Abstract
Z α1-antitrypsin (ZAAT) deficiency is a disease associated with emphysematous lung disease and also with liver disease. The liver disease of AAT deficiency is associated with endoplasmic reticulum (ER) stress. SEPS1 is a selenoprotein that, through a chaperone activity, decreases ER stress. To determine the effect of SEPS1 on ER stress in ZAAT deficiency, we measured activity of the grp78 promoter and levels of active ATF6 as markers of the unfolded protein response in HepG2 cells transfected with the mutant form of AAT, a ZAAT transgene. We evaluated levels of NFκB activity as a marker of the ER overload response. To determine the effect of selenium supplementation on the function of SEPS1, we investigated glutathione peroxidase activity, grp78 promoter activity, and NFκB activity in the presence or absence of selenium. SEPS1 reduced levels of active ATF6. Overexpression of SEPS1 also inhibited grp78 promoter and NFκB activity, and this effect was enhanced in the presence of selenium supplementation. This finding demonstrates a role for SEPS1 in ZAAT deficiency and suggests a possible therapeutic potential for selenium supplementation.
SEPS1 (selenoprotein S, VIMP, Tanis, or SelS) is a selenoprotein found in the endoplasmic reticulum (ER)3 membrane. SEPS1 participates in the processing and removal of misfolded proteins from the ER to the cytosol, where they are polyubiquitinated and degraded through the proteasome (1). SEPS1 can be induced by ER stress (2) and has been shown in macrophages to be protective from pharmacological ER stress agent-induced apoptosis (3).
The endoplasmic reticulum is one of the largest cell organelles. It serves many essential functions, including production of all components of cellular membranes, proteins, lipids, and sterols (4). Only correctly folded proteins are transported out of the ER, whereas incompletely folded proteins are retained in the organelle to complete the folding process or be targeted for destruction. ER stress is defined as an imbalance between the cellular demand for ER function and capacity of the organelle. It is characterized by a number of intracellular responses. These responses include the ER overload response (EOR), the unfolded protein response (UPR), and apoptosis.
α1-Antitrypsin (AAT) deficiency is a disease characterized by early onset emphysema and liver disease (5). The mutant Z form of this autosomal co-dominant disease occurs in >95% of all individuals with AAT deficiency (6). Liver disease occurs in ∼10% of all homozygous neonates who develop hepatitis and cholestasis. A proportion of these children progress to liver failure, requiring liver transplantation (7, 8). Cirrhosis can also occur in adults without a preceding history of childhood liver disease. The mutant Z AAT polymerizes and accumulates in the ER, leaving only 15% of ZAAT secreted (9, 10). This accumulation of abnormal protein in the ER gives rise to ER stress, which is believed to contribute to the liver disease that results from AAT deficiency. The cells respond to this perturbation by inducing the expression of novel genes whose products aim to restore normal ER function (11). SEPS1 is an example of a molecular chaperone that serves to augment the capacity of the ER for protein folding and degradation.
In this paper, we investigate the role of SEPS1 in regulating the cellular response to ER stress in HepG2 cells transfected with the ZAAT transgene. We investigate the effect of SEPS1 on the UPR component of this response by measuring grp78 promoter activity, a UPR-up-regulated gene that functions as a molecular chaperone, and by detecting activated ATF6, which occurs downstream to the activation of grp78. The EOR component of ER stress is investigated by measuring NFκB activation.
We study the effect of selenium supplementation on the action of SEPS1 to see if the function of this selenoprotein can be enhanced and, if so, through which pathways, looking specifically at grp78 promoter activity, ATF6 activation, and NFκB activation and also at glutathione peroxidase (GPx) activity and at the anti-inflammatory 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) pathway. GPx is a selenoprotein whose activity can be readily assayed. This was used as a measure of selenoprotein activity.
The role of SEPS1 in conformational diseases has not been evaluated. These diseases are caused by inherited or acquired modifications in protein structure, where specific proteins undergo a conformational rearrangement, causing deposition within cellular compartments, such as the ER. This can lead to devastating results. AAT deficiency is one such disease, but the group includes Alzheimer, Parkinson, and Huntington diseases and cystic fibrosis.
MATERIALS AND METHODS
Cell Culture, Treatments, and Transfections
HepG2 cells (3 × 105 cells/well) were grown in 24-well plates overnight in serum-free Eagle's minimum essential medium supplemented with l-glutamine (Glutamax) and 0.1 mm nonessential amino acids in preparation for transfection (Transfast; Promega). Cells were co-transfected with pCMV (empty vector), pSEPS1 (Origene), pZAAT, or pSEPS + pZAAT and an inducible grp78 promoter-linked (firefly) luciferase reporter plasmid or an inducible (NFκB)5 promoter (firefly) luciferase reporter plasmid and pRLSV40, a constitutive luciferase reporter plasmid for 24 h. Cells were treated with vehicle, tunicamycin (10 μg/ml) or IL-1β (10 ng/ml) for a further 24 h, as indicated. Supernatants or lysates were prepared for further analyses.
Western Blot Analysis
Cells (1 × 106) were grown in the absence or presence of selenium (as indicated) or in 6-well plates overnight in serum-free medium in preparation for transfection. Cells were transfected with appropriate plasmids and treated as indicated. Whole cell lysates were prepared. Immunoreactive proteins were detected using specific antibodies (SEPS1, ATF6; Abcam); signals were detected using horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technologies) and chemiluminescent substrate solution (Pierce). Equal protein loading was confirmed by Ponceau S staining.
Z Form of AAT Enzyme-linked Immunosorbent Assay
Detection of the Z form of AAT was carried out using the Wieslab AAT deficiency test according to the manufacturer's instructions. Duplicate samples of supernatants and lysates from pZAAT-transfected cells were used. Absorbance values were measured at 405 nm, and these were corrected for transfection efficiency.
Luciferase Assays
pRLSV40 luciferase activity was quantified, using coelenterazine as the luminescent enzyme substrate and using a Wallac Victor2 1420 multilabel counter. For (NFκB)5 and grp78 promoter luciferase assays, the Promega luciferase assay system was used according to the manufacturer's instructions. Values are expressed as (NFκB)5 or grp78 promoter activity per transfected cell.
Quantitative Reverse Transcription-PCR
Total RNA was isolated using TRI Reagent (Sigma). RNA (2 μg) was reverse-transcribed, and cDNAs were amplified using standard PCR conditions and primers for SEPS1 (sense, 5′-GAAACGGAAATCGGACAGAA-3′; antisense, 5′-GGCAATTGAATCGAGGGTTA-3′) and glyceraldehyde-3-phosphate dehydrogenase (sense, 5′-ACAGTCAGCCGCATCTTCTT-3′; antisense, 5′-GACAAGCTTCCCGTTCTCAG-3′). Amplifications were carried out for 35 cycles at 94 °C for 10 s, 55 °C for 10 s, and 72 °C for 7 s using a Roche Applied Science LightCycler (LC480). The expression of SEPS1 relative to glyceraldehyde-3-phosphate dehydrogenase was determined using the 2−ΔΔCT method.
Measurement of Serum Selenium Levels in Patients
Following informed consent under a protocol approved by the Beaumont Hospital Ethics Committee, serum samples were taken from 24 patients who had been screened for AAT deficiency and phenotyped. Twelve ZZ and 12 MM patients who were age- and sex-matched were compared. Serum selenium levels were measured in μg/liter by Trace Laboratories.
Selenium Supplementation of HepG2 Cells
Cells were cultured in the presence or absence of 40 nm or 150 μm seleno-dl-methionine (Sigma)-supplemented medium (12).
GPx Assay
GPx activity was quantified using the Sigma glutathione peroxidase cellular activity assay kit. This kit measures total GPx activity and does not differentiate different isoforms.
15-Deoxy-Δ12,14-Prostaglandin J2 Activity
HepG2 cells (9 × 105) transfected or treated appropriately were grown in 6-well plates in 3 ml of complete medium or selenium-supplemented medium (150 μm seleno-dl-methionine) for 48 h. 15-Deoxy-Δ12,14-prostaglandin J2 levels were quantified using an enzyme immunoassay kit (Correlate-EIA; Assay Designs).
Statistical Analysis
Data were analyzed with the GraphPad Prism 4 software package (GraphPad Software, San Diego, CA). Results are compared by t test or analysis of variance, as appropriate. Differences were considered significant at p values of less than 0.05.
RESULTS
Validation of Model System
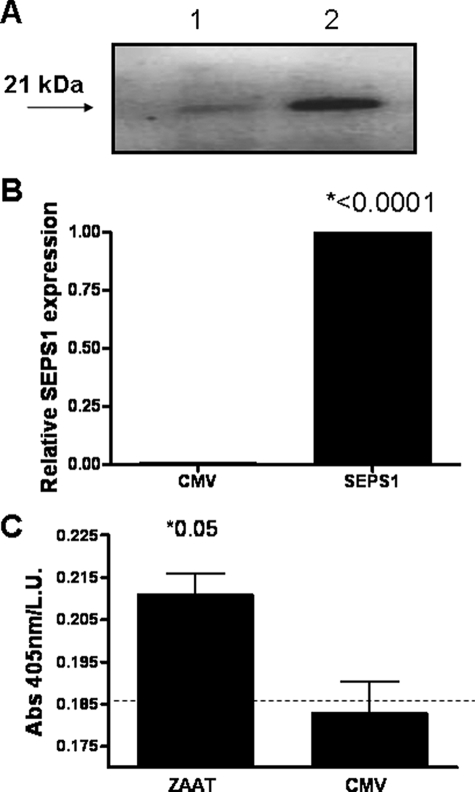
Fig. 1 shows that HepG2 cells transfected with pSEPS1 express higher SEPS1 protein (Fig. 1A) and mRNA (Fig. 1B) compared with empty vector-transfected cells. Cells transfected with pZAAT but not an empty vector produce ZAAT (Fig. 1C).
FIGURE 1.
Confirmation of expression of SEPS1 and ZAAT by HepG2 transfectants. A, HepG2 cells (1 × 106) were transfected with empty vector (lane 1) or pSEPS1 (lane 2). Protein extracts (40 μg) were analyzed by Western immunoblot. B, SEPS1 expression was quantified by quantitative reverse transcription-PCR. C, HepG2 cells were cotransfected with pRLSV40 and an empty vector (CMV) or pZAAT. ZAAT expression in cell lysates was quantified by an enzyme-linked immunosorbent assay. Absorbance at 405 nm per light unit (L.U.) indicates relative expression of ZAAT. The dashed line indicates background absorbance readings. Data shown are representative of three experiments.
Effect of SEPS1 Overexpression on ZAAT-induced grp78 Promoter Activity
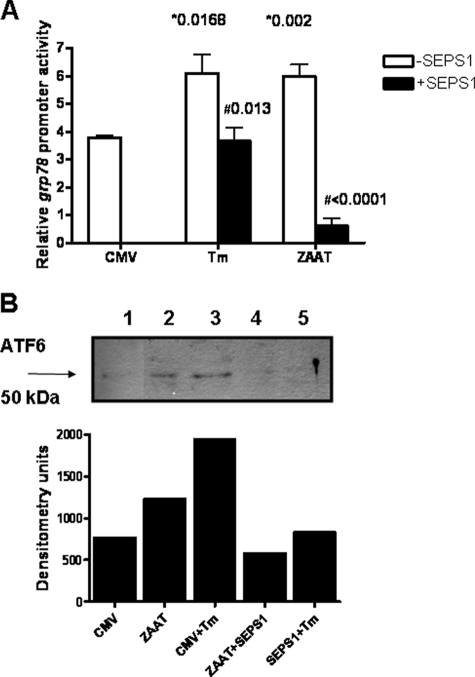
The unfolded protein response was examined by measuring grp78 promoter activity in HepG2 cells transfected with an empty vector (CMV) or with the ZAAT transgene (ZAAT in Fig. 2A). Cells transfected with an empty vector and treated with tunicamycin, a known ER stress agonist, had double the grp78 promoter activity of cells transfected with the empty vector alone. Overexpression of SEPS1 in these cells decreased grp78 promoter activity to the level of the empty vector alone. Transfection with pZAAT caused an increase in grp78 promoter activity to the level seen in the positive control with tunicamycin. When the cells were co-transfected with pSEPS1, grp78 promoter activity was abrogated (p < 0.0001).
FIGURE 2.
SEPS1 inhibits ZAAT-induced activation of UPR. A, triplicate samples of HepG2 cells (3 × 105) were cotransfected with an empty vector (pCMV) or pSEPSI; pZAAT; an inducible grp78 promoter-linked (firefly) luciferase reporter plasmid; and pRLSV40. Cells were treated with tunicamycin (Tm; 10 μg/ml) or DMSO for 16 h. Lysates were prepared, and luciferase production from both plasmids was quantified by luminometry using specific substrates. Relative grp78 promoter activity is shown (*, versus CMV; #, versus minus SEPS1). B, HepG2 cells (3 × 105) were transfected with pCMV or pZAAT, treated with tunicamycin (Tm; 10 μg/ml) or DMSO. Western immunoblotting for ATF6 was carried out on cell lysates, and densitometry of the resulting blot is also shown. Lane 1, empty vector (CMV); lane 2, pZAAT; lane 3, CMV + tunicamycin; lane 4, pZAAT + pSEPS1; lane 5, CMV + tunicamycin + pSEPS1. Data shown are representative of three experiments.
Effect of SEPS1 Overexpression on ZAAT-induced ATF6 Cleavage
The unfolded protein response was examined by Western immunoblotting for ATF6 (Fig. 2B). The active form of ATF6, occurring in ER stress, has a molecular mass of 50 kDa and is termed ATF6p50. There was no detectable ATF6p50 in empty vector-transfected cells (lane 1). Tunicamycin treatment and ZAAT overexpression lead to cleavage and activation of ATF6 (lanes 2 and 3, respectively). Co-transfection with SEPS1 inhibited generation of ATF6p50 (lanes 4 and 5).
Effect of SEPS1 Overexpression on NFκB Activity in HepG2 Cells
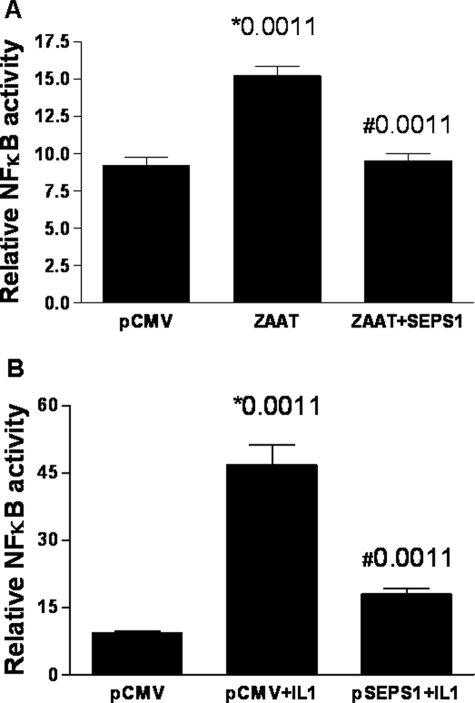
NFκB is a key transcription factor activated in ER stress. We examined the effect of SEPS1 expression on NFκB activation in HepG2 cells in response to ZAAT or another NFκB activator, IL-1β. HepG2 cells were co-transfected with an empty vector, CMV, or a plasmid carrying the ZAAT transgene and an (NFκB)5-luciferase reporter construct (Fig. 3). Overexpression of SEPS1 significantly reduced NFκB activation induced by ZAAT (Fig. 3A) or IL-1β (Fig. 3B).
FIGURE 3.
SEPS1 reverses the effect of ZAAT and IL-1β on NFκB activation in HepG2 cells. A, triplicate samples of HepG2 cells (3 × 105) were co-transfected with an empty vector (pCMV) or pZAAT, pSEPS1 (as indicated), an inducible NFκB (firefly) luciferase reporter plasmid, and pRLSV40. B, following overnight incubation, cells were treated with IL-1 (10 ng, 24 h). Lysates were prepared using reporter lysis buffer (Promega). Luciferase production from both plasmids was quantified by luminometry using specific substrates. Relative NFκB luciferase activity is shown (*, versus CMV; #, versus minus SEPS1). Data shown are representative of three experiments.
Serum Selenium Levels in α1-Antitrypsin-deficient Patients
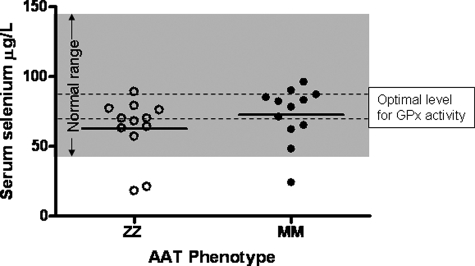
Patients were genotyped for the AAT gene. Serum selenium levels were measured in 12 Z homozygous and 12 M homozygous age- and sex-matched individuals. The levels of selenium in the ZZ patients were all in the lower normal range (Fig. 4). Although no significant difference was evident between the ZZ and MM individuals, mean serum selenium levels were lower than the optimal range for serum glutathione peroxidase activity (70–90 μg/liter) in 75% of the ZZ patients.
FIGURE 4.
Serum selenium levels in ZZ and MM AAT individuals. Individuals were screened to determine their AAT phenotype. Serum selenium levels (μg/liter) were measured in 12 ZZ and 12 MM individuals. The normal selenium range (shaded area) and optimal levels for serum glutathione peroxidase activity (dashed lines) are shown.
Glutathione Peroxidase Activity and SEPS1 Expression in Selenium-supplemented Cells
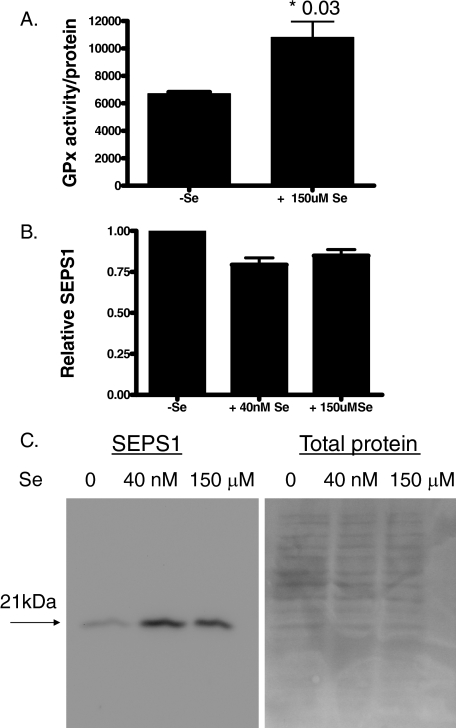
SEPS1 and GPx are selenoproteins. Selenocysteine is an essential component of selenoproteins and is often present in the active sites of these proteins. We investigated if supplementation with selenium would enhance the activity of SEPS1, using GPx activity as a surrogate marker of SEPS1 activity. HepG2 cells were grown in 150 μm seleno-dl-methionine-supplemented medium or routine serum-containing medium. Growth in selenium led to a decrease in overall cell number; however, once corrected for cell number, GPx activity in cell lysates was higher in cells grown in the presence of selenium (Fig. 5A). This showed an enhanced selenoprotein activity in these cells.
FIGURE 5.
Glutathione peroxidase activity and SEPS1 expression in control and selenium supplemented cells. HepG2 cells were grown in complete medium containing 0 nm, 40 nm, or 150 μm seleno-dl-methionine, as indicated, for 72 h. A, a glutathione peroxidase cellular activity assay kit (Sigma) was used to measure total GPx activity. The result shown is representative of three repeated experiments. B, RNA was isolated and used in quantitative reverse transcription-PCRs to detect SEPS1 and glyceraldehyde-3-phosphate dehydrogenase expression. Relative SEPS1 expression is shown. C, protein extracts were prepared, and samples (10 μg) were immunoblotted for SEPS1 or stained with Ponceau S for total protein loading.
We next assessed the status of SEPS1 mRNA and protein expression in selenium-deficient and a variety of selenium-sufficient conditions. Fig. 5B shows that compared with cells grown in the absence of selenium, incorporation of 40 nm or 150 μm selenium into the culture medium led to a small decrease in SEPS1 mRNA expression. Interestingly, SEPS1 protein expression was actually enhanced in cells grown in 40 nm or 150 μm selenium compared with selenium-starved cells (Fig. 5C).
15-Deoxy-Δ12,14-prostaglandin J2 Activity in Selenium-supplemented Cells
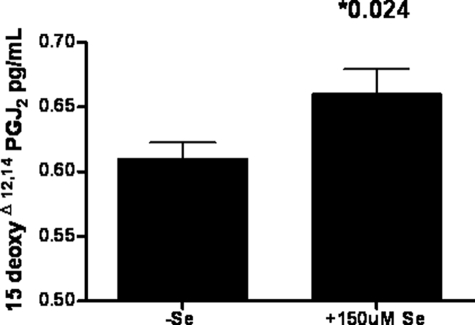
Selenium supplementation can have an anti-inflammatory effect through enhancing the activity of SEPS1 but also through its effect on 15d-PGJ2 formation, an endogenous inhibitor of IKKβ activity. HepG2 cells were grown in the presence or absence of 150 μm seleno-dl-methionine. Lysed cells were analyzed for 15d-PGJ2 concentration (Fig. 6). There was increased 15d-PGJ2 concentrations in the selenium-supplemented cells.
FIGURE 6.
Selenium increases 15-deoxy-Δ12,14-prostaglandin J2 production in HepG2 cells. HepG2 cells (9 × 105) were grown in complete medium (−Se) or medium supplemented with 150 μm seleno-dl-methionine (+Se) for 48 h. 15d-PGJ2 (pg/ml) was measured in whole cell lysates by an enzyme-linked immunosorbent assay. Data are representative of three experiments.
Effect of Selenium Supplementation on grp78 Promoter Activity and NFκB Activity
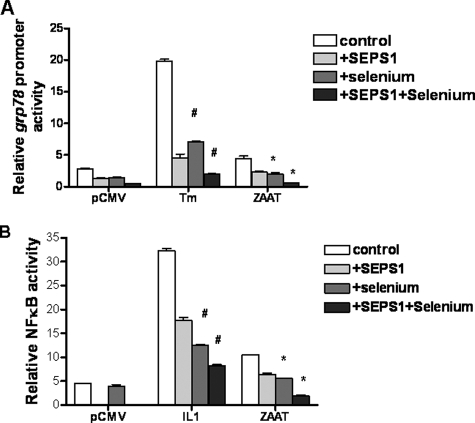
The effect of supplementation with 150 μm seleno-dl-methionine on the UPR component of ER stress was examined by measuring grp78 promoter activity in HepG2 cells transfected with pCMV, pSEPS1, pZAAT, or pSEPS1 + pZAAT. Fig. 7A shows that selenium enhances the effect of both endogenous and overexpressed SEPS1 on grp78 promoter activity.
FIGURE 7.
Selenium enhances the effect of endogenous and overexpressed SEPS1 on grp78 promoter activity and NFκB activity. A, triplicate samples of HepG2 cells (3 × 105) were transfected with an empty vector (pCMV or pZAAT) and co-transfected with an inducible grp78 promoter-linked (firefly) luciferase reporter plasmid and pRLSV40 and grown in the absence or presence (+Se) of 150 μm seleno-dl-methionine for 16 h and then left untreated or stimulated with tunicamycin (Tm; 10 μg/ml, 24 h) or DMSO (A) or IL-1β (10 ng/ml, 24 h) (B). Lysates were prepared, and luciferase production from both plasmids was quantified by luminometry. Relative grp78 promoter (#, versus tunicamycin; *, versus ZAAT) and NFκB activity (#, versus IL-1; #, versus ZAAT) are shown.
The effect on the EOR response was assessed by measuring NFκB activity in the presence of selenium supplementation (Fig. 7B). Selenium was also shown to enhance the effect of the endogenous and overexpressed SEPS1 on the activity of NFκB.
DISCUSSION
In this work, we have demonstrated a role for SEPS1 in modifying ER stress responses induced by ZAAT. ZAAT deficiency is a genetic disorder, the phenotype of which has varying degrees of severity and is likely to be affected by a number of modifying factors. Liver disease in this condition occurs in ∼10% of patients and results from the accumulation of polymers of the abnormal protein in the ER. This is exemplified by the fact that in the absence of the abnormal protein, as is the case in the null variant, liver disease does not occur (6). The in vitro model of the disease we have used is HepG2 cells transfected with a ZAAT transgene. HepG2 is a commonly used cell line in SEPS1 studies (2, 13). We validated our model prior to experiments by demonstrating the expression of SEPS1 and ZAAT in transfected cells.
The ER has a key role in protein folding and handling of misfolded proteins. Specific signaling pathways (14) and effector mechanisms have evolved to deal with the temporal and developmental variation in the ER load. The upstream signal that activates these pathways is referred to as ER stress and is defined functionally as an imbalance between the load of proteins facing the ER and the organelle's ability to process that load. The cellular response to ER stress has four main functional components: EOR, UPR (15), a decrease in protein synthesis, and programmed cell death (16). Here we investigated the effect of SEPS1 on different elements of the ER stress response.
GRP78/BiP is a major UPR-up-regulated target protein. This immunoglobulin heavy chain-binding protein/glucose-regulated protein has a molecular mass of 78 kDa and is one of the most highly expressed ER-resident chaperones. GRP78 is involved in many cellular processes, including translocating newly synthesized proteins, maintaining them in a state competent for subsequent folding and oligomerization, and regulating calcium homeostasis (17, 18). In addition to its chaperoning function, GRP78 is a key regulator of ER stress transducers. GRP78 binds and inhibits PERK, IRE1, and ATF6 activation in nonstressed cells (19). It senses the presence of misfolded proteins in the ER, dissociates from PERK, IRE1, and ATF6 and associates with the accumulating misfolded protein in the ER lumen. This titrating event leads to activation of UPR signaling. We demonstrate an increase in grp78 promoter activity in the setting of ER stress in the ZAAT-transfected cells. When SEPS1 is overexpressed, the effect of ZAAT is reduced, and the increase in grp78 promoter activity is decreased to a level seen in the control setting.
SEPS1 is a selenoprotein. Selenoproteins are a group of proteins that contain the rare amino acid selenocysteine (Sec). Sec is coded for by the dual function stop codon UGA when it is accompanied by the evolutionarily conserved cis- and trans-acting elements and protein factors dedicated to decoding of UGA as Sec (20). There are only 25 known selenoproteins in the human proteome (21). Selenium is an essential trace mineral of fundamental importance to human health and is known for its antioxidant activity and its chemopreventive, anti-inflammatory, and antiviral properties (22). Much of its beneficial influence on human health is attributed to its presence within the selenoproteins, but other pathways are also possible.
SEPS1 was initially cloned from diabetic rats, since it was observed that its expression was decreased in fasting diabetic animals. The protein was characterized under the name Tanis; however, the penultimate Sec residue was misinterpreted as a stop codon (23). The interaction of SEPS1 with serum amyloid (SAA), an acute phase protein, suggested a link between type 2 diabetes, inflammation, and cardiovascular disease. SEPS1 was later shown to be induced in response to ER stress (13). Of the 25 mammalian selenoproteins described, selenoproteins I, K, and S and the thioredoxin reductases 1, 2, and 3 share a penultimate rather then internal Sec residue (24). A hierarchy of selenium regulation exists whereby the position of Sec is thought to play a major role in susceptibility of selenoprotein mRNAs to nonsense-mediated decay. Although expression of GPx1 and thioredoxin reductases, for example, are known to be decreased by selenium deficiency, we and others found that SEPS1 displays only low susceptibility to regulation by selenium status (25). In contrast to the effect seen on SEPS1 mRNA, SEPS1 protein expression was enhanced by a growth in the presence of selenium. This is not surprising, given that Sec is cotranslationally incorporated into nascent polypeptides. Thus, in the presence of selenium, there are two levels of regulation: a small decrease in SEPS1 mRNA expression but a larger increase in Sec incorporation into SEPS1 protein.
We show in this paper, by demonstrating the effect on ATF6 and the grp78 promoter, that overexpression of SEPS1 inhibits UPR events triggered by ZAAT overexpression. The ER is an essential intracellular organelle for the synthesis and maturation of cell surface and secretory proteins and maintenance of calcium homeostasis. In hepatocytes in particular, it is extremely active, with an estimated synthesis of 13 million secretory proteins/min (26). Disruption of these physiological functions leads to accumulation of unfolded proteins and can induce UPR (4, 27). ATF6 is a type II transmembrane protein of the ER, which migrates to the Golgi compartment, where it is cleaved by site-1 protease and site-2 protease. The cleaved cytosolic N-terminal fragment of ATF6 migrates to the nucleus and acts as an active transcription factor, together with ATF4 and spliced XBP1, to increase the expression of the genes encoding proteins that function to augment the ER protein folding capacity. These gene targets include those encoding ER chaperones, such as BiP/GRP78 itself. ZAAT polymerizes and is retained in the ER (28). Here we link this event to ATF6 activation and show that that SEPS1 can reverse the effect.
ER stress also induces EOR (16), which leads to activation of the transcription factor NFκB (29). NFκB is normally found in an inactive cytoplasmic complex with IκB. Upon stimulation of cells with a number of agents, NFκB activity is induced. This involves phosphorylation and degradation of IκB and translocation of NFκB to the nucleus, where it activates transcription of proinflammatory target genes. In this study, we found that accumulation of ZAAT in the ER provided an NFκB-activating stimulus, which was decreased by the presence of increasing SEPS1. NFκB, although a regulator of proinflammatory gene expression, is also known to be cytoprotective. It remains to be determined if preventing excessive activation of NFκB represents a therapeutic benefit, since it would be important to avoid its complete suppression.
The anti-inflammatory effects of selenium are also mediated via an increased production of 15d-PGJ2 as an adaptive response to protect cells against oxidative stress-induced proinflammatory gene expression (30). The IKK complex is a key kinase in the NFκB cascade. IKKβ has the dominant role in signal-induced phosphorylation and degradation of the inhibitory and cytoplasmic retention protein of the NFκB complex, IκBα. IKKβ is also subject to control by cyclopentone prostaglandins, such as PGA1 and 15d-PGJ2. It has been shown that selenium supplementation of macrophages leads to an increase in 15d-PGJ2 via arachidonic acid oxidation by COX1. It has also been shown that some anti-inflammatory effects of 15d-PGJ2 were mediated by PPARγ-dependent mechanisms. We explored this mechanism of action of selenium by performing 15d-PGJ2 assays to demonstrate an increased concentration in cells supplemented with selenium.
In the experiments investigating grp78 promoter and NFκB activity in HepG2 cells, in the presence of selenium supplementation, we showed an additive effect to the inhibition when SEPS1 was overexpressed and selenium was supplemented. We had shown that SEPS1 exerted a direct effect, but the effect of selenium could potentially be through the effect on the selenoprotein, SEPS1, or via 15d-PGJ2. This in vitro work raises the potential of selenium supplementation as a therapeutic option in preventing liver disease in patients affected with Z variant AAT.
Interestingly, the SEPS1 gene is located on chromosome 15q26.3, a region associated with the conformational diseases diabetes and Alzheimer disease. SEPS1 is also a strong functional and positional candidate gene for various inflammation-related disorders, and genetic correlation studies have shown that the proinflammatory cytokines IL-1β, tumor necrosis factor α, and IL-6 may be affected by a gene that jointly influences their expression (31). Curran et al. (31) identified a key single nucleotide polymorphism in the promoter region of SEPS1 at position −105 that showed significant correlation to IL-1β, tumor necrosis factor α, and IL-6 plasma levels and demonstrated overwhelming support for a role of the SEPS1 −105G→A single nucleotide polymorphism as a functional variant for these proinflammatory biomarkers. This may be important in AAT deficiency, given that an aberrant inflammatory response is a probable factor determining the severity and outcome of disease. The SEPS1 promoter is GC-rich and contains one conserved and one potential ER stress response element II (ERSE), a consensus-binding site for transcription factors regulating ER stress responses. ERSEs are involved in the stress inducibility of ER proteins that assist protein folding in the ER and promote cell survival under stress conditions. The SEPS1 −105G→A single nucleotide polymorphism is located in the putative ERSEII. The A allele confers lower promoter activity than the G allele in response to stimulation with tunicamycin. Furthermore, the A allele is associated with higher cytokine levels. Thus, the phenotypes associated with the SEPS1 −105A allele are decreased SEPS1 expression and ERAD function and increased cytokine production.
Many countries in Europe and other parts of the world still have a dietary selenium intake below what is recommended by health regulatory bodies (32, 33). In the United States, where dietary intakes are higher than in many other countries, the current recommendation is 55 μg/day. This level is based on optimizing plasma glutathione peroxidase enzyme activity. Recommendations for intake are higher in European populations, where selenium intake is considerably lower. Suboptimal selenium status may increase susceptibility to various chronic disorders and is likely to lead to suboptimal functioning of the beneficial properties of SEPS1, which we have demonstrated. In our work, we measured serum selenium levels in ZZ patients and compared these with MM phenotypes. There was no difference between the two groups; however, we did find that the serum levels of selenium were in the lower range of normal and were not at the level likely to lead to optimal functioning of selenoproteins. Nonetheless, the sample size tested was very small, and any conclusions should be tested in an independent study with a larger sample size. Considerable variation in serum selenium levels is expected based on geographic location, and specific locations associated with low selenium may be important for individuals with AAT deficiency living there.
In view of these results, we believe that selenium supplementation has potential to be investigated as a therapeutic option for the liver disease associated with AAT deficiency. Taken together, these data point to the important role of selenium in the diet as an essential factor required to enhance the function of selenoproteins. Optimizing SEPS1 activity, particularly in cases where the −105A allele is present, could be of tremendous therapeutic benefit to an individual coping with intracellular accumulation of misfolded proteins, such as ZAAT.
In summary, we found that SEPS1 down-regulated a number of markers of the response to ER stress induced by ZAAT. We demonstrated that supplementation with selenium enhances the effect of this selenoprotein. We also showed effects of the anti-inflammatory selenium through the 15-deoxy-Δ12,14-prostaglandin J2 pathway. We demonstrated the selenium level to be in the low normal range for our population and below the optimal level for function of the antioxidant effect of selenoproteins. The potential therapies for conformational diseases, such as ZAAT deficiency, includes therapies directed at preventing polymerization of the aberrant protein, chemical chaperones, and anti-inflammatories. We have demonstrated how selenium can address two of these pathways. This is an essential trace element that is frequently low in adult populations and can be easily supplemented.
This work was supported by a Pilot and Feasibility grant from the Alpha One Foundation.
- ER
- endoplasmic reticulum
- EOR
- ER overload response
- UPR
- unfolded protein response
- AAT
- α1-antitrypsin
- ZAAT
- Z variant α1-antitrypsin
- GPx
- glutathione peroxidase
- CMV
- cytomegalovirus
- IL
- interleukin
- 15d-PGJ2
- 15-deoxy-Δ12,14-prostaglandin J2
- Sec
- selenocysteine
- ERSE
- ER stress response element II.
REFERENCES
- 1.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. ( 2004) Nature 429, 841– 847 [DOI] [PubMed] [Google Scholar]
- 2.Gao Y., Feng H. C., Walder K., Bolton K., Sunderland T., Bishara N., Quick M., Kantham L., Collier G. R. ( 2004) FEBS Lett. 563, 185– 190 [DOI] [PubMed] [Google Scholar]
- 3.Kim K. H., Gao Y., Walder K., Collier G. R., Skelton J., Kissebah A. H. ( 2007) Biochem. Biophys. Res. Commun. 354, 127– 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ron D., Walter P. ( 2007) Nat. Rev. Mol. Cell Biol. 8, 519– 529 [DOI] [PubMed] [Google Scholar]
- 5.Greene C. M., Miller S. D., Carroll T., McLean C., O'Mahony M., Lawless M. W., O'Neill S. J., Taggart C. C., McElvaney N. G. ( 2008) J. Inherit. Metab. Dis. 31, 21– 34 [DOI] [PubMed] [Google Scholar]
- 6.Brantly M., Nukiwa T., Crystal R. G. ( 1988) Am. J. Med. 84, 13– 31 [DOI] [PubMed] [Google Scholar]
- 7.Sveger T. ( 1976) N. Engl. J. Med. 294, 1316– 1321 [DOI] [PubMed] [Google Scholar]
- 8.Sveger T. ( 1988) Acta Paediatr. Scand. 77, 847– 851 [DOI] [PubMed] [Google Scholar]
- 9.Le A., Ferrell G. A., Dishon D. S., Le Q. Q., Sifers R. N. ( 1992) J. Biol. Chem. 267, 1072– 1080 [PubMed] [Google Scholar]
- 10.Le A., Graham K. S., Sifers R. N. ( 1990) J. Biol. Chem. 265, 14001– 14007 [PubMed] [Google Scholar]
- 11.Welihinda A. A., Tirasophon W., Kaufman R. J. ( 1999) Gene Expr. 7, 293– 300 [PMC free article] [PubMed] [Google Scholar]
- 12.Venkateswaran V., Klotz L. H., Fleshner N. E. ( 2002) Cancer Res. 62, 2540– 2545 [PubMed] [Google Scholar]
- 13.Gao Y., Hannan N. R., Wanyonyi S., Konstantopolous N., Pagnon J., Feng H. C., Jowett J. B., Kim K. H., Walder K., Collier G. R. ( 2006) Cytokine 33, 246– 251 [DOI] [PubMed] [Google Scholar]
- 14.Berridge M. J. ( 2002) Cell Calcium 32, 235– 249 [DOI] [PubMed] [Google Scholar]
- 15.Lawless M. W., Greene C. M., Mulgrew A., Taggart C. C., O'Neill S. J., McElvaney N. G. ( 2004) J. Immunol. 172, 5722– 5726 [DOI] [PubMed] [Google Scholar]
- 16.Ron D. ( 2002) J. Clin. Investig. 110, 1383– 1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendershot L. M. ( 2004) Mt. Sinai J. Med. 71, 289– 297 [PubMed] [Google Scholar]
- 18.Lee A. S. ( 2001) Trends Biochem. Sci. 26, 504– 510 [DOI] [PubMed] [Google Scholar]
- 19.Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. ( 2000) Nat. Cell Biol. 2, 326– 332 [DOI] [PubMed] [Google Scholar]
- 20.Chambers I., Frampton J., Goldfarb P., Affara N., McBain W., Harrison P. R. ( 1986) EMBO J. 5, 1221– 1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellano S., Lobanov A. V., Chapple C., Novoselov S. V., Albrecht M., Hua D., Lescure A., Lengauer T., Krol A., Gladyshev V. N., Guigó R. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 16188– 16193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papp L. V., Lu J., Holmgren A., Khanna K. K. ( 2007) Antioxid. Redox Signal. 9, 775– 806 [DOI] [PubMed] [Google Scholar]
- 23.Walder K., Kantham L., McMillan J. S., Trevaskis J., Kerr L., De Silva A., Sunderland T., Godde N., Gao Y., Bishara N., Windmill K., Tenne-Brown J., Augert G., Zimmet P. Z., Collier G. R. ( 2002) Diabetes 51, 1859– 1866 [DOI] [PubMed] [Google Scholar]
- 24.Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigó R., Gladyshev V. N. ( 2003) Science 300, 1439– 1443 [DOI] [PubMed] [Google Scholar]
- 25.Sunde R. A., Raines A. M., Barnes K. M., Evenson J. K. ( 2008) Biosci. Rep., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullins C. ( 2005) The Biogenesis of Cellular Organelles, pp. 63– 95, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 27.Wu J., Kaufman R. J. ( 2006) Cell Death Differ. 13, 374– 384 [DOI] [PubMed] [Google Scholar]
- 28.Lomas D. A., Evans D. L., Finch J. T., Carrell R. W. ( 1992) Nature 357, 605– 607 [DOI] [PubMed] [Google Scholar]
- 29.Pahl H. L., Baeuerle P. A. ( 1995) EMBO J. 14, 2580– 2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vunta H., Davis F., Palempalli U. D., Bhat D., Arner R. J., Thompson J. T., Peterson D. G., Reddy C. C., Prabhu K. S. ( 2007) J. Biol. Chem. 282, 17964– 17973 [DOI] [PubMed] [Google Scholar]
- 31.Curran J. E., Jowett J. B., Elliott K. S., Gao Y., Gluschenko K., Wang J., Abel Azim D. M., Cai G., Mahaney M. C., Comuzzie A. G., Dyer T. D., Walder K. R., Zimmet P., MacCluer J. W., Collier G. R., Kissebah A. H., Blangero J. ( 2005) Nat. Genet. 37, 1234– 1241 [DOI] [PubMed] [Google Scholar]
- 32.Combs G. F., Jr. ( 2001) Br. J. Nutr. 85, 517– 547 [DOI] [PubMed] [Google Scholar]
- 33.Rayman M. P. ( 2005) Proc. Nutr. Soc. 64, 527– 542 [DOI] [PubMed] [Google Scholar]