Abstract
Chloroplasts are the sites of photosynthesis in plants, and they contain their own multicopy, requisite genome. Chloroplasts are also major sites for production of reactive oxygen species, which can damage essential components of the chloroplast, including the chloroplast genome. Compared with mitochondria in animals, relatively little is known about the potential to repair oxidative DNA damage in chloroplasts. Here we provide evidence of DNA glycosylase-lyase/endonuclease activity involved in base excision repair of oxidized pyrimidines in chloroplast protein extracts of Arabidopsis thaliana. Three base excision repair components (two endonuclease III homologs and an apurinic/apyrimidinic endonuclease) that might account for this activity were identified by bioinformatics. Transient expression of protein-green fluorescent protein fusions showed that all three are targeted to the chloroplast and co-localized with chloroplast DNA in nucleoids. The glycosylase-lyase/endonuclease activity of one of the endonuclease III homologs, AtNTH2, which had not previously been characterized, was confirmed in vitro. T-DNA insertions in each of these genes were identified, and the physiological and biochemical phenotypes of the single, double, and triple mutants were analyzed. This mutant analysis revealed the presence of a third glycosylase activity and potentially another pathway for repair of oxidative DNA damage in chloroplasts.
Reactive oxygen species (ROS)2 are inevitable by-products of metabolism in all aerobic organisms (1). Plants and algae are especially prone to photo-oxidative stress because of ROS generated during oxygenic photosynthesis. Several types of ROS are generated at various sites in the photosynthetic electron transport chain in chloroplasts, and their production is enhanced by such factors as excess or varying light intensities and extremes of temperature, drought, nutrient deficiencies, and herbicides (2). These ROS can damage many chloroplast constituents, including lipids, proteins, pigments, and the multicopy genome.
Plants have evolved numerous mechanisms to deal with photo-oxidative stress, including dissipation of excess light energy, synthesis of antioxidant molecules and scavenging enzymes, and targeted repair (2). DNA repair of oxidized bases, such as thymine glycol (TG) or 8-oxoguanine, can be hypothesized as an important element of chloroplast photoprotection. Although there is considerable overlap in both the types of DNA lesions caused by different insults and the targeting of different DNA repair mechanisms, base excision repair (BER) is considered to be the main repair pathway for oxidative DNA damage, at least in the nucleus and mitochondrion (3, 4).
BER repairs single damaged bases (because of oxidation, deamination, alkylation, etc.) in DNA by removing them, breaking the phosphodiester backbone, excising the sugar residue at the abasic site, and filling the gap (reviewed in Refs. 5, 6). BER begins with a DNA glycosylase or glycosylase-lyase. There are many types of glycosylases in any given organism and across taxa, and they are distinguishable by their substrate specificity, whether they are monofunctional (glycosylase activity only) or bifunctional (glycosylase plus apurinic/apyrimidinic (AP) lyase activities; see below), by the phylogenetic family in which they reside, and/or by conserved structural characteristics (reviewed in Refs. 6–8). The glycosylases involved in BER of oxidative DNA damage can be roughly divided into those that target either oxidized purines or oxidized pyrimidines (4, 9). For example, TG is a common type of oxidized pyrimidine, which is removed primarily by endonuclease III (Nth), endonuclease VIII (Nei), or their homologs (10). TG is only poorly mutagenic, but it strongly blocks polymerases, inducing cell cycle arrest and potentially cell death if it is not removed.
After an appropriate glycosylase cleaves the N-glycosyl bond attaching a damaged base to deoxyribose, leaving an abasic site, the sugar-phosphate backbone is nicked. Bifunctional glycosylases also have an AP lyase activity that cleaves on the 3′ side of the AP site. However, the site still requires the function of a separate AP endonuclease that cuts on the 5′ side of the AP site to remove the 3′-deoxyribose residue at the nick site (11) before repair can continue. In the case of a monofunctional glycosylase, an AP endonuclease nicks the strand on the 5′ side of the AP site. Escherichia coli has two unrelated AP endonucleases, exonuclease III (Xth) and endonuclease IV (Nfo). In humans Ape1/Ref-1 is an Xth homolog, and in yeast Apn1p is an Nfo homolog (5, 12). Following generation of the AP site and nicking of the backbone, the gap is filled by a polymerase in either a short or long patch and then sealed by a ligase.
BER of oxidative DNA lesions such TG has been studied intensively in E. coli, yeast, and mammals, whereas comparatively little is known about BER in plants. For example, only two genes involved in BER of oxidized pyrimidines have been characterized previously in the model plant Arabidopsis thaliana (13, 14), and their localization within the plant cell is unknown. An Nth homolog in Arabidopsis, AtNTH1 (At2g31450), has the expected bifunctional glycosylase-lyase activity in vitro (14). The ARP gene (At2g41460) in Arabidopsis encodes an enzyme with AP endonuclease activity (13).
Here we present the results of experiments conducted to address whether there is BER of oxidized pyrimidines in the Arabidopsis chloroplast. Chloroplast protein extracts were assayed for glycosylase-lyase/endonuclease activity. The chloroplast localization of ARP, AtNTH1, and AtNTH2, a second Arabidopsis homolog of Nth, was tested experimentally, and the predicted activity of AtNTH2 was confirmed in vitro. In addition, an analysis of T-DNA insertion mutants affecting each of these three BER genes was performed.
EXPERIMENTAL PROCEDURES
Plant Materials
Arabidopsis lines (Col-0 strain background) were grown in growth chambers or in a greenhouse under standard conditions. Plants were grown under long days (16 h light/8 h dark; 50–100 μmol photons m−2 s−1; 18–23 °C) on either Sunshine Mix (Sun Gro) or Promix (Scott's) combined 3:1 with vermiculite. Plants were watered periodically with Hoagland's nutrient solution (15). T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center and backcrossed one to four times to wild type (Col-0). The lines used were SALK_054181 for AtNTH1, SALK_03155 for AtNTH2, and SAIL 866_H10 for ARP. The double mutants atnth1 atnth2 and atnth2 arp were crossed to generate the triple mutant. For phenotypic analysis of mutants, growth conditions were varied as described below.
Chloroplast Protein Preparation
Chloroplasts were isolated as described previously (16) with some modification. Arabidopsis plants were grown densely in whole flats or in randomly assorted 4-inch pots under short day (10 h light/14 h dark) conditions for 4–7 weeks. ∼50 g of above-ground tissue was harvested with a razor blade and washed quickly in cold water. All subsequent steps were carried out at 4 °C. Tissue was homogenized in 200–250 ml of fresh cold Xpl buffer, filtered through two layers of Miracloth (Calbiochem), and pelleted in 250-ml JA-14 tubes (Beckman) (1880 × g for 5 min at 4 °C). The pelleted material was gently resuspended in ∼4 ml of Xpl and carefully layered onto freshly prepared 40/80% Percoll gradients in 30-ml Corex tubes. The gradients were centrifuged (4740 × g for 18 min at 4 °C) with no braking in a swinging-bucket rotor (JS-13.1; Beckman). The interphase band was harvested into clean tubes, washed with 3 volumes of Xpl, and centrifuged for 5 min at 1880 × g. The pellet was transferred to microcentrifuge tubes and briefly repelleted at 7500 × g, and the supernatant was removed.
The resulting pellet of purified chloroplasts was then processed for total proteins (17). Chloroplasts were fractured by several passes through glass homogenizers and then resuspended and rinsed with ∼7 ml of homogenization buffer. In 10-ml beakers with stirring, 2 m KCl was added to a concentration of 450 mm, and the mixture was incubated for 30 min. This extract was centrifuged at 40,000 × g in a Beckman JA-20 rotor for 1 h. The supernatant was transferred to 10-ml beakers and stirred as ammonium sulfate was added to ∼70% saturation over the course of 1–2 h. The precipitate was centrifuged at 20,000 × g for 1 h; the supernatant was discarded, and the pellet was redissolved in dialysis buffer (25 mm HEPES-KOH, pH 7.8, 100 mm KCl, 12 mm MgCl2, 1 mm EDTA, 17% glycerol, and 2 mm dithiothreitol). The protein was dialyzed overnight against two changes of buffer. Protein was then quantified spectrophotometrically by Bradford assay (Bio-Rad), aliquoted, and frozen at −20 or −80 °C.
SDS-PAGE and Immunoblot Analysis
Proteins were analyzed by PAGE using standard protocols and precast Tris-glycine gels (Invitrogen). Gels were either analyzed by Coomassie Blue staining (for expressed protein) or by immunoblotting with anti-cytosolic (CSD1) and anti-chloroplastic (CSD2) Cu,Zn superoxide dismutase antibodies (18), anti-histone H3 (ab1791; Abcam), and anti-mitochondrial porin monoclonal antibodies (provided by Dr. Tom Elthon via Dr. Russell Jones). Equal protein amounts (25 μg) assessed by bicinchoninic acid assay (Sigma) of whole-cell or chloroplast extracts were loaded. For immunoblotting, proteins were transferred to a nitrocellulose membrane and blocked in TBS-T (20 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.1% (v/v) Tween) with 5% dry milk. Antibodies were detected with Supersignal West Femto electrochemical detection (Pierce).
DNA Glycosylase Activity Assay
Plasmid DNA template was prepared by incubating 5 μg of Midi Prep-prepared (Qiagen) pBluescript SK in 100 μl of distilled H2O with 0.04% OsO4 for 10 min at 75 °C to introduce TG lesions (19). Control plasmid was diluted identically, but not treated. ∼100 ng of DNA was incubated with a varied amount of protein with 200 μg of bovine serum albumin in 20-μl reactions in reaction buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.5 mm EDTA, and 0.5 mm dithiothreitol) for 7 or 15 min at 37 °C. Control reactions used cloned E. coli endonuclease III, human Ape1, or T4 endonuclease V (pyrimidine dimer glycosylase) (New England Biolabs) in the provided reaction buffers. Reactions were stopped with 2.2 μl of stopping buffer (40% (v/v) glycerol, 0.2% (w/v) bromphenol blue, and 1% (w/v) SDS) (19), and 10 μl of each reaction was analyzed on a 1% agarose gel.
Chloroplast Targeting Prediction
Candidates were tested for predicted chloroplast-targeting peptides with six on-line prediction algorithms: TargetP (20); ChloroP (21); iPSORT (22); Predotar (23); PSORT (24); and PCLR (25). PredictNLS, a nuclear localization sequence predictor, was also used (26).
Construction of GFP Fusion Plasmids
cDNA plasmids U13374 (At1g05900.1/AtNTH2) and U18037 (At2g31450/ AtNTH1) were obtained from the Arabidopsis Biological Resource Center. An ARP (At2g41460) cDNA was amplified by PCR from first-strand cDNA using primers BG1 and BG2 (see supplemental Table 3 for primers) with Pfu DNA polymerase (Stratagene). First-strand cDNA was generated with the Superscript II kit (Invitrogen) and oligo(dT) primer. cDNAs with NcoI and SalI restriction sites at the 5′ and 3′ ends, respectively, were amplified by PCR using primers BG3 and BG4 (AtNTH2), BG3 and BG5 (AtNTH1), and BG6 and BG7 (ARP). The same 5′ primer (BG3) was used for AtNTH1 and AtNTH2, because both cDNAs were provided in the same pUNI plasmid (27). Double digestion with NcoI and SalI allowed ligation of the cDNAs into the 35SΩ-sGFP(S65T) plasmid (28), in-frame with the GFP and under the control of the 35S promoter from cauliflower mosaic virus. For the 35S:AtNTH2:GFP fusion, the initiation codon of GFP was fused in-frame, one codon before the stop codon of the shorter (At1g05900.1) splice variant, thereby replacing a penultimate leucine with alanine and the terminal leucine with the methionine of GFP. In the 35S:AtNTH1:GFP construct, the initiating methionine of GFP replaced a lysine three residues from the C terminus of AtNTH1, and the preceding serine became a threonine as a result of the cloning. The 35S:ARP:GFP fusion omitted the C-terminal 54 amino acid residues of ARP, replacing a threonine with the GFP initial methionine and changing the preceding tyrosine to serine.
Transient Expression and Subcellular Localization of GFP Fusion Proteins
Protoplast isolation and transformation were done following the method of Abdel-Ghany et al. (29), with the following variations. Plants were grown on soil, and leaves were sliced into ∼1-mm wide strips in sterile water, then submerged, and briefly (∼1 min) vacuum-infiltrated in enzyme solution and incubated at room temperature with gentle shaking for 2–3 h. Protoplasts were pelleted at 200 × g for 2 min. Protoplasts (100 μl) were transformed with 10 μg of plasmid DNA with no salmon sperm DNA. Expression was allowed to occur overnight in 1 ml of W5 solution.
Expression in protoplasts was analyzed by confocal microscopy using a Zeiss 510 confocal laser scanning microscope. All micrographs were taken with a 100× oil immersion objective with a numerical aperture (NA) of 1.4, with the exception of Fig. 2F, which was taken with a 60× oil objective, NA of 1.35. GFP was excited with a 488 nm argon ion laser, and fluorescence was detected using 505–550 nm detector barrier filter. Chlorophyll was excited with a 543-nm helium-neon laser and detected using a 560–700-nm filter. DAPI was excited with a 364-nm Enterprise II laser and detected using a 370–420-nm filter. Images were analyzed and false-colored using Zeiss 510 software, version 2.8.
FIGURE 2.
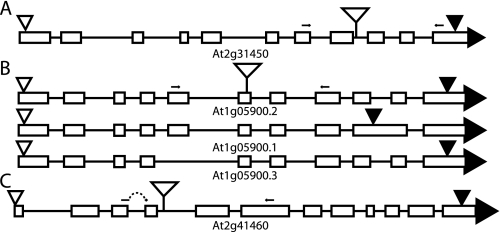
Schematic representations of BER gene models. Open triangles represent translation start sites; filled triangles represent translation termination sites; and stemmed open triangles represent sites of T-DNA insertions. Small arrows represent primers used to test mRNA expression by RT-PCR. A, AtNTH1/At2g31450 (T-DNA, SALK_054181/atnth1); B, AtNTH2/At1g05900 showing three expressed gene models (T-DNA, SALK_013055/atnth2); C, ARP/At2g41460 (T-DNA, SAIL_866_H10/arp). The left expression primer for ARP straddles the 3rd intron.
AtNTH2 Expression in E. coli
Total RNA was isolated from leaves of wild-type Arabidopsis using TRIzol reagent (Stratagene), beginning with 100 mg of leaf tissue ground in liquid nitrogen, and first-strand cDNA was amplified as described above. An At1g05900.2 cDNA was amplified in two rounds of PCR using PrimeSTAR high fidelity polymerase (Takara) and the nested primers BG8 and BG9 and then BG10 and BG11 (supplemental Table 3), which contained NdeI restriction sites. A 1130-bp NdeI product, corresponding to At1g05900.2 (Fig. 2B) without the N-terminal first 19 amino acid residues of the putative chloroplast transit peptide, was gel-purified, cloned in-frame (at M20) into the NdeI site of the pET-15b His6 vector (Novagen), and transformed first into a DH5α host strain for sequencing and subsequently into a Rosetta DE3 lysogen (Novagen) expression host. Protein was expressed (10) overnight at 18 °C. Cells were lysed with 100 μg/ml lysozyme for 20 min at 25 °C and sonicated in buffer A (50 mm sodium phosphate buffer, pH 7.5, 100 mm NaCl, 10 mm imidazole, and 5 mm β-mercaptoethanol). Cleared lysate was incubated in buffer A with pre-equilibrated nickel-nitrilotriacetic acid HisBind matrix (Novagen) for ∼3 h. The matrix/lysate was loaded into a 5-ml syringe blocked with glass wool and washed two times with 4 ml of buffer B (50 mm sodium phosphate buffer, pH 7.5, 300 mm NaCl, 30 mm imidazole, 0.1% Tween, and 5 mm β-mercaptoethanol). Protein was eluted six times with 0.5 ml of buffer C (50 mm sodium phosphate buffer, pH 7.5, 150 mm NaCl, 400 mm imidazole, and 5 mm β-mercaptoethanol) with 600 mm imidazole in the final two elutions, which were pooled and dialyzed overnight against 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm β-mercaptoethanol, and first 25% (v/v) glycerol and then 50% (v/v) glycerol. Dialyzed protein was aliquoted and stored at −20 or −80 °C.
Genotyping of T-DNA Insertion Mutants
Mutant lines were genotyped by PCR. DNA was isolated from young plant leaf samples as described previously (30). For identifying insertions in AtNTH1 and ARP, a three-primer PCR was carried out (30 cycles of 94 °C denaturation for 30 s, 57 °C annealing for 30 s, and 72 °C elongation for 1 min) with two gene-specific primers flanking the specific insertion site and one insert-specific primer. The genotyping for the T-DNA insertion in AtNTH2 (At1g05900) used two separate reactions, each containing the right-flanking primer and either the left-flanking primer or the insert primer. Primers were as follows: Salk insert left-border, BG12; AtNTH2-specific, BG13 and BG14; AtNTH1-specific, BG15 and BG16; SAIL insert left border, BG17; and ARP-specific, BG18 and BG19 (supplemental Table 3).
Analysis of RNA Expression
RNA expression was tested by RT-PCR of homozygous insertion lines. RNA was isolated from healthy leaf samples with TRIzol reagent (Stratagene). First-strand cDNA was generated as described above. Primers for each gene flanking an intron were then used in a standard high cycle PCR (36 cycles of 94 °C denaturation for 30 s, 58 °C annealing for 30 s, and 72 °C elongation for 1 min). Expression primers (supplemental Table 3) were BG20 to BG25, and reactions using primers BG26 and BG27, which efficiently amplified an unlinked control gene (At1g30480), were conducted to confirm the presence of cDNA.
RESULTS
Wild-type Arabidopsis Chloroplasts Contain DNA Glycosylase-Lyase/Endonuclease Activity
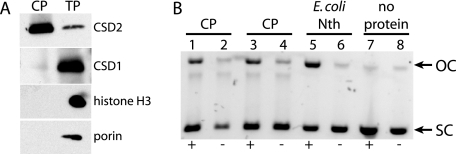
Chloroplasts were isolated from wild-type Arabidopsis leaves, and a soluble chloroplast protein extract was prepared by a method used previously to determine nucleotide excision repair and possible BER activity in Arabidopsis whole-cell extracts (17). Chloroplast protein preparations were enriched in a stromal marker protein, CSD2 (18), and were free of detectable contamination by cytosolic, nuclear, and mitochondrial proteins, as assessed by immunoblot analysis of marker proteins for each of these compartments (Fig. 1A).
FIGURE 1.
Chloroplast glycosylase-lyase/endonuclease activity. A, immunoblot analysis of chloroplast proteins from wild-type (Col-0). CP, chloroplast proteins; TP, total leaf proteins; CSD2, chloroplast stromal marker; CSD1, cytosolic marker; histone H3, nuclear marker; porin, mitochondrial marker. B, glycosylase-lyase/endonuclease activity assay. Supercoiled plasmid substrate was incubated for 7 min at 37 °C with replicate chloroplast protein preparations (lanes 1–4), E. coli Nth as a positive control (lanes 5 and 6), or no protein (lanes 7 and 8) and then run on a 1% agarose gel. +, lanes with OsO4-treated plasmid; −, untreated control plasmid; OC, relaxed open circular plasmid product; SC, supercoiled plasmid substrate.
BER enzymatic activities targeted to TG lesions were detected in the chloroplast protein preparation using a plasmid-nicking assay. A supercoiled plasmid DNA substrate, which had been treated with OsO4 to generate TG lesions, was incubated with the chloroplast protein extract, and nicking of the sugar-phosphate backbone by either bifunctional TG glycosylase-lyase or TG glycosylase plus AP endonuclease activities resulted in the appearance of a relaxed, open circular plasmid product of lower mobility in agarose gel electrophoresis (Fig. 1B). The activity was specific to DNA that contained TG lesions, because only a low level of nonspecific nicking activity was detected in the chloroplast extract using untreated control plasmid as substrate.
Identification of Candidate Genes Encoding Putative Chloroplast BER Enzymes
Candidate BER genes that might encode enzymes responsible for the activity detected in chloroplast extracts were initially selected by sequence similarity to known TG glycosylases. The Arabidopsis genome has two Nth homologs, AtNTH1 (At2g31450) and AtNTH2 (At1g05900), but no recognizable Nei homolog. AtNTH1 and AtNTH2 are homologous to their human and E. coli counterparts, and they have 65% identity and 80% similarity to each other at the protein level. AtNTH2 has two splice variants (At1g05900.1 and At1g05900.2) that are described in data bases (e.g. TAIR). The At1g05900.1 transcript retains a 3′ intron, which introduces a stop codon prior to the conserved C-terminal Fe-S domain (Fig. 2B). By RT-PCR, we identified a third splice variant that lacks the fifth exon (Fig. 2B).
Arabidopsis also has three genes that encode homologs of AP endonuclease enzymes from human (Ape1) and E. coli (Xth). One of these, ARP (At2g41460), has been shown (13) to have the predicted AP endonuclease activity in vitro. No homolog of the other E. coli AP endonuclease, Nfo, is present in Arabidopsis.
Subcellular localization of these candidate BER enzymes was predicted using six online prediction programs for chloroplast targeting (Table 1). Chloroplast targeting predicted by at least three programs was considered sufficient, and on this basis, AtNTH1, AtNTH2, and ARP were selected for further investigation.
TABLE 1.
Results of chloroplast targeting predictions and experiments
The following abbreviations are used: TP, TargetP; P, Predotar; CP, ChloroP; iP, iPSORT; NLS, PredictNLS; GFP, result of GFP fusion experiment (Fig. 3); C, chloroplast; M, mitochondria; N, nucleus; X, no predicted targeting sequence; ND, not determined.
| Protein | TP | P | CP | iP | PCLR | PSORT | NLS | GFP |
|---|---|---|---|---|---|---|---|---|
| AtNTH1 (At2g31450) | M | C | X | C | C | M | X | C |
| AtNTH2 (At1g05900) | C | M | C | C | C | M | X | C |
| ARP (At2g41460) | M | X | C | C | C | X | X | C |
| ARP-like (At4g36050) | M | X | X | X | X | M | X | ND |
| ARP-like (At3g48425) | X | X | X | X | X | N | X | ND |
Confirmation of Chloroplast Targeting Predictions
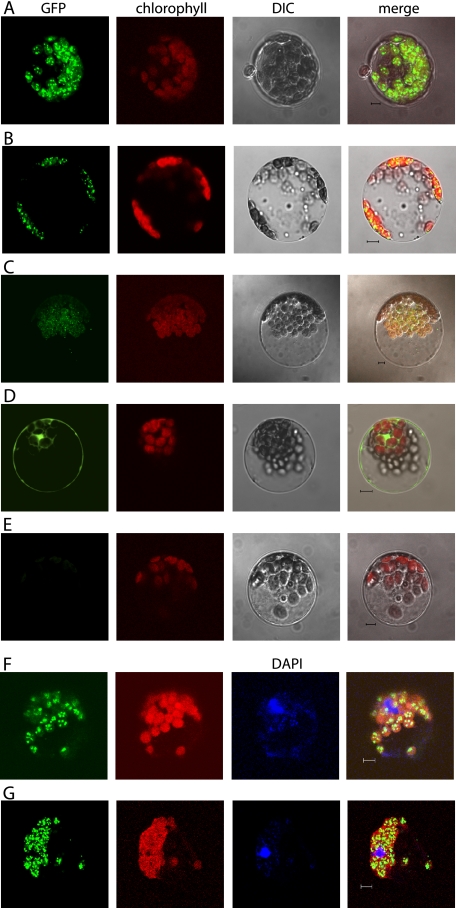
cDNAs of AtNTH1, AtNTH2, and ARP were cloned in-frame with the green fluorescent protein (GFP) under the control of the strong, constitutive 35S promoter from cauliflower mosaic virus. The GFP fusion plasmids were transformed into Arabidopsis protoplasts, which were examined by confocal microscopy after overnight expression. Transient expression of a 35S:GFP control plasmid showed GFP fluorescence in the cytosol and nucleus, but GFP was clearly excluded from the chloroplasts (Fig. 3D). All three fusion proteins expressed GFP in a pattern similar to each other. In each case the GFP fluorescence co-localized with the chlorophyll fluorescence and the visible structure of the chloroplast (Fig. 3, A–C). Moreover, the GFP fluorescence was seen in a distinct punctate pattern within the chloroplasts. DNA staining of these live cells with DAPI was difficult. However when the DAPI staining was successful, the GFP puncta clearly co-localized with the DAPI staining of chloroplast DNA, indicating that each of these proteins is not only chloroplast-targeted but is associated with the chloroplast nucleoids (Fig. 3, F and G).
FIGURE 3.
Subcellular localization of GFP fusion proteins. Confocal micrographs of individual Arabidopsis protoplasts expressing GFP fusion proteins. A, 35S:AtNTH1:GFP fusion; B, 35S:AtNTH2:GFP fusion; C, 35S:ARP:GFP fusion; D, 35S:GFP control; E, no DNA control; F, 35S:AtNTH2:GFP fusion; G, 35S:ARP:GFP fusion. A–E, columns from left are as follows: GFP fluorescence, chlorophyll autofluorescence, differential interference contrast transmission (DIC), and the merged image. F and G, columns from left are GFP fluorescence, chlorophyll autofluorescence, DAPI-DNA fluorescence, and the merged image.
Expression and Assay of AtNTH2 Activity in Vitro
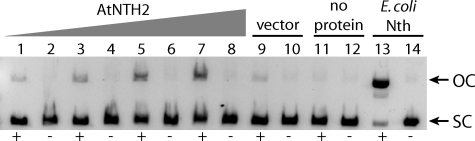
AtNTH1 was previously shown to have bifunctional glycosylase-lyase activity in vitro (14), but the enzymatic function of AtNTH2 has not been experimentally determined. To test whether AtNTH2 also has glycosylase-lyase activity, a cDNA of the At1g05900.2 splice variant (Fig. 2B), which includes the conserved C-terminal Fe-S domain in the open reading frame, was expressed in and purified from E. coli as a His6-tagged protein. Glycosylase-lyase activity of AtNTH2 was assayed using TG-containing (OsO4-treated) plasmid DNA as a substrate, as described above for chloroplast proteins. The expressed AtNTH2 protein exhibited significant nicking activity specifically on OsO4-treated substrate DNA, consistent with its predicted bifunctional TG glycosylase-lyase activity, whereas an eluate from an empty vector control showed essentially no activity (Fig. 4).
FIGURE 4.
Glycosylase-lyase activity assay of AtNTH2. Supercoiled plasmid substrate was incubated for 15 min at 37 °C with increasing volumes of His6-tagged AtNTH2 protein purified from E. coli (0.5 μl in lanes 1 and 2; 1 μl in lanes 3 and 4; 2 μl in lanes 5 and 6; and 4 μl in lanes 7 and 8), 4 μl of protein from an empty vector control strain (lanes 9 and 10), 4 μl of dialysis buffer (no protein) as a negative control (lanes 11 and 12), or E. coli Nth as a positive control (lanes 13 and 14), and then the substrate and product were separated on a 1% agarose gel. +, lanes with OsO4-treated plasmid; −, untreated control plasmid; OC, relaxed open circular plasmid product; SC, supercoiled plasmid substrate.
Identification and Characterization of T-DNA Insertion Mutants
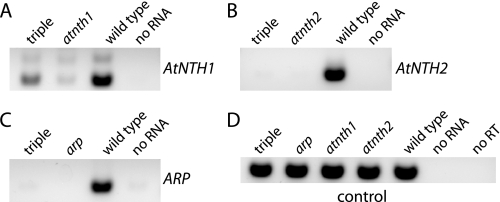
T-DNA insertional disruptions of AtNTH1, AtNTH2, and ARP (Fig. 2) were obtained from either the SALK collection (31) or the SAIL/GARLIC collection (32), and homozygous lines were identified by PCR. RT-PCR was conducted to examine mRNA accumulation in the homozygous mutants (Fig. 5). The atnth2 and arp mutant lines were confirmed to lack their respective transcripts, whereas the atnth1 mutant was found to express an AtNTH1 transcript at a reduced level (Fig. 5 and supplemental Fig. 7). The homozygous lines were crossed with each other to create double mutants and the triple mutant. The atnth1 atnth2 arp triple mutant lacked AtNTH2 and ARP mRNAs but accumulated an intermediate level of AtNTH1 transcript (Fig. 5).
FIGURE 5.
RT-PCR analysis of mRNA expression. Agarose-gel analysis of high cycle (36×) PCRs using first-strand cDNA template generated from total RNA of the indicated genotype and gene-specific primers. A, AtNTH1 mRNA; B, AtNTH2 mRNA; C, ARP mRNA; D, control mRNA (At1g30480).
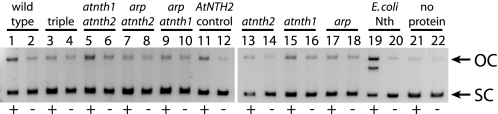
To determine whether the T-DNA insertion mutants affected the TG glycosylase-lyase/endonuclease activity detected in wild-type chloroplast extracts (Fig. 1B), the TG plasmid nicking assay was performed with chloroplast proteins isolated from the single, double, and triple mutant lines. The atnth1 and atnth2 mutants did not affect this glycosylase-lyase/endonuclease activity either independently or in their double mutant (Fig. 6). The arp mutation, however, eliminated the activity alone and in double and triple mutant combinations (Fig. 6).
FIGURE 6.
Glycosylase-lyase/endonuclease activity of T-DNA insertional mutant chloroplast extracts. Supercoiled plasmid substrate was incubated for 15 min at 37 °C with the following chloroplast proteins and controls: wild type (lanes 1 and 2); atnth1 atnth2 arp triple mutant (lanes 3 and 4); atnth1 atnth2 double mutant (lanes 5 and 6); arp atnth2 (lanes 7 and 8); arp atnth1 (lanes 9 and 10); His6-tagged AtNTH2 protein as a positive control (lanes 11 and 12); atnth2 (0.5×, lanes 13 and 14); atnth1 (lanes 15 and 16); arp (lanes 17 and 18); E. coli Nth as a positive control (lanes 19 and 20); and dialysis buffer (no protein) as a negative control (lanes 21 and 22). The substrate and product were then separated on a 1% agarose gel. +, lanes with OsO4-treated plasmid; −, untreated control plasmid; OC, relaxed open circular plasmid product; SC, supercoiled plasmid substrate.
All mutants were screened for growth differences and whole-plant phenotypes using a variety of measures and under a variety of growth conditions. Photo-oxidative stresses, including high light, hydrogen peroxide, methyl viologen, and UV light were imposed, and parameters such as plant size, weight, lifetime seed production (fitness), and photosynthetic efficiency were measured. No conditions yielded significant or reproducible differences in any measure, even in the triple mutant (supplemental Fig. 8 and supplemental Table 2).
DISCUSSION
BER Activity in the Chloroplast
The TG glycosylase-lyase/endonuclease activity observed in chloroplast extracts of wild-type Arabidopsis (Fig. 1B) represents the first time that BER activity has been reported targeting oxidative damage in the chloroplast. It is only the second time any glycosylase activity has been observed in chloroplasts, after the finding of uracil-DNA glycosylase in maize (33). AtNTH1 (14), AtNTH2 (Fig. 4), and ARP (13) have enzymatic activities in vitro that might be involved in TG glycosylase-lyase activity in vivo, and a T-DNA insertion in the ARP gene eliminated the activity in chloroplast extracts (Fig. 6). All three enzymes were co-localized to nucleoids within Arabidopsis chloroplasts (Fig. 3). Taken together, these results provide strong evidence for the occurrence of BER in chloroplasts.
Chloroplast Targeting of BER Enzymes
AtNTH1, AtNTH2, and ARP fusion proteins to GFP are not only targeted to chloroplasts, they are localized specifically in chloroplast nucleoids that stain with DAPI (Fig. 3). The observation of nucleoid co-localization supports the in vitro activity assays indicating that these proteins do in fact interact with and repair DNA. The punctate appearance is very similar to that seen in other studies of nucleoid proteins (34–36).
It is notable that there are two Nth homologs in the chloroplast, perhaps indicating an especially robust repair framework to cope with oxidative stress. However, the finding that both proteins are chloroplast-targeted also raises the question of what is catalyzing this type of repair in the nucleus. It is possible that there are splice or other variants of these genes that are also targeted to the nucleus. For example, there is expressed sequence tag support for transcription initiation variation in the AtNTH1 gene, which would allow for translation from a downstream ATG to generate a predicted protein lacking a chloroplast transit peptide. Splice-dependent localization of plant DNA repair enzymes has been postulated for one of the purine-specific glycosylases, Fpg (37, 38), and shown for DNA ligase I (38).
arp Mutant Uncovers an Additional TG Glycosylase Activity in Arabidopsis Chloroplasts
Although both AtNTH1 and AtNTH2 have Nth-type glycosylase-lyase activity in vitro, the T-DNA insertional mutants of each of these genes do not affect the activity detected in this qualitative assay of chloroplast extracts (Fig. 6). Because atnth1 does not appear to be a null mutant (Fig. 5), there might be residual AtNTH1 activity appearing in these lines. However, a T-DNA disruption of the ARP gene eliminated the TG glycosylase-lyase/endonuclease activity detected in wild-type Arabidopsis chloroplasts (Fig. 6), suggesting that AtNTH1 and AtNTH2 are not responsible for the major chloroplast glycosylase activity against TG and that there must be an additional TG glycosylase activity in Arabidopsis chloroplasts. Alternatively, this result could be explained by the presence of AP sites in the OsO4-treated substrate DNA; however, this possibility was eliminated by control experiments using the AP lyase activity of human Ape1 or T4 endonuclease V (pyrimidine dimer glycosylase) (supplemental Fig. 9). The additional TG glycosylase activity in chloroplasts might be a monofunctional glycosylase enzyme that requires the AP endonuclease activity of ARP.
The low apparent activity of AtNTH1 and AtNTH2 in chloroplast extracts might be explained in a number of ways. They might be present at much lower protein levels or have lower stability in our chloroplast protein extracts. Alternatively, they might have significantly slower kinetics than the ARP-dependent activity. In either case, it is intriguing that the recognized Nth homologs do not seem to be the only or even the most active enzymes in the chloroplast TG BER pathway. A more quantitative activity assay would be useful in exploring this further. Identification of the major TG glycosylase in the ARP-dependent pathway is an important future goal in analysis of BER in the chloroplast.
T-DNA Insertion Mutants in AtNTH1, AtNTH2, and ARP Lack an Obvious Whole-plant Phenotype
The lack of a growth or developmental phenotype in the T-DNA mutants was something of a surprise, given that the arp mutation blocked the major TG glycosylase pathway detected in chloroplasts (Fig. 6). In other organisms, DNA repair mutants often exhibit a detectable phenotype only when more than one gene is disrupted (39, 40), so the lack of a visible phenotype in the Arabidopsis single mutants was not unexpected. However, even the atnth1 atnth2 arp triple mutant did not exhibit a clear whole-plant phenotype, implying that chloroplast repair of TG lesions is unnecessary, that it is necessary on a time scale of generations, and/or that there is a redundant non-BER pathway masking the phenotype. A similar lack of phenotype was found for an ogg fpg double mutant in Arabidopsis apparently blocked in nuclear purine BER (41). The observed multiplicity of chloroplast enzymes for BER of TG lesions (two AtNTH proteins and a third activity) would seem to argue for the importance of this repair pathway. However, it is likely that there are other DNA repair pathways, such as nucleotide excision repair (42, 43), recombinational repair (44), or gene conversion (45) that are protecting chloroplast DNA in the absence of BER, at least in the short term. Over multiple generations, it is possible that phenotypes might emerge as mutations and/or damage accumulates in the plastome, similar to what has been observed for the nuclear genome in mismatch repair-deficient mutants (46).
Supplementary Material
Acknowledgments
We thank Steve Ruzin and Denise Schichnes and the University of California, Berkeley, CNR Biological Imaging Facility for use of and instruction in confocal microscopy; Marinus Pilon (Colorado State University) for the GFP vector and instruction; and Lillian Chan for technical assistance. We thank the Arabidopsis Biological Resource Center for DNA clones.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM071908 from NIGMS. This work was also supported by the United States Department of Agriculture.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 2 and 3 and Figs. 7–9.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 2 and 3 and Figs. 7–9.
Footnotes
- ROS
- reactive oxygen species
- AP
- apurinic/apyrimidinic
- BER
- base excision repair
- Nth
- endonuclease III
- RT
- reverse transcription
- GFP
- green fluorescent protein
- TG
- thymine glycol
- DAPI
- 4′,6-diamidino-2-phenylindole.
REFERENCES
- 1.Girard P. M., Boiteux S. ( 1997) Biochimie 79, 559– 566 [DOI] [PubMed] [Google Scholar]
- 2.Niyogi K. K. ( 1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333– 359 [DOI] [PubMed] [Google Scholar]
- 3.Cadet J., Bourdat A. G., D'Ham C., Duarte V., Gasparutto D., Romieu A., Ravanat J. L. ( 2000) Mutat. Res. 462, 121– 128 [DOI] [PubMed] [Google Scholar]
- 4.Dizdaroglu M. ( 2005) Mutat. Res. 591, 45– 59 [DOI] [PubMed] [Google Scholar]
- 5.Demple B., Harrison L. ( 1994) Annu. Rev. Biochem. 63, 915– 948 [DOI] [PubMed] [Google Scholar]
- 6.Huffman J. L., Sundheim O., Tainer J. A. ( 2005) Mutat. Res. 577, 55– 76 [DOI] [PubMed] [Google Scholar]
- 7.Krokan H. E., Standal R., Slupphaug G. ( 1997) Biochem. J. 325, 1– 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stivers J. T., Jiang Y. L. ( 2003) Chem. Rev. 103, 2729– 2759 [DOI] [PubMed] [Google Scholar]
- 9.Wallace S. S. ( 2002) Free Radic. Biol. Med. 33, 1– 14 [DOI] [PubMed] [Google Scholar]
- 10.Bandaru V., Sunkara S., Wallace S. S., Bond J. P. ( 2002) DNA Repair 1, 517– 529 [DOI] [PubMed] [Google Scholar]
- 11.Tell G., Damante G., Caldwell D., Kelley M. R. ( 2005) Antioxid. Redox. Signal. 7, 367– 384 [DOI] [PubMed] [Google Scholar]
- 12.Sung J. S., Demple B. ( 2006) FEBS J. 273, 1620– 1629 [DOI] [PubMed] [Google Scholar]
- 13.Babiychuk E., Kushnir S., Van Montagu M., Inzé D. ( 1994) Proc. Natl. Acad. Sci. U. S. A. 91, 3299– 3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roldán-Arjona T., García-Ortiz M. V., Ruiz-Rubio M., Ariza R. R. ( 2000) Plant Mol. Biol. 44, 43– 52 [DOI] [PubMed] [Google Scholar]
- 15.Taiz L., Zeiger E. ( 1991) Plant Physiology, pp. 111, Benjamin/Cummings, Redwood City, CA [Google Scholar]
- 16.Weigel D., Glazebrook J. ( 2002) Arabidopsis: A Laboratory Manual, pp. 217– 219, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17.Li A., Schuermann D., Gallego F., Kovalchuk I., Tinland B. ( 2002) Plant Cell 14, 263– 273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliebenstein D. J., Monde R. A., Last R. L. ( 1998) Plant Physiol. 118, 637– 650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda S., Biswas T., Roy R., Izumi T., Boldogh I., Kurosky A., Sarker A. H., Seki S., Mitra S. ( 1998) J. Biol. Chem. 273, 21585– 21593 [DOI] [PubMed] [Google Scholar]
- 20.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. ( 2000) J. Mol. Biol. 300, 1005– 1016 [DOI] [PubMed] [Google Scholar]
- 21.Emanuelsson O., Nielsen H., von Heijne G. ( 1999) Protein Sci. 8, 978– 984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. ( 2002) Bioinformatics 18, 298– 305 [DOI] [PubMed] [Google Scholar]
- 23.Small I., Peeters N., Legeai F., Lurin C. ( 2004) Proteomics 4, 1581– 1590 [DOI] [PubMed] [Google Scholar]
- 24.Nakai K., Horton P. ( 1999) Trends Biochem. Sci. 24, 34– 36 [DOI] [PubMed] [Google Scholar]
- 25.Schein A. I., Kissinger J. C., Ungar L. H. ( 2001) Nucleic Acids Res. 29, E82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cokol M., Nair R., Rost B. ( 2000) EMBO Rep. 1, 411– 415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada K., Lim J., Dale J. M., Chen H., Shinn P., Palm C. J., Southwick A. M., Wu H. C., Kim C., Nguyen M., Pham P., Cheuk R., Karlin-Newmann G., Liu S. X., Lam B., Sakano H., Wu T., Yu G., Miranda M., Quach H. L., Tripp M., Chang C. H., Lee J. M., Toriumi M., Chan M. M., Tang C. C., Onodera C. S., Deng J. M., Akiyama K., Ansari Y., Arakawa T., Banh J., Banno F., Bowser L., Brooks S., Carninci P., Chao Q., Choy N., Enju A., Goldsmith A. D., Gurjal M., Hansen N. F., Hayashizaki Y., Johnson-Hopson C., Hsuan V. W., Iida K., Karnes M., Khan S., Koesema E., Ishida J., Jiang P. X., Jones T., Kawai J., Kamiya A., Meyers C., Nakajima M., Narusaka M., Seki M., Sakurai T., Satou M., Tamse R., Vaysberg M., Wallender E. K., Wong C., Yamamura Y., Yuan S., Shinozaki K., Davis R. W., Theologis A., Ecker J. R. ( 2003) Science 302, 842– 846 [DOI] [PubMed] [Google Scholar]
- 28.Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. ( 1996) Curr. Biol. 6, 325– 330 [DOI] [PubMed] [Google Scholar]
- 29.Abdel-Ghany S. E., Müller-Moulé P., Niyogi K. K., Pilon M., Shikanai T. ( 2005) Plant Cell 17, 1233– 1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimyuk V. I., Carroll B. J., Thomas C. M., Jones J. D. ( 1993) Plant J. 3, 493– 494 [DOI] [PubMed] [Google Scholar]
- 31.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. ( 2003) Science 301, 653– 657 [DOI] [PubMed] [Google Scholar]
- 32.Sessions A., Burke E., Presting G., Aux G., McElver J., Patton D., Dietrich B., Ho P., Bacwaden J., Ko C., Clarke J. D., Cotton D., Bullis D., Snell J., Miguel T., Hutchison D., Kimmerly B., Mitzel T., Katagiri F., Glazebrook J., Law M., Goff S. A. ( 2002) Plant Cell 14, 2985– 2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bensen R. J., Warner H. R. ( 1987) Plant Physiol. 84, 1102– 1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi-Ham C. L., Keaton M. A., Cannon G. C., Heinhorst S. ( 2002) Plant Mol. Biol. 49, 621– 631 [DOI] [PubMed] [Google Scholar]
- 35.Cho H. S., Lee S. S., Kim K. D., Hwang I., Lim J. S., Park Y. I., Pai H. S. ( 2004) Plant Cell 16, 2665– 2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato N., Nakayama M., Hase T. ( 2001) FEBS Lett. 487, 347– 350 [DOI] [PubMed] [Google Scholar]
- 37.Murphy T. M., Gao M. J. ( 2001) J. Photochem. Photobiol. B 61, 87– 93 [DOI] [PubMed] [Google Scholar]
- 38.Sunderland P. A., West C. E., Waterworth W. M., Bray C. M. ( 2006) Plant J. 47, 356– 367 [DOI] [PubMed] [Google Scholar]
- 39.Jiang D., Hatahet Z., Blaisdell J. O., Melamede R. J., Wallace S. S. ( 1997) J. Bacteriol. 179, 3773– 3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito Y., Uraki F., Nakajima S., Asaeda A., Ono K., Kubo K., Yamamoto K. ( 1997) J. Bacteriol. 179, 3783– 3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy T. M. ( 2005) Physiol. Plant. 123, 227– 232 [Google Scholar]
- 42.Hays J. B. ( 2002) DNA Repair 1, 579– 600 [DOI] [PubMed] [Google Scholar]
- 43.Netrawali M. S., Nair K. A. ( 1984) Environ. Exp. Bot. 24, 63– 70 [Google Scholar]
- 44.Cerutti H., Osman M., Grandoni P., Jagendorf A. T. ( 1992) Proc. Natl. Acad. Sci. U. S. A. 89, 8068– 8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khakhlova O., Bock R. ( 2006) Plant J. 46, 85– 94 [DOI] [PubMed] [Google Scholar]
- 46.Hoffman P. D., Leonard J. M., Lindberg G. E., Bollmann S. R., Hays J. B. ( 2004) Genes Dev. 18, 2676– 2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.