Abstract
NSC-741909 is a recently identified novel anticancer agent that suppresses the growth of several NCI-60 cancer cell lines with a unique anticancer spectrum. However, its molecular mechanisms remain unknown. To determine the molecular mechanisms of NSC-741909-induced antitumor activity, we analyzed the changes of 77 protein biomarkers in a sensitive lung cancer cell line after treatment with this compound by using reverse-phase protein microarray. The results showed that phosphorylation of mitogen-activated protein (MAP) kinases (P38 MAPK, ERK, and JNK) were persistently elevated by the treatment with NSC-741909. However, only the JNK-specific inhibitor SP600125 effectively blocked the apoptosis induced by NSC-741909. Moreover, NSC-741909-mediated apoptosis was also blocked by a dominant-negative JNK construct, suggesting that sustained activation of JNK is critical for the apoptosis induction. Further studies revealed that treatment with NSC-741909 suppressed dephosphorylation of JNK and the expression of MAPK phosphatase-1. Thus, NSC-741909-mediated inhibition of JNK dephosphorylation results in sustained JNK activation, which leads to apoptosis in cancer cells.
Because of genetic and epigenetic changes in cancer cells, it is possible to identify tumor-selective cytotoxic agents by synthetic lethality screening for compounds that kill isogenic cancer cells but not their normal counterparts (1). The term synthetic lethality was originally used to describe a lethal phenotype caused by mutations of two genes (2), i.e. mutations of the two genes are lethal if they occur together but viable if they occur separately. A synthetically lethal phenotype often indicates that the two genes or two related pathways affect a common essential biologic function. Unfortunately, our current knowledge of molecular networks in normal or cancer cells is not adequate for us to predict what genes are synthetically lethal partners to an oncogene or a mutated tumor suppressor gene. Nevertheless, synthetic lethality screening allows us to identify cytotoxic agents specific for certain cancer cells because a compound targeting to such a partner can be identified by their lethality when administered to cancer cells with elevated activities of a particular oncogene.
Using synthetic lethality screening, we recently identified an indole compound (designated oncrasin-1) that kills immortalized and tumorigenic human ovarian epithelial cells expressing mutant K-Ras but not cells expressing wild-type Ras genes (3). Furthermore, this compound effectively induced apoptosis at low micromolar or nanomolar concentrations in a variety of lung cancer cells with K-Ras mutations but did not kill cells with wild-type Ras genes. Molecular characterization revealed that oncrasin-1 can induce abnormal aggregation of protein kinase C-ι in the nucleus of oncrasin-sensitive cells but not in oncrasin-resistant cells and that oncrasin-1-induced apoptosis was blocked by siRNA3 of K-Ras or protein kinase C-ι (3), demonstrating that oncrasin-1 is synthetically lethal for K-Ras and protein kinase C-ι, one of the downstream effectors of Ras signaling pathways (4). Our search for oncrasin-1 analogues identified several active compounds with similar chemical structures. Testing of one of the oncrasin-1 analogues, oncrasin-60 (NSC-741909), on NCI-60 cancer cell lines showed that it is highly active against several cell lines derived from lung, colon, breast, ovary, and kidney cancers and that it lies outside the category of adequately studied classes of antitumor agents, suggesting that those compounds could be novel anticancer agents. However, the mechanisms of apoptosis induction by oncrasin compounds remain to be characterized. Here, we used reverse-phase protein array to determine molecular changes induced by NSC-741909 in a sensitive cell line. Our results indicated that sustained c-Jun N-terminal protein kinase (JNK) activation caused by suppression of JNK dephosphorylation contributes to NSC-741909-induced apoptosis.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
The human non-small cell lung carcinoma H460 and H157 cell lines were routinely grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 mg/ml penicillin-streptomycin (all from Invitrogen). Cells were cultured at 37 °C in a humidified incubator containing 5% CO2. We also used human ovarian surface epithelial cells immortalized with the catalytic subunit of human telomerase reverse transcriptase and the SV40 early genomic region (designated T29) and its tumorigenic derivatives transformed with mutant K-Ras (T29Kt1) (5). The culture conditions were the same as above.
Chemicals and Antibodies
NSC-741909 was synthesized by Zhejiang Yuancheng MST Inc. (Hangzhou, China). The purity of this compound as determined by high performance liquid chromatography-mass spectrum analysis is 98.5%. The chemical structure was confirmed by nuclear magnetic resonance spectrum analysis. U0126, SB203580, and JNK inhibitor II (SP600125) were purchased from Calbiochem. Antibodies to the following proteins were used for Western blot analysis: JNK, phospho-JNK, phospho-c-Jun, P38, phospho-P38, phospho-ERK, phospho-MKK7 (Cell Signaling), MKP1 (c-19) (Santa Cruz Biotechnology), poly(ADP-ribose) polymerase (BD Pharmingen), caspase-8 (ALEXIS Biochemicals), β-actin, and hemagglutinin (HA) (Sigma).
Reverse-phase Protein Array
The cells were washed twice in ice-cold phosphate-buffered saline (PBS) and then lysed in reverse-phase protein microarrays (RPPA) lysis buffer (1% Triton X-100, 50 mmol/liter HEPES (pH 7.4), 150 mmol/liter NaCl, 1.5 mmol/liter MgCl2, 1 mmol/liter EGTA, 100 mmol/liter NaF, 10 mmol/liter NaPPi, 10% glycerol, 1 mmol/liter Na3VO4, 1 mmol/liter phenylmethylsulfonyl fluoride, and aprotinin 10 μg/ml; alternatively, whole proteinase inhibitor tablets could be used (Roche Applied Science)) for 30 min with frequent vortexing on ice. The resultant solution was centrifuged for 15 min at 14,000 rpm, the supernatant was collected, and the protein concentration was determined by bicinchoninic acid protein assay kit (Thermo Scientific) assays and then adjusted to 1–1.5 mg/ml by lysis buffer. Cell lysates were mixed with one-quarter volume of 4× SDS sample buffer containing 40% glycerol, 8% SDS, 0.25 m Tris-HCl, pH 6.8, and 10% (v/v) 2-mercaptoethanol (freshly added).
RPPA assay was performed at the Functional Proteomics Reverse-Phase Protein Array Core facility at our institution. Serial diluted cell lysates were printed on nitrocellulose-coated slides (Whatman, Inc) by using a G3 arrayer (Genomic Solutions). A total of 960 array spots could be arranged on each slide, including 96 spots corresponding to positive and negative controls prepared from dilution buffer. A total of 80–90 slides were made for each sample. Each slide was probed with a validated primary antibody plus a biotin-conjugated secondary antibody. The signal was amplified using a DakoCytomation-catalyzed system (Dako) and visualized by a 3,3′-diaminobenzidine tetrahydrochloride colorimetric reaction. The slides were scanned, analyzed, and quantified using customized software, Microvigene (VigeneTech, Inc.), to generate spot intensity. Signals from each dilution were fitted with the logistic model developed by the Department of Bioinformatics and Computational Biology at the M. D. Anderson Cancer Center. The protein concentrations of each set of slides were then normalized and corrected across samples by the linear expression values, using the median expression levels of all antibody experiments to calculate a loading correction factor for each sample.
Cell Viability Assay
Cells were seeded at a density of 1 × 104 cells/well in 96-well plates. After overnight incubation, the cells were treated with NSC-741909 (0.03–10 μm) alone or in combination with different compounds for 24 h. The inhibitory effects of NSC-741909 alone or in combination with other agents on cell growth were determined using the sulforhodamine B assay, as described previously (6). Each experiment was performed in quadruplicate and repeated for a total of at least three times.
Flow Cytometric Assay
The flow cytometric assay was performed as described previously (7). In brief, cells were seeded at a density of 2.5 × 105 cells/well in six-well plates and allowed to grow overnight. The cells were treated with NSC-741909 (1 μm) alone or in combination with different inhibitors for 24 h. The inhibitors were added 30 min before NSC-741909. After treatment, cells were harvested, washed with PBS, and fixed by 70% ethanol overnight at 4 °C. Before analysis, cells were stained with propidium iodide for 30 min. Then the flow cytometric assay was used to measure the sub-G0/G1 cellular DNA content using Cell Quest software (BD Biosciences). The flow cytometric assays were performed in the Core Laboratory of the M. D. Anderson Cancer Center.
Western Blot
Cells were harvested and subjected to lysis in Laemmli lysis buffer. Equal amounts of lysates (40 μg) were separated by 10% SDS-PAGE and then transferred to Hybond-enhanced chemiluminescence membranes (Amersham Biosciences). Membranes were then blocked with PBS buffer containing 5% low fat milk and 0.05% Tween (PBST) for 1 h and then incubated with primary antibodies overnight at 4 °C. After being washed three times with PBST, membranes were incubated with peroxidase-conjugated secondary antibodies for 1 h at room temperature. The membranes were washed with PBST again and developed with a chemiluminescence detection kit (ECL kit; Amersham Biosciences). β-Actin was used as a loading control.
Plasmid and siRNA Transfection
H157 cells were transfected with the plasmid pLNCX-3X/HA-p46JNK1α (dnJNK1 (dominant-negative JNK1 mutant construct)) (kindly provided by Dr. L. E. Heasley, University of Colorado Health Sciences Center, Denver, CO) (8), which encoded an HA-tagged dominant-negative JNK1 mutant whose phosphorylation site, Thr-Pro-Tyr (TPY), is altered to Ala-Pro-Phe (APF). The retroviral vector encoding dnJNK1 was then prepared from 293/Phoenix cells and used to infect target cells. Stable transfectants were selected for growth in the presence of 500 μg/ml Geneticin (G418). Plasmid or siRNA transfection was performed with FuGENE6 reagent (Roche Diagnostics) or ON-TARGETplus siRNA reagents (Dharmacon).
Total JNK Phosphatase Activity
Total JNK phosphatase activity was determined as reported earlier (9). Briefly, cells were homogenized in 50 mm Tris-HCl (pH 7.4), 250 mm NaCl, 3 mm EDTA, 3 mm EGTA, 1% Triton X-100, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and proteinase inhibitor complex. After 20 min of centrifugation at 16,000 × g at 4 °C, JNK1/2 were removed from the supernatants by immunoprecipitation, and the immunodepleted lysates were mixed with phosphorylated JNK1/2 isolated from H460 cells 4 h after treatment with 1 μm NSC-741909 at 37 °C. The reactions were terminated by the addition of 2% SDS and separated by SDS-PAGE. Dephosphorylation of activated JNK was then detected with an anti-phospho-JNK antibody.
Real-time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). A 500-ng aliquot of each RNA sample was reverse-transcribed in a 20-μl reaction volume using the Taqman reverse transcription reagents (Applied Biosystems). The 10-fold dilutions of the cDNA product were used in real-time PCR. Real-time PCR was carried out in 20 μl of a reaction mixture containing 2 μl of diluted cDNA, 10 μl of 2× Absolute Blue QPCR SYBR Green mix buffer (Thermo Fisher Scientific), 5.2 μl of double-distilled water, and 1.4 μl each of sense and antisense primers (70 nm final concentration). Real-time PCR assays were performed in triplicate using a CRF96TM real-time system (Bio-Rad) with the following conditions: 95 °C for 15 min, 40 cycles at 95 °C for 10 s, 58 °C for 15 s, and 72 °C for 30 s. The following primer sequences for the genes were used: MKP1, sense 5′-CCAGTACAAGAGCATCCCTGT-3′, and antisense 5′- AGTGGACAAACACCCTTCCTC-3′; GAPDH, sense 5′- GGCTCTCCAGAACATCATCC-3′, and antisense 5′-TAGCCCAGGATGCCCTT-3′. The sets of gene primer for the target gene MKP1 were confirmed to have amplification efficiency equal to that of the reference gene GAPDH. The relative RNA expression was calculated automatically by the installed software of the instrument with the ΔΔCt method, using GAPDH as a reference gene.
Data Analysis
Differences between treatment groups were assessed using the unpaired Student's t test at a significance level of p < 0.05.
RESULTS
Antitumor Activity of NSC-741909 in NCI-60 Cancer Cell Lines
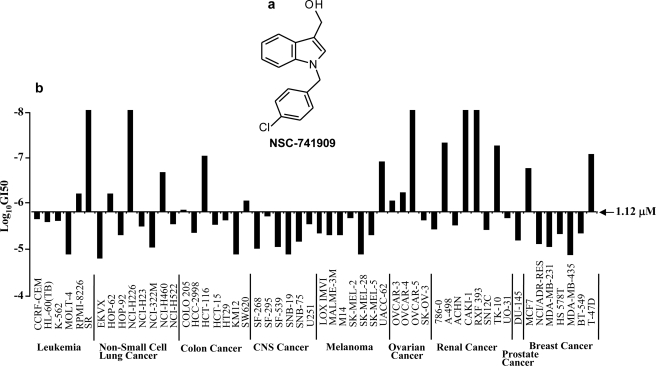
After we identified oncrasin-1 through synthetic lethality screening (3), we tested a number of analogues with similar chemical structure and found that several were very active in T29Kt1 and H460 cells that harbor mutant K-Ras but were inactive in T29 cells with wild-type Ras genes. We submitted a few highly active compounds, including NSC-741909 (oncrasin-60, Fig. 1a), to the NCI Developmental Therapeutics Program (National Institutes of Health) for testing their effect on a panel of 60 cancer cell lines derived from various tissues and organs. The test performed at the NCI (National Institutes of Health) showed that NSC-741909 was active against a number of cancer cell lines derived from non-small-cell lung cancer, colon cancer, melanoma, ovarian cancer, renal cancer, and breast cancer. Among 54 of the cancer cell lines tested, the median 50% growth inhibitory concentration (GI50) for NSC-741909 was 1.12 μm. For five of the most sensitive cell lines, the GI50 was <10−8 m or 10 nm, the lowest concentration tested by the NCI (National Institutes of Health) (Fig. 1b). The analysis performed by the Developmental Therapeutics Program at the NCI (National Institutes of Health) also showed that NSC-741909 lies outside the category of adequately studied classes of antitumor agents, suggesting that it has novel anticancer mechanisms. This compound is currently under preclinical evaluation for its in vivo efficacy and toxicity by the Rapid Access to Intervention Development (RAID) Program at the NCI (National Institutes of Health).
FIGURE 1.
Antitumor activity of NSC-741909. a, the structure of NSC-741909. b, antitumor activity of NSC-741909 in NCI-60 cancer cell lines. NSC-741909 was submitted to the NCI Developmental Therapeutics Program (National Institutes of Health) for testing its effect on a panel of 60 cancer cell lines derived from various tissues and organs. The 50% growth inhibitory concentrations (GI50) at logarithmic scale were calculated and are shown in the bar graph. The middle line represents the median 50% growth inhibitory concentration for NSC-741909 (i.e. −5.95 for the logarithmic concentration). CNS Cancer, central nervous system cancer.
Sustained Activation of MAPK Signaling Pathways by NSC-741909
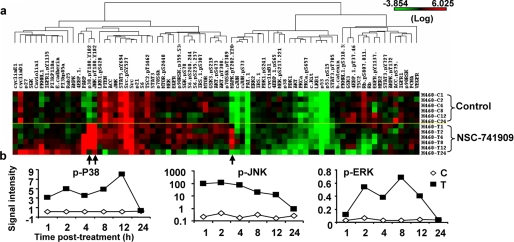
To characterize molecular changes induced by NSC-741909, we treated H460 cells with 1 μm NSC-741909 (a concentration around IC80) and harvested the cells at various time points after the treatment; cells treated with solvent alone (DMSO) were used as controls. The samples were then immobilized in RPPA, and each array was tested with a validated antibody specific to a particular protein or a particular post-translational modification of a protein. Seventy-seven antibodies specific for proteins or their post-translational modifications that are involved in various signaling pathways were used in the RPPA analysis (Fig. 2a). The result showed that treatment with NSC-741909 dramatically increased the signal intensity of phosphorylated P38 (p38.pT180.Y182), phosphorylated JNK (JNK.pT180.Y182), and phosphorylated ERK (MAPK.pT202.Y204) when compared with results for control cells. The increase of those phosphorylation signals occurred as early as 1 h after treatment and were as high as >100-fold for JNK (Fig. 2b). In comparison, the signal changes for other proteins or post-translational modifications were insignificant.
FIGURE 2.
Changes of protein detected by RPPA. a, heat map of protein levels detected by RPPA. The biomarkers, a protein or a phosphorylated isotype detected with specific antibodies, are arrayed on the horizontal axis, and the control and NSC-741909-treated samples are arrayed on the vertical axis. The levels of each biomarker are presented with colors, with red for highest and green for lowest. b, the time-dependent changes of phosphorylated P38 (p-P38), JNK (p-JNK), and ERK (p-ERK). The values represent signal intensity detected at each time point as indicated. C, control sample; T, NSC-741909-treated sample.
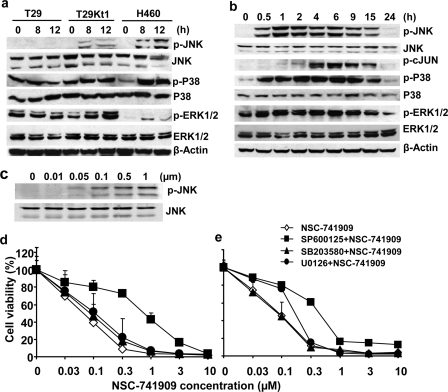
To verify the results of the RPPA assay, we treated two oncrasin-sensitive cell lines, H460 (1 μm) and T29Kt1 (10 μm, also around IC80 for this cell line), and one oncrasin-resistant cell line, T29 (10 μm), with NSC-741909 and harvested the cells at 0, 8, and 12 h after the treatment. Western blot analysis showed that treatment with NSC-741909 induced dramatic up-regulation of phospho-JNK, phospho-P38, and phospho-ERK at 8 and 12 h after treatment in the two sensitive cell lines but not in the resistant cell line (Fig. 3a). We then evaluated the time-dependent changes of these signal molecule alterations after NSC-741909 treatment in H460 cells. The results showed that the phosphorylations of JNK, P38 MAPK, and p44/p42 ERK were induced as early as 30 min after NSC-741909 treatment, whereas the expression of basal JNK or basal P38 MAPK was not affected (Fig. 3b). The elevation of phosphorylated JNK, P38, and ERK lasted for at least 15 h after treatment (Fig. 3b). We also evaluated the dose response of JNK phosphorylation after treatment with NSC-741909 for 6 h. The result showed that induction of JNK phosphorylation by NSC-741909 was dose-dependent and occurred at the dose of 50 nm (Fig. 3c). Moreover, there is no apparent cell viability change(s) or apoptosis induction at 6 h after treatment with all the concentrations tested (up to 1 μm, data not shown). Together, these results suggest that treatment with NSC-741909 induced activation of JNK, which occurred before cell growth suppression and apoptosis induction.
FIGURE 3.
NSC-741909-induced activation of MAP kinases. a, cells were treated with NSC-741909 (1 μm for H460; 10 μm for T29 and T29Kt1) for 0, 8, and 12 h. The whole-cell lysates were subjected to Western blot analysis to detect basal and phosphorylated JNK (p-JNK), ERK (p-ERK1/2), and P38 (p-P38). b, H460 cells were treated with 1 μm NSC-741909 for the indicated times. Cell lysates were then subjected to Western blot analysis. β-Actin was used as the loading control. c, H460 cells were treated for 6 h with the indicated concentrations of NSC-741909. Levels of phospho-JNK and JNK were detected by Western blot analysis. d and e, H460 cells (d) and H157 cells (e) were treated with different concentrations of NSC-741909 in the presence or absence of 10 μm SP600125 (JNK-specific inhibitor), 20 μm U0126 (MEK-specific inhibitor), or 20 μm SB203580 (P38 inhibitor). Cell viability was determined at 24 h after treatment. Cells treated with DMSO were used as a control, with viability set at 100%. Each data point represents the means ± S.D. of three independent experiments.
JNK Inhibitor but Not P38 or ERK Inhibitor Diminished NSC-741909-induced Cell Killing
To investigate whether any MAP kinases activated by NSC-741909 were involved in NSC-741909-mediated cell killing, we evaluated combination effects of NSC-741909 with kinase inhibitors for JNK (SP600125, also called JNK inhibitor II), P38 MAPK (SB203580), and MEK (U0126). We treated the sensitive human lung cancer cell lines H460 (Fig. 3d) and H157 (Fig. 3e) with 0.03–10 μm NSC-741909 alone or in combination with different inhibitors for 24 h. Cell viability was then determined by using the sulforhodamine B assay. The results showed that the presence of the JNK inhibitor SP600125 (10 μm) significantly blocked the cell death induced by NSC-741909 (1 μm) in both H460 and H157 cell lines. The IC50 for NSC-741909 in the presence of SP600125 was about 10-fold greater than that for NSC-741909 alone. The MEK-specific inhibitor U0126 (20 μm) and the P38 inhibitor SB203580 (20 μm) had either no effects or only minor effects on NSC-741909-induced suppression of cell growth. These data suggest that JNK activation was critical in NSC-741909-mediated cell killing.
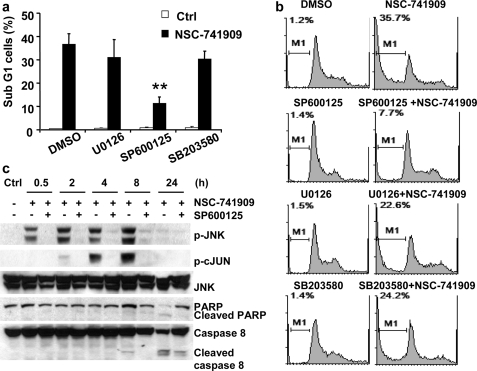
To further evaluate the role of MAPK activation in NSC-741909-mediated cell killing, we tested the effects of three MAP kinase inhibitors on apoptosis induction by NSC-741909. H460 cells were treated with 1 μm NSC-741909 for 24 h, and the percentage of apoptotic cells was determined by flow cytometric analysis. The result showed that treatment with NSC-741909 led to a significantly greater proportion of cells in sub-G1 phase when compared with cells treated with solvent (Fig. 4a). Treatment of H460 cells with SP600125 (10 μm), U0126 (20 μm), or SB203580 (20 μm) alone did not affect the cell growth cycle or induce apoptosis. The presence of SP600125 (10 μm) markedly diminished NSC-741909-induced apoptosis. The proportion of apoptotic cells was 35.7% in the cells treated with NSC-741909 alone but only 7.7% in the cells treated with NSC-741909 plus SP600125. In contrast, the presence of either U0126 or SB203580 had only a minor effect on NSC-741909-induced apoptosis (Fig. 4b).
FIGURE 4.
Apoptosis induction by NSC-741909. Cells were treated 1 μm NSC-741909 in the presence or absence of 10 μm SP600125, 20 μm U0126, or 20 μm SB203580. Cells treated with DMSO or the MAP kinase inhibitors alone were used as controls (Ctrl). Apoptotic cells were determined by flow cytometric analysis at 24 h after treatment. a, the percentages of apoptotic cells. The values represent the means + S.D. of three analyses. **, p < 0.01, when compared with cells treated with NSC-741909 alone. b, an example of fluorescence-activated cell sorter histograms. c, H460 cells were treated with 1 μm NSC-741909 in the presence or absence of 10 μm SP600125. Whole-cell lysates were harvested for Western blot analysis of JNK activation and apoptosis at the indicated time points. p-JNK, phospho-JNK; p-cJUN, phospho-c-Jun; PARP, poly(ADP-ribose) polymerase.
The effect of SP600125 on NSC-741909-induced apoptosis was further verified by Western blot analysis. The presence of SP600125 (10 μm) markedly blocked the phosphorylation of JNK and c-Jun induced by NSC-741909 (1 μm) but had no obvious effects on the basal JNK level (Fig. 4c), suggesting that SP600125 (10 μm) was sufficient to block activation of JNK by NSC-741909. Cleavages of poly(ADP-ribose) polymerase and caspase-8 after treatment with NSC-741909 were also attenuated in the presence of SP600125 (Fig. 4c). Together, these results indicate that JNK activation contributes to NSC-741909-mediated apoptosis, although NSC-741909 activated all three MAPK signaling pathways.
Dominant-negative JNK-1 Suppressed Cell Killing and Apoptosis Induced by NSC-741909
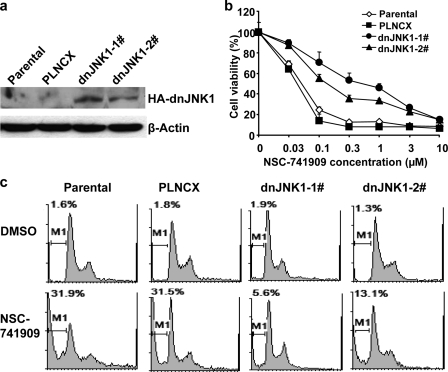
To further evaluate the role of JNK activation in NSC-741909-induced apoptosis, we determined cell growth suppression and apoptosis induction by NSC-741909 in cells of the human lung cancer cell line H157 stably transfected with a HA-tagged dnJNK1. H157 cells were transfected with a plasmid expressing dnJNK1 or with a control plasmid. After selection with G418, H157 clones expressing dnJNK1 were identified by Western blot analysis with anti-HA antibody (Fig. 5a). Parental, vector-transfected, or dnJNK1-transfected H157 cells were then analyzed for their susceptibility to NSC-741909. Dose-response analysis showed that H157 cells expressing dnJNK1 had a dramatic shift of their dose-response curves to the right (Fig. 5b). The IC50 for NSC-741909 in two dnJNK1 stably transfected H157 clones increased about 10-fold when compared with parental and vector-transfected cells. Similarly, flow cytometric analysis indicated that the introduction of dnJNK1 to H157 protected cells from NSC-741909-induced apoptosis (Fig. 5c) when compared with parental H157 cells or H157 infected with control vector.
FIGURE 5.
Effect of dominant-negative JNK-1 on NSC-741909-induced cell killing. H157 cells were transfected with a plasmid expressing dnJNK1 or a control vector. After selection with G418, two H157 clones expressing dnJNK1 were identified by Western blot analysis with anti-HA (an epitope tag from influenza hemagglutinin) antibody. a, parental, vector-transfected, or dnJNK1-transfected H157 cells were then analyzed for their susceptibility to NSC-741909. b, dose response to NSC-741909 as determined by the sulforhodamine B assay. Values represent the means ± S.D. from three independent experiments. c, apoptosis induction as determined by the flow cytometric assay. Cells were treated with 1 μm NSC-741909, and apoptosis was determined at 24 h after treatment. The numbers indicate the proportions of apoptotic cells.
NSC-741909 Suppressed JNK Dephosphorylation
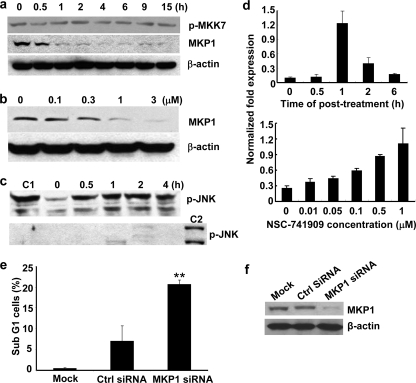
JNKs are activated by dual phosphorylation on the Thr-Pro-Tyr motif in the activation loop through mitogen-activated protein kinase kinase 4 (MKK4) and 7 (MKK7) and inactivated by dephosphorylation through a group of MAP kinase phosphatases (10, 11). To investigate whether NSC-741909-induced JNK activation is caused by elevated activity of upstream kinases or by decreased activity of MAP kinase phosphatases, we tested the changes of phospho-MKK4, phospho-MKK7, and MAP kinase phosphatase 1 (MKP1) in H460 cells after treatment with NSC-741909. The activity of MKK7 is regulated by phosphorylation of Ser/Thr in a conserved region of the kinase domain through its upstream kinases such as MAPK kinase kinase (MAP3K) MEK kinase 1 (MEKK1) and the Ser/Thr kinase STE20-like protein kinases (12). H460 cells treated with 1 μm NSC-741909 were harvested at various time points. Western blot analysis showed that treatment with NSC-741909 had no obvious effect on phosphorylation of MKK7 (Ser-271/Thr-275). Analysis on phospho-MKK4 was not informative because no signal was detected in all samples. In contrast, treatment with NSC-741909 resulted in a dramatic reduction of MKP1, which correlated with the time course of JNK activation (Fig. 6a). The NSC-741909-induced suppression of MKP1 is also dose-dependent when evaluated at doses ranging from 0.1 to 3 μm. Suppression of MKP1 was detectable at 0.1 μm and became more obvious at 0.3–3 μm, in a dose-dependent manner (Fig. 6b).
FIGURE 6.
Effects of NSC-741909 on MAP kinase phosphatase and JNK dephosphorylation. a and b, H460 cells were treated with 1 μm NSC-741909 for the indicated times (a) or treated for 1 h with the indicated concentrations of NSC-741909 (b). Levels of phospho-MKK7 (p-MKK7) (Ser-271/Thr-275) and MKP1 were detected by Western blot analysis. c, lysates from cells treated as in A were depleted of active JNK and then incubated with equal amount of active JNK at 37 °C for 30 min (top panel). The levels of remaining phospho-JNK (p-JNK) were detected by Western blot analysis. Active JNK added to a boiled cell lysate was used as a control for the initial amount of active JNK added (C1). The efficiency of JNK depletion in the cell lysates before adding active JNK was shown in the bottom panel. The cell lysate from H460 treated with 1 μm NSC-741909 for 4 h was used as a positive control (C2). d, H460 cells were treated with 1 μm NSC-741909 for the indicated times (top panel) or treated for 1 h with the indicated concentrations of NSC-741909 (bottom panel). Levels of MKP1 mRNA were detected by real-time PCR. The values were normalized with GAPDH mRNA levels. e, mock, control siRNA (Ctrl siRNA), or MKP1 siRNA-treated H460 cells were analyzed for cell apoptosis at 72 h after treatment. The values represent the means + S.D. of three analyses. **, p < 0.01, when compared with cells treated with control siRNA. f, Western blot analysis for MKP1 expression after knock-down by siRNA transfection.
To test whether the NSC-741909-induced decrease of MKP1 level was caused by inhibition of transcription, we tested the changes of MKP1 mRNA after NSC-741909 treatment with real-time PCR. The results showed that NSC-741909 induced the increase of MKP1 mRNA expression in both a time-dependent and a dose-dependent manner (Fig. 6d). The peak occurred at 1 h after treatment, which had a 5–10-fold increase when compared with DMSO-treated control. This result indicates that NSC-741909 may suppress MKP1 expression at the post-transcriptional level. Increased MKP1 mRNA expression might reflect a negative feedback to its protein level decrease caused by NSC-741909.
Reduced MKP1 protein levels by NSC-741909 suggested that increased JNK phosphorylation may be caused by suppression of JNK dephosphorylation. To investigate whether JNK dephosphorylation was inhibited by NSC-741909 treatment, we incubated activated JNK with active JNK-depleted lysates of untreated and NSC-741909-treated cells. Activated JNK was isolated by immunoprecipitation from H460 cells treated with 1 μm NSC-741909 for 4 h. Similarly, we depleted active JNK from the lysates of untreated and NSC-741909-treated cells by immunoprecipitation. The cell lysates after depletion were divided into two equal parts. Half of them were used for verification of the depletion efficiency, and the other half were incubated with an equal amount of activated JNK in a phosphatase buffer. Dephosphorylation of activated JNK was then detected with an anti-phospho-JNK antibody. The phospho-JNK was dramatically reduced when incubated with lysates from control cells but remained unchanged when incubated with lysates from cells treated with NSC-741909 for 0.5–4 h (Fig. 6c). Together, the above results demonstrated that NSC-741909-mediated sustained JNK activation is caused by the inhibition of JNK dephosphorylation.
We also tested whether inhibition of MKP1 expression itself is sufficient to induce apoptosis. For this purpose, H460 cells were treated with MKP1 siRNA or control siRNA. Apoptosis was determined at 72 h after the treatment. The result showed that transfection with MKP1 siRNA led to a dramatic increase of apoptotic (sub-G1) cells when compared with mock-treated or control siRNA-treated cells The proportion of apoptotic cells was 20.7% in the MKP1 siRNA-treated cells but only 0.45% in mock-treated and 6.97% in the control siRNA-treated cells (Fig. 6, e and f), suggesting that suppression of MKP1 is sufficient to induce apoptosis in some cancer cells.
DISCUSSION
In the current study, we characterized the antitumor activity mechanism of NSC-741909, an analogue of oncrasin-1 that we recently identified as inducing synthetic lethality with oncogenic K-Ras and protein kinase C-ι (3). Tests on NCI-60 cancer cell lines found that NSC-741909 was highly active against several cell lines derived from lung, colon, breast, ovary, and kidney cancers. Here, we used RPPA to detect molecular changes induced by NSC-741909 treatment. The advantage of RPPA is that a single test probe (antibody) is used for each array so that the testing condition will be consistent for each antibody, thereby providing better reproducibility and sensitivity when compared with other protein array techniques. Our results showed that treatment with NSC-741909 induced activation of MAP kinases, including JNK, P38, and ERK, that activation of JNK contributed to NSC-741909-mediated apoptosis, and that pan-activation of MAP kinases is likely caused by suppression of MAP kinase phosphatases.
It is not clear whether Ras signaling pathways facilitate NSC-741909-induced activation of MAP kinases. Nevertheless, MAP kinases are known downstream effectors of Ras signaling pathways. Among these pathways, the Ras/Raf/MEK/ERK pathway is the most well characterized in Ras-induced proliferation and transformation (13). Indeed, a compound named erastin was recently reported to induce non-apoptotic cell death in oncogenic Ras-transfected cells through a mechanism involved in the Ras-Raf-Mek pathway (14). Nevertheless, we have evaluated the effects of oncrasin compounds on the Ras-Raf-Mek pathway and did not find a direct association with this pathway. It is noteworthy that, in addition to the Ras-Raf-Mek pathway, many other signaling pathways were also regulated by Ras, including phosphatidylinositol 3-kinase and guanine nucleotide exchange factors for the Ral family of GTPases. Phosphatidylinositol 3-kinase has been implicated in activation of the small GTPase Rac (15). Interestingly, Rac functions as an activator of a number of the MAP kinases, including JNK (16, 17).
Evidence has also indicated that JNK and Jun proteins complexed with Ras proteins and mediated Ras signaling transductions (18). JNK was originally identified as oncogenic Ras and UV light-responsive protein kinase that binds and phosphorylates the c-Jun activation domain (19, 20). The Ras proteins can directly bind to JNK and c-Jun and regulate their function in a dose-dependent manner (18). Studies of the effect of Jnk gene disruption on Ras-induced transformation of murine fibroblasts indicate that JNK may act as a suppressor of Ras transformation and that the JNK signaling pathway contributes to the apoptotic elimination of Ras-transformed cells in vivo (22, 23). In fact, agents such as capsaicin and anisomycin can selectively induce apoptosis in Ras-transformed cells but not in their normal cell counterparts through JNK-dependent pathway (24, 25). Those results collectively suggested that JNK signaling pathways can also be explored for induction of apoptosis in Ras transformed cells.
JNKs are a group of MAP kinases that play crucial roles in several physiological processes, including proliferation, apoptosis, and differentiation (10). JNKs are activated by dual phosphorylation on the Thr-Pro-Tyr motif in the activation loop through MKK4 and MKK7. Dephosphorylation by a group of MAP kinase phosphatases ultimately leads to inactivation of JNK (10, 11). The biological consequences of JNK activation may depend on the cell type, the nature of the stimulus, the duration of JNK activation, and the activity of other signaling pathways. Evidences have found that JNK activation is required for apoptosis induced by various stress stimuli (11, 26–29); however, most forms of environmental stress that are sufficient for JNK activation do not cause apoptosis (10), suggesting that JNK activation is necessary but not sufficient for apoptosis. Moreover, evidence suggests either that a JNK-dependent pathway is required for TNF-α-induced apoptosis (26) or that JNK activation is not involved in the induction of apoptosis by TNF-α (30). These inconsistencies are presumably due to the difference in cellular contexts and duration of JNK activation. TNF-α-induced JNK activation was transient in TNF-α-resistant human breast carcinoma MCF-7-R cells but was prolonged in TNF-α-sensitive MCF-7 cells. Restoration of transient JNK activation in the MCF-7 cells blocked TNF-α-induced apoptosis (31), indicating that prolonged JNK activation is crucial for the proapoptotic function of JNK.
Our data showed that NSC-741909-mediated increase of phospho-JNK is primarily caused by suppression of JNK dephosphorylation but not by the activation of its upstream kinases. Reduced expression of MKP1 may contribute to NSC-741909-mediated JNK dephosphorylation. However, involvement of other MAP kinase phosphatases is also possible. As a group of dual specificity phosphatases, MAPK phosphatases can effectively dephosphorylate both phospho-tyrosine and phospho-threonine residues on MAPKs (32). Suppression of MAPK phosphatase expressions or their activities is expected to break the balances between the phosphorylating kinases and dephosphorylating phosphatases, thereby dramatically altering MAPK signaling activities. Interestingly, a combination of computational and experimental analysis had revealed that MAPK phosphatases, but not the kinases, dictate the extent of MAPK phosphorylation (21). Because treatment with NSC-741909 also activated P38 and ERK, it is possible that several of the MAPK phosphatases were suppressed by NSC-741909. Together, our results suggested that NSC-741909, as a novel antitumor agent, has selective cell killing activity in a subgroup of cancer cell lines and that sustained JNK activation due to inhibition of JNK dephosphorylation is one of primary effects of the anticancer activity of NSC-741909.
Acknowledgments
We thank Michael Worley for editorial review of the manuscript, the Developmental Therapeutics Program of the NCI (National Institutes of Health) for testing on NCI-60 cancer cell panels, and Henry Peng and Ji Wang for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA 092487 (to B. F.) through the NCI. This work was also supported by Lockton Grant matching funds and by Cancer Center Support Grant CA 3P30CA016672 (J. Mendelsohn).
- siRNA
- small interfering RNA
- JNK
- c-Jun N-terminal kinase
- MAP
- mitogen-activated protein
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- MEK
- MAPK/ERK kinase
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- TNF
- tumor necrosis factor
- DMSO
- dimethyl sulfoxide
- dn
- dominant-negative.
REFERENCES
- 1.Kaelin W. G., Jr. ( 2005) Nat. Rev. Cancer 5, 689– 698 [DOI] [PubMed] [Google Scholar]
- 2.Dobzhansky T. H. ( 1946) Genetics 31, 269– 290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo W., Wu S., Liu J., Fang B. ( 2008) Cancer Res. 68, 7403– 7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray N. R., Jamieson L., Yu W., Zhang J., Gökmen-Polar Y., Sier D., Anastasiadis P., Gatalica Z., Thompson E. A., Fields A. P. ( 2004) J. Cell Biol. 164, 797– 802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Yang G., Thompson-Lanza J. A., Glassman A., Hayes K., Patterson A., Marquez R. T., Auersperg N., Yu Y., Hahn W. C., Mills G. B., Bast R. C., Jr. ( 2004) Cancer Res. 64, 1655– 1663 [DOI] [PubMed] [Google Scholar]
- 6.Rubinstein L. V., Shoemaker R. H., Paull K. D., Simon R. M., Tosini S., Skehan P., Scudiero D. A., Monks A., Boyd M. R. ( 1990) J. Natl. Cancer Inst. 82, 1113– 1118 [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Gu J., Lin T., Huang X., Roth J. A., Fang B. ( 2002) Gene Ther. 9, 1262– 1270 [DOI] [PubMed] [Google Scholar]
- 8.Wojtaszek P. A., Heasley L. E., Siriwardana G., Berl T. ( 1998) J. Biol. Chem. 273, 800– 804 [DOI] [PubMed] [Google Scholar]
- 9.Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. ( 2005) Cell 120, 649– 661 [DOI] [PubMed] [Google Scholar]
- 10.Davis R. J. ( 2000) Cell 103, 239– 252 [DOI] [PubMed] [Google Scholar]
- 11.Lin A. ( 2003) BioEssays 25, 17– 24 [DOI] [PubMed] [Google Scholar]
- 12.Tournier C., Whitmarsh A. J., Cavanagh J., Barrett T., Davis R. J. ( 1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7337– 7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joneson T., Bar-Sagi D. ( 1997) J. Mol. Med. 75, 587– 593 [DOI] [PubMed] [Google Scholar]
- 14.Yagoda N., von Rechenberg M., Zaganjor E., Bauer A. J., Yang W. S., Fridman D. J., Wolpaw A. J., Smukste I., Peltier J. M., Boniface J. J., Smith R., Lessnick S. L., Sahasrabudhe S., Stockwell B. R. ( 2007) Nature 447, 864– 868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Viciana P., Warne P. H., Khwaja A., Marte B. M., Pappin D., Das P., Waterfield M. D., Ridley A., Downward J. ( 1997) Cell 89, 457– 467 [DOI] [PubMed] [Google Scholar]
- 16.Coso O. A., Chiariello M., Yu J. C., Teramoto H., Crespo P., Xu N., Miki T., Gutkind J. S. ( 1995) Cell 81, 1137– 1146 [DOI] [PubMed] [Google Scholar]
- 17.Minden A., Lin A., Claret F. X., Abo A., Karin M. ( 1995) Cell 81, 1147– 1157 [DOI] [PubMed] [Google Scholar]
- 18.Adler V., Pincus M. R., Brandt-Rauf P. W., Ronai Z. ( 1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10585– 10589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibi M., Lin A., Smeal T., Minden A., Karin M. ( 1993) Genes Dev. 7, 2135– 2148 [DOI] [PubMed] [Google Scholar]
- 20.Dérijard B., Hibi M., Wu I. H., Barrett T., Su B., Deng T., Karin M., Davis R. J. ( 1994) Cell 76, 1025– 1037 [DOI] [PubMed] [Google Scholar]
- 21.Bhalla U. S., Ram P. T., Iyengar R. ( 2002) Science 297, 1018– 1023 [DOI] [PubMed] [Google Scholar]
- 22.Kennedy N. J., Davis R. J. ( 2003) Cell Cycle 2, 199– 201 [PubMed] [Google Scholar]
- 23.Kennedy N. J., Sluss H. K., Jones S. N., Bar-Sagi D., Flavell R. A., Davis R. J. ( 2003) Genes Dev. 17, 629– 637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang H. J., Soh Y., Kim M. S., Lee E. J., Surh Y. J., Kim H. R., Kim S. H., Moon A. ( 2003) Int. J. Cancer 103, 475– 482 [DOI] [PubMed] [Google Scholar]
- 25.Santibañez J. F., Hurtado C. ( 2005) FEBS Lett. 579, 6459– 6464 [DOI] [PubMed] [Google Scholar]
- 26.Deng Y., Ren X., Yang L., Lin Y., Wu X. ( 2003) Cell 115, 61– 70 [DOI] [PubMed] [Google Scholar]
- 27.Tournier C., Hess P., Yang D. D., Xu J., Turner T. K., Nimnual A., Bar-Sagi D., Jones S. N., Flavell R. A., Davis R. J. ( 2000) Science 288, 870– 874 [DOI] [PubMed] [Google Scholar]
- 28.Verheij M., Bose R., Lin X. H., Yao B., Jarvis W. D., Grant S., Birrer M. J., Szabo E., Zon L. I., Kyriakis J. M., Haimovitz-Friedman A., Fuks Z., Kolesnick R. N. ( 1996) Nature 380, 75– 79 [DOI] [PubMed] [Google Scholar]
- 29.Xia Z., Dickens M., Raingeaud J., Davis R. J., Greenberg M. E. ( 1995) Science 270, 1326– 1331 [DOI] [PubMed] [Google Scholar]
- 30.Liu Z. G., Hsu H., Goeddel D. V., Karin M. ( 1996) Cell 87, 565– 576 [DOI] [PubMed] [Google Scholar]
- 31.Tang F., Tang G., Xiang J., Dai Q., Rosner M. R., Lin A. ( 2002) Mol. Cell. Biol. 22, 8571– 8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeffrey K. L., Camps M., Rommel C., Mackay C. R. ( 2007) Nat. Rev. Drug Discov. 6, 391– 403 [DOI] [PubMed] [Google Scholar]