Abstract
The x-ray crystal structure of the thiostrepton resistance RNA methyltransferase (Tsr)·S-adenosyl-l-methionine (AdoMet) complex was determined at 2.45-Å resolution. Tsr is definitively confirmed as a Class IV methyltransferase of the SpoU family with an N-terminal “L30-like” putative target recognition domain. The structure and our in vitro analysis of the interaction of Tsr with its target domain from 23 S ribosomal RNA (rRNA) demonstrate that the active biological unit is a Tsr homodimer. In vitro methylation assays show that Tsr activity is optimal against a 29-nucleotide hairpin rRNA though the full 58-nucleotide L11-binding domain and intact 23 S rRNA are also effective substrates. Molecular docking experiments predict that Tsr·rRNA binding is dictated entirely by the sequence and structure of the rRNA hairpin containing the A1067 target nucleotide and is most likely driven primarily by large complementary electrostatic surfaces. One L30-like domain is predicted to bind the target loop and the other is near an internal loop more distant from the target site where a nucleotide change (U1061 to A) also decreases methylation by Tsr. Furthermore, a predicted interaction with this internal loop by Tsr amino acid Phe-88 was confirmed by mutagenesis and RNA binding experiments. We therefore propose that Tsr achieves its absolute target specificity using the N-terminal domains of each monomer in combination to recognize the two distinct structural elements of the target rRNA hairpin such that both Tsr subunits contribute directly to the positioning of the target nucleotide on the enzyme.
RNA modifications and the enzymes that catalyze their formation are critical for cellular viability. Certain RNA modifications are extremely well characterized, such as CCA addition and amino acylation of the 3′-ends of tRNA, and the contributions of some nucleotide modifications to the creation of specific functional tRNA structures (1–3). Although the single most common nucleotide modification is pseudouridine, by far the most abundant type of RNA chemical modification is methylation (4). A vast array of unique mono-, di-, and trimethylations of each RNA base and/or ribose sugar 2′-OH is possible, and important new functions for these modifications continue to emerge. In ribosomal RNA (rRNA),2 for example, modifications cluster in functionally critical regions where methylation may act as a checkpoint in ribosome subunit assembly (5), influence the process of translation (6), and alter resistance to certain antibiotics (7, 8).
RNA methylation is catalyzed by members of two classes (I and IV) of S-adenosyl-l-methionine (AdoMet)-dependent RNA methyltransferase (MTase) enzymes (9). In bacteria, rRNA methylations are incorporated by both “housekeeping” MTases and those that confer resistance to antibiotics. Although members of the former group are often highly conserved, the latter are generally only found in the antibiotic-producing strain as one mechanism of defense against self-intoxication (10). However, several instances of antibiotic resistance MTase genes in non-producer strains, including pathogenic bacteria, have been identified, and it is clear that these genes are mobile resistance determinants, usually obtained by lateral gene transfer.
Several classes of antibiotics target the conserved centers on the ribosome, altering or blocking critical steps in translation such as decoding and peptidyl transfer, to exert their bactericidal effect (11). RNA MTases have been identified as clinically significant resistance determinants to a number of these, including the aminoglycoside (Arm MTase) and erythromycin (Erm MTase) antibiotics (12, 13). Another functionally critical ribosome domain, the factor binding site (or “GTPase” center), is also the target for a family of thiazole-containing peptide antibiotics (14), which includes thiostrepton. These antibiotics have been important biochemical tools for studies of ribosome function but are of limited clinical use due to their poor aqueous solubility. Thiostrepton is, however, used in veterinary medicine, and recent studies suggest it may have application in development of novel antimalarial and anticancer strategies (15, 16). The minimal rRNA sequence for interaction of thiostrepton is a highly conserved, independently folded 58-nucleotide rRNA domain that is also bound by ribosomal protein L11. Resistance to thiostrepton can result from mutations in the N-terminal domain of L11 or its entire absence, whereas mutation of the target nucleoside (A1067) confers far greater resistance (17–19). In the thiostrepton producer Streptomyces azureus the thiostrepton resistance MTase (Tsr) catalyzes the 2′-O-methylation of A1067 resulting in specific and total resistance to thiostrepton (20).
Here we present the crystal structure of Tsr in complex with AdoMet. The structure definitively places Tsr into the SpoU/TrmD (SPOUT) family of enzymes and provides the basis for modeling the Tsr·rRNA recognition process.
EXPERIMENTAL PROCEDURES
Tsr Protein Expression, Purification, and Crystallization
Tsr from S. azureus was PCR-amplified from pUC-TSR and ligated into pET28a (Novagen) to generate a Tsr expression plasmid with an N-terminal hexahistidine tag and thrombin cleavage site for its removal. Tsr expression was induced by isopropyl 1-thio-β-d-galactopyranoside in Escherichia coli BL21(λDE3)/pLysS cultures grown at 37 °C in LB medium supplemented with kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml). The protein was purified to homogeneity by HisTrapTM HP Ni2+-affinity chromatography followed by thrombin protease cleavage of the His6 tag and further purification over HiTrap Benzamidine FF and HisTrapTM HP columns, and Mono Q anion exchange chromatography. Tsr-Ala-88 was created by QuikChange Lightning Site-directed Mutagenesis (Stratagene) and expressed and purified under identical conditions to the wild-type protein.
The purified protein was dialyzed against 50 mm Tris buffer, pH 7.0, containing 75 mm KCl, 10 mm β-mercaptoethanol, and 10% glycerol, and concentrated to ∼8.5 mg/ml. Crystallization conditions were identified using the JBScreen HTS1 screen (JenaBioscience) on an Innovadyne 96 crystallization robot. Crystals used in diffraction experiments were obtained by hanging drop vapor diffusion in 4-μl drops containing equal volumes of protein and a solution containing 27% polyethylene glycol 4000, 0.2 m sodium acetate, and 0.1 m Tris buffer, pH 8.0. Crystals appeared in 2–3 days and reached their maximum dimensions (∼800 × 150 × 80 μm) after a further 5 days. Fully grown crystals were derivatized with AdoMet by soaking in the same solution supplemented with 5 mm AdoMet for 24 h.
X-ray Diffraction Data Collection, Structure Determination, and Refinement
Crystals were cryoprotected by dragging through neat perfluoropolyether and flash cooled by immersion in liquid nitrogen. Diffraction data were collected on ID23-1 at the European Synchrotron Radiation Facility and processed with the XDS program package (21). The Tsr structure was solved by molecular replacement with the CCP4 program MOLREP (22, 23), using the RNA 2′-O-ribose MTase RrmA (1ipa.pdb) as a starting model. As RrmA only has an overall 27% sequence homology to Tsr, the highly variable NTD was removed to leave only the conserved CTD and improve the starting model quality. All residues were replaced to produce a 160-residue polyserine molecular replacement search model. MOLREP successfully placed two molecules in the asymmetric unit, consistent with the observed Mathews coefficient, VM, of 2.12 Å3/Da and a solvent content of 42%.
The initial electron density for the missing NTDs was poorly defined making manual building difficult. Initial automated builds using RESOLVE (24) could only place ∼280 amino acids, all of which were within the conserved C-terminal domains. The dual-space molecular replacement model completion method (25) was therefore used to improve the quality of the model. The initial molecular replacement solution was subjected to cycles of phase calculation in OASIS06 (25) and density modification with DM (23, 26), followed by automated model building with RESOLVE and refinement with CNS (27). After seven rounds, RESOLVE was able to successfully place over 470 residues, including side chains, into the electron density. The remaining amino acids were built manually in Coot (28) before a final refinement with Phenix.refine (29) incorporating translation libration screw refinement. The final model contains amino acids 8–268 of each Tsr monomer in the asymmetric unit. Full data collection, processing, and refinement statistics are shown in Table 1.
TABLE 1.
X-ray data collection and refinement statistics for the Tsr·AdoMet complex
| Tsr-AdoMet complex | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Resolution (Å) | 19.6-2.45 (2.51-2.45) |
| Unit cell: a,b,c (Å) | 40.80, 56.20, 213.90 |
| Redundancy | 4.4 (4.5) |
| Total observations | 85,388 |
| Unique observations (khl) | 18,568 |
| Completeness (%) | 95.8 (90.0) |
| Rsyma (%) | 10.7 (36.5) |
| 〈I/s(I)〉 | 10.2 (2.85) |
| Refinement | |
| Rworkb (%) | 21.9 (26.9) |
| Rfreec (%) | 27.3 (32.3) |
| Number of atoms | |
| Protein | 3,802 |
| Solvent | 90 |
| AdoMet | 52 |
| B-factor (Å2), overall | 60.5 |
| Protein | 60.9 |
| Solvent | 34.7 |
| AdoMet | 57.7 |
| Ramachandran plot (%) | |
| Favorable | 84.6 |
| Allowed | 11.8 |
| Generous | 3.6 |
| Disallowed | 0 |
| Room mean square deviation | |
| Bond lengths (Å) | 0.022 |
| Bond angles (°) | 2.007 |
aRsym = Σhkl Σi|Ii (hkl) − 〈I(hkl)〉|/Σhkl Σi Ii(hkl).
b Rwork = Σhkl|Fo(hkl) − Fc(hkl)|/Σhkl|Fo(hkl), where Fo and Fc are observed and calculated structure factors, respectively.
c For the calculation of Rfree, 5% of reflections were chosen at random to constitute a test set. Values in parenthesis are for the highest resolution shell.
Gel Electrophoresis Mobility Shift Assays
The 29-nucleotide RNA-encoding plasmids were created by ligation of pairs of annealed chemically synthesized DNA oligonucleotides into a pUC plasmid modified for RNA in vitro transcription (30). The 58-nucleotide RNA-encoding plasmids, methods for RNA in vitro transcription from linearized plasmid templates using T7 RNA polymerase and purification by denaturing PAGE were described previously (30–32). RNA samples (6 μm final concentration) were annealed at 65 °C in 100 mm MOPS buffer, pH 7.0, containing 5 mm MgCl2 and 175 mm KCl. Tsr·RNA complexes were prepared by adding Tsr (0, 3, 6, 12, 20, and 40 μm final concentration) and incubating for 30 min at room temperature. For each sample, free RNA and complexes were separated on a 10% acrylamide native gel run for 1 h at 120 V and visualized by ethidium bromide staining.
Gel Filtration Chromatography
The 58-nucleotide RNA samples were annealed at 65 °C in 50 mm Tris buffer, pH 8.0, containing 5 mm MgCl2, 75 mm KCl, and 10% glycerol. Gel filtration chromatography was performed in the same buffer using a Superdex 200 10/300 GL column (Amersham Biosciences) on an ÄKTApurifier10 system. Complexes were prepared by mixing approximately stoichiometric quantities of 58-nucleotide RNAs (3 μm) and Tsr (6 μm; 3 μm dimer) and incubating at room temperature for 30 min before applying to the column. Elution of Tsr·RNA complexes and free components was monitored by UV absorbance at 260 nm and 230 nm.
Tsr in Vitro Methylation Assays
RNA samples were resuspended in 10 mm Hepes buffer, pH 8.0, containing 10 mm MgCl2, 50 mm KCl, 10 mm NH4Cl, and 6 mm β-mercaptoethanol, and annealed at 65 °C. Methylation assays were performed at 37 °C in the same buffer with a final volume of 50 μl containing 1 μm RNA, 100 μm AdoMet, 1 μCi of [methyl-3H]AdoMet, and 165 nm Tsr. Samples were removed at 10 and 30 min, and the RNAs were recovered using G-25 spin columns (Amersham Biosciences). Assays were performed without RNA to measure unincorporated [3H]AdoMet recovered from the spin columns bound to Tsr. Incorporation of 3H was determined by liquid scintillation counting in EcoscintTM scintillation fluid (National Diagnostics). Each assay was performed at least three times.
Molecular Modeling Experiments
The coordinates for the 58-nucleotide L11-binding domain rRNA were taken from the protein data bank (1HC8.pdb) and modified at position 1061 to the wild-type E. coli sequence (U1061). Docking experiments were performed with the program Hex first applying a rigid-body prediction with the RNA (the “ligand”) oriented toward the cleft formed by the conserved CTDs of the Tsr dimer (the “receptor”). Subsequently both shape-only and shape-electrostatics correlation algorithms were used with a search radius of n = 30, and the top 10 docking solutions were inspected visually in Coot.
RESULTS AND DISCUSSION
Crystal Structure of Tsr, a SpoU MTase
Our x-ray crystal structure (Fig. 1) definitively confirms Tsr is a member of the SpoU family of MTases as hypothesized from sequence analysis (33, 34). Tsr is composed of two structural domains, with amino acids 1–102 and 108–269 forming the N-terminal domain (NTD) and C-terminal domain (CTD), respectively. The latter contains all of the common SpoU sequence and structural motifs that define the global structure of these enzymes (supplemental Fig. S1).
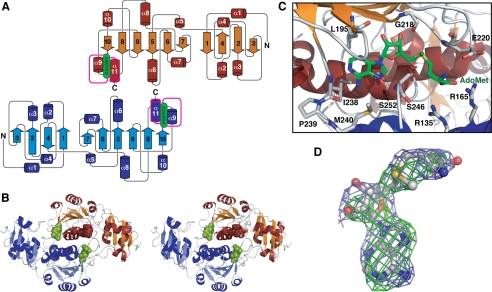
FIGURE 1.
X-ray crystal structure of the Tsr-AdoMet complex. A, Tsr dimer protein secondary structure topology diagram. The knotted sequence is colored magenta. B, stereo view schematic of the Tsr dimer with bound AdoMet (green). C, the AdoMet binding pocket with key Tsr amino acids indicated. D, AdoMet in an extended conformation shown with omit Fo − Fcc (green) and omit 2Fo − 1Fc (light blue) maps contoured at 3.5 and 1.0 σ, respectively.
In Tsr, the core of the conserved C-terminal catalytic domain is a twisted six-stranded parallel β-sheet with a central topological switch point, flanked on its outer edges by loops and sandwiched between seven α-helices. Three of these α-helices are on the outer surface of the CTD and four on the dimer interface (Fig. 1 and supplemental Fig. S2). The linker between the two domains ends in a short α-helix (α5) that is packed against the two end strands (β7–β6) of the β-sheet and the adjacent helix α8. In addition to those conserved in all SpoU MTases, Tsr contains an additional α-helix between β8 and β9. As a result, the CTD maintains a strict alternating α/β secondary structure (Fig. 1).
SPOUT MTases exist as tightly bound homodimers mediated by interaction of their CTDs (34). This feature is also observed in the crystal structure of Tsr and is supported by our in vitro and in silico analyses of the Tsr·RNA interaction (see below) indicating that Tsr dimerization is functionally critical. Dimerization is driven by the extensive interaction of two α-helices (α6 and α11) on the inner face of each Tsr CTD and an extended loop formed by amino acids 238–245. Dimer formation buries ∼3500-Å2 surface of each protein (30% of the total surface) and involves many hydrophobic interactions. Leu-247 and Val-251 form reciprocal hydrophobic interactions with Thr-136′ and Leu-140′, respectively (where primes denote amino acids from the other Tsr protomer). Uniquely to Tsr, the amino acid at position 258, which is typically conserved as Tyr or Phe (supplemental Fig. S1), is replaced by a His that interacts across the dimer interface with the highly conserved Glu-259′. His-258 and His-258′ are located close together in the interface and sandwich the two glutamates. The arrangement thus results in a reciprocal ionic interaction network between the two pairs of residues that is further supplemented by the adjacent Lys-204 and Lys-204′ that cap the dimer interface with two salt bridges.
AdoMet Binding Pocket
The Tsr dimer binds two AdoMet molecules and, although each binding pocket is formed predominantly by one monomer, amino acids from both proteins contribute to binding of each. One of the defining characteristics of the SPOUT class of MTases is the presence of a deep trefoil knot at the C-terminal end of the protein that forms the AdoMet binding pocket. In Tsr this knot is created by the threading of amino acids 237–269 through the short untwisted loop of amino acids 195–203 containing the SpoU Motif 3 (35). The highly conserved IPM—–SLN sequence intimately sequesters the adenosine moiety of AdoMet in a deep pocket of hydrophobic residues. The adenosine ring is positioned by hydrogen bonds from N1 and N6 to the main-chain amino and carbonyl groups, respectively, of the conserved Ile-238, and sits on a hydrophobic surface created by the Ile-238 and Ser-252 side chains (Fig. 1C). Further protein backbone-AdoMet contacts hold the ribose moiety in place, with Leu-195 and the highly conserved Gly-218 hydrogen bonding to the 2′-OH and 3′-OH, respectively. The methionine group is positioned on either side by packing against the Cβ of Glu-220 and Ser-246, and Val-249 beneath, and through electrostatic interactions with Arg-165′ and the highly conserved Arg-135′ (Fig. 1C).
Although the electron density for the end of the methionine tail is less well defined, indicating some degree of flexibility or potentially degradation of the molecule, AdoMet is clearly bound in an extended conformation (Fig. 1D and supplemental Fig. S2B) with O4′-C4′-C5′-Sδ and C4′-C5′-Sδ-Cγ dihedral angles of 151° and 152°, respectively. Extended AdoMet conformations have been observed in numerous structures of Class I and Class II MTases (9), but those of Class IV SPOUT enzymes have invariably shown bound AdoMet, or the reaction product S-adenosyl-l-homocysteine, in a tightly bent conformation (36–38). Tsr is unique in having a basic amino acid at 165 (supplemental Fig. S1), and this may directly influence the different AdoMet conformation observed in this Class IV MTase.
Structure of the Target Specificity Domain
The secondary structure of the smaller NTD of Tsr has an alternating α/β structure with four strands forming a central β-sheet sandwiched between two α-helices on each side. The central β-sheet motif is of mixed polarity with a β3↑-β2↑-β4↓-β1↑ topology, whereas the α-helices on each side are parallel, with the pairs of helices oriented orthogonally with respect to each other. The NTD has no sequence conservation with other SpoU MTases (supplemental Fig. S1). However, a Tsr NTD structure search using the DALI server (39) identified several proteins, including yeast ribosomal proteins L30e and L7e, the eukaryotic release factor (eRF) 1, and two related SpoU MTases, AviRb and RlmB (38, 40), that also contain extended “L30-like” NTDs (supplemental Fig. S1).
The SpoU subfamily of MTases can be subdivided into three distinct subclasses: single domain MTases (i.e. conserved catalytic/AdoMet binding domain only) and those containing extended N-terminal regions with either an L5- or L30-like domain (38). We have now shown that Tsr contains an L30-like N-terminal domain. Although there is a high degree of structural conservation between these domains in Tsr and other MTases such as AviRb and RlmB, there is comparatively little sequence similarity. Presumably the variation in target specificity and selectivity these MTases achieve for their target sites must arise from their unique NTD primary sequences and their relative orientation to the catalytic CTDs.
The two domains in Tsr are joined by an elongated loop formed by amino acids 101–118 that contains a short helix in the center (α5). Two lines of evidence suggest there is inherent flexibility within Tsr through this long linker. First, for one monomer where there are fewer crystal packing contacts made by the NTD the electron density is more poorly defined for the entire domain. Second, the crystallographic B factors are correspondingly higher on average for both NTDs (61 and 95) compared with the two CTDs (38 and 52). This suggests there is dynamic freedom between the two domains, which may be necessary for correctly positioning these domains on the rRNA target for methyl transfer.
rRNA Binding and Methylation by Tsr
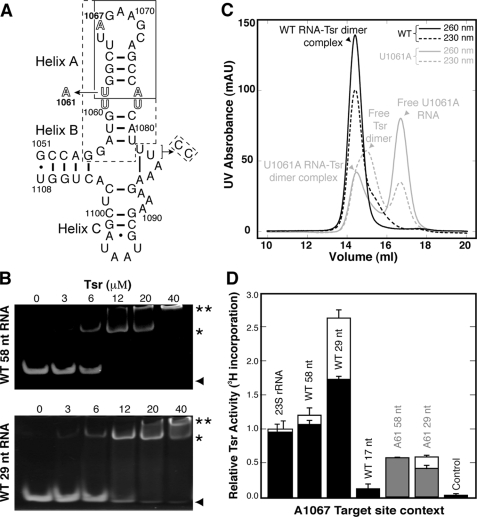
Gel mobility shift assays were used to examine the Tsr-rRNA interaction using both the entire 58-nucleotide L11-binding domain RNA and a 29-nucleotide hairpin containing the A1067 target site (Fig. 2). As expected for a SpoU MTase, we observed a major complex with a 2:1 protein:rRNA stoichiometry for both the 58- and 29-nucleotide rRNA fragments. Higher molecular weight complexes were also seen with increasing protein concentration that have been attributed to tetramer enzyme·RNA complexes for another SpoU MTase (41). These results show that for both RNAs the functional complex is a Tsr dimer bound to one target rRNA. We also examined a 58-nucleotide rRNA containing a U1061 to A mutation (U1061A RNA) within the internal bulge of Helix A, the only site distant from the target loop where mutation dramatically reduces Tsr methylation activity (18). Binding of the U1061A rRNA was significantly weaker with higher molecular weight species dominating at higher Tsr concentrations (supplemental Fig. S3).
FIGURE 2.
rRNA binding and methylation. A, the 58-nucleotide L11 rRNA binding domain containing the Tsr target nucleotide A1067. Boxed regions correspond to smaller RNA hairpins: 17 nucleotides (solid line) and 29 nucleotides (dashed). A modification of the 3′-UU end for the latter RNA to generate Watson-Crick pairing is indicated. The mutation U1061 to A is indicated, and the internal loop within Helix A is shown in outline type. B, gel electrophoresis shift assays with wild-type 58-nucleotide (upper panel) and 29-nucleotide (lower panel) RNAs at a constant concentration of 6 μm RNA per assay and Tsr input at the concentrations indicated above each gel. Free RNA (▴), RNA-Tsr dimer 1:1 complex (*) and higher molecular weight complexes (**) are indicated on the right hand side. C, gel filtration chromatography of 1:1 mixtures of wild-type (black) and U1061A mutant (gray) 58-nucleotide RNAs (3 μm) and Tsr dimer (3 μm). Elution from the column was monitored at 260 (solid line) and 230 nm (dashed line). The content of each peak is identified as indicated on the basis of apparent molecular weight and relative intensity of absorbance at each wavelength. D, methylation activity was measured for 23 S rRNA and three wild-type (58, 29, and 17 nucleotides) and two U1061A mutant (58 and 29 nucleotides) L11-binding domain RNAs by 3H incorporation. Solid bars represent data at the 10-min time point, and where present open bars represent the 30-min time point (both were measured for all RNAs but for some maximum methylation was reached by the earlier time point). Control experiments used an unrelated 54-nucleotide RNA (see supplemental Fig. S4). Error bars are the standard deviation from at least three independent experiments.
We next examined stoichiometric complexes of Tsr and the wild-type and U1061A 58-nucleotide RNAs by gel filtration chromatography. The Tsr-wild-type 58-nucleotide L11-binding domain RNA complex eluted as a single peak corresponding to a complex of Tsr dimer and one RNA (Fig. 2C). In contrast, the U1061A mutant RNA was eluted as a mixture of complex and free Tsr and RNA. These results confirm that the mutation reduces binding affinity presumably by disrupting important Tsr·RNA contacts.
Tsr in vitro methylation assays were conducted with E. coli 23 S rRNA, three substrates of 58, 29, and 17 nucleotides in length derived from the L11 rRNA target domain (Fig. 2A), and an unrelated 54-nucleotide domain from ribosomal protein L10 mRNA (supplemental Fig. S4) as control. For the L11 rRNA domain transcripts of 58 and 29 nucleotides, we examined both the wild-type and U1061A mutant RNAs. 23 S rRNA and wild-type 58-nucleotide RNA were methylated with similar efficiency and were ∼2-fold poorer substrates than the 29-nucleotide hairpin (Fig. 2D). In contrast, the 17-nucleotide RNA containing only the target loop and the first four base pairs of the stem was a much poorer substrate with methylation reduced ∼20-fold compared with the larger hairpin RNA. Methylation of the 17-nucleotide RNA was however still significantly above that of the non-methylated control RNA. The U1061A mutation also significantly reduced methylation in both the 58- and 29-nucleotide RNA contexts, although in our assay the effect was less pronounced than previously observed (18). Each RNA was methylated to a similar extent, corresponding to ∼2-fold and 4.5-fold, respectively, lower activity compared with the equivalent wild-type RNAs. Together, these results demonstrate Tsr to be a hairpin-binding protein that requires two distinct structural features of the target RNA for recognition.
Modeling of Tsr·RNA Interactions
The target site for Tsr lies within the structurally well characterized L11-binding domain of 23 S rRNA (42, 43), allowing us to conduct molecular docking experiments. We first examined the electrostatic potential of the protein dimer surface. One face of the protein has a large stripe of positive surface surrounding the cleft between the two Tsr protomers (Fig. 3), with the NTD of each positioned on either side. In contrast, the reverse side is predominantly negatively charged across the center and is therefore very unlikely to have significant affinity for RNA. Rigid body docking was performed beginning with the 58-nucleotide rRNA domain oriented to face the positively lined cleft of the Tsr dimer. Docking experiments were performed using both shape-only and shape-electrostatics correlations, and each produced an extremely similar final docked orientation as the clear top solution (Fig. 3). The interactions predicted between Tsr and the rRNA extend over extensive complimentary surfaces and are largely electrostatic in nature. However, the exposed nature of the target loop and unusual structure of the internal bulge within Helix A leave open the possibility of direct recognition of base edges.
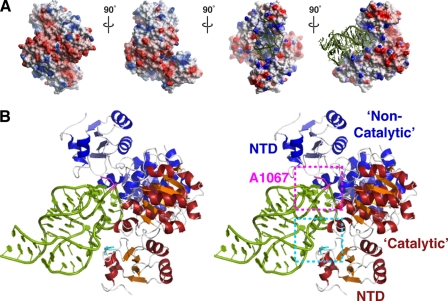
FIGURE 3.
Molecular modeling of Tsr-rRNA interactions. A, four orthogonal views around the vertical axis of the Tsr dimer with electrostatic surface potential indicated in red (negative) and blue (positive). Docked RNA is shown in the two orientations on the right only. B, stereo view schematic of the docked Tsr·rRNA complex. Regions encompassing the A1067 target loop (magenta) recognized by the non-catalytic Tsr and the internal loop (cyan) recognized by the catalytic Tsr, including Phe-88 (also see Fig. 4), are shown in dashed boxes.
The docked structure provides additional insight into the catalytic “asymmetry” of the Tsr dimer. The backbone of the 58-nucleotide RNA domain Helix A in this docked structure lies predominantly along the surface of a single Tsr protein with the A1067 target loop placed deep into the cleft formed by the CTDs. The cleft can accommodate only a single RNA, and its orientation defines one Tsr as “catalytic,” i.e. bound to the AdoMet molecule that will provide the methyl group, and the other as “non-catalytic” (amino acids denoted with a prime). The NTDs of each Tsr make extensive contacts with the RNA: the domain of the non-catalytic Tsr is placed against the target loop on the opposite side to the modeled active site, while the catalytic Tsr NTD contacts the internal bulge loop in the center of Helix A. Although the non-catalytic Tsr NTD is located near the RNA Helix C, no direct contacts are predicted. Thus Tsr recognition of the 58-nucleotide domain appears to be dictated entirely by the Helix A hairpin in good agreement with our observation that the hairpin RNA is the optimal target for in vitro methylation. Most significantly, the model predicts that direct recognition of two regions of unusual backbone geometry and their relative positions within this RNA domain are likely to be the critical determinants of specific target site recognition.
In this model, recognition of the 58-nucleotide domain by the catalytic Tsr extends some distance from the target loop with contacts made by both protein NTD and CTD to the RNA backbone around the internal bulge within Helix A (Fig. 3). The unusual geometry of the RNA is probed by a collection of basic residues, including Arg-17, Lys-23, Arg-26, and Lys-89 of the N-terminal domain, and Arg-158, Arg-159, and Arg-162 of the C-terminal domain. Of these, only Arg-26 and Arg-162 are conserved in other L30-like SpoU enzymes, with the latter moderately conserved across the wider SPOUT family (supplemental Fig. S1), predicting that this large collection of basic residues is organized to specifically recognize the A1067 hairpin of 23 S rRNA.
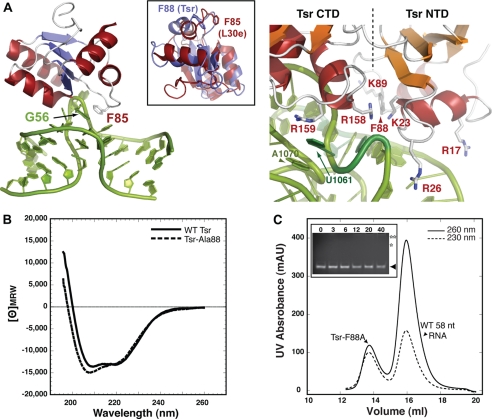
In the yeast L30e·mRNA autoregulatory complex (44, 45) a critical determinant of binding is an aromatic ring stacking interaction of Phe-85 with the first unpaired nucleotide (G56) of a kink-turn motif in the RNA (Fig. 4A). Mutation of this amino acid to Ala causes a 20-fold reduction in binding (45). Unlike the other SpoU enzymes with L30-like domains (supplemental Fig. S1), the equivalent position in Tsr is also phenylalanine (Phe-88) and is positioned close to the open RNA minor groove at the RNA internal loop in the docked structure (Fig. 4A). Although Phe-88 is oriented into a hydrophobic pocket within the free protein, a small rotation toward the RNA would allow it to probe the RNA internal loop structure. We therefore mutated Phe-88 to alanine (Tsr-Ala-88) to examine its contribution to recognition in the Tsr·rRNA complex. The Tsr-Ala-88 was expressed as a soluble protein and purified in an identical manner to wild-type Tsr. Although slightly different around the 208 nm peak, the CD spectrum of Tsr-Ala-88 was consistent with a folded protein of similar structure to the wild-type protein (Fig. 4B). Most significantly however, Tsr-Ala-88 was defective in RNA binding as monitored by the gel mobility shift and gel filtration assays (Fig. 4C). Thus, the prediction of involvement of Tsr-Phe-88 in recognition of the RNA internal loop is confirmed, providing further experimental validation of the model.
FIGURE 4.
Tsr Phe-88 is a critical determinant of recognition. A, interaction of L30e Phe-85 with the L30e mRNA kink turn (44, 45) (left) and close-up view of the modeled Tsr-rRNA interaction in the region of Phe-88 (right). The inset shows an alignment of the Tsr L30-like NTD and L30e. B, CD spectra of wild-type and Ala-88 Tsr proteins. C, the Tsr-Ala-88 protein is defective in RNA binding as monitored by gel filtration chromatography and gel mobility shift assay (inset; compare with data of Fig. 2, B and C). Protein concentrations in the gel shift assay are indicated above each lane, and bands corresponding to free RNA (▴) and approximate positions of absent shifted bands (asterisks) are indicated.
On the opposite side of the internal loop, further interactions are predicted where the 152–157 loop of the catalytic Tsr approaches U1061. This base is turned out of Helix A to form a unique tertiary stacking interaction on the surface of the RNA with A1070 from the target loop. It is thus possible Tsr also directly recognizes these bases, because mutation of either dramatically reduces methyl transfer (18). Mutation of U1061 to A also significantly increases the stability of the RNA tertiary structure suggesting that the reduction in Tsr activity might correspond to an increased energetic cost to unfold the RNA tertiary structure. Although this may be partly true, our methylation data argue against this, because the U1061 to A mutation is equally detrimental in the context of either the 58- or 29-nucleotide RNA. In the latter case, there is no RNA tertiary structure to unfold, and the mutation actually decreases the thermostability of the hairpin (data not shown). Because the mutation has a direct detrimental effect on RNA binding by Tsr in our gel filtration assay, together these data support the direct recognition of this nucleotide by Tsr.
Positioning of the Target Nucleotide Loop
Our molecular model indicates that the non-catalytic Tsr subunit also plays a major role in the recognition and positioning of the A1067 target loop. The CTD of the catalytic Tsr subunit makes extensive interactions with the opposite side of the A1067 target loop from the catalytic center. Gly-128, Arg-162, and Arg-158 lie close to the RNA making contacts to the backbone and potentially recognizing base edges of residues U1066, G1068, and A1069, where mutation is known to eliminate methyl transfer (18).
The target nucleotide A1067 is at the apex of the RNA loop with base and ribose exposed on the surface of the RNA available for direct recognition (42, 43). However, despite being placed deep into the cleft made by the Tsr dimer interface, simple docking of the RNA against the Tsr dimer could not position the target ribose 2′-oxygen closer than ∼10 Å from the bound AdoMet. Residues from the non-catalytic Tsr, including Lys-89′ and Arg-92′ of the NTD and Lys-125′, Arg-158′, Arg-159′, and Lys-221′ of the CTD, are in close proximity to the RNA backbone and appear to collectively recognize the unusual conformation of the RNA loop and could potentially alter it to move the target atom into the catalytic center. Lys-89′ in particular extends directly toward A1067 suggesting it could directly influence the loop conformation and thus positioning of the target nucleotide. Also in favor of this hypothesis, we note that there is a small pocket on the catalytic Tsr surface adjacent to the loop that contains several conserved amino acids proposed to be important for both RNA and AdoMet binding in other SPOUT MTases (33, 34, 38, 41) into which A1067 could be positioned by such changes. However, the molecular fine details of this component of Tsr·rRNA recognition will require a high resolution structure of the protein·RNA complex.
CONCLUSION
The results of molecular docking experiments based on our Tsr-AdoMet complex crystal structure provide a structural rationalization for the findings of our in vitro binding and methylation assays with RNA and mutant Tsr protein. Tsr directly binds a single hairpin loop structure within the ribosomal L11-binding domain but uses each of its L30-like NTDs to recognize two distinct components of its structure: the A1067 target loop and a more distant internal bulge. Undoubtedly Tsr employs a distinct set of recognition strategies compared with L11 (32) to bind the same rRNA domain. Like L11, however, Tsr almost certainly exploits the unique conformations in this rRNA domain to achieve absolute specificity of target selection. With the major contemporary clinical challenge of combating resistant bacterial strains, a deeper molecular understanding of the specific recognition mechanisms of rRNA resistance methyltransferases will be an essential platform for producing new designer antibiotics.
Supplementary Material
Acknowledgments
The tsr gene from S. azureus (as pUC-TSR) and the 17-nucleotide RNA were generous gifts from Dr. G. Wright (McMaster University, Hamilton, Ontario, Canada) and Prof. D. E. Draper (Johns Hopkins University), respectively. We are grateful to Dr. A. Fordham-Skelton for use of the Innovadyne crystallization robot (Synchrotron Radiation Source, Daresbury Laboratories), and to Dr. C. M. Dunham for comments on the manuscript. Dr. E. Daub, R. Kinach, S. Tanner, and G. Couture are gratefully acknowledged for early contributions to this research.
This work was supported by The Wellcome Trust (Grant 79242 to G. L. C.) and National Sciences and Engineering Council (Canada) (to J. F. H.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- rRNA
- ribosomal RNA
- Tsr
- thiostrepton resistance methyltransferase
- AdoMet
- S-adenosyl-l-methionine
- MTase
- methyltransferase
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- A1067
- 23 S rRNA nucleotide 1067 (E. coli numbering)
- SPOUT
- SpoU/TrmD methyltransferase family
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Gustilo E. M., Vendeix F. A., Agris P. F. (2008) Curr. Opin. Microbiol. 11, 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clouet-d'Orval B., Gaspin C., Mougin A. (2005) Biochimie (Paris) 87, 889–895 [DOI] [PubMed] [Google Scholar]
- 3.Ferré-D'Amaré A. R. (2003) Curr. Opin. Struct. Biol. 13, 49–55 [DOI] [PubMed] [Google Scholar]
- 4.Rozenski J., Crain P. F., McCloskey J. A. (1999) Nucleic Acids Res. 27, 196–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z., O'Farrell H. C., Rife J. P., Culver G. M. (2008) Nat. Struct. Mol. Biol. 15, 534–536 [DOI] [PubMed] [Google Scholar]
- 6.Decatur W. A., Fournier M. J. (2002) Trends Biochem. Sci. 27, 344–351 [DOI] [PubMed] [Google Scholar]
- 7.Conn G. L., Savic M., Macmaster R. (2009) in DNA and RNA Modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution ( Grosjean H. ed) Landes Bioscience, Austin, TX [Google Scholar]
- 8.Long K. S., Vester B. (2009) in DNA and RNA Modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution ( Grosjean H. ed) Landes Bioscience, Austin, TX [Google Scholar]
- 9.Schubert H. L., Blumenthal R. M., Cheng X. D. (2003) Trends Biochem. Sci. 28, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cundliffe E. (1989) Annu. Rev. Microbiol. 43, 207–233 [DOI] [PubMed] [Google Scholar]
- 11.Poehlsgaard J., Douthwaite S. (2005) Nat. Rev. Microbiol. 3, 870–881 [DOI] [PubMed] [Google Scholar]
- 12.Doi Y., Arakawa Y. (2007) Clin. Infect. Dis. 45, 88–94 [DOI] [PubMed] [Google Scholar]
- 13.Gaynor M., Mankin A. S. (2003) Curr. Top. Med. Chem. 3, 949–961 [DOI] [PubMed] [Google Scholar]
- 14.Harms J. M., Wilson D. N., Schluenzen F., Connell S. R., Stachelhaus T., Zaborowska Z., Spahn C. M., Fucini P. (2008) Mol. Cell. 30, 26–38 [DOI] [PubMed] [Google Scholar]
- 15.Goodman C. D., Su V., McFadden G. I. (2007) Mol. Biochem. Parasitol. 152, 181–191 [DOI] [PubMed] [Google Scholar]
- 16.Liao Z., Thibaut L., Jobson A., Pommier Y. (2006) Mol. Pharmacol. 70, 366–372 [DOI] [PubMed] [Google Scholar]
- 17.Wienen B., Ehrlich R., Stöffler-Meilicke M., Stöffler G., Smith I., Weiss D., Vince R., Pestka S. (1979) J. Biol. Chem. 254, 8031–8041 [PubMed] [Google Scholar]
- 18.Bechthold A., Floss H. G. (1994) Eur. J. Biochem. 224, 431–437 [DOI] [PubMed] [Google Scholar]
- 19.Cameron D. M., Thompson J., Gregory S. T., March P. E., Dahlberg A. E. (2004) Nucleic Acids Res. 32, 3220–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson J., Schmidt F., Cundliffe E. (1982) J. Biol. Chem. 257, 7915–7917 [PubMed] [Google Scholar]
- 21.Kabsch W. (1988) J. Appl. Crystallogr. 21, 916–924 [Google Scholar]
- 22.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 23.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 24.Terwilliger T. C. (2003) Acta Crystallogr. Sect. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y., Yao D. Q., Gu Y. X., Lin Z. J., Zheng C. D., Fan H. F. (2007) Acta Crystallogr. Sect. D Biol. Crystallogr. 63, 793–799 [DOI] [PubMed] [Google Scholar]
- 26.Cowtan K. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 [Google Scholar]
- 27.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 28.Emsley P., Cowtan K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 30.Walker S. C., Avis J. M., Conn G. L. (2003) NAR Methods 31, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conn G. L., Gutell R. R., Draper D. E. (1998) Biochemistry 37, 11980–11988 [DOI] [PubMed] [Google Scholar]
- 32.Dunstan M. S., Guhathakurta D., Draper D. E., Conn G. L. (2005) Chem. Biol. 12, 201–206 [DOI] [PubMed] [Google Scholar]
- 33.Anantharaman V., Koonin E. V., Aravind L. (2002) Nucleic Acids Res. 30, 1427–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tkaczuk K. L., Dunin-Horkawicz S., Purta E., Bujnicki J. M. (2007) BMC Bioinf. 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gustafsson C., Reid R., Greene P. J., Santi D. V. (1996) Nucleic Acids Res. 24, 3756–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor A. B., Meyer B., Leal B. Z., Kötter P., Schirf V., Demeler B., Hart P. J., Entian K. D., Wöhnert J. (2008) Nucleic Acids Res. 36, 1542–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahn H. J., Kim H. W., Yoon H. J., Lee B. I., Suh S. W., Yang J. K. (2003) EMBO J. 22, 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosbacher T. G., Bechthold A., Schulz G. E. (2005) J. Mol. Biol. 345, 535–545 [DOI] [PubMed] [Google Scholar]
- 39.Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 40.Michel G., Sauvé V., Larocque R., Li Y., Matte A., Cygler M. (2002) Structure 10, 1303–1315 [DOI] [PubMed] [Google Scholar]
- 41.Watanabe K., Nureki O., Fukai S., Ishii R., Okamoto H., Yokoyama S., Endo Y., Hori H. (2005) J. Biol. Chem. 280, 10368–10377 [DOI] [PubMed] [Google Scholar]
- 42.Conn G. L., Draper D. E., Lattman E. E., Gittis A. G. (1999) Science 284, 1171–1174 [DOI] [PubMed] [Google Scholar]
- 43.Wimberly B. T., Guymon R., McCutcheon J. P., White S. W., Ramakrishnan V. (1999) Cell 97, 491–502 [DOI] [PubMed] [Google Scholar]
- 44.Chao J. A., Williamson J. R. (2004) Structure 12, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 45.Mao H., White S. A., Williamson J. R. (1999) Nat. Struct. Biol. 6, 1139–1147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.