to fibulin-2, fibulin-4, and fibulin-5.
Cell cultures established from fibulin-2 null and fibulin-4 null mice were tested by immunofluorescence to determine whether loss of either of these fibulins affected matrix incorporation of LTBPs or fibrillin-1. Fibrillin-1 (Fig. 7, A and B), LTBP-1 (Fig. 7, C and D), and LTBP-4 (Fig. 7, E–H) were assembled into the matrix by cells lacking either fibulin-2 (Fig. 7, A, C, E, and G) or fibulin-4 (Fig. 7, B, D, F, and G). LTBP-4 matrix deposition occurred more slowly than either fibrillin-1 or LTBP-1 (compare Fig. 7, E and F with G and H) in these cells, as well as in wild type and Fbn2 null cells (data not shown). Immunofluorescence of tissues from embryonic day 18.5 fibulin-4 null mouse limbs showed no differences in matrix incorporation of fibrillin-1, fibrillin-2, LTBP-1, or LTBP-4 (data not shown).
FIGURE 7.
Matrix deposition of fibrillin-1, LTBP-1 and LTBP-4 in fibulin-2 and fibulin-4 null cultures. Immunofluorescence, using antibodies specific for fibrillin-1 (pAb 9543) (A and B), LTBP-1 (pAb 8579) (C and D), and LTBP-4 (pAb 2101) (E–H), demonstrated incorporation of all three proteins into fibrillar matrices in both fibulin-2 null (A, C, E, and G) and fibulin-4 null (B, D, F, and H) cells. Cells were grown for 4 (A–F) and 9 days (G and H). Bar, 20 μm.
Secondary Structures of the Mutated Fibrillin-1 Recombinant Polypeptides Were Not Significantly Altered
Mutations generated in the rF23-like peptides could have affected the LTBP or fibulin binding sites by altering the conformation of the whole peptide or of adjacent binding sites. To test this possibility, CD spectra of rF68, rF79, and rF23mut1 were analyzed and compared with rF23 (supplemental Fig. S2). Secondary structures were not significantly altered by the mutations present in these polypeptides. Therefore, we conclude that gross conformational changes in the mutated peptides were not the cause of the reduced binding affinities that were measured using surface plasmon resonance.
DISCUSSION
We showed previously that LTBP-1 and LTBP-4 interact with fibrillin-1 in vitro and that LTBP-1 is associated with fibrillin microfibrils in vivo (9). Subsequent studies in Fbn1 mutant mice reported abnormal activation of TGFβ signaling (21–23). Therefore, we first investigated whether loss of fibrillin-1 affected matrix deposition of LTBPs, because this might be a mechanism responsible for aberrant TGFβ signaling. Previous studies have focused on the domains in LTBP-1 that are required for matrix deposition (8, 35) and for TGFβ activation (36). However, no previous study has determined matrix requirements for the incorporation of LTBPs. Here we showed for the first time that fibrillin-1 is required for matrix deposition of LTBPs in cultured fibroblasts. A similar requirement for fibrillin-1 was also found using vascular smooth muscle cells obtained from Fbn1 null mouse aorta. These cells also failed to assemble LTBP-1 and LTBP-4, whereas vascular smooth muscle cell cultures from wild type littermates incorporated fibrillin-1, LTBP-1, and LTBP-4 into fibrils.5 Our results are novel because it was previously thought that matrix deposition of LTBPs was dependent on fibronectin, based on coimmunolocalization studies (8, 37). In our experiments, fibronectin fibrils were clearly assembled in Fbn1 null fibroblast cultures, but LTBPs were still not incorporated into the matrix.
To test whether lack of matrix incorporation of LTBPs in Fbn1 mutant mice may play a role in the abnormal activation of TGFβ signaling, we immunolocalized LTBPs in tissues from Fbn1 null and Fbn2 null mice. We found that LTBPs are normally distributed in fibrillar patterns in skin, perichondrium, and tendon in Fbn2 null mice, in which fibrillin-1 appears to be normal and similarly distributed. However, in Fbn1 null mice, some LTBP-4 was clearly present in fibrillar staining patterns similar to fibrillin immunolocalization patterns, but in low magnification fields, LTBP-4 staining was grossly reduced, both in the skin and perichondrium. It was more difficult to determine by this method whether LTBP-1 staining was reduced in Fbn1 null tissues, because LTBP-1 appears to be abundant in the tissues where it is expressed (perichondrium and tendon). In Fbn1 null mice, fibrillin-2 microfibrils are assembled (31) and are apparently abundant in the early postnatal tissues that we examined here. Therefore, incorporation of LTBP-4 and LTBP-1 in the matrix of Fbn1 null tissues may be mediated by fibrillin-2. Further studies are required to determine the role of fibrillin-2 in mediating matrix incorporation of LTBPs in Fbn1 null mice.
Our interaction studies suggest that the Hyb1 domain contributes residues to the binding sites for LTBP-1 and LTBP-4. However, these two binding sites appear to be different, because deletion of Hyb1 abolishes binding of fibrillin-1 to LTBP-4, but only reduces binding to LTBP-1. Moreover, all three mutations in EGF3 affected binding to LTBP-4, but did not perturb binding to LTBP-1. Because both rF38N and rF80 bind to LTBP-1, albeit with lower affinities, we conclude that both EGF2/EGF3 and Hyb1 contribute to the LTBP-1 binding site, but it was not possible, with the mutants that we utilized, to precisely determine contributions of residues within EGF2/EGF3 and Hyb1 to the LTBP-1 binding site. In contrast, we conclude that the LTBP-4 binding site is primarily contained within Hyb1, because deletion of Hyb1 in rF80 abolishes binding. It is possible that the mutations generated in EGF3 may have perturbed the conformation of the binding site in Hyb1. However, because CD spectra did not indicate substantial conformational changes in rF23mut1, rF68, and rF79, these perturbations must have been subtle and yet sufficient to lower the KD values of these interactions with LTBP-4. Therefore, it is also possible that residues in EGF3, as well as residues in Hyb1, contribute to the LTBP-4 binding site in fibrillin-1.
A recent study demonstrated that binding of fibulin-2, fibulin-4, and fibulin-5 to fibrillin-1 requires Hyb1, but that Hyb1 is not sufficient to mediate these interactions within the context of the domains flanking Hyb2 (11). Furthermore, results from this previously published study were interpreted to indicate that additional synergy sites mediating fibulin binding were located downstream of cbEGF2 (11). Our results confirm the importance of Hyb1 to these interactions. However, by probing the role of EGF3 with our panel of mutants, our results suggest that residues in EGF3 may also contribute to the binding sites for these fibulins.
Probing the binding sites in fibrillin-1 for LTBP-1, LTBP-4, fibulin-2, fibulin-4, and fibulin-5 with our panel of mutated and deleted polypeptides revealed exquisite specificities, a phrase that has been used to describe interactions between monoclonal antibodies and their specific epitopes. We use this phrase to indicate that this concept of exquisite specificities may be applied more broadly to protein-protein interactions. Each of these five proteins bind to closely related sites, and interactions between these proteins are competitive. The structure of EGF3/Hyb1 has not been determined. Identification of the exact interaction sites in fibrillin-1 with these five proteins will require additional structural studies.
In fibulin-2 and fibulin-4 null cell cultures, LTBPs and fibrillins were incorporated into fibrillar matrices, and no differences in immunolocalization of LTBPs or fibrillins were observed in tissues from fibulin-4 null mice. These data indicate that individual fibulins are not required for LTBP or fibrillin-1 matrix assembly. However, because fibulins can compete with LTBPs for binding to fibrillin-1, the function of these fibulins may be to modulate the sequestration of LTBPs. Together with regulation of gene expression in tissue-specific spatial and temporal patterns, competitive protein-protein interactions may also play important roles in certain contexts. These contexts may vary according to factors that modulate the concentrations of these proteins.
Fibulin-2 null mice are viable and fertile (32). Fibulin-4 null mice die during the perinatal period with elastic fiber abnormalities in multiple tissues (38). Fibulin-5 null mice are also viable and fertile, and they also demonstrate elastic fiber abnormalities (39, 40). The fates of LTBPs and fibrillins in these mice have not been reported.
LTBP-2, which has been shown to bind to fibrillin-1 (41), also interacts with fibulin-5 and has been suggested to function as a molecular switch that determines on which microfibrils fibulin-5 will be deposited (42). The N-terminal end of LTBP-2 was found to bind to fibulin-5 (42), and the C-terminal end of LTBP-2 interacted with fibrillin-1 (41). In our studies, the C-terminal ends of LTBP-1 and LTBP-4 interacted with fibrillin-1, but not to fibulins. The fibrillin-1 binding sites within the fibulins were not determined. Future additional in vitro and in vivo studies are required to fully understand the significance of the interactions between LTBPs, fibulins, and fibrillin-1 for microfibril structure, function, and biogenesis. In addition, future studies will address the importance of these interactions to TGFβ signaling.
The FBN1 mutation, N164S in EGF3, identified in a family with dominant ectopia lentis (34), was modeled in the recombinant polypeptide rF23mut1. It is not clear from sequence analysis why this mutation should cause disease, because serine residues occur in this position in other EGF domains. In Marfan syndrome, missense mutations are predicted to cause domain misfolding or destabilization of the fibrillin-1 molecule. N164S does not appear to be this type of mutation. However, our studies show that this mutation substantially reduced binding of fibrillin-1 to LTBP-4, fibulin-2, fibulin-4, and fibulin-5. Interestingly, the mutation did not affect binding of fibrillin-1 to LTBP-1. Therefore, it may be that modulation of these interactions is sufficient to cause ectopia lentis. In contrast, mutations in FBN1 that destabilize the molecule and affect these interactions as well as other interactions, including interactions between fibrillin-1 and LTBP-1, cause Marfan syndrome. In other words, modulation of the interaction between fibrillin-1 and LTBP-1 may be required to cause Marfan syndrome. This hypothesis will be tested in future studies.
Supplementary Material
Acknowledgments
We thank Dr. Kerry Maddox and the Analytical Core Facility of the Portland Shriners Hospital for protein sequencing and amino acid analysis, and Glen Corson for help with engineering some of the fibrillin recombinant peptides used in this study.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 AR049698 (to F. R., D. B. R., and L. Y. S.). This work was also supported a grant from the Shriners Hospitals for Children (to H. P. B. and L. Y. S.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
4 M. L. Chu, unpublished data.
5 L. Zilberberg and D. B. Rifkin, unpublished data.
- LTBP
- latent transforming growth factor-β-binding protein
- TGF-β
- transforming growth factor β
- EGF
- epidermal growth factor like
- cbEGF
- calcium-binding epidermal growth factor like
- Hyb
- hybrid domain
- Fbn
- fibrillin gene
- Fbln
- fibulin gene
- pAb
- polyclonal antibody
- rF
- recombinant fibrillin
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Charbonneau N. L., Ono R. N., Corson G. M., Keene D. R., Sakai L. Y. ( 2004) Birth Defects Res. C Embryo Today 72, 37– 50 [DOI] [PubMed] [Google Scholar]
- 2.Sengle G., Charbonneau N. L., Ono R. N., Sakai L. Y. ( 2008) Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism, 7th Edition, pp. 27– 32, American Society for Bone and Mineral Research, Washington, DC [Google Scholar]
- 3.Sakai L. Y., Keene D. R., Engvall E. ( 1986) J. Cell Biol. 103, 2499– 2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H., Hu W., Ramirez F. ( 1995) J. Cell Biol. 129, 1165– 1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charbonneau N. L., Dzamba B. J., Ono R. N., Keene D. R., Corson G. M., Reinhardt D. P., Sakai L. Y. ( 2003) J. Biol. Chem. 278, 2740– 2749 [DOI] [PubMed] [Google Scholar]
- 6.Corson G. M., Chalberg S. C., Dietz H. C., Charbonneau N. L., Sakai L. Y. ( 1993) Genomics 17, 476– 484 [DOI] [PubMed] [Google Scholar]
- 7.Dallas S. L., Miyazono K., Skerry T. M., Mundy G. R., Bonewald L. F. ( 1995) J. Cell Biol., 131, 539– 549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dallas S. L., Keene D. R., Bruder S. P., Saharinen J., Sakai L. Y., Mundy G. R., Bonewald L. F. ( 2000) J. Bone Miner. Res. 15, 68– 81 [DOI] [PubMed] [Google Scholar]
- 9.Isogai Z., Ono R. N., Ushiro S., Keene D. R., Chen Y., Mazzieri R., Charbonneau N. L., Reinhardt D. P., Rifkin D. B., Sakai L. Y. ( 2003) J. Biol. Chem. 278, 2750– 2757 [DOI] [PubMed] [Google Scholar]
- 10.Reinhardt D. P., Sasaki T., Dzamba B. J., Keene D. R., Chu M. L., Göhring W., Timpl R., Sakai L. Y. ( 1996) J. Biol. Chem. 271, 19489– 19496 [DOI] [PubMed] [Google Scholar]
- 11.El-Hallous E., Sasaki T., Hubmacher D., Getie M., Tiedemann K., Brinckmann J., Bätge B., Davis E. C., Reinhardt D. P. ( 2007) J. Biol. Chem. 282, 8935– 8946 [DOI] [PubMed] [Google Scholar]
- 12.Gibson M. A., Hughes J. L., Fanning J. C., Cleary E. G. ( 1986) J. Biol. Chem. 261, 11429– 11436 [PubMed] [Google Scholar]
- 13.Hanssen E., Hew F. H., Moore E., Gibson M. A. ( 2004) J. Biol. Chem. 279, 29185– 29194 [DOI] [PubMed] [Google Scholar]
- 14.Trask B. C., Trask T. M., Broekelmann T., Mecham R. P. ( 2000) Mol. Biol. Cell 11, 1499– 1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinboth B., Hanssen E., Cleary E. G., Gibson M. A. ( 2002) J. Biol. Chem. 277, 3950– 3957 [DOI] [PubMed] [Google Scholar]
- 16.Isogai Z., Aspberg A., Keene D. R., Ono R. N., Reinhardt D. P., Sakai L. Y. ( 2002) J. Biol. Chem. 277, 4565– 4572 [DOI] [PubMed] [Google Scholar]
- 17.Tiedemann K., Sasaki T., Gustafsson E., Göhring W., Bätge B., Notbohm H., Timpl R., Wedel T., Schlötzer-Schrehardt U., Reinhardt D. P. ( 2005) J. Biol. Chem. 280, 11404– 11412 [DOI] [PubMed] [Google Scholar]
- 18.Gregory K. E., Ono R. N., Charbonneau N. L., Kuo C. L., Keene D. R., Bächinger H. P., Sakai L. Y. ( 2005) J. Biol. Chem. 280, 27970– 27980 [DOI] [PubMed] [Google Scholar]
- 19.Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., Sakai L. Y. ( 2008) J. Biol. Chem. 283, 13874– 13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rifkin D. B. ( 2005) J. Biol. Chem. 280, 7409– 7412 [DOI] [PubMed] [Google Scholar]
- 21.Neptune E. R., Frischmeyer P. A., Arking D. E., Myers L., Bunton T. E., Gayraud B., Ramirez F., Sakai L. Y., Dietz H. C. ( 2003) Nat. Genet. 33, 407– 411 [DOI] [PubMed] [Google Scholar]
- 22.Habashi J. P., Judge D. P., Holm T. M., Cohn R. D., Loeys B. L., Cooper T. K., Myers L., Klein E. C., Liu G., Calvi C., Podowski M., Neptune E. R., Halushka M. K., Bedja D., Gabrielson K., Rifkin D. B., Carta L., Ramirez F., Huso D. L., Dietz H. C. ( 2006) Science 312, 117– 121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohn R. D., van Erp C., Habashi J. P., Soleimani A. A., Klein E. C., Lisi M. T., Gamradt M., ap Rhys C. M., Holm T. M., Loeys B. L., Ramirez F., Judge D. P., Ward C. W., Dietz H. C. ( 2007) Nat. Med. 13, 204– 210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arteaga-Solis E., Gayraud B., Lee S. Y., Shum L., Sakai L., Ramirez F. ( 2001) J. Cell Biol. 154, 275– 281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinbaum J. S., Broekelmann T. J., Pierce R. A., Werneck C. C., Segade F., Craft C. S., Knutsen R. H., Mecham R. P. ( 2008) J. Biol. Chem. 283, 25533– 25543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keene D. R., Jordan C. D., Reinhardt D. P., Ridgway C. C., Ono R. N., Corson G. M., Fairhurst M., Sussman M. D., Memoli V. A., Sakai L. Y. ( 1997) J. Histochem. Cytochem. 45, 1069– 1082 [DOI] [PubMed] [Google Scholar]
- 27.Sasaki T., Göhring W., Pan T. C., Chu M. L., Timpl R. ( 1995) J. Mol. Biol. 254, 892– 899 [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi N., Kostka G., Garbe J. H., Keene D. R., Bächinger H. P., Hanisch F. G., Markova D., Tsuda T., Timpl R., Chu M. L., Sasaki T. ( 2007) J. Biol. Chem. 282, 11805– 11816 [DOI] [PubMed] [Google Scholar]
- 29.Giltay R., Timpl R., Kostka G. ( 1999) Matrix Biol. 18, 469– 480 [DOI] [PubMed] [Google Scholar]
- 30.Wuttke M., Müller S., Nitsche D. P., Paulsson M., Hanisch F. G., Maurer P. ( 2001) J. Biol. Chem. 276, 36839– 36848 [DOI] [PubMed] [Google Scholar]
- 31.Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S. Y., Lenart B., Starcher B., Merkel C. A., Sukoyan M., Kerkis A., Hazeki N., Keene D. R., Sakai L. Y., Ramirez F. ( 2006) J. Biol. Chem. 281, 8016– 8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sicot F. X., Tsuda T., Markova D., Klement J. F., Arita M., Zhang R. Z., Pan T. C., Mecham R. P., Birk D. E., Chu M. L. ( 2008) Mol. Cell. Biol. 28, 1061– 1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak K. J., Schmittgen T. D. ( 2001) Methods 25, 402– 408 [DOI] [PubMed] [Google Scholar]
- 34.Comeglio P., Evans A. L., Brice G., Cooling R. J., Child A. H. ( 2002) Br. J. Ophthalmol. 86, 1359– 1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unsöld C., Hyytiäinen M., Bruckner-Tuderman L., Keski-Oja J. ( 2001) J. Cell Sci. 114, 187– 197 [DOI] [PubMed] [Google Scholar]
- 36.Fontana L., Chen Y., Prijatelj P., Sakai T., Fässler R., Sakai L. Y., Rifkin D. B. ( 2005) FASEB J. 19, 1798– 1808 [DOI] [PubMed] [Google Scholar]
- 37.Taipale J., Saharinen J., Hedman K., Keski-Oja J. ( 1996) J. Histochem. Cytochem. 44, 875– 889 [DOI] [PubMed] [Google Scholar]
- 38.McLaughlin P. J., Chen Q., Horiguchi M., Starcher B. C., Stanton J. B., Broekelmann T. J., Marmorstein A. D., McKay B., Mecham R., Nakamura T., Marmorstein L. Y. ( 2006) Mol. Cell. Biol. 26, 1700– 1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura T., Lozano P. R., Ikeda Y., Iwanaga Y., Hinek A., Minamisawa S., Cheng C. F., Kobuke K., Dalton N., Takada Y., Tashiro K., Ross J., Jr., Honjo T., Chien K. R. ( 2002) Nature 415, 171– 175 [DOI] [PubMed] [Google Scholar]
- 40.Yanagisawa H., Davis E. C., Starcher B. C., Ouchi T., Yanagisawa M., Richardson J. A., Olson E. N. ( 2002) Nature 415, 168– 171 [DOI] [PubMed] [Google Scholar]
- 41.Hirani R., Hanssen E., Gibson M. A. ( 2007) Matrix Biol. 26, 213– 223 [DOI] [PubMed] [Google Scholar]
- 42.Hirai M., Horiguchi M., Ohbayashi T., Kita T., Chien K. R., Nakamura T. ( 2007) EMBO J. 26, 3283– 3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.