Abstract
Recently, we identified Txnip (thioredoxin-interacting protein) as a mediator of glucotoxic beta cell death and discovered that lack of Txnip protects against streptozotocin- and obesity-induced diabetes by preventing beta cell apoptosis and preserving endogenous beta cell mass. Txnip has therefore become an attractive target for diabetes therapy, but although we have found that txnip transcription is highly induced by glucose through a unique carbohydrate response element, the factors controlling this effect have remained unknown. Using transient transfection experiments, we now show that overexpression of the carbohydrate response element-binding protein (ChREBP) transactivates the txnip promoter, whereas ChREBP knockdown by small interfering RNA completely blunts glucose-induced txnip transcription. Moreover, chromatin immunoprecipitation demonstrated that glucose leads to a dose- and time-dependent recruitment of ChREBP to the txnip promoter in vivo in INS-1 beta cells as well as human islets. Furthermore, we found that the co-activator and histone acetyltransferase p300 co-immunoprecipitates with ChREBP and also binds to the txnip promoter in response to glucose. Interestingly, this is associated with specific acetylation of histone H4 and recruitment of RNA polymerase II as measured by chromatin immunoprecipitation. Thus, with this study we have identified ChREBP as the transcription factor that mediates glucose-induced txnip expression in human islets and INS-1 beta cells and have characterized the chromatin modification associated with glucose-induced txnip transcription. In addition, the results reveal for the first time that ChREBP interacts with p300. This may explain how ChREBP induces H4 acetylation and sheds new light on glucose-mediated regulation of chromatin structure and transcription.
Txnip (thioredoxin-interacting protein) is a ubiquitously expressed protein involved in cellular redox control and apoptosis (1–3). In particular, we have previously demonstrated that Txnip levels are significantly up-regulated in pancreatic islets of different mouse models of type 1 and type 2 diabetes and that increased Txnip expression induces pancreatic beta cell apoptosis (4). Loss of pancreatic beta cell mass by apoptosis is a key factor in the pathogenesis of both type 1 and type 2 diabetes (5–8), raising the possibility that Txnip may play an important role in this process.
In fact, using Txnip-deficient mice harboring a naturally occurring nonsense mutation in their txnip gene (HcB-19) as well as our beta cell-specific txnip knock-out mice (bTKO), we discovered that lack of txnip protects against diabetes induced by multiple low dose injections of streptozotocin (a classical model of type 1 diabetes) (9). In addition, Txnip deficiency was also able to rescue severely obese and insulin-resistant BTBRob/ob mice lacking leptin from diabetes, as demonstrated by our double-mutant congenic BTBRlepob/obtxniphcb/hcb (BTBR.ob/hcb) mice (9). BTBRob/ob mice are considered one of the most stringent models of type 2 diabetes, since, unlike B6ob/ob BTBRob/ob mice typically develop severe diabetes already at 6 weeks of age and completely decompensate at around 10 weeks (10). In contrast, Txnip-deficient BTBR.ob/hcb mice remained normoglycemic for over 6 months. Interestingly, the main mechanism responsible for this Txnip-mediated protection against streptozotocin- and obesity-induced diabetes was a dramatic ∼50-fold reduction in beta cell apoptosis and preservation of endogenous beta cell mass (9). In addition, we found that Txnip represents a critical link between glucose toxicity and beta cell death (11). Based on these combined findings, Txnip has become an attractive novel target for diabetes therapy, but still very little is known about its regulation.
Txnip expression is highly induced by glucose in different tissues, and we originally identified it as the top glucose-induced gene in our human islet oligonucleotide microarray study (12). Quantitative RT2-PCR and immunoblotting further confirmed these findings in INS-1 beta cells as well as in isolated mouse islets (4, 11). We further established that this regulation occurs at the transcriptional level, and a detailed promoter deletion analysis revealed that a short proximal promoter region was necessary and sufficient for glucose-induced transactivation (4). More specifically, a unique nonpalindromic 17-bp E-box repeat was found to be highly conserved between species and function as the cis-acting carbohydrate response element (ChoRE) for glucose-induced txnip transcription (4), but the trans-acting factors remained unknown.
The present study was therefore aimed at identifying the transcription factor(s) as well as potentially associated co-activators and chromatin modifications that mediate glucose-induced txnip expression.
EXPERIMENTAL PROCEDURES
Cell Culture
INS-1 and HIT-T15 cells were maintained as described previously (4). INS-1 cells were incubated at 5 mm glucose for 30 h prior to being used for any experiments.
Human islets were obtained through the Islet Cell Resources Basic Science Islet Distribution Program and incubated overnight in RPMI medium containing 5 mm glucose prior to a 24-h incubation at low (1.67 mm) or high (16.7 mm) glucose. (We used these conditions to reproduce the experimental design of our original human islet oligonucleotide microarray and quantitative RT-PCR studies that identified TXNIP as a highly glucose-induced gene (12).)
Transient Transfection Assays and RNA Interference
Carbohydrate response element-binding protein (ChREBP) was cloned from mouse liver, and NeuroD was cloned from human islets using the Elongase amplification system (Invitrogen) and primers 13 and 14 and primers 17 and 18, respectively (Table S1). Amplicons where then subcloned into the pcDNA3.1/V5-His TOPO vector (Invitrogen) under the control of the CMV promoter. All other plasmids have been described previously (4). HIT-T15 cells were grown in 12-well plates and transfected with CMV-ChREBP, CMV-NeuroD, or CMV-LacZ together with the D4 or mut D4 txnip reporter constructs at a 3:1 ratio (0.5 μg of DNA/well) using Fugene 6 (Roche Applied Science) in serum-free RPMI (Invitrogen) containing 2.5 mm glucose. 72 h after transfection, cells were harvested, and luciferase activities were determined by a dual luciferase assay (Promega). The pRL-TK control plasmid (Promega), co-transfected at 5 ng/well, was used to correct for transfection efficiency.
INS-1 cells were grown in 6-well plates and transfected with a txnip promoter-driven luciferase reporter plasmid (FL-TXNIP) or an SV40-driven pGL3 control plasmid (1 μg/ml) and ChREBP-specific small interfering RNA oligonucleotide (siChREBP) (0.1 μm) (RSS301430; Invitrogen) or scrambled oligonucleotides (0.1 μm) (D-001810-01-20; Dharmacon) using DharmaFECT Duo transfection reagent (Dharmacon/Thermo Scientific, Chicago, IL) (5 μl/well) and incubated at low (5 mm) or high (25 mm) glucose prior to analysis by the dual luciferase assay. To confirm ChREBP knockdown, cells transfected with siChREBP or scrambled oligonucleotide and grown at 25 mm glucose were harvested for RNA extraction with the RNeasy minikit (Qiagen, Valencia, CA) and for protein extraction using 150 μl of whole cell extract lysis buffer (11).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as described previously (13). 500 μg of protein (by BCA protein assay kit) were incubated overnight at 4 °C with 4 μg of ChREBP (sc-21189), Mlx (sc-14705), USF2 (sc-862X), RNA polymerase II (Pol II) (sc-899X) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies or specific antibodies against acetylated histone H3 (06-599), acetylated histone H4 (06-866) (Upstate/Millipore, Billerica, MA), lysine 4-trimethylated histone H3 (ab8580; Abcam, Cambridge, MA), or an affinity-purified polyclonal antibody as a negative control (β-galactosidase) (Abcam). Human islets were cross-linked by adding formaldehyde directly to the cell media to a final concentration of 1%. Sonication was performed with a Misonix cup horn sonicator for 10 30-s pulses at output 4. Immunoprecipitation was performed with anti-human ChREBP (sc-21189) or p300 antibodies (sc-584X) (Santa Cruz Biotechnology). Immune complexes were captured with 50 μl of 50% protein A-Sepharose or protein G-agarose slurry. DNA fragments were purified using a Qiagen PCR purification kit and quantified by real time PCR with primers listed in Table S1. Although histone acetylation occurs at the promoter and 5′-coding region, methylation is enriched in the coding region (14–16). Since we only saw modest changes in acetylation in the txnip promoter region (data not shown), we used the 5′-coding region primers 7 and 8 (Table S1) for modified histone ChIP analysis.
Co-immunoprecipitation Studies
Human islet nuclear extracts were prepared as described previously (17). Nuclear proteins (250 μg) were incubated with 2 μg of polyclonal rabbit p300 (sc-584) or goat ChREBP (sc-21189) antibodies in 200 μl of immunoprecipitation buffer (20 mm HEPES, pH 7.9, 0.15 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture) for 4 h at 4 °C followed by precipitation with protein A- or protein G-Sepharose. Pellets were resuspended in NuPAGE LDS electrophoresis sample buffer (Invitrogen) and boiled for 5 min at 95 °C. Samples were analyzed by SDS-PAGE and immunoblotting with polyclonal mouse anti-p300 (1:300; 05-257; Upstate/Millipore), horseradish peroxidase-conjugated rabbit anti-ChREBP (1:500; NB400–135H; Novus Biologicals) or goat anti-Mlx (1:300; sc-14705; Santa Cruz Biotechnology) followed by secondary anti-mouse or anti-goat antibodies (1:5000; Santa Cruz Biotechnology). Bands were visualized by ECL Plus detection reagent (GE Healthcare).
Statistical Analysis
p values were calculated by Student's t test or by one-way analysis of variance for data sets of more than two groups.
RESULTS
The txnip Promoter Contains a Putative ChREBP Binding Site
Glucose-induced txnip transcription depends on a short proximal region of the txnip promoter and especially on a well conserved ChoRE consisting of two E-boxes (4). Analysis of this critical 17-bp ChoRE sequence of the human, rat, and mouse txnip promoter using MatInspector (Genomatix) predicted ChREBP as the only conserved putative trans-acting factor binding to this region (Fig. 1A). In fact, ChREBP has been recognized as the key transcription factor mediating glucose-induced gene expression especially in liver (18). However, although ChREBP belongs to the B group of basic helix-loop-helix transcription factors that recognize E-boxes containing CACGTG or CATGTG sequences, the E-boxes in the txnip promoter are nonpalindromic and consist of a repeat of CACGAG sequences (also referred to as N-boxes (19)).
FIGURE 1.
ChREBP effects on txnip promoter activity. A, conserved ChREBP binding site (gray) in the txnip promoter of mouse, rat, and human. The core sequence of the four highest conserved consecutive positions is shown in capital letters (MatInspector), and E-boxes are shown in boldface type. B, mutation of the core sequence and first E-box. Shown are the effects of ChREBP (C) and NeuroD (D) overexpression on txnip promoter activity, as measured by luciferase activity. Bars, means ± S.E. of three independent experiments performed in duplicates.
ChREBP Overexpression Activates txnip Transcription in Beta Cells
To test experimentally if ChREBP might induce txnip transcription, we performed transfection experiments using luciferase reporter constructs driven by the proximal txnip promoter region containing the intact ChoRE (D4) or a mutation of the first E-box (mut D4) (Fig. 1B) and overexpressing ChREBP or LacZ. As a negative control, we also performed parallel experiments overexpressing NeuroD, which is involved in glucose-induced transcription of the insulin gene. Indeed, our results demonstrated that ChREBP overexpression stimulates transcription from the txnip promoter (Fig. 1C), whereas NeuroD overexpression had no effect (Fig. 1D). Moreover, mutation of the core sequence of the txnip-ChoRE in the mut D4 completely abolished ChREBP-mediated transactivation, indicating that this cis-acting element is critical for the ChREBP effect.
Glucose Induces ChREBP Binding to the txnip-ChoRE Promoter Sequence in Vivo
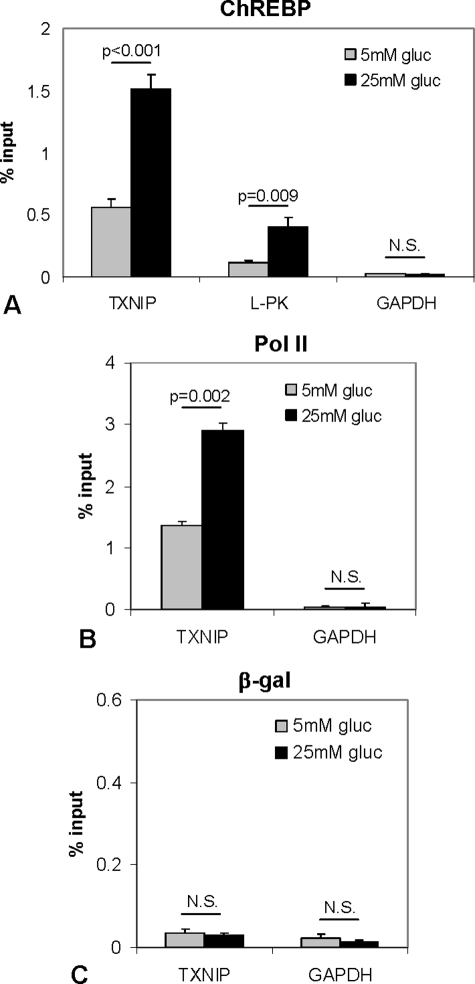
To examine whether ChREBP binds the txnip promoter in vivo, we performed ChIP assays using INS-1 beta cells incubated at low (5 mm) or high (25 mm) glucose for 6 h and utilized PCR primers flanking the txnip-ChoRE. A moderate level of ChREBP binding to the txnip promoter was observed even at low glucose, whereas upon high glucose, ChREBP recruitment to the txnip promoter increased ∼3-fold (Fig. 2A). Very similar effects were also observed with another primer pair (data not shown). ChREBP is known to mediate glucose-induced transactivation of liver-type pyruvate kinase (LPK) in the liver as well as in beta cells (18, 20) and was therefore used as a positive control. Indeed, glucose led to a similar induction of ChREBP binding to the LPK promoter, although the level of ChREBP binding to the txnip promoter was higher overall (Fig. 2A). Moreover, the glucose-induced increase in ChREBP binding to the txnip-ChoRE was also associated with a 2-fold increase in Pol II recruitment to the txnip promoter (Fig. 2B), consistent with the role of ChREBP as a transcriptional activator. In contrast, the IgG/β-galactosidase control showed no enrichment of txnip-ChoRE (Fig. 2C), and binding to the GAPDH internal control was negligible, confirming the specificity of these ChIP assays. In addition, we also studied the role of USF2, another B group basic helix-loop-helix transcription factor that recognizes E-boxes. Although overexpression of USF2 increased txnip promoter activity in transfection experiments and ChIP assays demonstrated some USF2 binding to the txnip promoter in INS-1 beta cells, no increase in USF2 binding was observed in response to glucose (data not shown), suggesting that USF2 is not mediating the observed glucose-induced activation of txnip transcription.
FIGURE 2.
In vivo binding of ChREBP to txnip promoter in INS-1 beta cells by ChIP. INS-1 beta cells were incubated at 5 or 25 mm glucose for 6 h, and ChIP assays were performed using ChREBP (A), Pol II (B), and β-galactosidase (as IgG control) (C) antibodies. The ChoRE regions of rat txnip and LPK as well as the GAPDH coding region were amplified by quantitative real time RT-PCR, and the percentage of bound promoter was calculated. Bars, means ± S.E. of at least three independent ChIP assays.
Glucose-induced ChREBP Binding to the txnip-ChoRE Is Dose-dependent and Follows a Time Course
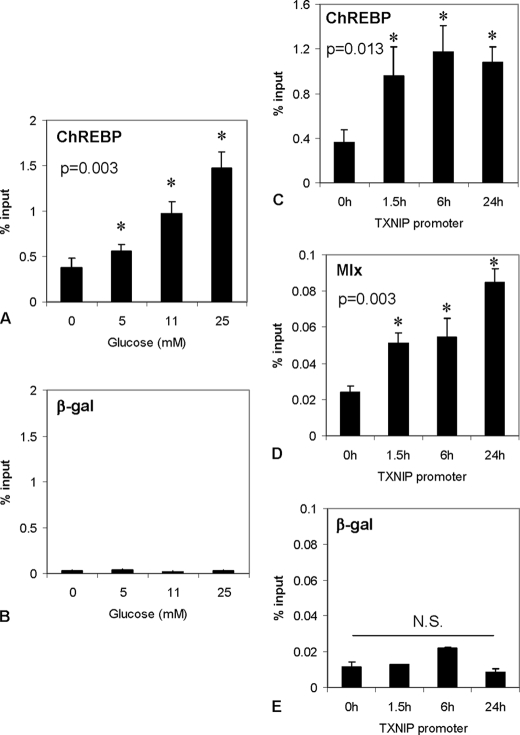
We also performed dose response and time course experiments using INS-1 beta cells treated with increasing concentrations of glucose (0–25 mm) for 6 h or treated with 25 mm glucose for various periods of time (0–24 h). ChREBP binding to txnip-ChoRE increased in a dose-dependent manner, showing the biggest effect at 25 mm glucose (Fig. 3A). Interestingly, we found that ChREBP binding occurs as early as 15 min in response to glucose (Fig. S1) and persists for at least 24 h (Fig. 3C). Since active ChREBP forms heterodimers with Mlx in the nucleus, we again employed ChIP analysis to examine whether there is also Mlx binding to the txnip-ChoRE in vivo. Indeed, Mlx not only bound to the txnip-ChoRE, but its DNA binding increased in a time-dependent manner in response to high glucose (Fig. 3D), further supporting the role of ChREBP/Mlx in glucose-induced txnip transcription.
FIGURE 3.
Dose response and time course of glucose-induced recruitment of ChREBP to txnip promoter. A and B, dose response. INS-1 cells were treated with increasing glucose concentrations (0, 5, 11, and 25 mm) for 6 h prior to analysis by ChIP. C–E, time course. INS-1 cells were incubated at 25 mm glucose for 0, 1.5, 6, and 24 h prior to ChIP. Bars, means ± S.E. of at least three independent ChIP assays.
ChREBP Knockdown Blunts Glucose-induced txnip Transcription in Beta Cells
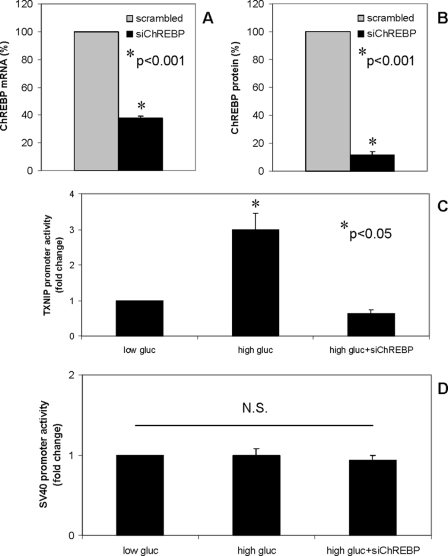
To further confirm that ChREBP is the main regulator of glucose-induced txnip gene expression, we knocked down ChREBP by RNA interference (Fig. 4, A and B). While transfection of INS-1 beta cells with scrambled oligonucleotide did not affect glucose-induced txnip promoter activity, siChREBP completely blunted the glucose effect (Fig. 4C), indicating that ChREBP is critical for glucose-induced txnip transcription. In contrast, siChREBP had no effect on the SV40 promoter activity run as a negative control (Fig. 4D).
FIGURE 4.
Effects of ChREBP knockdown on glucose-induced txnip promoter activity. INS-1 cells were transfected with siChREBP or scrambled oligonucleotide, and successful ChREBP knockdown was confirmed at the mRNA (A) and at the protein level (B) by quantitative RT-PCR and immunoblotting, respectively. INS-1 cells were co-transfected with the txnip promoter-driven luciferase reporter plasmid (C) (or the SV40-driven pGL3 control plasmid (D)) and with scrambled oligonucleotide or siChREBP and incubated at low (5 mm) or high (25 mm) glucose for 24 h. Bars, -fold changes in luciferase activity; means ± S.E. of three independent experiments performed in triplicates.
Beta Cell txnip Gene Expression Is Induced by Histone Deacetylase (HDAC) Inhibition
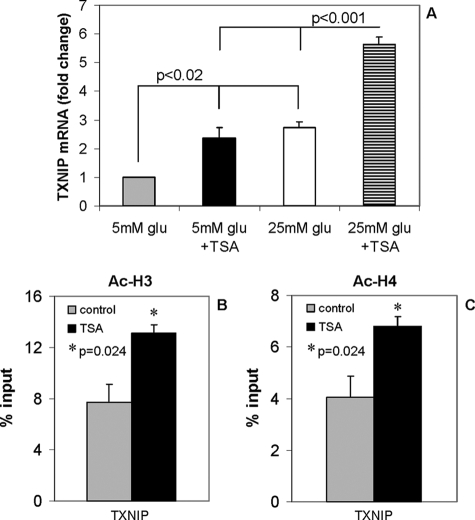
Chromatin modification and, in particular, histone deacetylation by HDACs has been shown to be associated with inhibition of gene transcription. To investigate whether histone deacetylation may play a role in the inhibition of txnip gene expression at low glucose, INS-1 cells incubated at low (5 mm) glucose were treated with the HDAC inhibitor trichostatin A (TSA) (60 nm) (Sigma) or with high (25 mm) glucose with or without TSA for 6 h, and txnip mRNA levels were measured by quantitative RT-PCR. Interestingly, TSA induced txnip gene expression significantly in the presence of low or high glucose (Fig. 5A), suggesting that histone acetylation/deacetylation plays an important role in the control of txnip transcription. Furthermore, this TSA-mediated txnip induction was as potent as the one conferred by high glucose, and together they resulted in an additive effect. To further define the specific histone modifications conferred by TSA, we performed ChIP assays using specific antibodies detecting acetylated H3 and H4 representing common activation marks. The results revealed that TSA induces both H3 and H4 acetylation of the txnip gene (Fig. 5, B and C).
FIGURE 5.
TSA effects on txnip mRNA expression and histone H3/H4 acetylation. A, INS-1 cells were incubated at 5 mm glucose (gray), 5 mm glucose with 60 nm HDAC inhibitor TSA (black), 25 mm glucose (white), or 25 mm glucose with 60 nm of TSA (shaded) for 6 h, and RNA was isolated and analyzed for txnip expression by quantitative RT-PCR. Bars, mean -fold changes ± S.E. of three independent experiments. INS-1 cells were again incubated at 5 mm glucose in the presence or absence of TSA (60 nm) for 6 h, and ChIP assays were performed using antibodies directed against acetylated histone H3 (Ac-H3) (B) and H4 (Ac-H4) (C). Bars, means ± S.E. of at least three independent ChIP experiments.
Glucose Specifically Induces Histone H4 Acetylation of the txnip Gene
To examine the possible role of histone acetylation in glucose-induced txnip gene expression, we performed modified histone ChIP assays using INS-1 beta cells incubated again at low or high glucose for various periods of time, focusing specifically on H4 and H3, which are frequently hyperacetylated in actively transcribed genes. In addition, we investigated H3 lysine 4 trimethylation as another classical chromatin activation mark. Interestingly, glucose led to a significant increase in H4 acetylation of the txnip gene (Fig. 6A), whereas H3 acetylation (Fig. 6B) and H3 lysine 4 trimethylation (Fig. 6C) or dimethylation (data not shown) were not affected by glucose.
FIGURE 6.
Glucose-induced histone modification of the txnip gene. INS-1 cells were treated with 25 mm glucose for different times, as indicated in the figure. ChIP assays were performed using antibodies directed against acetylated histone H4 (Ac-H4) (A), H3 (Ac-H3) (B), and lysine 4-trimethylated H3 (H3-K4me3) (C). Bars, means ± S.E. of at least three independent ChIP experiments.
Glucose Induces ChREBP and p300 Recruitment to the TXNIP Gene Promoter in Human Islets
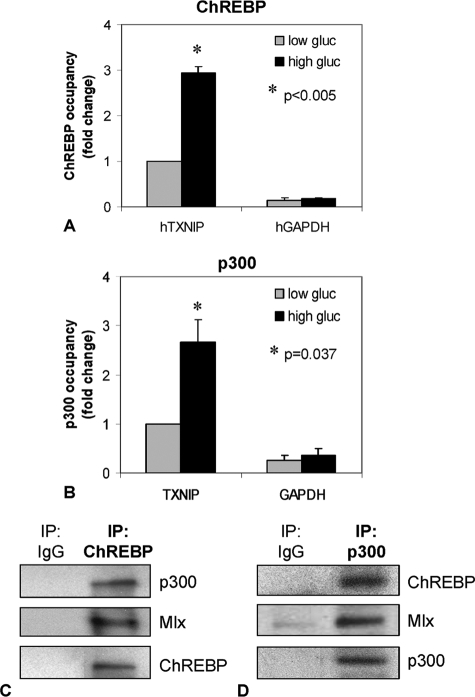
To further confirm our findings in human islets, ChIP analysis was performed using isolated human islets incubated again at low or high glucose. Similar to our findings in INS-1 beta cells, we observed a significant 3-fold increase in ChREBP binding to the human TXNIP promoter in response to high glucose (Fig. 7A).
FIGURE 7.
ChREBP and p300 interaction and recruitment to the txnip promoter in human islets. Human islets were incubated at low or high glucose for 24 h, and ChIP assays were performed using antibodies directed against ChREBP (A) and p300 (B). Bars, means ± S.E. of at least three independent ChIP assays using islets of different donors, whereby each donor served as its own control. Co-immunoprecipitation experiments were performed using nuclear extracts of human islets incubated for 24 h at high glucose and immunoprecipitating with ChREBP (C) or p300 (D) and immunoblotting for p300, ChREBP, and Mlx. One representative of at least three independent experiments is shown.
To further study potential cofactors involved in this glucose-induced activation of TXNIP transcription, we also examined the possible role of p300. The transcriptional co-activator and histone acetyltransferase (HAT) p300 has been shown to regulate the expression of other glucose-responsive genes, such as insulin and fibronectin (21–23), and to physically interact with other E-box-binding basic helix-loop-helix proteins, such as NeuroD (21), USF2 (24), MyoD (25), SREBP (26), and Myc (27). Moreover, p300 is one of the major HATs (in addition to HAT1 and TIP 60) that acetylate H4. As a co-activator, p300 directly associates with Pol II (28–31), suggesting that it could confer the Pol II recruitment observed in our studies. Taken together with our observation that glucose induces txnip H4 acetylation, p300 seemed to represent an attractive candidate. Indeed, ChIP analysis again using isolated human islets revealed that p300 was recruited to the txnip promoter at high glucose (Fig. 7B).
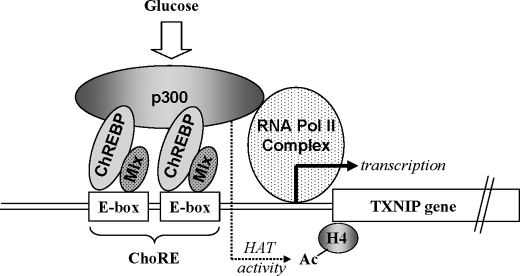
Furthermore, using nuclear extracts of human islets incubated at high glucose and co-immunoprecipitation experiments, we were able to demonstrate that p300 physically interacts with ChREBP/Mlx (Fig. 7C). Together, these findings suggest that glucose exposure of beta cells leads within minutes to binding of a complex containing ChREBP/Mlx and p300 to the txnip promoter, which results in H4 acetylation, opening of the chromatin structure, recruitment of Pol II, and activation of transcription (Fig. 8).
FIGURE 8.
Schematic of glucose-induced transactivating complex formation on the txnip promoter. Glucose induces the binding of ChREBP/Mlx heterodimers to the ChoRE, which in turn recruits p300 to the promoter via direct (or indirect) physical interaction between ChREBP and p300. The recruited p300 stimulates acetylation of histone H4 through its HAT activity and promotes Pol II promoter occupancy, resulting in increased txnip transcription.
DISCUSSION
Increased beta cell Txnip expression, especially induced by high glucose, plays a major role in the pathogenesis of pancreatic beta cell loss and diabetes. In this study, we have now identified ChREBP as the critical transcription factor, p300 as the co-activator/HAT, and H4 acetylation as the specific chromatin modification mediating this glucose-induced transcription of beta cell txnip.
Glucose-regulated transcription has been shown to be often mediated by ChoREs consisting of a palindromic E-box repeat, such as CACGTG, present in the LPK-ChoRE (18, 32). However, in the case of txnip, the ChoRE is not composed of a canonical E-box sequence but rather of two nonpalindromic (CACGAG) E-boxes or N-boxes. Nevertheless, this txnip-ChoRE was able to confer strong transactivation induced by ChREBP, as shown in our transfection experiments (Fig. 1), and appears to have a high affinity for ChREBP, as demonstrated by in vivo ChREBP binding in our ChIP experiments using INS-1 beta cells as well as human islets (Figs. 2, 3, and 7). Some ChREBP binding to the txnip promoter was even observed at low glucose, but upon stimulation with high glucose, ChREBP binding increased 3-fold, which was also associated with a 2-fold increase in Pol II occupancy. These results are consistent with the finding that ChREBP shuttling from the cytoplasm to the nucleus occurs at both low and high glucose in beta cells but that high glucose leads to a 3-fold increase in the rate of its nuclear entry (33) and results in the required dephosphorylation steps (34). Glucose-induced ChREBP recruitment displayed a dose- and time-dependent pattern, and the very rapid increase of ChREBP binding to the txnip promoter observed within 15 min of exposure to high glucose (Fig. S1) further suggests a direct effect and is consistent with the known role of txnip as an early response gene and its rapid changes in expression (35).
Other E-box-binding proteins, such as NeuroD, were not able to mimic the ChREBP effects (Fig. 1D), and although USF2 may be involved in general modulation of txnip transcription (data not shown), ChIP analysis failed to reveal any increase in USF2 recruitment to the txnip promoter in response to glucose, suggesting that USF2 is not mediating the glucose effects. This is consistent with previous finding in regard to the regulation of LPK in beta cells (20). In contrast, Mlx, the heterodimeric ChREBP partner, also bound to the txnip promoter, and its binding activity was increased in response to high glucose (Fig. 3D). Mlx appears to be required for glucose-activated transcription, since overexpression of a dominant-negative form of Mlx in hepatocytes abolished glucose-induced txnip mRNA expression (36). However, since Mlx is known to lack intrinsic transcriptional activity (37, 38), it might exert its function via stabilizing the binding of two ChREBP/Mlx heterodimers to the ChoRE (39).
In HA1ER cells, a cell line derived from normal human renal epithelium, MondoA, a paralog of ChREBP, has recently been reported to regulate glucose-stimulated txnip gene expression (40). However, as mentioned by the authors, this role of MondoA/Mlx seems to be tissue-specific and restricted to skeletal muscle and the particular cell lines used. In this regard, it is also surprising that the subcellular localization of MondoA was not regulated by glucose in either primary human skeletal muscle cells or in C2C12 myoblasts (41). On the other hand, even important components of the core transcription machinery were recently found to be different in skeletal muscle as compared with other tissues (42, 43), which may help to explain some of these tissue-specific differences in the mechanisms mediating glucose-induced txnip expression. Indeed, overexpression of ChREBP, but not MondoA, rescued LPK luciferase reporter expression when a dominant-negative form of Mlx was expressed in primary hepatocytes, suggesting that ChREBP is the critical transcription factor in liver (36). Furthermore, a very recent study using transient reporter gene transfection also demonstrated that ChREBP regulates glucose-induced txnip transcription in human prostate carcinoma cells. In regard to pancreatic beta cells, our knockdown experiments using ChREBP small interfering RNA demonstrated that glucose-induced txnip transcription was completely abolished (Fig. 4), proving that ChREBP is essential for glucose-activated txnip transcription in beta cells. Together with our ChIP findings in INS-1 beta cells as well as human islets demonstrating for the first time in vivo binding of ChREBP to the txnip promoter, these results provide strong evidence for ChREBP/Mlx being the key regulatory complex of glucose-stimulated txnip gene expression in pancreatic beta cells.
Although ChREBP has been studied extensively as a mediator of glucose-induced transcription of glycolytic and lipogenic genes, especially in the liver (18), our current results suggest that the effects of ChREBP may reach far beyond regulation of hepatic enzymes and also include control of prodiabetic Txnip and pancreatic beta cells as a major site of action.
In this regard, our observation of ChREBP-p300 interaction is of particular interest (Fig. 7). The transcriptional co-activator p300 also functions as a HAT for H3/H4 and has been shown to interact with other E-box-binding proteins (21, 24–27) and to be involved in the regulation of glucose-regulated genes (21–23). Its interaction with ChREBP may therefore not only explain how ChREBP induces H4 acetylation and recruitment of Pol II to the txnip promoter but may also be key for the molecular understanding of the regulation of other ChREBP target genes and their potential chromatin modification and transactivation.
The importance of chromatin modification as a powerful mechanism of transcriptional control has gained more recognition over recent years, and especially histone acetylation of H3 and/or H4 has been found to effectively open the chromatin structure, thereby enabling Pol II binding and transcriptional activation (44). The balance of histone acetylation/deacetylation is tightly regulated by a variety of HATs and HDACs. Interestingly, although glucose-induced LPK expression has been found to be associated with H4 as well as H3 acetylation (45) and the same was true for TSA-induced txnip expression (Fig. 5, B and C), our data indicate that glucose specifically induces H4 but not H3 acetylation of the txnip gene (Fig. 6). These findings suggests that glucose and HDAC inhibitors mediate their effects on txnip transcription at least in part by independent molecular pathways, which is consistent with their additive effects that we observed on txnip mRNA expression (Fig. 5A). Although TSA is obviously only an experimental stimulus, our findings of increased txnip expression in response to this HDAC inhibitor suggest that histone deacetylation by HDACs might play an important physiological role in the inhibition of txnip gene expression at low glucose.
In summary, the results of the present study provide new insight into the molecular mechanisms governing pancreatic beta cell txnip expression in particular and glucose-regulated gene transcription and chromatin modification in general. Since Txnip has emerged as a novel target for diabetes therapy, these findings are of special relevance and reveal new approaches to specifically inhibit glucose-induced txnip transcription. This may help to interrupt the vicious cycle of glucose inducing beta cell Txnip expression, resulting in beta cell apoptosis, worsening of the hyperglycemia, and diabetes progression.
Supplementary Material
Acknowledgments
We thank the Islet Cell Resources Basic Science Islet Distribution Program for providing the isolated human islets used in this study.
This work was supported, in whole or in part, by National Institutes of Health, NIDDK, Grant R01DK-078752 and National Institutes of Health, NHLBI, Grant R21HL-089205. This work was also supported by American Diabetes Association Grant 7-07-CD-22 and Juvenile Diabetes Research Foundation Grant 1-2007-790.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
Footnotes
- RT
- reverse transcription
- ChORE
- carbohydrate response element
- ChREBP
- carbohydrate response element-binding protein
- CMV
- cytomegalovirus
- siChREBP
- ChREBP-specific small interfering RNA oligonucleotide
- ChIP
- chromatin immunoprecipitation
- Pol II
- polymerase II
- TSA
- trichostatin A
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- LPK
- liver-type pyruvate kinase.
REFERENCES
- 1.Junn E., Han S. H., Im J. Y., Yang Y., Cho E. W., Um H. D., Kim D. K., Lee K. W., Han P. L., Rhee S. G., Choi I. ( 2000) J. Immunol. 164, 6287– 6295 [DOI] [PubMed] [Google Scholar]
- 2.Nishiyama A., Masutani H., Nakamura H., Nishinaka Y., Yodoi J. ( 2001) IUBMB Life 52, 29– 33 [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., De Keulenaer G. W., Lee R. T. ( 2002) J. Biol. Chem. 277, 26496– 26500 [DOI] [PubMed] [Google Scholar]
- 4.Minn A. H., Hafele C., Shalev A. ( 2005) Endocrinology 146, 2397– 2405 [DOI] [PubMed] [Google Scholar]
- 5.Eizirik D. L., Kutlu B., Rasschaert J., Darville M., Cardozo A. K. ( 2003) Ann. N.Y. Acad. Sci. 1005, 55– 74 [DOI] [PubMed] [Google Scholar]
- 6.Poitout V., Robertson R. P. ( 2002) Endocrinology 143, 339– 342 [DOI] [PubMed] [Google Scholar]
- 7.Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., Butler P. C. ( 2003) Diabetes 52, 102– 110 [DOI] [PubMed] [Google Scholar]
- 8.Rhodes C. J. ( 2005) Science 307, 380– 384 [DOI] [PubMed] [Google Scholar]
- 9.Chen J., Hui S. T., Couto F. M., Mungrue I. N., Davis D. B., Attie A. D., Lusis A. J., Davis R. A., Shalev A. ( 2008) FASEB J. 22, 3581– 3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan H., Rabaglia M. E., Stoehr J. P., Nadler S. T., Schueler K. L., Zou F., Yandell B. S., Attie A. D. ( 2003) Diabetes 52, 688– 700 [DOI] [PubMed] [Google Scholar]
- 11.Chen J., Saxena G., Mungrue I. N., Lusis A. J., Shalev A. ( 2008) Diabetes 57, 938– 944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalev A., Pise-Masison C. A., Radonovich M., Hoffmann S. C., Hirshberg B., Brady J. N., Harlan D. M. ( 2002) Endocrinology 143, 3695– 3698 [DOI] [PubMed] [Google Scholar]
- 13.Cha-Molstad H., Keller D. M., Yochum G. S., Impey S., Goodman R. H. ( 2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13572– 13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurdistani S. K., Grunstein M. ( 2003) Nat. Rev. Mol. Cell Biol. 4, 276– 284 [DOI] [PubMed] [Google Scholar]
- 15.Espinoza C. R., Feeney A. J. ( 2005) J. Immunol. 175, 6668– 6675 [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T. ( 2007) Cell 128, 693– 705 [DOI] [PubMed] [Google Scholar]
- 17.Schreiber E., Matthias P., Müller M. M., Schaffner W. ( 1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita H., Takenoshita M., Sakurai M., Bruick R. K., Henzel W. J., Shillinglaw W., Arnot D., Uyeda K. ( 2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9116– 9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akazawa C., Sasai Y., Nakanishi S., Kageyama R. ( 1992) J. Biol. Chem. 267, 21879– 21885 [PubMed] [Google Scholar]
- 20.Wang H., Wollheim C. B. ( 2002) J. Biol. Chem. 277, 32746– 32752 [DOI] [PubMed] [Google Scholar]
- 21.Qiu Y., Guo M., Huang S., Stein R. ( 2002) Mol. Cell. Biol. 22, 412– 420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosley A. L., Corbett J. A., Ozcan S. ( 2004) Mol. Endocrinol. 18, 2279– 2290 [DOI] [PubMed] [Google Scholar]
- 23.Kaur H., Chen S., Xin X., Chiu J., Khan Z. A., Chakrabarti S. ( 2006) Diabetes 55, 3104– 3111 [DOI] [PubMed] [Google Scholar]
- 24.Breen G. A., Jordan E. M. ( 1999) Biochim. Biophys. Acta 1428, 169– 176 [DOI] [PubMed] [Google Scholar]
- 25.Eckner R., Yao T. P., Oldread E., Livingston D. M. ( 1996) Genes Dev. 10, 2478– 2490 [DOI] [PubMed] [Google Scholar]
- 26.Yang F., Vought B. W., Satterlee J. S., Walker A. K., Jim Sun Z. Y., Watts J. L., DeBeaumont R., Saito R. M., Hyberts S. G., Yang S., Macol C., Iyer L., Tjian R., van den Heuvel S., Hart A. C., Wagner G., Näär A. M. ( 2006) Nature 442, 700– 704 [DOI] [PubMed] [Google Scholar]
- 27.Faiola F., Liu X., Lo S., Pan S., Zhang K., Lymar E., Farina A., Martinez E. ( 2005) Mol. Cell. Biol. 25, 10220– 10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kee B. L., Arias J., Montminy M. R. ( 1996) J. Biol. Chem. 271, 2373– 2375 [DOI] [PubMed] [Google Scholar]
- 29.Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. ( 2000) Cell 103, 667– 678 [DOI] [PubMed] [Google Scholar]
- 30.Kim M. Y., Hsiao S. J., Kraus W. L. ( 2001) EMBO J. 20, 6084– 6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosma M. P. ( 2002) Mol. Cell 10, 227– 236 [DOI] [PubMed] [Google Scholar]
- 32.Rufo C., Teran-Garcia M., Nakamura M. T., Koo S. H., Towle H. C., Clarke S. D. ( 2001) J. Biol. Chem. 276, 21969– 21975 [DOI] [PubMed] [Google Scholar]
- 33.Davies M. N., O'Callaghan B. L., Towle H. C. ( 2008) J. Biol. Chem. 283, 24029– 24038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawaguchi T., Takenoshita M., Kabashima T., Uyeda K. ( 2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13710– 13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitoh T., Tanaka S., Koike T. ( 2001) J. Neurochem. 78, 1267– 1276 [DOI] [PubMed] [Google Scholar]
- 36.Ma L., Tsatsos N. G., Towle H. C. ( 2005) J. Biol. Chem. 280, 12019– 12027 [DOI] [PubMed] [Google Scholar]
- 37.Billin A. N., Eilers A. L., Coulter K. L., Logan J. S., Ayer D. E. ( 2000) Mol. Cell. Biol. 20, 8845– 8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meroni G., Cairo S., Merla G., Messali S., Brent R., Ballabio A., Reymond A. ( 2000) Oncogene 19, 3266– 3277 [DOI] [PubMed] [Google Scholar]
- 39.Ma L., Sham Y. Y., Walters K. J., Towle H. C. ( 2007) Nucleic Acids Res. 35, 35– 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoltzman C. A., Peterson C. W., Breen K. T., Muoio D. M., Billin A. N., Ayer D. E. ( 2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6912– 6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sans C. L., Satterwhite D. J., Stoltzman C. A., Breen K. T., Ayer D. E. ( 2006) Mol. Cell. Biol. 26, 4863– 4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deato M. D., Marr M. T., Sottero T., Inouye C., Hu P., Tjian R. ( 2008) Mol. Cell 32, 96– 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deato M. D., Tjian R. ( 2007) Genes Dev. 21, 2137– 2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grunstein M. ( 1997) Nature 389, 349– 352 [DOI] [PubMed] [Google Scholar]
- 45.Eckert D. T., Zhang P., Collier J. J., O'Doherty R. M., Scott D. K. ( 2008) Biochem. Biophys. Res. Commun. 372, 131– 136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.